Abstract
Type III secretion (TTS) is an essential virulence function for Shigella flexneri that delivers effector proteins that are responsible for bacterial invasion of intestinal epithelial cells. The Shigella TTS apparatus (TTSA) consists of a basal body that spans the bacterial inner and outer membranes and a needle exposed at the pathogen surface. At the distal end of the needle is a “tip complex” composed of invasion plasmid antigen D (IpaD). IpaD not only regulates TTS, but is required for the recruitment and stable association of the translocator protein IpaB at the TTSA needle tip in the presence of deoxycholate or other bile salts. This phenomenon is not accompanied by induction of TTS or the recruitment of IpaC to the Shigella surface. We now show that IpaD specifically binds fluorescein-labeled deoxycholate and, based on energy transfer measurements and docking simulations, this interaction appears to occur where the N-terminal domain of IpaD meets its central coiled-coil, a region that may also be involved in needle-tip interactions. TTS is initiated as a series of distinct steps and that small molecules present in the bacterial milieu are capable of inducing the first step of TSS through interactions with the needle tip protein IpaD. Furthermore, the amino acids proposed to be important for deoxycholate binding by IpaD appear to have significant roles in regulating tip complex composition and pathogen entry into host cells.
Shigella flexneri is the etiologic agent of shigellosis, a potentially life-threatening bacillary dysentery in humans. Although shigellosis is typically considered a disease of the developing world, it is an underreported problem in industrialized nations where it is an infectious agent in child daycare centers, nursing homes, and in any situation where sanitation procedures become compromised (1). Shigellosis is spread by the fecal-oral route with larger outbreaks linked to contaminated water, which makes it a serious public health problem anywhere treatment regimens are inadequate. Following ingestion, Shigella travels to the colon where it crosses M cells and kills macrophages to gain access to the basal side of the colonic epithelium. S. flexneri then promotes its own uptake into epithelial cells by inducing membrane ruffling at the site of pathogen contact.
Shigella invasiveness is the product of a 31-kb segment on its large virulence plasmid, which encodes the components of a type III secretion system (TTSS).4 The Shigella TTSS is used to subvert the normal host cell mechanisms that control the actin cytoskeleton to promote invasion (2, 3). Within this genetic region, the mxi/spa operons encode the type III secretion apparatus (TTSA) and the ipa/ipg operon encodes the type III secreted protein effectors/translocators, IpaA-D, and IpgC, the cytoplasmic chaperone for IpaB and IpaC (4). The TTSA is a nanomachine that provides a conduit for secretion from the bacterial cytoplasm to the membrane and cytoplasm of target cells. It is composed of two main parts: an external needle that allows host cell contact and a basal body that traverses the bacterial inner and outer membranes and cell wall and is in many respects similar to the flagellar basal body (5, 6). In S. flexneri the exposed needle is a homopolymer of MxiH that is ∼45 nm in length, 7.0 nm in diameter, and with a central channel that is about 2.5 nm in diameter (7). Of the TTSS-secreted proteins, the three translocators, IpaB, IpaC, and IpaD, are essential for invasion (8).
We recently showed that IpaD resides at the tip of the S. flexneri TTSA needle (9). IpaD has an overall dumbbell-shape with an intramolecular coiled-coil providing the handle (10). IpaD most likely resides at the needle tip with its N-terminal globular domain and the C-terminal tail near the needle tip and the second globular domain positioned away from the needle (10, 11). The C-terminal end of IpaD, which contributes to formation of one of the stabilizing coiled-coil helices, appears to be involved with anchoring it at the tip of the MxiH needle (9). When S. flexneri is grown in tryptic soy broth (TSB), IpaD is the only Ipa protein found exposed at the tip of the needle (9). When the bacteria are grown in the presence of the bile salt deoxycholate, however, IpaB is recruited to the needle tip to form a ternary complex consisting of MxiH, IpaD, and IpaB (11). This step may serve to prepare the S. flexneri TTSA for host cell contact because it is IpaB that has been shown to bind to the host cell components involved in the entry process (12–14).
