Abstract
Coagulation factor XI (FXI) is a covalent homodimer consisting of two identical subunits of 80 kDa linked by a disulfide bond formed by Cys-321 within the Apple 4 domain of each subunit. Because FXIC321S is a noncovalent dimer, residues within the interface between the two subunits must mediate its homodimeric structure. The crystal structure of FXI demonstrates formation of salt bridges between Lys-331 of one subunit and Glu-287 of the other subunit and hydrophobic interactions at the interface of the Apple 4 domains involving Ile-290, Leu-284, and Tyr-329. FXIC321S, FXIC321S,K331A, FXIC321S,E287A, FXIC321S,I290A, FXIC321S,Y329A, FXIC321S,L284A, FXIC321S,K331R, and FXIC321S,H343A were expressed in HEK293 cells and characterized using size exclusion chromatography, analytical ultracentrifugation, electron microscopy, and functional assays. Whereas FXIC321S and FXIC321S,H343A existed in monomer/dimer equilibrium (Kd ∼ 40 nm), all other mutants were predominantly monomers with impaired dimer formation by analytical ultracentrifugation (Kd = 3–38 μm). When converted to the active enzyme, FXIa, all the monomeric mutants activated FIX similarly to wild-type dimeric FXIa. In contrast, these monomeric mutants could not be activated efficiently by FXIIa, thrombin, or autoactivation in the presence of dextran sulfate. We conclude that salt bridges formed between Lys-331 of one subunit and Glu-287 of the other together with hydrophobic interactions at the interface, involving residues Ile-290, Leu-284, and Tyr-329, are essential for homodimer formation. The dimeric structure of FXI is essential for normal proteolytic activation of FXI by FXIIa, thrombin, or FXIa either in solution or on an anionic surface but not for FIX activation by FXIa in solution.
Factor XI (FXI),3 the zymogen form of a serine protease coagulation enzyme that is essential for normal hemostasis, is activated either by FXIIa or by thrombin or by autoactivation (1, 2). Once converted to FXIa, the enzyme recognizes its natural macromolecular substrate, FIX, the Ca2+-dependent activation of which requires the exposure of a substrate-binding site within the Apple 2 (A2) and/or Apple 3 (A3) domains of FXIa and the γ-carboxyglutamic acid domain of FIX, as well as an extended, macromolecular substrate-binding exosite in the protease domain of FXIa (3–9). The activation of FIX to FIXaβ involves two cleavages by FXIa, one after Arg-145 and the other after Arg-180, thereby releasing an 11-kDa activation peptide (3, 4, 10). FIX is also activated to FIXaβ by the tissue factor-FVIIa complex (11).
FXI and plasma prekallikrein (PK) are 58% identical in their amino acid sequences, and the domain structures of the two molecules are very similar, with each molecule containing four homologous apple (A1–A4) domains (12). The high homology between the heavy chain of FXI and PK indicates a common origin of these two zymogens, in contrast to FXII and other coagulation factors (13, 14). However, FXI is a homodimer of two identical subunits joined by a disulfide bond formed by Cys-321 within the A4 domain of each subunit, whereas PK exists as a monomer (15, 16). Cys-321 in PK forms an intrachain disulfide bond with Cys-326, whereas Cys-326 in FXI is a Gly (12). If the interchain disulfide bond formed by Cys-321 in FXI is the only site responsible for homodimer formation, then FXIC321S or FXIC321A should be monomeric; however, both of these mutants exist predominantly as dimers (15, 17), strongly suggesting that other noncovalent interactions are also important for maintaining the dimeric structure of FXI. The fact that FXI is a homodimer composed of two identical subunits, whereas PK is a monomer, also suggests that among three possible pathways for the evolution of a dimer (18), the FXI dimer might have evolved from mutations of surface residues of an ancestral monomer. Thus it is reasonable to speculate that it would be possible to reconstitute stable monomeric FXI by replacing the residues that, according to the crystal structure (19), are present at the dimer interface.
We therefore aimed to evaluate the relative contributions of selected residues within the A4 domain to dimer formation and to understand the importance of the dimeric structure of FXI to its normal function. After examining the crystal structure of FXI (19) and comparing the amino acid sequence of FXI and PK (12), we were able to predict the candidate residues (Leu-284, Glu-287, Ile-290, Tyr-329, and Lys-331) within the A4 domain that may be involved in homodimer formation. These FXI A4 domain residues were mutated and the mutant proteins expressed in HEK293 cells, and the purified proteins were examined by size exclusion chromatography, analytical ultracentrifugation, and electron microscopy for their capacity to mediate homodimer formation and for their functional properties.
EXPERIMENTAL PROCEDURES
Expression of Factor XI Mutants—All the mutants (FXIC321S, FXIC321S,K331A, FXIC321S,E287A, FXIC321S,I290A, FXIC321S,Y329A, FXIC321S,L284A, FXIC321S,K331R, and FXIC321S,H343A) were constructed using the QuikChange® site-directed mutagenesis kit II (Stratagene, La Jolla, CA). Wild-type (WT) FXI cDNA inserted into pJVCMV vector (kindly provided by Dr. David Gailani, Vanderbilt University, Nashville, TN) served as the template in all our reactions. The plasmid constructs were cotransfected into HEK293 cells (ATCC, Manassas, VA) with pCMV neo vector (which conferred G418 resistance) by Lipofectamine 2000 (Invitrogen), and the clones selected against 800 μg/ml G418 were further screened using a FXI enzyme-linked immunosorbent assay kit (Enzyme Research Laboratories Inc, South Bend, IN) to confirm FXI expression by these clones. The clones with highest protein yield were transferred to 2-liter roller bottles and maintained in Dulbecco's modified Eagle's medium (Cellgro, Herndon, VA) containing 10% fetal bovine serum, 0.5 units/ml penicillin, and 0.5 mg/ml streptomycin until cells reached 90% confluence. For the expression of recombinant proteins, the serum-containing medium was replaced by serum-free Dulbecco's modified Eagle's medium supplemented with 10 μg/ml lima bean trypsin inhibitor, 10 μg/ml soybean trypsin inhibitor (Sigma), insulin (10 mg/liter), transferrin (5.5 mg/liter), and selenium (6.7 μg/ml) (the latter three from Invitrogen). The medium was collected and filtered through 0.22-μm membranes before loading onto an affinity column packed with FXI monoclonal (5F7) antibody conjugated to cyanogen bromide-activated Sepharose beads. The column was washed with TBS buffer (100 mm NaCl, 25 mm Tris-HCl, pH 7.4) containing 5 mm benzamidine, and the bound recombinant proteins were eluted with 2 m potassium thiocyanate buffer. The eluted proteins were concentrated with Centricon columns (molecular weight cutoff of 10,000, Millipore, Billerica, MA), and dialyzed in Slide-A-Lyzer dialysis cassettes (molecular weight cutoff of 10,000, Pierce) against TBS buffer (NaCl 150 mm, Tris-HCl 50 mm, pH 7.4) overnight. Protein concentrations of purified proteins were determined by BCA assay (Pierce), and the proteins were assessed for purity by size fractionation on SDS-PAGE and visualizing the bands using Coomassie Brilliant Blue.
