Abstract
This study describes a new method for analyzing microcirculatory videos. It introduces algorithms for quantitative assessment of vessel length, diameter, the functional microcirculatory density distribution and red blood-cell (RBC) velocity in individual vessels as well as its distribution. The technique was validated and compared to commercial software. The method was applied to the sublingual microcirculation in a healthy volunteer and in a patient during cardiac surgery. Analysis time was reduced from hours to minutes compared to previous methods requiring manual vessel identification. Vessel diameter was detected with high accuracy (>80%, d > 3 pixels). Capillary length was estimated within 5 pixels accuracy. Velocity estimation was very accurate (>95%) in the range [2.5, 1,000] pixels/s. RBC velocity was reduced by 70% during the first 10 s of cardiac luxation. The present method has been shown to be fast and accurate and provides increased insight into the functional properties of the microcirculation.
Keywords: Orthogonal polarized spectral (OPS) imaging, Side-stream dark field (SDF) imaging, Vessel density, Blood velocity, Space–time diagram
Introduction
Sublingual orthogonal polarization spectral (OPS) imaging [2, 4, 23, 25, 27–29, 34] and side-stream dark field (SDF) imaging [17] are currently being used extensively in clinical microcirculatory research, especially in surgery and intensive care medicine. This research has gained clinical importance by the finding in several centers that microcirculatory alterations nonresponsive to therapy predict a poor outcome in critically ill patients [32, 39]. This predictive value of microcirculatory images was not found in systemic hemodynamic or oxygen-derived parameters measured conventionally at the bedside. Furthermore, clinical investigations have shown that the impact of standard as well as innovative therapies could best be demonstrated by their effect on the sublingual microcirculation [7, 10, 34, 35]. In demonstrating their effects, OPS and SDF images have been analyzed manually by semi-quantitative scoring methods [4, 8, 9, 34]. Although these methods have been validated and prove sensitive and specific in identifying the severity of disease in critically ill patients they are cumbersome, very time consuming and semi-quantitative. Klyscz et al. [22] described an early quantitative method for estimating red blood-cell (RBC) velocity, limited to straight vessel segments selected manually by the user [11, 13, 18–20, 22, 24, 28]. Local vessel width is determined manually with an on-screen caliper; vessel length is obtained using a drawing tool that allows manual tracing of vessels. Although the program is unique in its field, it requires a large amount of user interaction, which increases observer bias and analysis time.
The image analysis techniques proposed in the current paper provide a high degree of automation and yield quantitative measures of vessel length, vessel diameter, the functional capillary density distribution, RBC velocity in individual vessel segments and the RBC velocity distribution. Space–time diagrams [20] are used for velocity estimation and the new technique, in contrast to earlier similar image analysis software, is able to generate space–time diagrams of curved vessels. We introduce automatic detection of the line orientation in space–time diagrams for automated velocity estimation. The accuracy of the present method was validated using video simulations and compared to commercially available software (CapiScope [6, 14, 33]). Finally, in order to illustrate the application of the software we present analyses of sublingual video recordings from a healthy volunteer and from a patient during cardiac luxation in open-heart surgery.
Methods
With currently available imaging techniques, such as capillaroscopy, OPS or SDF imaging, “vessels” are only observed in the presence of RBCs. The RBCs contain haemoglobin, which highly absorbs the incident wavelength used in these techniques, in contrast to the background medium. The capillary vessel wall is basically invisible to these imaging techniques. Videos of the microcirculation therefore show structures of red blood cells that are bounded by vessel walls. These structures are referred to as “vessels” in this paper.
The analysis techniques described involve vessel segmentation (the operation that extracts vessel segments from an image) and RBC velocity estimation using space–time diagrams, and require a comprehensive series of image processing steps as indicated by Fig. 1. The performance of the image analysis algorithms depends on a series of parameters that are listed in Table 1. These parameters are adjusted for optimal performance, considering the utilized image scale, and need only be adapted when changing optical magnification.
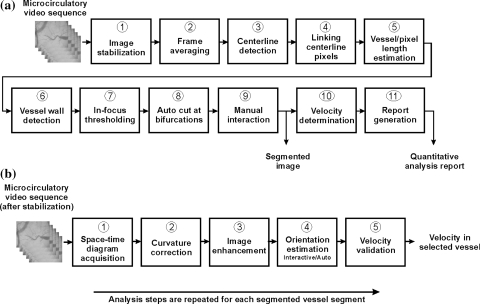
Fig. 1.
a Analysis steps for vessel segmentation and for assessment of quantitative analysis parameters. Step 10, velocity detection, is detailed in the second diagram. b Analysis steps for quantifying RBC velocity per vessel segment
Table 1.
Parameter settings for automated microcirculatory analysis
| Symbol | Description | Setting | Motivation |
|---|---|---|---|
| r max | Search range for linking pixels | 5 pixels | If the search range is set too large, spurious vessel segments are linked together |
| α | Search angle for linking pixels | 90° | ±45° allows strong curvature yet rejects perpendicular continuation of a vessel |
| σ | Standard deviation of Gaussian derivatives for centerline detection; Many other filter segmentation parameters are derived from this scale parameter | 3 pixels | Pragmatically determined |
| σ cross | Edge detection; standard deviation of highest derivative filter in direction normal to vessel orientation | 1/3 × σ | This filter setting gives no considerable overestimation of capillary diameter (>4 μm) |
| σ long | Edge detection; standard deviation for averaging distance to vessel wall in longitudinal direction | 3 × σ | This filter settings spans small plasma gaps |
| s min | Minimum vessel segment length | 5 × σ a | Pragmatically determined |
| σ edge | Standard deviation of edge distance smoothing | 3 × σ | Pragmatically determined |
| ε thr | Centerline detection threshold | 0.7b | Pragmatically determined |
| σ H | Standard deviation for smoothing Hough score diagram (Fig. 7a) | 2° | Pragmatically determined |
| v min | Lower limit for velocity assessment | 2 μm/s | Pragmatically determined |
aSet to 2σ for interactive assessment
bSet to 0 for interactive assessment
Movement of the subject or the hand-held imaging device can result in unstable images that hamper vessel recognition and velocity measurements. In order to stabilize images 2D cross correlation was used [1]. During this stabilization process, image enhancement is optionally performed in two ways. First, intensity variations in the background are reduced for each frame by subtracting the quadratic polynomial surface that best-fits the image, and by adding the average intensity of the original image. Secondly, contrast improvement is achieved by manipulating the image gray-scale histogram, by mapping each gray-level of the input image to a gray-level of the output image using a so-called transfer function, as described by Pries [30]. The latter method may affect the vessel geometry and is therefore not recommended before performing spatial measurements. It is convenient, however, to evaluate space–time diagrams.