Bile salts have been implicated as agents that modulate the virulence capacity of S. flexneri (15). Although they are not inducers of type III secretion (11), bile salts do increase the adherence and invasion properties of S. flexneri (15). Here we present experimental and computational evidence that deoxycholate binds directly to IpaD at a position that may be at the interface between IpaD and MxiH at the needle tip. This information will be of value in the rational design of compounds that may block known steps in TTSA needle tip maturation and thus serve as TTSS-specific anti-infective agents.
EXPERIMENTAL PROCEDURES
Materials—Antibodies to IpaB, IpaC, and IpaD were provided by E. V. Oaks (Walter Reed Army Institute for Research, Silver Spring, MD). The S. flexneri ipaD null strain (SF622) was from P. J. Sansonetti (Institute Pasteur, Paris, France) and grown at 37 °C on trypticase soy agar containing 0.025% Congo red to ensure the selection of bacteria harboring the virulence plasmid. Fluorescent probes and Alexa Fluor-labeled secondary antibodies were from Invitrogen. Escherichia coli Tuner(DE3), E. coli NovaBlue, pET15b, and ligation reagents were from Novagen (Madison, WI). Restriction enzymes were from New England BioLabs (Tozer, MA). Oligonucleotide primers were from IDT (Coralville, IA). IMAC affinity resin was from Sigma. Other chemicals were reagent grade.
Growth of S. flexneri in the Presence of Deoxycholate—S. flexneri is routinely grown to early or mid log phase in TSB after inoculation from a trypticase soy agar plate containing Congo red. These growth conditions do not lead to detectable levels of IpaB on the bacterial surface. To induce mobilization of IpaB to the needle tip, the S. flexneri was grown for 30 min in TSB containing 2.5 mm (0.1%, w/v) deoxycholate. None of the conditions used here promoted the recruitment of IpaC to the Shigella surface or led to induction of type III secretion (data not shown).
Immunofluoresence Microscopy—S. flexneri were grown to early log phase in TSB with or without 2.5 mm deoxycholate. Bacteria were collected by centrifugation, washed twice with PBS, and resuspended in 4% (v/v) formaldehyde in 10 mm phosphate, pH 7.2, 150 mm NaCl (phosphate-buffered saline). The bacteria were fixed to slides and blocked with 1% bovine serum albumin in PBS. IpaD was detected using monoclonal mouse anti-IpaD antibodies and Alexa Fluor 488 goat anti-mouse IgG. IpaB was detected using rabbit anti-IpaB antiserum and Alexa Fluor 568 goat anti-rabbit IgG. Imaging was performed using a Yokugawa-type spinning disk confocal attached to an Olympus IX-81 microscope, alternately using either the 473-nm laser line with a 505–530 band-pass emission filter (Alexa 488) or a 561-nm laser line with a 580–520 band-pass emission filter (Alexa 568). Transmission images were also obtained. To determine the importance of IpaD-bile salt interactions on the presence of IpaD and IpaB on the bacterial surface, mutations were introduced into IpaD and the number of bacteria labeled with anti-IpaD and/or anti-IpaB antibodies was determined. Multiple fields (n > 10) were viewed following fluorescence labeling and the number of bacteria with IpaD on their surface was counted as a function of total bacteria in the field. The same was then done for IpaB. Greater than 90% of bacteria with a given protein on their surfaces was considered positive (+), whereas fewer than 5% of the bacteria with the protein of interest labeled on their surfaces was considered negative (–) for the presence of that protein.