Size Exclusion Chromatography—Size exclusion chromatography was performed using a Superose 12 column (GE Healthcare) fitted to an AKTA UPC-90 FPLC workstation (GE Healthcare). Each sample (100 μl, at a concentration of 200 μg/ml) was loaded on the column and fractionated at a flow rate of 0.5 ml/min. Elution profiles and retention times were obtained from the absorbance values at 280 nm. Plasma FXI and PK served as references for dimeric and monomeric proteins, respectively.
Analytical Ultracentrifugation Analysis—WTFXI, PK, and the expressed mutants were subjected to equilibrium sedimentation analysis on a Beckman XLA ultracentrifuge utilizing six-sector cells in an eight-cell rotor. In each run the plasma FXI and PK in TBS served as reference samples. The sample cell was loaded with 100 μl of FXI, PK, or purified mutants and the reference cell with TBS buffer. Three rotor speeds (12,000, 15,000, and 20,000 rpm) and a total run time of 20–24 h were employed for each sample. Molar extinction values used for FXI subunit and PK were 83,780 and 92,030, respectively. Partial specific volumes (v̄) were calculated from the sequences using “Sednterp” (20) and adjusted (21) for glycosylation levels of 5% for FXI and FXI mutants and 15.5% for PK (12). The pKd value is defined as the negative base 10 logarithm of the monomer-dimer dissociation constant in molar units.
Electron Microscopy—Samples were prepared by conventional negative staining with 0.75% (w/v) uranyl formate as described before (22). Images were collected with a Tecnai T12 electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 filament and operated at an acceleration voltage of 120 kV. Images were recorded on imaging plates at a nominal magnification of ×67,000 and a defocus value of –1.5 μm using low dose procedures. Imaging plates were read out with a Ditabis micron imaging plate scanner (DITABIS Digital Biomedical Imaging System AG, Pforzheim, Germany) using a step size of 15 μm, a gain setting of 20,000, and a laser power setting of 30%, 2 × 2 pixels were averaged to yield a pixel size of 4.5 Å on the specimen level.
Image Processing—Using BOXER, the display program associated with the EMAN software package (23), 4,269 particles were interactively selected from 29 images of wild-type FXI (WTFXI), 4,150 particles from 17 images of the mutant FXIC321S,K331A, and 4,601 particles from 12 images of PK. The WTFXI particles were windowed into 64 × 64 pixel images and the FXIC321S,K331A and PK particles into 50 × 50 pixel images. Using the SPIDER software package (24), the particles were rotationally and translationally aligned and subjected to 10 cycles of multireference alignment. Each round of multireference alignment was followed by k means classification specifying 50 output classes. The references used for the first multireference alignment were randomly chosen from the raw images.
Clotting Assay—The specific clotting activities of plasma FXI and all the mutants were determined based on an activated partial thromboplastin time (APTT) assay utilizing FXI-deficient plasma and commercial APTT reagent (Sigma). The mutants were mixed with FXI-deficient plasma and incubated with APTT reagent for 2 min, and the clotting time was obtained after the addition of 5 mm CaCl2. Standard curves were generated with serially diluted normal pooled plasma.
Activation of Factor XI—Wild-type and plasma FXI and the mutants (30 nm, determined according to the molecular weight of wild-type dimer) were activated by FXIIa (3 nm, Enzyme Research Laboratories Inc., South Bend, IN) in a molar ratio of 10:1 at 37 °C in TBSA buffer (NaCl 150 mm, Tris-HCl 50 mm, pH 7.4, 0.1% BSA) either in the absence or presence of dextran sulfate (500 kDa, 1 μg/ml). At different time points, the aliquots of samples were taken into the 96-well plates, and the FXIIa was inhibited by corn trypsin inhibitor (30 nm, Enzyme Research Laboratories Inc, South Bend, IN). The generation of FXIa was determined by its capacity to cleave the synthetic chromogenic substrate S2366 (330 μm, Chromogenix, Milano, Italy), and the standard curve was plotted against serially diluted purified FXIa (Enzyme Research Laboratories Inc.). The activation of FXI mutants by higher concentrations of FXIIa (6 and 30 nm) was performed in similar manner.
The activation of FXI by α-thrombin was measured at 37 °C in the presence of dextran sulfate (500 kDa, 1 μg/ml) with a molar ratio of 30:1, FXI to thrombin. The samples were aliquoted at different time points, and the remaining thrombin was inhibited by hirudin (5 nm). The conversion of FXI to FXIa was measured by its ability to cleave S2366 (330 μm). The autoactivation of FXI and the mutants in the presence of a negatively charged surface was investigated by incubating them with dextran sulfate (500 kDa, 1 μg/ml) for 0–60 min at 37 °C, and conversion to FXIa was measured as described above by cleavage of S2366 (330 μm).
Activation of Factor IX by Factor XIa and the Activated Factor XI Mutants—The WTFXI and FXI mutants were activated by incubating them with FXIIa in a molar ratio of 1:2 in TBS buffer (NaCl 50 mm, Tris-HCl 50 mm, pH 7.4) at 37 °C for 1 h, and the FXIIa was removed by corn trypsin inhibitor-conjugated Sepharose-4 beads. The activated proteins were examined by SDS-PAGE under reducing conditions and visualized by Western blot using anti-FXI polyclonal antibody (Enzyme Research Laboratories Inc.). FIX (400 nm) was incubated with FXIa or activated mutants (2 nm, i.e. molar ratio 200:1) in TBS buffer at 37 °C, and the reaction was stopped at different time points by boiling (3 min) in SDS buffer containing 10% β-mercaptoethanol. These samples were then analyzed by SDS-PAGE, and the gels were stained with Coomassie Blue.