After stabilization, video frames are time-averaged to fill up interruptions in capillaries that exist due to the presence of plasma gaps or white blood cells. Averaging causes capillaries to be detected as a continuous structure, irrespective of interrupted cell flow. Averaging also reduces the contribution of noise, which is beneficial for the vessel segmentation process.
The remaining analysis steps rely on the scale parameter (σ). Analyzing at larger values of the scale parameter detects larger vessels whereas smaller values of this parameter detects smaller structures. The scale parameter is explained in greater detail in the Appendix. The pre-processed image is subjected to vessel segmentation as detailed in the Appendix.
The vessel diameter may be overestimated, especially in small vessels, if the microcirculatory image is not in focus. It is, therefore, important to exclude those vessels that are out of focus. In the present study the average gradient magnitude at all edge points of a vessel is used to determine a focus score per vessel. This focus score [12] is made less sensitive to background variations by normalization to the background intensity, local (200 × 200 pixels2) to each edge pixel. The user is able to exclude vessels with a focus score below a manually adjusted limit.
Blood flow splits into two branches at a bifurcation, causing RBC velocity to change. For accurate RBC velocity assessment, space–time diagrams have to be determined from vessel segments between bifurcations. The process of cutting vessels at bifurcations is automated by determining the distance between the end of a blood vessel segment and the wall of neighboring vessels. If a vessel approaches a neighbor within less than 1½ × the neighbor’s radius, the neighbor is cut in two at the point of approach. The 1½ × factor allows cutting of vessels that bifurcate or intersect, yet prevents cutting vessels that run parallel. This cutting procedure is repeated for all available vessel segments.
Following the above-described automatic segmentation, the user is able to manipulate these intermediate results by deleting, cutting, or connecting vessel segments. Undetected vessel segments can be manually drawn in where the software suggests a present vessel segment, given a user-selectable scale (i.e. σ = 1.5, σ = 3.0, σ = 6.0 or σ = 12.0). If computer-assisted vessel detection fails, one can add remaining vessels by manual tracing with a user-selected diameter.
RBC velocity is determined using space–time diagrams [20], which are obtained by automatically tiling the centerline intensity of a vessel as vertical lines (corrected for vessel curvature, see the Appendix) for a number of consecutive frames. Moving cells and plasma gaps cause tilted lines to appear in these diagrams (see Fig. 7b for an example). The line orientation is indicative for RBC velocity. Acquiring the space–time diagram from curved vessels is an improvement on previous methods which only allow velocity estimation from straight vessel segments where the user draws a straight centerline. Image histogram equalization [31] is utilized to automatically improve visibility of the line structure in space–time diagrams. RBC velocity is estimated automatically using gray-scale Hough transform [15, 26], detailed in the Appendix. The user is allowed to overrule the result of automatic analysis by tracing lines in the space–time diagram interactively. When interactively tracing lines, the average orientation is used for further processing. Finally, the acquired orientation is converted to an actual velocity value (see Appendix).
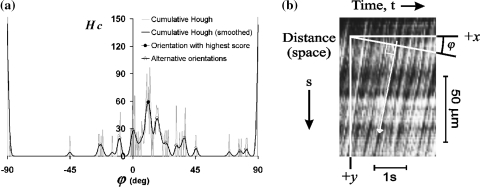
Fig. 7.
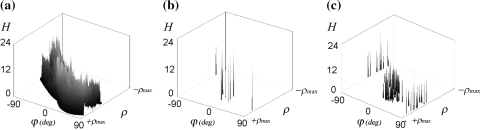
a Adding accumulator cells from Fig. 6c that have the same orientation, results in the Hough score diagram (gray curve). Spikes are removed by Gaussian smoothing (black curve). The highest peak identifies the global orientation of lines in the space–time diagram. b Original space–time diagram and the definition of orientation parameter φ. The white arrow indicates the resulting orientation
Some of the above described techniques, numbered in Fig. 1a, b, are new in microcirculatory image analysis and are therefore explained in the Appendix in greater detail. These include: Fig. 1a-(3) centerline detection, Fig. 1a-(6) vessel wall detection in the presence of interrupted cell flow, Fig. 1b-(2) curvature correction, Fig. 1b-(4) automatic orientation and velocity estimation and Fig. 1b-(5) theoretical range of velocity assessment.
Experiments
In all experiments detailed below, the algorithms were configured according to the settings given in Table 1. Contrast enhancements were not used in any of the experiments.
Validation
To validate the performance of vessel length, diameter and RBC velocity, simulation videos were created. The main advantage of simulation videos is that the actual vessel and flow characteristics are fully known and the ability of the software to measure it can accurately be determined. In addition, such simulation videos exclude optical effects, such as, scattering of light in surrounding tissue and widening of vessels due to point spread effects [38].
The simulation video for length and diameter validation (500 × 500 pixels) contains five lines of different length (50, 100, 150, 200 and 250 pixels) with a Gaussian cross-sectional profile (with standard deviation σ l). The vessel wall of these simulated vessels is marked by the points where the maximum gradient is found, i.e. at ± σ l, yielding d = 2σ l, where d is the line diameter. The background and centerline intensity were set to 200 and 50 au (au = arbitrary units). The effect of vessel orientation was incorporated by including frames with different line orientation in the range [0, 90]° with 15° increments.