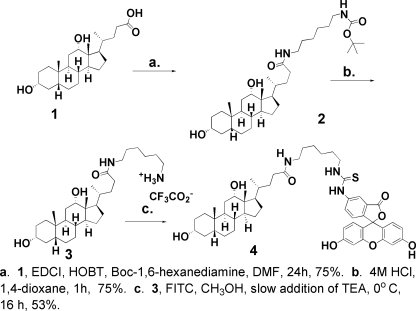
Preparation of FITC-Deoxycholate—The three-step synthesis of fluorescein 4-isothiocyanate-labeled deoxycholate (FITC-deoxycholate, 4) is shown in Fig. 1. Additional experimental details for each step and the spectral data are given under supplementary Materials. FITC-deoxycholate 4 is prepared by first converting deoxycholic acid (1) into a hexylamino amide 3 by treatment of a N,N-dimethylformamide solution of 1 with hydroxybenzotriazole and 1-ethyl-3-(3-dimethyl aminopropyl)carbodiimide hydrochloride followed by addition of tert-butyl 6-aminohexylcarbamate. After stirring for 22 h at room temperature, the solution was diluted with ethyl acetate, extracted with water, and the organic phase dried with anhydrous magnesium sulfate. The solvent was removed under vacuum and 15 ml of 4 m HCl in 1,4-dioxane was added and the solution stirred for 1 h. Removal of the solvent under reduced pressure gave a white precipitate of the trifluoroacetate salt of 3. The salt (3) was mixed with FITC in methanol and stirred for 30 min to dissolve the reactants. Triethylamine was then slowly added and the reaction mixture stirred for an additional 16 h. The solvent was then removed under reduced pressure to yield a reddish brown precipitate that was purified by flash column chromatography to give a yellowish waxy precipitate of fluorescein 4-isothiocyanate-labeled deoxycholate (FITC-deoxycholate, 4) as shown in Fig. 1. The structure of 4 was confirmed by mass spectrometry (see supplemental materials Fig. S1).
FIGURE 1.
Synthesis of FITC-deoxycholate (4). A schematic of the synthetic sequence for labeling deoxycholic acid (1) with FITC through a 1,6-hexanediamine tether, 2, 3, is shown. The synthetic details and the MS analysis of the final FITC-deoxycholate product 4 are given under supplemental Materials and Fig. S1.
Protein Expression and Purification—Recombinant IpaD, SipD, and BipD have been prepared by standard methods as previously described (16, 17). Briefly, Tuner(DE3) containing D/pET15b, SipD/pET15b, or BipD/pET15b were grown to 0.5 A600 units, induced to express protein with 1.0 mm isopropyl 1-thio-β-d-galactopyranoside and incubated a further 3 h. The bacteria were collected by centrifugation, lysed by sonication, and the HisTag protein from the clarified supernatant was purified by standard nickel affinity chromatography.
For fluorescence spectroscopic analysis, IpaD was labeled with 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM) at its single Cys residue (Cys-322). Briefly, IpaD at >2 mg/ml in PBS was diluted with an equal volume of N,N-dimethylformamide. CPM was added from a stock solution prepared in N,N-dimethylformamide to give a final concentration of 2 mm and the mixture was incubated for at least 1 h on ice. Any precipitate that formed was removed by centrifugation and the labeled protein was separated from free dye by gel filtration using a Sephadex G-50 column equilibrated with PBS.
Fluorescence Polarization—The interaction of IpaD with FITC-deoxycholate was measured using fluorescence polarization (FP) spectroscopy (18). FP provides a measure of the rotational diffusion of a fluorescent molecule in solution. The association of the small fluorescent FITC-deoxycholate with a larger nonfluorescent partner like IpaD results in an increase in the molecular volume of the fluorescent species, thus giving rise to an increased mP value. FP measurements for FITC-deoxycholate were obtained using a Beacon fluorescence polarimeter (Panvera Corp.) as described (18). Briefly, a constant concentration of FITC-deoxycholate (30–50 nm) in PBS was mixed with increasing concentrations of IpaD (micromolar range). The change in FP was monitored as a change in millipolarization units (ΔmP). The ability for non-fluorescent bile salts or their derivatives to compete for FITC-deoxycholate binding to IpaD was tested using unlabeled sodium salts of deoxycholate, cholate hydrate, taurodeoxycholate, chenodeoxycholate, and dehydrocholate. A negative control used here was CAPS buffered to neutral pH. In these experiments, IpaD (5 μm) was simultaneously incubated with 30 nm FITC-deoxycholate and increasing concentrations of the nonfluorescent competitor for 1 h. At this point, the mP value of the FITC-deoxycholate was determined and plotted as a function of competitor concentration.