RESULTS
Interactions at the Interface of the Two Subunits of FXI—The residues identified from the FXI crystal structure (19) as potential candidates to mediate FXI homodimer formation, other than Cys-321 forming a disulfide bond in WTFXI, are shown in Fig. 1 (A–C). Glu-287 of one subunit appears to form a salt bridge with Lys-331 of the other subunit (Fig. 1A), suggesting that two salt bridges formed between the two subunits may mediate dimer formation. Glu-287 is conserved in PK, whereas the residue at position 331 is an Arg. Although both Lys and Arg are positively charged, and the distance between Glu-287 of one subunit and Arg-331 of the other may not allow formation of a salt bridge. At the interface of the two subunits, hydrophobic interactions also appear to mediate homodimer interaction, including interactions between Ile-290 (Leu-290 in PK) of one subunit and Leu-284 (Gly-284 in PK) of the other (Fig. 1B) and between the two Tyr-329 (Phe-329 in PK) residues of each subunit (Fig. 1C). Leu-290 and Phe-329 in PK are also hydrophobic, but a Gly at position 284 may orient the backbone in a direction unfavorable for hydrophobic interaction at the interface of the two PK A4 subunits. Based on these structural observations, we have expressed in HEK293 cells and purified the following mutant proteins (in which Cys-321 was also mutated to serine to prevent covalent disulfide bond formation between the two A4 subunits): FXIC321S,K331A, FXIC321S,E287A, FXIC321S,I290A, FXIC321S,Y329A, FXIC321S,L284A, and FXIC321S,K331R. As an additional control, the amino acid residue His-343, which is near the FXI dimer interface in the crystal structure, but does not appear to interact with any residue in the opposite subunit, was also mutated to alanine to construct the double mutant FXIC321S,H343A. Each mutant was found to migrate as a single band of ∼80 kDa on SDS-PAGE under nonreducing conditions, whereas WTFXI and plasma FXI migrated with a molecular mass of ∼160 kDa (data not shown). In enzyme-linked immunosorbent assays using both monoclonal and polyclonal antibodies WTFXI, plasma FXI, and the mutants were indistinguishable (data not shown)
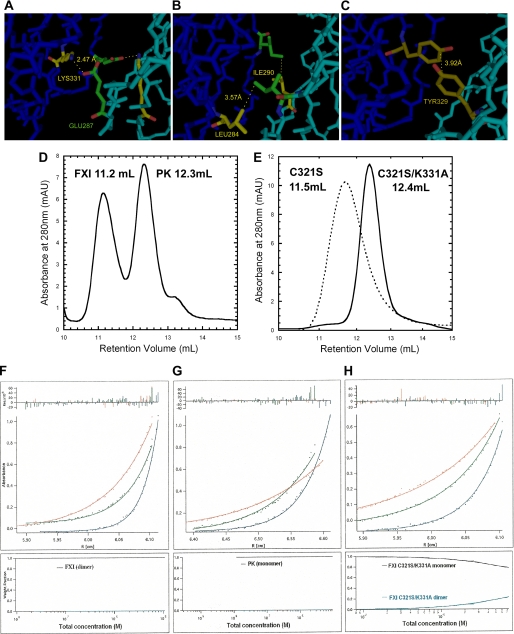
FIGURE 1.
Structure of FXI dimer interface and characterization of FXI mutants by size exclusion chromatography and analytical ultracentrifugation. A–C, structure of the FXI dimer interface based on the crystal structure of FXI (19) predicting a salt bridge between the positively charged Lys-331 residue of one subunit 2.47 Å away from the negatively charged Glu-287 residue on the opposite subunit (A); a hydrophobic interaction (3.57 Å) between Leu-284 and Ile-290 (B); and a hydrophobic interaction (3.92 Å) between the two Tyr-329 residues of the two A4 domains (C). D and E, size exclusion chromatography elution profiles, carried out as described under the “Experimental Procedures.” The retention volumes of purified plasma FXI and PK, 11.2 and 12.3 ml, were adopted as references for dimer and monomer, respectively (D). The mutant FXIC321S migrated with only one peak that had a retention volume, 11.5 ml, similar to that of the FXI dimer, whereas the FXIC321S,K331A mutant displayed a retention volume of 12.4 ml (E), i.e. similar to PK (D). All the other monomeric double mutants also migrated with one predominant peak that had the retention volume around that of PK even at a concentration of 200 μg/ml (data not shown). F–H, equilibrium sedimentation analysis, performed as described under the “Experimental Procedures” at 12,000 rpm (upper curve, red), 15,000 rpm (middle curve, green), and 20,000 rpm (lower curve, blue) for 20–24 h. The radial absorbance profiles (points) and fits (lines) to equations describing equilibrium sedimentation of FXI (F) and PK (G) are shown. The FXIC321S,K331A mutant was selected as a representative of all the mutants as shown in H. The bottom part of each panel showed the species plot of weight fraction against concentration. For FXI and PK, there was only one species (upper black curves); however, the mutant FXIC321S,K331A existed in the equilibrium of dimer (upper black curve) and monomer (lower blue curves), and the dimer species became dominant with the increment of concentration.
Size Exclusion Chromatography—Elution profiles of plasma FXI, PK, FXIC321S, and FXIC321S,K331A are shown in Fig. 1, D and E. Whereas the retention volume of FXIC321S (Fig. 1E) is very close to that of plasma FXI (Fig. 1D), the mutant FXIC321S,K331A (Fig. 1E) migrated as a monomer, with a retention volume similar to that of PK (Fig. 1D). The other five mutants FXIC321S,E287A, FXIC321S,I290A, FXIC321S,Y329A, FXIC321S,L284A, and FXIC321S,K331R migrated with retention volumes similar to that of FXIC321S,K331A (data not shown), i.e. as monomers, whereas the double mutant FXIC321S,H343A exhibited one predominant peak close to the single mutant FXIC321S, suggesting its existence as a noncovalent dimer (data not shown).
Analytical Ultracentrifugation Analysis—The equilibrium concentration distributions of all the mutant macromolecules as well as purified plasma FXI and WTFXI and PK were determined by sedimentation equilibrium experiments as shown in Fig. 1, F–H. Each panel of Fig. 1, F–H, shows radial absorbance profiles (points) and fits (lines) to equations describing equilibrium sedimentation (25). Molecular weights from curve fits were consistent with 100% dimer for WTFXI (Mr = 142,850, v̄ 0.736, see Fig. 1F), and 100% monomer for PK (Mr = 79,928, v̄ = 0.726, see Fig. 1G). All the mutants were in monomer-dimer equilibrium with different values of pKd. The equilibrium sedimentation profiles of mutant FXIC321S,K331A are shown in Fig. 1H; all the other monomeric mutants displayed similar equilibrium sedimentation profiles (data not shown). Consistent with the gel filtration data, the double mutants had much higher Kd values than the single mutant FXIC321S (Kd ∼ 40 nm, see Table 1). Among all the double mutants, the values of Kd, ranged from 3.4 to 37 μm, representing a loss of 1.36–2.78 kcal/mol of binding energy, compared with that of FXIC321S. Collectively, these data suggest that, at the physiological concentration of FXI, e.g. 30 nm, all these mutant proteins would exist almost entirely in monomeric form.
TABLE 1.
Analysis of analytical ultracentrifugation data for FXI mutants and APTT results
| Species | pKd | Kd | ΔGa | DΔGb | Clotting activity |
|---|---|---|---|---|---|
| μm | kcal/mol | kcal/mol | % | ||
| C321S | 7.46 | 0.04 | 8.81 | 0 | 85 ± 10 |
| C321S/L284A | 4.82 | 15 | 6.57 | -2.24 | 81 ± 6 |
| C321S/I290A | 5.03 | 9 | 6.86 | -1.95 | 80 ± 1 |
| C321S/Y329A | 5.46 | 3.4 | 7.45 | -1.36 | 83 ± 5 |
| C321S/E287A | 4.69 | 20 | 6.39 | -2.42 | 78 ± 13 |
| C321S/K331A | 4.41 | 38.6 | 6.01 | -2.8 | 72 ± 5 |
| C321S/K331R | 4.42 | 37 | 6.03 | -2.78 | 74 ± 8 |
ΔG is the Gibbs free energy of monomer-monomer binding, calculated from the values of Kd (ΔG = —RTln Kd), whereas DΔG refers to the difference in free energy between double mutants versus the single mutant FXI C321S.