A second simulation video (250 × 250 pixels) was created for validating velocity assessment. Each video frame shows a simulated vessel containing “cells” being circular blobs with a Gaussian cross-sectional intensity profile (σ cell = 3 pixels). These cells (approximately 1 cell per 5 pixels of vessel length, background intensity 200 au, center at 50 au) were drawn at random locations but within the boundaries of an imaginary vessel of 10 pixels wide that extends to the edges of each video frame. The accuracy of interactive and automatic velocity assessment was tested in a vessel oriented at 0° in the velocity range [2.5, 2,000] pixels/s. The lower limit of this range was chosen pragmatically while the upper limit is in accordance with the physical limit of detection (∼v max, see Eq. 2 in the Appendix, L = 250 pixels, f = 25 frames/s). Velocity results were obtained interactively, by tracing up to five available lines in the space–time diagram, and automatically (see Appendix).
Each video fragment covered 100 frames. Gaussian noise was added to each frame with σ noise = 10 au, which is approximately twice that of a typical SDF image. The validation experiments were made independent of optical magnification, by expressing the accuracy of assessment in terms of pixels/s. The two simulation videos have been put on the Internet (http://www.sdfimaging.net) as information for the reader and for use in validation of other software developments.
Comparison
To evaluate the utility and accuracy of our software we compared its performance to that of a commercially available microcirculation image analysis package. In this context, CapImage [22] and CapiScope [14] are commercially available software packages used to analyze microcirculatory video sequences. CapImage represents one of the few software packages that has been described and evaluated in the literature in any detail. To our knowledge this package is no longer available. Instead a new software package was developed with similar modalities but using improved technology, called CapiScope. A validation study [6, 33] showed that CapiScope provides comparable values for microcirculatory parameters, such as, vessel diameter and RBC velocity, to those obtained with CapImage. In the present study we compared the performance of our software to that of CapiScope (version 3.6.4.0) (KK-Technology, Bridleways Holyford, Devon, England).
In the comparison study the ability of the software to measure the average vessel diameter was compared to that measured by CapiScope. In the CapiScope method an average of five determinations at different locations along the vessel was taken as the average diameter and in the present software the diameter was averaged over the entire vessel segment. RBC velocity was measured in a simulation video as well as in an SDF imaging recording of the sublingual microcirculation in a healthy male volunteer (see http://www.sdfimaging.net). In the latter experiment the interactively obtained velocity results were most accurate and served as reference for determining the error in automatic analysis. For automatic analysis, a velocity error level up to 20% compared to interactive assessment, was considered acceptable within the framework of the experiment.
For comparing the vessel length estimation, the vessel density (VD) was also determined by both programs by analyzing ten sublingual recordings of healthy individuals. The VD is defined as the functional capillary density (FCD) [16, 22] and includes thick vessels as well as capillaries.
To evaluate the time saved by the present method two experienced analysis researchers applied the two methods to the analysis of the simulation as described before and to SDF image recordings of the sublingual microcirculation.
Clinical application
Sublingual video recordings were made using a MicroScan SDF system [17] (MicroScan B.V., Amsterdam, The Netherlands) with a standard 5× optical magnification, which results in microcirculation images with a pixel spacing of approximately (h × w) 1.5 × 1.4 μm. The disposable microscope tip is held gently against the tissue and guarantees a fixed distance (∼1 mm) and no perspective between specimen and lens over the entire field of view. The hardware features a point spread function [38] similar to a Gaussian distribution with a standard deviation of approximately 1 pixel in the x and y direction. Capillaries, having a diameter of about 4-5 μm, are therefore approximately 3 pixels wide in standard SDF images.
A sublingual video recording from a healthy male volunteer was selected with high contrast and moderate RBC velocity, which allowed us to evaluate the feasibility of automatically analyzing space–time diagrams of clinical observations. Another sublingual recording was made during cardiac luxation in a patient who underwent cardiac bypass surgery using off-pump coronary artery grafting (OPCAB). Cardiac luxation is a procedure that is used during cardiac surgery where the heart is lifted and repositioned causing an immediate decrease of cardiac output and thereby sublingual microcirculation. During this procedure sublingual SDF imaging was applied and cessation of the microcirculation was observed during luxation-induced hypotension. The luxation videos have also been put on the Internet (http://www.sdfimaging.net) as information for the reader.
Results
Validation experiments
Vessel length
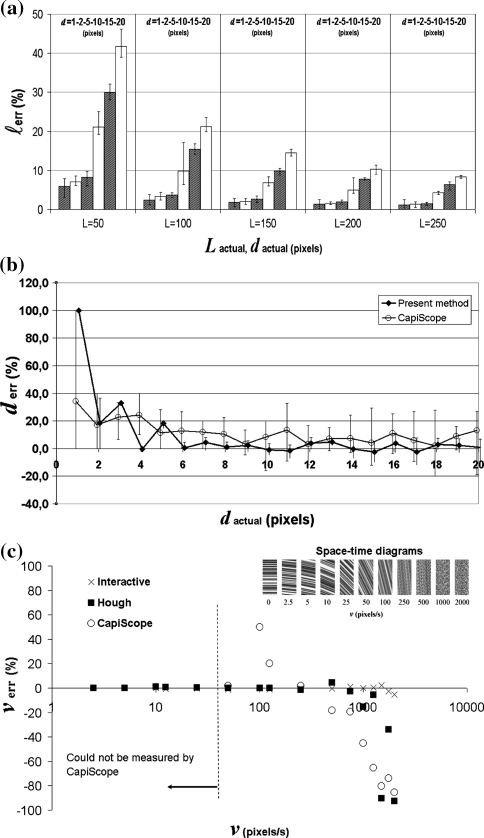
In two out of 35 measurements at a diameter d = 1 pixel automatic vessel detection failed due to the presence of noise. The bars in Fig. 2a show the average length deviation of lines at different orientation. The error bars indicate the small error range due to line orientation and image noise (<5% for capillaries with L > 100 pixels and d < 5 pixels). The graph shows that the accuracy of length assessment strongly depends on the diameter of the simulated vessel (due to the scale of analysis). CapiScope could not measure vessel length automatically.
Fig. 2.