Förster Resonance Energy Transfer (FRET) Analyses—FRET measurements were obtained for CPM-labeled IpaD or CPM-IpaD (donor) and FITC-deoxycholate (acceptor). CPM-IpaD was used at a final concentration of 50 nm and increasing concentrations of either deoxycholate (to give Fd in which the CPM-IpaD donor is associated with nonfluorescent ligand) or FITC-deoxycholate (to give Fda in which the CPM-IpaD is associated with FITC-deoxycholate acceptor). Fluorescence emission was scanned from 400 to 520 nm using an excitation of 385 nm, which allowed for monitoring changes in coumarin emission upon addition of deoxycholate or FITC-deoxycholate. For each concentration of ligand, the energy transfer efficiency was determined according to E = 1 – Fda/Fd (Equation 1). The Fd term involved the use of nonfluorescent deoxycholate to account for possible changes in coumarin emission that might occur upon ligand binding, however, no change in CPM-IpaD emission was observed upon the addition of deoxycholate (data not shown). A RO value (the theoretical distance that would give 50% energy transfer efficiency) was determined to be 52 Å (19), which was used with the calculated energy transfer maximum to determine the distance between the coumarin on Cys-322 of CPM-IpaD to the fluorescein on FITC-deoxycholate according to E = RO6/(RO6 + RO6) (Equation 2), where R is the calculated distance separating the donor and acceptor fluorophores (20). In determining the RO value, the relative orientation of donor and acceptor dipoles was assumed to be random (κ2 = 2/3). Although this assumption can introduce potential error to the measurement of an absolute distance (21), the error introduced is typically minimal (not more than about 10%) (22) and this is not expected to distract from the localization of IpaD's deoxycholate binding site as described here.
Molecular Modeling—Structures of all ligands were sketched in SYBYL (23) and were optimized to default levels of structural convergence via Tripos Molecular Force Field (24) and Gasteiger-Marsili electrostatics (25). Receptor structures were imported into SYBYL from their native PDB format (i.e. 1HGU in the case of IpaD (26)). These structures were then protonated in SYBYL, crystallographic waters were removed, and Gasteiger-Marsili were added to all atoms. All Asp and Glu residues were assumed to be anionic, whereas Lys and Arg residues were rendered as cations. One hundred docked complexes for each ligand-receptor pair were then generated via AutoDock (27) using the Lamarckian Genetic Algorithm search protocol (ligand torsional, positional, and rotational coordinates all varied; receptor held fixed) according to default settings. Results were then parsed according to clustering techniques provided within the AutoDock suite, and the most plausible complex structure was considered to be the lowest free energy structure (as quantified by the native AutoDock score function) of the dominant cluster.
Preparation of ipaD Mutants for Expression in S. flexneri SF622 or E. coli—D/pwpsf4 containing the ipaD gene has been described (28). ipaD mutants were made by inverse PCR using D/pwpsf4 as template, a primer composed of GAGAGA, a restriction site, and 18 nucleotides flanking the nucleotides to be mutated and a primer going the opposite direction encoding GAGAGA, a restriction site, and 18 nucleotides (28). The primers used to introduce mutations into D/pwpsf4 are given in supplemental materials Table S1. The resulting PCR products were digested with the appropriate restriction enzyme, intramolecularly ligated, and used to transform E. coli NovaBlue. Each plasmid was electroporated into S. flexneri SF622. Ampicillin selection ensured the presence of the ipaD-expressing plasmid and kanamycin resistance with Congo red differentiation ensured that the Shigella virulence plasmid was present.