Clotting activity of the mutants based on the APTT assay is expressed as the percentage of normal plasma FXI.
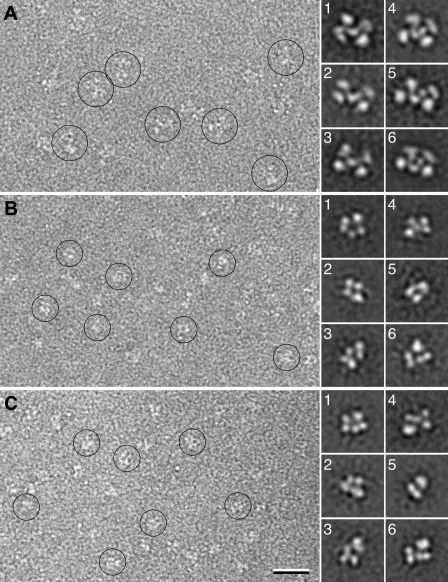
Electron Microscopy—To confirm the oligomeric state of wild-type FXI and FXIC321S,K331A, negatively stained samples were prepared and imaged in the electron microscope. The images of WTFXI showed mono-dispersed particles homogeneous in size (some particles circled in Fig. 2A), of which 4,269 particles were selected and classified into 50 classes (supplemental Fig. 1A). All the class averages have a very similar appearance, suggesting that the particles adsorbed to the EM grid in a single preferred orientation. The class averages revealed X-shaped particles, about 8.5 × 14 nm in size, consisting of four peripheral domains connected by a central domain (Fig. 2A, averages 1–3). In some cases the central domain was resolved into two smaller domains (Fig. 2A, averages 4–6). The small variations between the class averages may be due to slightly different stain embedding, subtle differences in the orientation of the particles on the grid, or somewhat flexible linkers between the various domains of the FXI molecule. Nevertheless, all class averages are consistent in size and shape with the FXI dimer seen in the x-ray structure (19).
FIGURE 2.
Electron microscopy of WTFXI, FXIC321S,K331A, and PK. A, raw image (left) and representative class averages (right, numbered 1–6) of WTFXI, showing the wild-type protein forming a dimer. B, raw image (left) and representative class averages (right, numbered 1–6) of FXIC321S,K331A, showing that the mutations prevent the protein from dimerizing. C, raw image (left) and representative class averages (right, numbered 1–6) of PK, revealing its structural similarity to FXIC321S,K331A. The scale bar represents 25 nm, and the side length of the panels showing individual class averages is 22.5 nm.
Images of negatively stained FXIC321S,K331A also showed a homogeneous particle population (some particles circled in Fig. 2B), but the particles appeared smaller than those seen in the images of WTFX1. Class averages obtained by classification of 4,150 particles into 50 classes (supplemental Fig. 1B) confirmed that the particles were indeed smaller, about 7 × 11.5 nm, indicating that the FXIC321S,K331A mutant does not dimerize. The class averages of FXIC321S,K331A (Fig. 2B, averages 1–6) show more structural variability compared with those of wild-type FXI. The most likely reason for this observation is that FXIC321S,K331A adsorbs to the grid in different orientations. The dimeric WTFXI molecule has an extended, flat shape, which causes it to adsorb to the grid in a preferred orientation, in which it has the largest interaction surface with the carbon film. In contrast, the monomeric FXIC321S,K331A molecule has a more globular shape and can thus adsorb to the grid in more orientations, giving rise to the different projection averages. This interpretation is supported by images of negatively stained PK (Fig. 2C), a molecule with high sequence homology to FXI but existing as a monomer. Fifty class averages obtained with 4,601 particles (supplemental Fig. 1C) show particles similar in size and shape to those observed with FXIC321S,K331A (compare averages 1–6 in Fig. 2C with the corresponding averages in Fig. 2B). These results thus support the notion that WTFXI is dimeric, whereas FXIC321S,K331A, like PK, is monomeric in solution.
Clotting Assay—All the mutants showed relatively normal clotting activity, ranging from 70 to 85% of WTFXI, with mutant FXIC321S,K331A having the lowest clotting activity, ∼70% (Table 1).
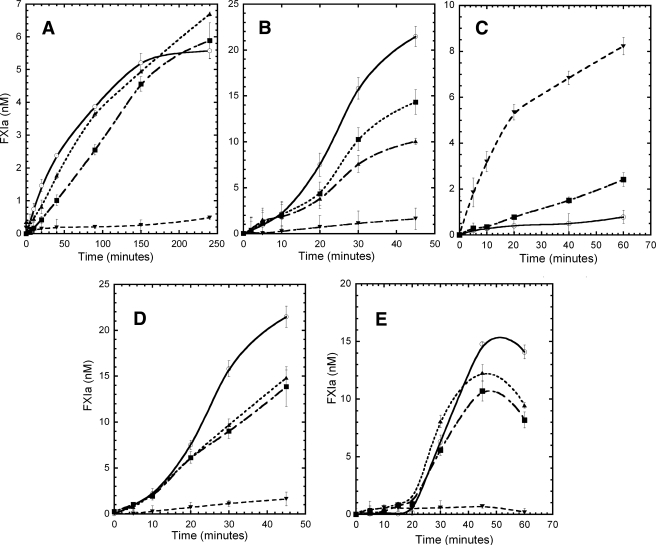
Activation of WTFXI and Monomeric Mutants—The monomeric mutants displayed markedly decreased rates of activation by FXIIa (Fig. 3, A and B), by thrombin (Fig. 3C), and by autoactivation in the presence of dextran sulfate (Fig. 3D). When FXI activation by FXIIa was examined in solution (i.e. the absence of dextran sulfate, Fig. 3A), the FXIC321S mutant (▪), which at 30 nm was at monomer-dimer equilibrium (Kd ∼ 40 nm), displayed initial activation rates approximately half of that observed for WTFXIa (○), a result expected for a protein that is 50% monomer and 50% dimer, if the dimeric structure is important for activation. The double mutant, FXIC321S,H343A (Fig. 3A, ▴), which displayed one predominant peak close to the single mutant FXIC321S (suggesting its existence as a protein in a noncovalent monomer-dimer equilibrium), was also activated at rates very similar to FXIC321S, suggesting that the mutation of His-343 to alanine did not disrupt either the dimeric structure or the functional properties of FXI. In contrast, the monomeric mutant FXIC321S,K331A (Fig. 3A, ▾) displayed markedly reduced rates of activation by FXIIa (Fig. 3A), as did all the other monomeric mutants, FXIC321S,E287A, FXIC321S,I290A, FXIC321S,Y329A, FXIC321S,L284A, and FXIC321S,K331R (data not shown). When the activation by FXIIa of WTFXI and the FXI mutants was examined in the presence of dextran sulfate, the rate of activation of WTFXI was increased by ∼10-fold, compared with the absence of dextran sulfate, but all the monomeric mutants displayed markedly impaired activation rates as they did in the absence of dextran sulfate (Fig. 3B).