Results of evaluation using simulation video. a Average vessel length estimation error (l err) versus actual length (L, pixels) and diameter (d, pixels). b Average diameter estimation error (d err) versus actual vessel diameter (d in pixels). Measurements at whole pixel intervals are slightly shifted apart for clarity. The error bars in both figures indicate the error range due to image noise and vessel orientation as tested in the range [0, 90], with 15° increments. c Accuracy of interactive, automatic (Hough) and CapiScope velocity estimation. The inset shows the space–time diagrams that yielded these results
Vessel diameter
Figure 2b shows the relative diameter-estimation error of simulated vessels as obtained by the present method (σ = 3, σ cross = 1) and by CapiScope. The error bars indicate the range as a result of vessel orientation and image noise. CapiScope tends to overestimate vessel diameter slightly over the entire range. The present method performs better for vessels wider than 5 pixels. It also shows a smaller diameter variation due to orientation and image noise. With the present method, vessels with a diameter in the range [2σ cross, 13] pixels show an absolute diameter error <1 pixel. This results in a relative error that drops below 20% for vessels wider than 3 pixels (Fig. 2b).
Velocity
RBC velocity was estimated by the present method using the acquired space–time diagrams shown in Fig. 2c (inset). These diagrams show that a line structure is clearly visible at low velocities while the images turn rather noisy at high velocities. Figure 2c shows the deviation in velocity assessment as obtained with the new method by drawing lines in the space–time diagram manually, automatically using the Hough method, and using CapiScope. Interactively tracing lines in the space–time diagram gives the best results and appears feasible up to 1,000 pixels/s in this simulation experiment (accuracy >95%). Automatic velocity assessment performs excellently up to 750 pixels/s (>95% accurate). At higher velocities (>1250 pixels/s), the method fails and selects an alternative orientation that results in a large velocity error. CapiScope was not able to measure velocities below 50 pixels/s. Higher velocities showed a relatively large error (Fig. 2c).
Comparison experiments
This section compares the analysis results of the present method with CapiScope in finding vessel density, vessel diameter and RBC velocity. For these experiments SDF image recordings were used that show the sublingual microcirculation of healthy volunteers.
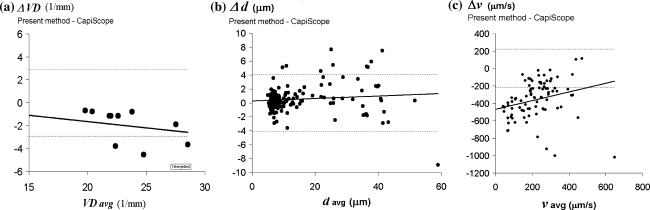
The Bland–Altman plots [3] in Fig. 3a, b illustrate the similarity between the present method and CapiScope in measuring vessel density and the diameter of sublingual vessels. With the present method, VD measurements were performed in 67% of the time required by using CapiScope (10 recordings in 56 min with the present method compared to 84 min with CapiScope). The vessel diameter measurement using CapiScope took approximately 4 h while the present method provided the same data in approximately 10 min.
Fig. 3.
Agreement of present method and CapiScope, represented by Bland–Altman plots [3] showing, a difference against average vessel density (VD), b difference against average diameter, c difference against average velocity. Data was obtained from sublingual microcirculatory video recordings. All curves are drawn with 95% limits of agreement (dashed lines) and regression line
Velocity results obtained by the present method do not correspond with those obtained by CapiScope (as illustrated by the graph in Fig. 3c). With the present method, line orientation in the space–time diagram was analyzed automatically and if the presented orientation failed, lines were traced manually. Visual inspection of the video fragment, together with the many space–time diagrams, confirmed the presence of relatively low velocities in this experiment (<200 μm/s) that were largely overestimated by CapiScope. This finding is similar to the results of the simulation experiment shown in Fig. 2c which illustrates the disparity between the two methods. Manual velocity analysis with CapiScope took 3 h in this experiment compared to 20 min using the present automated method.
Clinical application
This section describes the application of the present method in analyzing microcirculatory images from a healthy volunteer and from a patient during cardiac surgery.
Healthy volunteer
The video recording of a healthy volunteer was analyzed after averaging frames within a 2 s interval. In this experiment 31% of the total vessel length required manual interaction. The functional microcirculatory density distribution is given in Fig. 4a. It shows the presence of a bimodal distribution with a large portion of the image area being occupied by capillaries in the range 5-10 μm.
Fig. 4.
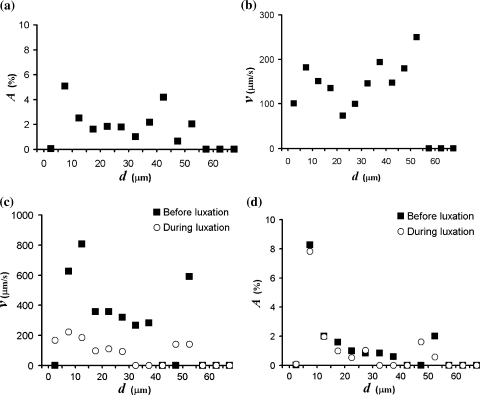
Distributions a Sublingual microcirculatory density distribution [A represents the relative image area occupied by vessels in the given diameter (d) range] and, b velocity distribution, both of a healthy male individual. c Velocity distribution, and d functional microcirculatory density distribution, both showing the results before and during the first 10 s of cardiac luxation (see also Fig. 5)
In this analysis a total of 207 vessel segments was analyzed. In 99 segments (48%) the space–time diagrams did not reveal a visible line structure. In some of these cases vessel segments were too short to allow velocity analysis (see Eq. 2 in the Appendix). The space–time diagrams of the remaining 108 vessel segments (52%) showed a line structure that was analyzed both interactively, by tracing lines, and automatically. The velocity distribution in Fig. 4b illustrates the result of interactive analysis and shows that RBC velocity is in the same order of magnitude for all vessels in the given diameter range d = [0, 60] μm. With automatic analysis 29 segments (27%) fell within the 20% error level of acceptance.