To produce recombinant mutant IpaD protein, mutant ipaD genes were subcloned into pET15b as previously described (29, 30). The resulting plasmids were used to transform Tuner(DE3) cells and the recombinant protein purified as described above.
Gentamycin Protection Assay—Newly designed SF622 strains expressing mutant forms of ipaD were assessed for their ability to invade Henle 407 cells by a standard gentamycin protection assay as previously described (18, 28) with one minor modification. In previous use of this assay, bacterial-host cell contact was facilitated by centrifugation, however, this step was found to interfere with assessing subtle differences in the invasion capacity of these strains. Therefore, the centrifugation step normally used for monitoring Shigella invasiveness was omitted.
RESULTS
Deoxycholate-induced Recruitment of IpaB to the Shigella Surface Appears to Correlate with Deoxycholate Binding by IpaD—We have previously shown that S. flexneri grown to early log phase in TSB has IpaD present on its surface. This population of IpaD is located at the tip of TTSA needles (11, 29), presumably due to direct interaction between IpaD and MxiH (11, 31). IpaB and IpaC were not observed on the surface of these bacteria. When 2.5 mm deoxycholate was included in this growth medium, however, IpaB (but not IpaC) was seen on the bacterial surface (11). This new population of IpaB was associated with IpaD at the tip of the TTSA needle (11) (see supplemental materials Fig. S2). The same result was achieved for S. flexneri grown to early log phase and then exposed to chenodeoxycholate or taurodeoxycholate (11). Furthermore, deoxycholate triggered IpaB surface localization at micromolar concentrations (data not shown).
To determine whether the observed deoxycholate effects are due to bile salt recognition by IpaD, a fluorescent derivative of deoxycholate was synthesized so that the IpaD-deoxycholate interaction could be monitored in vitro by fluorescence spectroscopic methods. Previously, Mills et al. (32) derivatized cholate and other bile salts (33, 34) at the single available carboxyl group with fluorescein using a lysine linker. These fluorescent bile salt derivatives were found to maintain native properties such as lecithin solubilization, hepatic uptake, biliary secretion, and stimulation of biliary lipid secretion (32–36). Similarly, we labeled deoxycholate on the carboxyl group via an amide linkage with the α-amino group of boc-1,6-diaminohexane. After deprotection, the resulting primary amine was labeled with FITC (Fig. 1 and supplementary materials). S. flexneri was then grown in TSB containing FITC-deoxycholate to determine whether the addition of the tether and fluorophore affected the biological activity of the deoxycholate. FITC-deoxycholate was able to induce the mobilization of IpaB to the needle tip complex at the same micromolar concentrations as deoxycholate alone (data not shown).
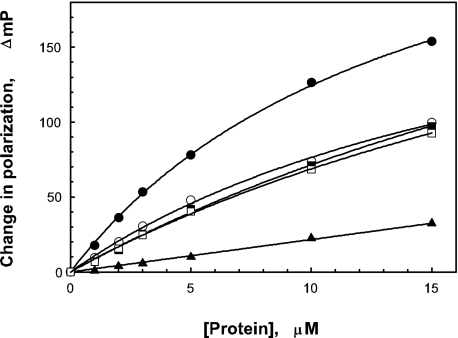
Using FITC-deoxycholate as a ligand, FP was used to determine whether deoxycholate directly interacts with IpaD. FP measures the rate of molecular rotation for a fluorescent molecule in solution (see “Experimental Procedures”). Small molecules rotate faster in solution, thus giving low polarization values, whereas larger molecules rotate slower, resulting in higher polarizations. The increase in size seen when a small fluorescent molecule binds to a larger nonfluorescent moiety gives rise to a characteristic increase in polarization. The initial polarization value (in mP units) for FITC-deoxycholate was 27.5 mP. This small value was expected based on the size of FITC-deoxycholate and the flexibility of the chosen hexyl tether. When FITC-deoxycholate associates with IpaD and SipD, the IpaD homologue from Salmonella typhimurium, an increase in mP was seen. The maximum increase for FITC-deoxycholate with IpaD was about 250 mP units with an apparent dissociation constant (kd) in the micromolar range (varying between 5 and 15 μm). With SipD, FITC-deoxycholate had a maximum increase of about 160 mP with a larger apparent kd (see Fig. 2). In contrast, the putative needle tip protein from Burkholderia pseudomallei BipD did not give rise to an increase in FITC-deoxycholate polarization. The negative control in these experiments was IpgC, the cytoplasmic chaperone for IpaB and IpaC (Fig. 2). These findings link the ability for deoxycholate to induce in vivo changes in S. flexneri with its ability to bind to IpaD in vitro.