FIGURE 3.
FXI activation by FXIIa and thrombin and autoactivation. A, activation of FXI and the mutants (30 nm) by FXIIa (3 nm) in solution were carried out in TBSA buffer (NaCl 150 mm, Tris-HCl 50 mm, pH 7.4, 0.1% BSA). The incubation mixtures were sampled at the indicated time points; the activity of FXIIa was inhibited by CTI, and the generation of FXIa was determined by its capacity to cleave the synthetic substrate S2366 (330 μm). B, activation of FXI and the mutants (30 nm) by FXIIa (3 nm) was carried out in the presence of 1 μg/ml of dextran sulfate (500 kDa) in TBSA buffer. C, FXIC321S,K331A mutant (30 nm) was activated by FXIIa at different enzyme:substrate molar ratios, 1:10 (○), 1:5 (▪), and 1:1 (▾). The reactions were carried out in TBSA buffer (NaCl 150 mm, Tris-HCl 50 mm,pH 7.4, 0.1% BSA). The FXIIa activity was inhibited by CTI as described under “Experimental Procedures,” and the generation of FXIa was determined by its capacity to cleave the synthetic substrate S2366 (330 μm). D, activation of WTFXI and mutants (30 nm) by thrombin (1 nm) was carried out in the presence of 1 μg/ml of dextran sulfate (500 kDa) in TBSA buffer. The thrombin was inhibited by addition of hirudin (5 nm) as described under “Experimental Procedures,” and the generation of FXIa was determined by its capacity to cleave the synthetic substrate S2366 (330 μm). E, autoactivation of FXI and mutants (30 nm) on dextran sulfate (1 μg/ml, 500 kDa) surface in TBSA buffer (NaCl 150 mm, Tris-HCl 50 mm, pH 7.4, 0.1% BSA). The incubation mixtures were sampled at the indicated time points, and the generation of FXIa was determined by its capacity to cleave the synthetic substrate S2366 (330 μm). Data are shown for activation of WTFXI (○), FXIC321S (▪), FXIC321S,H343A (▴), and FXIC321S,K331A (▾). Results (not shown) for the monomeric mutants FXIC321S,E287A, FXIC321S,I290A, FXIC321S,Y329A, and FXIC321S,L284A were virtually identical to those for FXIC321S,K331A.
When the monomeric mutant, FXIC321S,K331A, was incubated in the absence of dextran sulfate with higher concentrations of FXIIa, the results showed that the mutant could be activated by FXIIa in solution at rates (Fig. 3C) similar to those observed with WTFXI (Fig. 3A), provided a high enough concentration of FXIIa was employed (i.e. at a sufficiently high molar ratio of enzyme to substrate). Thus the rate of activation of FXIC321S,K331A by 30 nm FXIIa (Fig. 3C) was similar to the rate of activation of WTFXI by 3 nm FXIIa (Fig. 3A) in solution. This result may help to explain the relatively normal results in the clotting assay of the monomeric mutants, because it has been estimated that during the preincubation phase of the APTT assay, almost all plasma FXII (300 nm) is converted to activated FXII (FXIIa) (26).
When FXI activation by thrombin in the presence of dextran sulfate was examined (Fig. 3D), the dissociable mutant FXIC321S displayed activation rates ∼55% of those of the wild-type protein as did the control protein FXIC321S,H343A, whereas the monomeric mutant FXIC321S,K331A was activated by thrombin at a rate only ∼4% that of WTFXI (Fig. 3D). Almost identical results were obtained with the other monomeric mutants (FXIC321S,E287A, FXIC321S,I290A, FXIC321S,Y329A, FXIC321S,L284A, and FXIC321S,K331R; data not shown). Finally, when autoactivation experiments were carried out to examine FXI activation in the presence of dextran sulfate and in the absence of any exogenously added protease, after a lag time of ∼20 min, WTFXI was rapidly activated to generate fully activated FXIa over the subsequent ∼20 min (Fig. 3E) as demonstrated previously (1, 2). Similar rates of activation of FXIC321S and FXIC321S,H343A were observed, whereas in marked contrast, the monomeric mutant FXIC321S,K331A (Fig. 3E) remained virtually unactivated over a 60-min incubation as did all the other monomeric mutants (FXIC321S,E287A, FXIC321S,I290A, FXIC321S,Y329A, FXIC321S,L284A, and FXIC321S,K331R; data not shown).
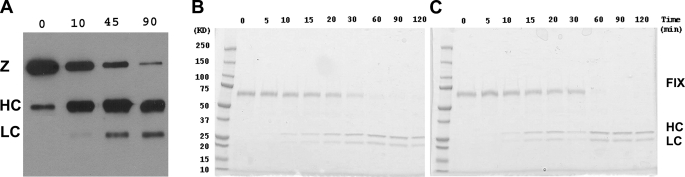
Factor IX Activation by Wild-type Factor XIa and Factor XIa Mutants—The activation of the monomeric mutant FXIC321S,K331A (200 nm) by FXIIa (100 nm) was monitored with SDS-PAGE and Western blot analysis, demonstrating >90% proteolytic cleavage of the monomeric mutant provided a sufficiently high molar ratio of enzyme to substrate was utilized (Fig. 4A), confirming the results depicted in Fig. 3C. Utilizing fully activated WTFXIa and monomeric mutant FXIa molecules, rates of activation of FIX by plasma or WTFXIa (molar ratio 200:1) and by each of the activated mutants were assessed by examining time courses of proteolytic cleavage of FIX by SDS-PAGE in the presence of 10% β-mercaptoethanol. Very similar rates of FIX activation were observed for WTFXIa (Fig. 4B) and for the monomeric mutant, FXIaC321S,K331A (Fig. 4C), reflecting similar rates of cleavage of FIX at both scissile bonds (at Arg-145 and at Arg-180) by the dimeric and monomeric proteins. Each of the other mutant proteins (FXIaC321S,K331A, FXIaC321S,E287A, FXIaC321S,I290A, FXIaC321S,Y329A, and FXIaC321S,L284A) activated FIX at similar rates and cleavage patterns (data not shown), strongly suggesting that all mutant proteins were properly folded. These data also suggest that normal rates of FIX activation by FXIa in solution do not require the dimeric structure of FXIa.
FIGURE 4.