Cardiac luxation
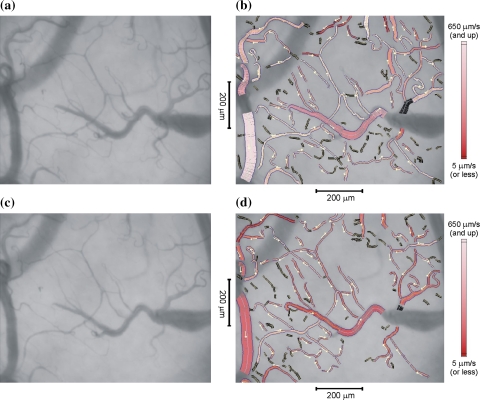
The present method was used to measure the changes which occur when the heart is repositioned during off-pump cardiac surgery. Figure 5a, c indicate the average of 250 frames (10 s) from the sublingual video recording before and during cardiac luxation. The figures at the right (Fig. 5b, d) show the same video data with the results of analysis superimposed. Vessel segments with a diameter larger than 60 μm were excluded. In these two experiments 95% (before luxation) and 80% (during luxation) of the total vessel length were segmented automatically, the remaining vessels were added interactively. We traced up to 20 lines in each space–time diagram (10 s interval) to get an impression of the average velocity in each vessel segment during that interval. Space–time diagrams showed a clear line structure in 44% of the vessel segments before, and 48% during luxation. These represent approximately 75% of the segmented vessel length in both cases. RBC velocity is color-coded in the vessels of Fig. 5b, d. Dark colors in Fig. 5d clearly show that RBC velocity is reduced during cardiac luxation. Figure 4c demonstrates the velocity distribution.
Fig. 5.
a Average frame out of a video sequence showing sublingual microcirculation before a luxation procedure. b Same image as (a) with analysis results superimposed. c Average of frames obtained during a luxation procedure, d with analysis results superimposed. The magnitude of red-blood cell velocity is color-coded in the range [5 (dark), 650 (bright)] μm/s. Vessel segments with space–time diagrams that could not be analysed are marked black. The small arrows indicate the direction of blood flow
The observed image area that was occupied by vessels in the cardiac luxation example changed from 17.1 to 14.6%, which is a 15% reduction. The reduction of vessels was confirmed by visual inspection of the images of Fig. 5. It shows that some small vessels are not visible, i.e. the presence of red blood cells is lacking or is reduced, during cardiac luxation. The density distributions of Fig. 4d illustrate that a slight reduction of small vessels (d < 45 μm) occurs during luxation.
Discussion
This present study has introduced advanced image analysis techniques for the analysis of microcirculatory video sequences which allow determination of vessel length, diameter and RBC velocity, from curved vessels, quantitatively. The method combines automatic vessel identification with manually tracing vessels. It further provides the microcirculatory density distribution and the RBC velocity distribution. A first step towards automatic detection of RBC velocity from space–time diagrams has been made. The method was validated using simulation video sequences and was compared with commercially available software (CapiScope). Finally, clinical application of the software was demonstrated by analyzing microcirculatory images from a healthy volunteer and from a patient during cardiac surgery.
The measurements performed on sublingual recordings showed that 69–95% of the total vessel length was detected automatically at a single scale of analysis. Compared to CapiScope, the present method reduced analysis time from hours to minutes. It can therefore be concluded that computer-assisted vessel segmentation drastically reduces user interaction although visual inspection of the superimposed results and possible interaction at selectable scales, remain necessary.
The validation experiments were all performed at the same small scale of automatic analysis, which focuses on small image features, such as small vessels. This explains why vessel length and diameter estimation were less accurate for large-diameter vessels. In addition, the eigenvalue |λ n| (see “centerline detection” in the Appendix) reduces with vessel diameter. In the simulation experiments, where vessels end in a step edge (i.e., |λ t| is fixed), less pixels are consequently marked as being centerline pixels. This explains the underestimation of vessel length for large-diameter vessels. Considering the length of actual vessels in OPS and SDF images (L ≈ 100 pixels on average), it can be concluded that length estimation is very accurate (>95%) for capillaries up to 5 pixels wide. Vessel diameter could accurately be determined (>80%) for vessels wider than 3 pixels as in standard SDF images. Velocity estimation with the present method was very accurate (>95%) for both interactive velocity estimation (range [2.5, 1,000] μm/s) and automatic analysis of space–time diagrams (range [2.5, 750] pixels/s). CapiScope on the other hand, was not able to identify vessels automatically. It also could not measure vessel length automatically, could not measure velocities in curved vessels and was not able to measure velocities below 50 pixels/s while higher velocities were relatively inaccurate.
Clinical application of the present method illustrated the use of the functional microcirculatory distribution. Its bimodal behavior demonstrates the presence of microcapillaries as well as larger vessels. The velocity distribution was also bimodal and showed that velocities are of the same order of magnitude in vessels ranging [3, 60] μm. It was also demonstrated that RBC velocity reduced to approximately one third in all vessels ranging [3, 60] μm, during episodes of shock with severe hypotension caused by cardiac luxation. The present method of analysis was able to detect the microcirculatory alterations adequately and this illustrates its potential use in clinical microcirculation research.
The clinical experiments confirmed the feasibility of analyzing space–time diagrams automatically using the Hough transform (∼25% of the vessels with a visible line structure in the space–time diagram). The performance of automatic velocity analysis strongly depends on the quality of microcirculatory video recordings. In this respect, RBC velocity measurements benefit from higher frame rates, which increases the velocity range (limited by v max, see Eq. 2 in the Appendix), and from stroboscopic illumination with very short exposure times, which reduce longitudinal motion blur in vessels, thereby yielding high-contrast space–time diagrams. This would increase the performance of automatic orientation detection of space–time diagrams and may ultimately render velocity detection fully automatic.
The image analysis technique described in this study drastically reduce analysis time. It further reduces user interaction and observer bias. The method proved to be fast and accurate. It enables determination of vascular density and RBC velocity distributions that were otherwise impossible to obtain. We expect that the present method will allow much more widespread analysis of microcirculatory images which currently is very time consuming and thereby prohibiting. It is expected that the use of the present method will encourage microcirculation research and will increase our insight into the central role of the microcirculation in health and disease.
A full-featured version of the software that includes all analysis algorithms in this paper can be downloaded free of charge for evaluation at http://www.sdfimaging.net.
Acknowledgments
We gratefully acknowledge Keshen Mathura and Peter Goedhart (AMC) for helpful discussions during software development.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Appendix
Vessel segmentation
Scale of analysis. The image analysis techniques described in this paper require determination of spatial intensity derivatives. These image derivatives are noise sensitive if calculated as the difference between adjacent pixels. Gaussian derivatives are therefore used [37, 38] that include image data within the working distance of the Gaussian kernel. The Gaussian derivatives are obtained by convolving the image with the corresponding derivative of a Gaussian. The standard deviation (σ) of the Gaussian filter serves as the scale parameter. Many distance related analysis parameters are based on this scale parameter as indicated in Table 1.