FIGURE 2.
FITC-deoxycholate binds to IpaD. FITC-deoxycholate (30 nm) incubated with nonfluorescent IpaD (closed circles), SipD (open circles), BipD (closed triangles), and IpgC (open triangles). IpgC is a Shigella cytoplasmic chaperone that was used here as a negative control for FITC-deoxycholate binding. The starting mP for FITC-deoxycholate is 27.5. For IpaD binding the apparent ΔmPmax is 248 and the apparent kd is in the micromolar range between about 5 to 15 μm.
When different nonfluorescent bile salts or bile salt derivatives were tested for the ability to compete with FITC-deoxycholate for binding to IpaD, deoxycholate was the most efficient competitor, however, cholate hydrate, taurodeoxycholate, and chenodeoxycholate also competed with FITC-deoxycholate for binding to IpaD (Fig. 3). In contrast, dehydrocholate was not found to be competitive for FITC-deoxycholate binding (Fig. 3) and it failed to promote IpaB recruitment to the Shigella surface (data not shown). This suggests that placement of functional groups on these bile salt derivatives may have a role in contributing to their recognition by IpaD.
FIGURE 3.
Competition for FITC-deoxycholate binding to IpaD by nonfluorescent molecules. The ability for nonfluorescent deoxycholate (open circles), cholate hydrate (closed circles) chenodeoxycholate (open squares), taurodeoxycholate (closed squares), and dehydrocholate (open triangles). The negative control used here was CAPS (closed triangles) as described under “Experimental Procedures.” All of these compounds were used below their recognized critical micelle concentrations.
Localization of the IpaD Region That Binds Deoxycholate Based on FRET Measurements—To provide a starting point for identifying the deoxycholate binding pocket on IpaD, FRET (nonradiative) was carried out using a coumarin (CPM) donor covalently linked to Cys-322 of IpaD and the fluorescein acceptor attached to FITC-deoxycholate. To estimate maximum FRET efficiency, CPM-labeled IpaD was titrated with increasing concentrations of FITC-deoxycholate (data not shown). Based on these findings, FITC-deoxycholate appeared to bind to full-length IpaD with a low micromolar kd to give a theoretical maximum FRET efficiency of 78%, suggesting that the two probes are relatively close to one another. Using approximately a 30-fold excess of FITC-deoxycholate, the level of energy transfer efficiency can be seen in the typical FRET spectrum shown in Fig. 4. In all of these experiments, the concentration of FITC-deoxycholate was kept as low as possible to avoid artifacts that could be introduced by a fluorescein-dependent inner filter affect. An RO value of 52 Å was determined here, which is largely in agreement with coumarin to fluorescein distances determined previously (19) and for standard RO values published elsewhere (37). These calculations assume a random orientation for the component dipoles, which is reasonable considering the nature of the tether linking FITC to deoxycholate. Ultimately, the observed 78% energy transfer efficiency translates to a distance of 42.1 Å (about ±5 Å) from the coumarin probe to the fluorescein moiety on deoxycholate. Based on the published crystal structure of IpaD, this places the binding site of the FITC-deoxycholate near the midpoint of the central coiled-coil of IpaD.