FIX activation by FXIa mutants. A, activation of the monomeric mutant FXIC321,K331A (200 nm) by FXIIa (100 nm) was carried out in TBS buffer (NaCl 150 mm, Tris-HCl 50 mm, pH 7.4), and the conversion of zymogen to enzyme at different time points was examined by SDS-PAGE under reducing conditions. The conversion of FXI (80 kDa) to FXIa (50 kDa heavy chain, HC, and 30 kDa light chain, LC) was detected by Western blot with anti FXI polyclonal antibody. B and C, activation of FIX (400 nm) by FXIa or activated monomeric mutant (2 nm) was carried out in TBS buffer (NaCl 150 mm, Tris-HCl 50 mm, pH 7.4) in the presence of 2 mm CaCl2. At the indicated time point samples were removed, and the reaction was stopped by adding 10% SDS buffer containing 10% β-mercaptoethanol. The samples were subjected to SDS-PAGE after boiling for 3 min. Representative data are shown for WTFXIa (B) and the activated mutant FXIaC321S,K331A (C). Very similar results were obtained with all other mutants, including FXIaC321S,H343A, FXIaC321S,E287A, FXIaC321S/I290A, FXIaC321S,Y329A, and FXIaC321S,L284A.
DISCUSSION
It is well established that the A4 domain of FXI mediates dimer formation between the two identical polypeptide chains comprising FXI through disulfide bond formation at Cys-321 (12, 15, 27–30). The FXI dimer interface of each subunit is 567 Å2, which is about 2.24% of the overall surface area of the FXI monomer (25,312 Å2) and considerably smaller than that of most other dimeric proteins (3–44%) (31). Interestingly, the C321A mutant is apparently dimeric, indicating that other noncovalent interactions taking place at the interface are sufficient to promote dimer formation in the endoplasmic reticulum and the Golgi. The FXI crystal structure shows both electrostatic and hydrophobic interactions in the dimer interface. The studies presented here demonstrate that the salt bridges formed by Lys-331 of one A4 subunit and Glu-287 of the other, together with the hydrophobic interactions of residues Ile-290, Tyr-329, and Leu-284, are essential for FXI homodimer formation (Fig. 1). These observations are consistent with the results of a survey of 136 homodimeric proteins, which demonstrated that the majority of homodimeric proteins are “stabilized by a combination of small hydrophobic patches, polar interactions, and a considerable number of bridging water molecules” (32). They are also consistent with the results of studies of 122 dimer interfaces (31), which showed that four residues (Leu, Ile, Val, and Met) together contribute ∼25% of dimer interface interactions. Thus the hydrophobic interactions between the FXI subunits are shown here to be mediated predominantly by interactions between Leu-284 of one monomer and Ile-290 of the other, in addition to interactions of Tyr-329 of one subunit with the corresponding residue of the other subunit. Previous studies of the noncovalent dimerization of the isolated A4 domain showed that when the disulfide bond between the two monomers was eliminated by mutation of Cys-321, dimerization is highly dependent upon the salt concentration (27), suggesting that electrostatic interactions between charged residues such as Lys-331 and Glu-287 might play a relatively important role in noncovalent dimerization of FXI. Although the charged residues Lys and Glu are less frequently involved in other dimer subunit interactions (31), our present studies (Fig. 1 and Table 1) demonstrate the importance of interactions of Lys-331 with Glu-287 on the opposite subunit, because the two residues are within 2.47 Å distance from one another, consistent with the formation of a salt bridge across the dimer interface between the A4 subunits. The introduction of a K331A mutation on the C321S mutant leads to a dramatic decrease of affinity between the two subunits of FXI equal to 2.8 kcal/mol of binding energy (Table 1), indicating its important role in noncovalent dimer formation.
The conclusion that the mutant FXIC321S,K331A exists in a monomeric form is strongly supported by the EM images (Fig. 2 and supplemental Fig. 1), which are very similar to those of (monomeric) PK and distinctly different from the wild-type (dimeric) FXI. Moreover, it is especially interesting to compare these ultrastructural images of dimeric WTFXI with the x-ray crystal structure of the full-length FXI zymogen (19), because the domain organization and overall shape of the molecule are remarkably similar in these two depictions, both of which show a central V-shaped domain clearly (crystal structure) or likely (EM averages) representing the two A4 domains exhibiting 2-fold molecular symmetry. Associated with each A4 domain in the EM images are two globular domains, which by reference to the crystal structure (19) most likely represent the remaining apple (A1–A3) domains and the catalytic domains of each monomer. Although it is not possible to distinguish which of these two globular domains represents the catalytic domain and which represent the apple domains, the EM images are consistent with the conclusion that the two catalytic domains of the dimer are oriented in opposite directions.
The fact that rabbit FXI circulates in plasma as a noncovalent dimer is explained by the fact that a histidine residue in rabbit FXI replaces the Cys-321 that forms the interchain disulfide linkage in human FXI (17, 33, 34). Moreover, rabbit FXI has functional properties that are identical to those of human FXI, and the two proteins are 87% identical in sequence, with complete conservation of all the A4 domain residues (Leu-284, Glu-287, Ile-290, Tyr-329, and Lys-331) identified in this study to mediate noncovalent dimer formation. This strongly supports the view (15) that covalent stabilization of the FXI dimer via the disulfide bond mediated by the Cys-321 residues of the two A4 domains is not a prerequisite either for FXI dimerization or for normal FXI function. The studies reported here demonstrate that when covalent dimer formation is precluded by mutation of Cys-321 to serine, the resulting noncovalent dimeric FXI molecule dissociates to monomer with a Kd value of ∼40 nm (Table 1). This value is very close to that previously determined (Kd ∼ 36 nm) for an FXI mutant (G326C) in which an intrachain disulfide bond was formed between Cys-321 and Cys-326 as in PK, a monomeric protein with 58% sequence identity to FXI, thereby forming a dissociable dimer (35). This value is also very close to that previously determined (Kd ∼ 45 nm) for the isolated A4 domain of FXI (29). These results provide the basis for a high level of confidence that the totality of the binding energy mediating noncovalent dimer formation of FXI resides within the A4 domain, as indicated by x-ray crystallography (19, 29, 36).
The fact that FXI is unique in its homodimeric structure among coagulation proteins has led to studies of the functional relevance of dimerization. For example, it has been suggested that impaired intracellular dimerization of FXI can lead to impaired secretion, because a common mutant FXI (F283L results in a defect in dimerization of the molecule (rather than a decrease in the rate of mRNA transcription), leading to accumulation of non-native forms of FXI and decreased secretory rates (30). Because our structural studies (19, 29) indicate that the Phe-283 side chain does not directly participate in the dimer interface, and because the F283L mutation leads to a 4-fold increase in the Kd value for dimerization and a 1 kcal/mol stabilization of the monomer (29), we have suggested that the F283L mutation “leads to increased dimer dissociation by stabilizing a monomeric state with altered side chain packing that is unfavorable for homodimer formation.”