Centerline detection is based on the method described by Steger [37]. In short, this method calculates the eigenvectors of the local Hessian matrix [21] and results in a vector that points in the vessel direction (t) and a vector in the perpendicular direction (n). Image pixels are considered candidate centerline pixels if the second order spatial intensity derivative in the direction of n, represented by eigenvalue λ n, is markedly higher than in the perpendicular direction t, represented by λ t. This condition is tested by evaluating |λ n |/(|λ n |+|λ t |) ≥ ε thr, with ε thr a given threshold value (Table 1). Candidate pixels are identified as centerline pixels if the cross-intensity profile, i.e. the intensity profile in the direction of n, is locally at its extremum.
Centerline pixels are subsequently grouped into vessel segments. Grouping starts at the centerline pixel with the highest value of λ n, and includes neighboring pixels that are on the same centerline (and do not belong to a vessel that runs parallel), and have similar eigenvalue directions. These conditions are tested as described by Staal et al. [36]. In the present paper, the search area for neighboring pixels is illustrated by a triangle, defined by an opening angle, with bisector towards tangent vector (t), and a perpendicular bisector (towards n) at a distance r in a given search range ([1, r max], Table 1). The orientation that resulted from Hessian analysis helps to calculate the length contribution per centerline pixel and the total length of a vessel segment. Vessel segments that exist due to the presence of noise are usually small, which justifies removing segments with a limited length (<s min, Table 1).
Vessel wall detection. The vessel wall is detected at each centerline pixel and is marked by the points where the cross-sectional intensity profile shows its maximum steepness (in the direction of the normal vector n, described above). With this information a vessel’s cross-sectional intensity profile is determined by sampling the image at sub-pixel level (by linear interpolation) in the normal direction. This process is repeated for each centerline pixel to obtain straightened vessels. Gaps may occur in these straightened vessels if plasma gaps or white blood cells interrupt the continuous flow of red blood cells. For this reason an anisotropic diffusion kernel is used [38] with a Gaussian response (σ long, Table 1) that largely extends in the longitudinal direction of a vessel. It effectively closes possible interruptions and detects vessels as a whole. Horizontally, the first derivative of a Gaussian filter kernel is used as a maximum gradient detector, with a small extent in the cross direction (σ cross, Table 1) to preserve well localized edge detection [5]. Convolution with the edge detection kernel may bias vessel diameter estimation, especially for small vessels. This is caused by the filter’s own pulse response, which shows its highest gradient (G″(x) = 0) at x = ±σ cross. Vessels are therefore detected as being at least 2σ cross pixels wide.
Misinterpreted edge points (artifacts) largely deviate by their mean distance to the centerline. This property is used to remove artifacts iteratively by excluding the most-distant edge point that exceeds two standard deviations from the mean distance in each iteration pass. This process is repeated until all remaining distance samples are within two standard deviations from the mean distance. The resulting mean distance is assigned to all artifact locations. Finally, Gaussian filtering (σ edge, Table 1) is performed to smooth the acquired centerline-wall distance in the longitudinal direction. The above described procedure is repeated for the opposite vessel wall and yields estimates of the local and average vessel diameter.
Velocity determination
The slope of the line structure in space–time diagrams [20] is a measure of RBC velocity, which is calculated as: v = Δs/Δt = tan φ with Δs the longitudinal displacement along the vessel centerline in time fragment Δt (see Fig. 7b).
Curvature correction. The time axis of space–time diagrams is a multiple of the frame interval, which is very accurate in CCD cameras. The space axis, on the other hand, is not uniformly distributed since the length contribution per pixel depends on the local vessel orientation. This orientation dependence is compensated for, by mapping the randomly spaced centerline pixels onto the equidistant intervals of the space–time diagram using linear interpolation. In this study the number of distance pixels of the space–time diagram was taken equal to the number of centerline pixels that describes the corresponding vessel.
Automatic velocity determination. A possible way for automatic determination of RBC velocity from space–time diagrams is by orientation estimation using the Hough transform [15]. The conventional Hough transform is a method for detecting straight lines (or curves) in images. It is basically a point-to-curve transformation that detects the parameters of straight lines in images. The technique considers the polar representation of a line:
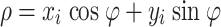 |
1 |
with (x
i
, y
i
) the coordinate of each line pixel in the space–time diagram, φ the orientation of the vector normal to the line and starting at the origin (top-left image position as in Fig. 7b), and ρ the length of this vector, which is equal to the line distance to the origin. Each line pixel is mapped to a sinusoidal curve in parameter space, ρ(φ). The discrete image of parameter space consists of accumulator cells, H(φ, ρ), that are incremented for each sinusoidal curve that passes the cell. By converting all line pixels of the space–time diagram into sinusoidal curves, the accumulator cells increment to the line length (L, in pixel units). An accumulator cell therefore yields the characteristic parameters (φ, ρ, L) of a line. With the space–time diagram as input image, a high response (count) is expected at a specified orientation (φ) and for multiple lines with a different distance to the origin (ρ). The response is also used to reject small line structures (artifacts in space–time diagrams) by thresholding accumulator cells with relatively low counts (<90% of the largest Hough count, see Fig. 6b). By performing this procedure for all (or a selection of) gray levels, as in gray-scale Hough transformation [26], and by adding the responses, we obtain a result that is less sensitive to the noisy character of space–time diagrams (Fig. 6c, named the “long-line” Hough space). Since the global line orientation is required, the total count of accumulator cells representing the same orientation
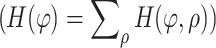 finally serves as a “score” per orientation (Fig. 7a). This curve is finally smoothed using a Gaussian filter kernel (σ
H, Table 1). The highest peak in the filtered curve, Fig. 7a, gives the best estimation of the global line orientation that we seek and represents equally oriented long lines at different gray levels.
finally serves as a “score” per orientation (Fig. 7a). This curve is finally smoothed using a Gaussian filter kernel (σ
H, Table 1). The highest peak in the filtered curve, Fig. 7a, gives the best estimation of the global line orientation that we seek and represents equally oriented long lines at different gray levels.
Fig. 6.
a Hough transform of pixels in a space–time diagram (see Fig. 7b) having the same gray level. The Hough count (H) is the number of pixels on a line with orientation angle φ at distance ρ from the origin. The orientation (φ) represents the angle between the vector normal to the line and the positive x-axis (Fig. 7b). b Result of thresholding, which accepts long lines exceeding 90% of the maximum Hough count in (a). c is obtained by adding the responses, as in (b), for all gray levels. The obtained result is less sensitive to noise or artifacts in the space–time diagram and preserves long lines out of the space–time diagram
Theoretical range of velocity assessment. The physical upper limit of velocity assessment depends on vessel length (L in μm) and video frame rate (f) of the CCD camera. Velocity measurements from slowly sampled scenes may be hampered by aliasing. It is theoretically possible to calculate RBC velocity from the space–time diagram if an object travels at constant velocity and is visible in only two successive frames. However, it is not possible to tell with certainty whether the cell object in the first frame is the same as in the second frame. If additional video frames show that the object moves with a rather constant displacement between successive frames, it is “more likely” that one and the same object is being observed. Therefore, a minimum of three frame intervals is chosen for determining the maximum physical velocity limit (vmax):
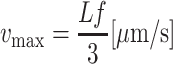 |
2 |
Space–time diagrams are often marked by horizontal lines as a result of dark spots at fixed locations, e.g. due to intersecting vessels (see peaks at φ = ±90° in Fig. 7a), or by vertical lines, e.g. due to periodic variations in illumination. Orientations that correspond to velocities above the physical limit, as described above, or below a given lower limit (v min, Table 1) are therefore rejected and require manual assessment, i.e. by tracing lines in the space–time diagram interactively.
References
- 1.Altman DG. Correlation—mathematics and worked examples. In: Altman DG, editor. Practical statistics for medical research. USA: Chapman & Hall; 1999. p. 293. [Google Scholar]
- 2.Biberthaler P, Langer S. Comparison of the new OPS imaging technique with intravital microscopy: analysis of the colon microcirculation. Eur Surg Res. 2002;24:124–128. doi: 10.1159/000048898. [DOI] [PubMed] [Google Scholar]
- 3.Bland JM, Altman DG. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet. 1995;346:1085–1087. doi: 10.1016/S0140-6736(95)91748-9. [DOI] [PubMed] [Google Scholar]
- 4.Boerma EC, Mathura KR, Van der Voort PHJ, Spronk PE, Ince C. Quantifying bedside-derived imaging of microcirculatory abnormalities in septic patients: a prospective validation study. Crit Car. 2005;9(6):R601–R606. doi: 10.1186/cc3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canny J. A computational approach to edge detection. IEEE Trans Pattern Anal PAMI. 1986;8(6):679–698. [PubMed] [Google Scholar]
- 6.Dadasch B, Harris AG, Kellam KR, Christ F (2001) Validation of an analysis software for intravital microscopy with OPS imaging applied to the hamster skinfold chamber. Proceedings of the 7th World Congress for Microcirculation, pp 3–16
- 7.De Backer D, Creteur J, Dubois MJ, Sakr Y, Koch M, Verdant C, Vincent JL. The effects of dobutamine on microcirculatory alterations in patients with septic shock are independent of its systemic effects. Crit Care Med. 2006;34(2):403–408. doi: 10.1097/01.CCM.0000198107.61493.5A. [DOI] [PubMed] [Google Scholar]
- 8.De Backer D, Creteur J, Preiser JC, Dubois MJ, Vincent JL. Microvascular blood flow is altered in patients with sepsis. Am J Respir Crit Care Med. 2002;166:98–104. doi: 10.1164/rccm.200109-016OC. [DOI] [PubMed] [Google Scholar]
- 9.De Backer D, Hollenberg S, Boerma C, Goedhart P, Buchele G, Ospina-Tascon G, Dobbe I, Ince C (2007) How to evaluate the microcirculation? Report of a round table conference. Crit Care 11:R101 (in press) [DOI] [PMC free article] [PubMed]
- 10.De Backer D, Verdant C, Chierego M, Koch M, Gullo A, Vincent JL. Effects of drotrecogin alfa activated on microcirculatory alterations in patients with severe sepsis. Crit Care Med. 2006;34(7):1918–1924. doi: 10.1097/01.CCM.0000220498.48773.3C. [DOI] [PubMed] [Google Scholar]
- 11.De Vriese AS, Verbeuren RJ, Vallez MO, Lameire MH, De Buyzere M, Vanhoutte PM. Off-line analysis of red blood cell velocity in renal arterioles. J Vasc Res. 2002;37:26–31. doi: 10.1159/000025710. [DOI] [PubMed] [Google Scholar]
- 12.Dobbe JGG, Streekstra GJ, Hardeman MR, Ince C, Grimbergen CA. Measurement of the distribution of red blood cell deformability using an automated rheoscope. Clin Cytom. 2002;50:313–325. doi: 10.1002/cyto.10171. [DOI] [PubMed] [Google Scholar]
- 13.Ellis CG, Ellsworth ML, Pittman RN, Burgess WL. Application of image analysis for evaluation of red blood cell dynamics in capillaries. Microvasc Res. 1992;44:214–225. doi: 10.1016/0026-2862(92)90081-Y. [DOI] [PubMed] [Google Scholar]
- 14.Genzel-Boroviczény O, Christ F, Glas V. Blood transfusion increases functional capillary density in de skin of anemic preterm infants. Pediatr Res. 2004;56(5):751–755. doi: 10.1203/01.PDR.0000141982.38959.10. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez RC, Woods RE. Global processing via the Hough Transform. In: Gonzalez RC, Woods RE, editors. Digital image processing. Massachusetts: Addison-Wesley; 1992. pp. 432–439. [Google Scholar]