This study provides the tools required for addressing interesting questions concerning the functional significance of the homodimeric structure of FXI, unique among coagulation proteins. For example, studies of the metabotropic glutamate receptor, mGlu5, which is also a dimer mediated by both covalent and noncovalent interactions, demonstrated that the noncovalent dimeric mutant was fully functional compared with the wild-type dimer, suggesting that the covalent interaction is not required for the normal function of this dimeric protein (37). Thus, in addition to determining the major A4 domain residues that mediate noncovalent homodimer formation, this study provides us with mutant FXI molecules that exist mainly in monomeric form at physiological concentrations. The observation that all the monomeric FXIa molecules studied were able to cleave the two scissile bonds of FIX at rates equivalent to those cleaved by WTFXIa (Fig. 4B) strongly suggests that the mutations introduced did not result in protein misfolding. It has been suggested that since the proteolytic activation of FIX by FXIa occurs without the release of an intermediate, dimeric FXIa is required for positioning of both its active sites for simultaneous cleavage of the two scissile bonds in FIX (38). In conflict with this possibility is the observation in this study that monomeric FXIa activates FIX by cleavage of both scissile bonds at the same rate as dimeric FXIa (Fig. 4B). These results are similar to those previously reported (28) in which a monomeric FXIa chimera (with the A4 domain of PK in place of the FXI A4 domain) activated FIX in solution at rates similar to those observed for dimeric WTFXIa. These studies utilizing a monomeric version of FXI (FXI/PKA4) compared with a dimeric version of FXI/PKA4 (FXI/PKA4-Gly-326) as a control also demonstrated impaired rates of FIX activation in the presence of activated platelets, suggesting a novel model for FIX activation in which FXIa binds to activated platelets by one chain of the dimer, while binding to FIX through the other (28). Subsequently, it was shown that a dissociable dimeric FXIa mutant (G326C) activated FIX by cleavage of both its scissile bonds at rates similar to those observed with WTFXIa both in solution and in the presence of activated platelets (35), demonstrating that a preformed FXIa dimer is not required for normal FIX activation. Very recent studies by Smith et al. (39) are in agreement with these results and demonstrate conclusively that each monomeric subunit of the FXIa dimer behaves as an independent enzyme in FIX activation. Similar conclusions have been drawn for other dimeric proteins, such as Drosophila 14-3-3ζ (D14-3-3ζ), a dimeric G-protein-coupled receptor protein (40), and the Ca2+ receptor (41). In both cases, the monomeric subunit of the constitutive dimer could function as well as the wild-type dimer. Similar results have been obtained with the dimeric enzyme alkaline phosphatase, in which amino acids substitutions at the interface disrupt dimerization but have no significant effect on the kinetics of substrate under physiological conditions (42). In contrast the dimeric structure of the enzyme, ornithine decarboxylase, is required for normal function because constituents from both subunits are required for formation of the enzyme active site at the dimer interface (43).
The monomeric proteins generated in this study were also utilized to address important questions concerning the role of FXI dimeric structure in the mechanism of activation of FXI by FXIIa, thrombin, and FXIa in solution and on a dextran sulfate surface. The results of these studies, presented in Fig. 3, demonstrate that all the monomeric mutant FXI molecules prepared and characterized in this study demonstrated profound decreases in rates of activation by FXIIa both in the absence (Fig. 3A) and in the presence of a dextran sulfate surface (Fig. 3B), and by thrombin (studied only in the presence of dextran sulfate, Fig. 3D). In addition, whereas WTFXI, after a 20-min lag, underwent rapid rates of autoactivation, as reported previously (1, 2), the monomeric mutant, FXIC321S,K331A (Fig. 3E), and all the other monomeric mutants (not shown), were refractory to autoactivation. By comparison, the dissociable noncovalent dimer, FXIC321S, and the control (normally functioning and noncovalent dimer) protein, FXIC321S,H343A, were activated by FXIIa and thrombin at rates ∼50% of those observed with WTFXI, possibly reflecting their concentrations (30 nm), which were close to the equilibrium dissociation constant for noncovalent dimer-monomer equilibrium (∼40 nm) as determined herein, at which ∼50% of the protein would be monomeric.
These interesting results are possibly subject to rational interpretation based on the crystal structure of FXI (19), from which it was suggested that FXI activation might proceed via a transactivation mechanism, whereby thrombin bound through exosite I to the FXI A1 domain of one subunit (44, 45) cleaves the scissile bond of the opposite subunit (i.e. in trans). Thus, the results of this study could be rationalized if the enzymes that activate FXI, FXIIa, which binds to the A4 domain (46), and thrombin, which binds to the A1 domain (44), utilize one subunit to bind to the substrate, FXI, and cleave the scissile bond in the opposite subunit. A similar transactivation mechanism could explain autoactivation, whereby the binding of FXI to dextran sulfate, after a lag period, results in the formation of FXIa in one subunit that can then cleave the scissile bond in the opposite subunit (1, 2). In support of this possibility is the recent demonstration that the conversion of FXI to FXIa proceeds through an intermediate with a single active site present in one of the two subunits (39). Although this transactivation hypothesis is speculative, it would explain our present observation that the monomeric mutants are refractory to autoactivation and activation by thrombin and FXIIa, because the enzymes required for proteolytic cleavage of FXI (thrombin, FXIIa, and FXIa) are positioned on a single monomer and may fail to position their active sites in close proximity to the scissile bond, because the opposite monomer is missing.
Supplementary Material
Acknowledgments
The molecular EM facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained by National Institutes of Health Grant GM62580 (to S. C. Harrison).
This work was supported, in whole or in part, by National Institutes of Health Grants HL46213 and HL74124 (to P. N. W.) and HL40387 (to J. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: FXI, factor XI; An, Apple n; PK, prekallikrein; WT, wild-type; APTT, activated partial thromboplastin time; BSA, bovine serum albumin.
References
- 1.Naito, K., and Fujikawa, K. (1991) J. Biol. Chem. 266 7353–7358 [PubMed] [Google Scholar]
- 2.Gailani, D., and Broze, G. J., Jr. (1991) Science 253 909–912 [DOI] [PubMed] [Google Scholar]
- 3.Fujikawa, K., Legaz, M. E., Kato, H., and Davie, E. W. (1974) Biochemistry 13 4508–4516 [DOI] [PubMed] [Google Scholar]
- 4.Di Scipio, R. G., Kurachi, K., and Davie, E. W. (1978) J. Clin. Investig. 61 1528–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha, D., Seaman, F. S., and Walsh, P. N. (1987) Biochemistry 26 3768–3775 [DOI] [PubMed] [Google Scholar]
- 6.Baglia, F. A., Jameson, B. A., and Walsh, P. N. (1991) J. Biol. Chem. 266 24190–24197 [PubMed] [Google Scholar]
- 7.Sun, Y., and Gailani, D. (1996) J. Biol. Chem. 271 29023–29028 [DOI] [PubMed] [Google Scholar]
- 8.Sun, M. F., Zhao, M., and Gailani, D. (1999) J. Biol. Chem. 274 36373–36378 [DOI] [PubMed] [Google Scholar]
- 9.Sinha, D., Marcinkiewicz, M., Navaneetham, D., and Walsh, P. N. (2007) Biochemistry 46 9830–9839 [DOI] [PubMed] [Google Scholar]
- 10.Lindquist, P. A., Fujikawa, K., and Davie, E. W. (1978) J. Biol. Chem. 253 1902–1909 [PubMed] [Google Scholar]
- 11.Osterud, B., and Rapaport, S. I. (1977) Proc. Natl. Acad. Sci. U. S. A. 74 5260–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujikawa, K., Chung, D. W., Hendrickson, L. E., and Davie, E. W. (1986) Biochemistry 25 2417–2424 [DOI] [PubMed] [Google Scholar]
- 13.Veloso, D., Shilling, J., Shine, J., Fitch, W. M., and Colman, R. W. (1986) Thromb. Res. 43 153–160 [DOI] [PubMed] [Google Scholar]
- 14.Jiang, Y., and Doolittle, R. F. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMullen, B. A., Fujikawa, K., and Davie, E. W. (1991) Biochemistry 30 2056–2060 [DOI] [PubMed] [Google Scholar]
- 16.Meijers, J. C., Mulvihill, E. R., Davie, E. W., and Chung, D. W. (1992) Biochemistry 31 4680–4684 [DOI] [PubMed] [Google Scholar]
- 17.Sinha, D., Marcinkiewicz, M., Gailani, D., and Walsh, P. N. (2002) Biochem. J. 367 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu, D., Tsai, C. J., and Nussinov, R. (1998) Protein Sci. 7 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papagrigoriou, E., McEwan, P. A., Walsh, P. N., and Emsley, J. (2006) Nat. Struct. Mol. Biol. 13 557–558 [DOI] [PubMed] [Google Scholar]
- 20.Laue, T., Shaw, B. D., Ridgeway, T. M., and Pelletier, S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding, S. E., Rowe, A. J., and Horton, J. C., eds) pp. 90–125, Royal Society of Chemistry, Cambridge, UK
- 21.Fairman, R., Fenderson, W., Hail, M. E., Wu, Y., and Shaw, S. Y. (1999) Anal. Biochem. 270 286–295 [DOI] [PubMed] [Google Scholar]
- 22.Ohi, M., Li, Y., Cheng, Y., and Walz, T. (2004) Biol. Proced. Online 6 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludtke, S. J., Baldwin, P. R., and Chiu, W. (1999) J. Struct. Biol. 128 82–97 [DOI] [PubMed] [Google Scholar]
- 24.Frank, J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M., and Leith, A. (1996) J. Struct. Biol. 116 190–199 [DOI] [PubMed] [Google Scholar]
- 25.Minton, A. P. (1990) Anal. Biochem. 190 1–6 [DOI] [PubMed] [Google Scholar]
- 26.Kogan, A. E., Kardakov, D. V., and Khanin, M. A. (2001) Thromb. Res. 101 299–310 [DOI] [PubMed] [Google Scholar]
- 27.Dorfman, R., and Walsh, P. N. (2001) J. Biol. Chem. 276 6429–6438 [DOI] [PubMed] [Google Scholar]
- 28.Gailani, D., Ho, D., Sun, M. F., Cheng, Q., and Walsh, P. N. (2001) Blood 97 3117–3122 [DOI] [PubMed] [Google Scholar]
- 29.Riley, P. W., Cheng, H., Samuel, D., Roder, H., and Walsh, P. N. (2007) J. Mol. Biol. 367 558–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meijers, J. C., Davie, E. W., and Chung, D. W. (1992) Blood 79 1435–1440 [PubMed] [Google Scholar]
- 31.Jones, S., and Thornton, J. M. (1995) Prog. Biophys. Mol. Biol. 63 31–65 [DOI] [PubMed] [Google Scholar]
- 32.Larsen, T. A., Olson, A. J., and Goodsell, D. S. (1998) Structure (Lond.) 6 421–427 [DOI] [PubMed] [Google Scholar]
- 33.Wiggins, R. C., Cochrane, C. G., and Griffin, J. H. (1979) Thromb. Res. 15 475–486 [DOI] [PubMed] [Google Scholar]
- 34.Wiggins, R. C., Cochrane, C. G., and Griffin, J. H. (1979) Thromb. Res. 15 487–495 [DOI] [PubMed] [Google Scholar]
- 35.Sinha, D., Marcinkiewicz, M., Lear, J. D., and Walsh, P. N. (2005) Biochemistry 44 10416–10422 [DOI] [PubMed] [Google Scholar]
- 36.Samuel, D., Cheng, H., Riley, P. W., Canutescu, A. A., Nagaswami, C., Weisel, J. W., Bu, Z., Walsh, P. N., and Roder, H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 15693–15698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano, C., Miller, J. K., Hyrc, K., Dikranian, S., Mennerick, S., Takeuchi, Y., Goldberg, M. P., and O'Malley, K. L. (2001) Mol. Pharmacol. 59 46–53 [PubMed] [Google Scholar]
- 38.Wolberg, A. S., Morris, D. P., and Stafford, D. W. (1997) Biochemistry 36 4074–4079 [DOI] [PubMed] [Google Scholar]
- 39.Smith, S. B., Verhamme, I. M., Sun, M. F., Bock, P. E., and Gailani, D. (2008) J. Biol. Chem. 283 6696–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, Y., Reddy, S., Murrey, H., Fei, H., and Levitan, I. B. (2003) J. Biol. Chem. 278 10073–10080 [DOI] [PubMed] [Google Scholar]
- 41.Ray, K., Hauschild, B. C., Steinbach, P. J., Goldsmith, P. K., Hauache, O., and Spiegel, A. M. (1999) J. Biol. Chem. 274 27642–27650 [DOI] [PubMed] [Google Scholar]
- 42.Martin, D. C., Pastra-Landis, S. C., and Kantrowitz, E. R. (1999) Protein Sci. 8 1152–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers, D. P., Jackson, L. K., Ipe, V. G., Murphy, G. E., and Phillips, M. A. (2001) Biochemistry 40 13230–13236 [DOI] [PubMed] [Google Scholar]
- 44.Baglia, F. A., and Walsh, P. N. (1996) J. Biol. Chem. 271 3652–3658 [DOI] [PubMed] [Google Scholar]
- 45.Yun, T. H., Baglia, F. A., Myles, T., Navaneetham, D., Lopez, J. A., Walsh, P. N., and Leung, L. L. (2003) J. Biol. Chem. 278 48112–48119 [DOI] [PubMed] [Google Scholar]
- 46.Baglia, F. A., Jameson, B. A., and Walsh, P. N. (1993) J. Biol. Chem. 268 3838–3844 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.