- 16.Groner W, Winkelman JW, Harris A, Ince C, Bouma GJ, Messmer K, Nadeau RG. Orthogonal polarization spectral imaging: a new method for study of the microcirculation. Nature. 1999;5(10):1209–1213. doi: 10.1038/13529. [DOI] [PubMed] [Google Scholar]
- 17.Ince C. The microcirculation is the motor of sepsis. Crit Car. 2005;9(suppl 4):S13–S19. doi: 10.1186/cc3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Japee SA, Ellis CG, Pittman RN. Flow visualization tools for image analysis of capillary networks. Microcirculation. 2004;11:39–54. doi: 10.1080/10739680490266171. [DOI] [PubMed] [Google Scholar]
- 19.Japee SA, Pittman RN, Ellis CG. A new video image analysis system to study red blood cell dynamics and oxygenation in capillary networks. Microcirculation. 2005;12:489–506. doi: 10.1080/10739680591003332. [DOI] [PubMed] [Google Scholar]
- 20.Jähne B. Motion as orientation in space-time images. In: Jähne B, editor. Digital image processing. 6th revised and extended edn. Berlin: Springer; 2005. pp. 403–406. [Google Scholar]
- 21.Jähne B. The structure tensor. In: Jähne B, editor. Digital image processing. 6th revised and extended edn. Berlin: Springer; 2005. pp. 364–368. [Google Scholar]
- 22.Klyscz T, Jünger M, Jung F, Zeintl H. Cap Image—ein neuartiges computerunterstütztes Videobildanalysesystem für die dynamische Kapillarmikroskopie. Biomedizinische Technik Band 42 Heft. 1997;6:168–175. doi: 10.1515/bmte.1997.42.6.168. [DOI] [PubMed] [Google Scholar]
- 23.Langer S, Biberthaler P, Harris AG, Steinau HU, Messmer K. In vivo monitoring of microvessels in skin flaps: introduction of a novel technique. Microsurgery. 2001;21:317–324. doi: 10.1002/micr.1058. [DOI] [PubMed] [Google Scholar]
- 24.Lentner A, Berger F, Wienert V. Das “spatial shift alignment (SSA)”—eine neue Methode zur Bestimmung der Blutflussgeschwindigkeit in de Video-Kapillarmikroskopie. Biomedizinische Techniek Band 39 heft. 2002;7–8:170–175. doi: 10.1515/bmte.1994.39.7-8.170. [DOI] [PubMed] [Google Scholar]
- 25.Lindert J, Werner J, Redlin M, Kuppe H, Habazettl H, Pries AR. OPS imaging op human microcirculation: a short technical report. J Vasc Res. 2002;39:368–372. doi: 10.1159/000065549. [DOI] [PubMed] [Google Scholar]
- 26.Lo RC, Tsai WH. Gray-scale Hough transform for thick line detection in gray-scale images. Int J Pattern Recognit. 1995;28(5):647–661. doi: 10.1016/0031-3203(94)00127-8. [DOI] [Google Scholar]
- 27.Mathura KR, Bouma GJ, Ince C. Abnormal microcirculation in brain tumours during surgery. Lancet. 2001;358:1698–1699. doi: 10.1016/S0140-6736(01)06722-8. [DOI] [PubMed] [Google Scholar]
- 28.Mathura KR, Vollebregt KC, Boer K, De Graaff JC, Ubbink DT, Ince C. Comparison of OPS imaging and conventional capillary microscopy to study the human microcirculation. J Appl Physiol. 2001;91:74–78. doi: 10.1152/jappl.2001.91.1.74. [DOI] [PubMed] [Google Scholar]
- 29.Pennings FA, Bouma GJ, Ince C. Direct observation of the human cerebral microcirculation during aneurysm surgery reveals increased arteriolar contractility. Stroke. 2004;35:1284–1288. doi: 10.1161/01.STR.0000126039.91400.cb. [DOI] [PubMed] [Google Scholar]
- 30.Pries AR. A versatile video image analysis system for microcirculatory research. Int J Microcirc Clin Exp. 1988;7:327–345. [PubMed] [Google Scholar]
- 31.Russ JC. Histogram equalization. In: Russ JC, editor. The image processing handbook. 4th edn. Florida: CRC Press LLC; 2002. pp. 211–219. [Google Scholar]
- 32.Sakr Y, Dubois MJ, De Backer D, Creteur J, Vincent JL. Persistent microcirculatory alterations are associated with organ failure and death in patients with septic shock. Crit Care Med. 2004;32(9):1825–1831. doi: 10.1097/01.CCM.0000138558.16257.3F. [DOI] [PubMed] [Google Scholar]
- 33.Schaudig S, Dadasch B, Kellam KR, Christ F (2001) Validation of an analysis software for OPS-imaging used in humans. Proceedings of the 7th World Congress for Microcirculation, pp 2–59
- 34.Spronk PE, Ince C, Gardien MJ, Mathura KR, Oudemans-Van straaten HM, Zandstra DF. Nitroglycerin in septic shock after intravascular volume resuscitation. Lancet. 2002;360:1395–1396. doi: 10.1016/S0140-6736(02)11393-6. [DOI] [PubMed] [Google Scholar]
- 35.Spronk PE, Rommes JH, Schaar C, Ince C. Thrombolysis in fulminant purpura: observations on changes in microcirculatory perfusion during successful treatment. Thromb Haemost. 2006;95(3):576–578. doi: 10.1160/TH05-11-0746. [DOI] [PubMed] [Google Scholar]
- 36.Staal J, Abràmoff MD, Viergever MA, Van Ginneken B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans Med Imaging. 2004;23(4):501–509. doi: 10.1109/TMI.2004.825627. [DOI] [PubMed] [Google Scholar]
- 37.Steger C. An unbiased detector of curvilinear structures. IEEE Trans Pattern Anal. 1998;20(2):113–125. doi: 10.1109/34.659930. [DOI] [Google Scholar]
- 38.Ter Haar Romeny BM (2003) Anisotropy. In: Ter Haar Romeny BM (ed) Front-end vision and multi-scale image analysis: multi-scale computer vision theory and applications, written in Mathematica, Kluwer, Dordrecht, pp 48-49 and 74–48. ISBN: 1-4020-1507-0
- 39.Trzeciak S, Dellinger RP, Parrillo JE, Guglielmi M, Bajaj J, Abate NL, Arnold RC, Colilla S, Zanotti S, Hollenberg SM (2006) Early microcirculatory perfusion derangements in patients with severe sepsis and septic shock: relationship to hemodynamics, oxygen transport, and survival. Ann Emerg Med (in press) [DOI] [PubMed]