FIGURE 4.
Determining the distance from the coumarin from CPM-Cys-322 to the fluorescein of FITC-deoxycholate. FRET from the donor CPM-Cys-322 residue of IpaD (30 nm) to the acceptor fluorescein on FITC-deoxycholate (in a 30-fold or more excess of acceptor) is seen in the fluorescence emission spectrum depicted by the dashed line. The same conditions using deoxycholate that was not labeled with FITC is shown by the solid line. In this particular experiment, nearly 70% energy transfer efficiency was observed, however, titration experiments were used to determine that the maximum energy transfer from the CPM to FITC probes is 78%. The data shown are from a representative experiment that was performed six times.
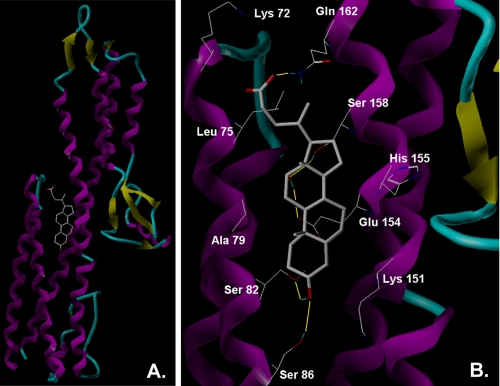
Molecular Modeling Tentatively Defines the Composition of the Deoxycholate Binding Site—Although the FRET provides a tentative distance measurement from the Cys-322 of the FITC-deoxycholate binding site, it does not provide details on the amino acid residues involved in the binding event. Therefore, computational analyses were used to dock deoxycholate onto the published crystal structure of IpaD to determine whether a tentative binding pocket for this bile salt could be identified. As shown in Fig. 5, the consensus binding site for deoxycholate on IpaD is at a site on the stabilizing coiled-coil of the protein where it interacts with the N-terminal helix-turn-helix globular domain. This position is consistent with the FRET data presented above. Docking simulations using full-length IpaD implicate residues within the central coiled-coil of the protein (e.g. Ser-158) in deoxycholate binding, as well as N-terminal domain residues like Leu-75. Docking analyses using FITC-deoxycholate as the ligand suggest that it also binds at this site in the same way as deoxycholate (not shown). Furthermore, additional docking simulations suggested that while the IpaD N-terminal domain may contribute to deoxycholate binding, it is not essential for this event because docking could also be accomplished using a truncated form of IpaD (IpaDΔ1–120) and FP analyses using recombinant IpaDΔ1–120 showed that this protein was still able to bind FITC-deoxycholate (data not shown). The identification of a putative deoxycholate binding site on IpaD now provides the basis for conducting molecular analyses to confirm whether this is indeed an important functional site within IpaD.
FIGURE 5.
Simulated docking of deoxycholate onto the crystal structure of IpaD is shown. A, deoxycholate docked onto the ribbon structure of IpaD. B, enlarged version of the binding pocket showing key contacts. Structures of all ligands were sketched in SYBYL and the protein was imported into SYBYL from its native PDB format. One hundred docked complexes for each ligand-receptor pair were generated via AutoDock as described under “Experimental Procedures.”
Mutagenesis of the IpaD Residues Suggested to Interact with Deoxycholate in Docking Analyses—To explore the role of the IpaD residues predicted to be involved in deoxycholate binding, site-directed mutagenesis was employed as a means to probe how mutations might alter bile salt binding with IpaD and influence the function of IpaD in host cell invasion. Not surprisingly, mutation of residues within the N-terminal domain of IpaD that might interact with deoxycholate (e.g. residues Lys-72, Leu-75, or Ala-79), do not have an obvious effect on the invasion function of IpaD (Table 1). Because the N-terminal domain (here considered to be residues 31–120) is not required for IpaB recruitment to the bacterial surface upon addition of deoxycholate, these residues were not expected to have a major role in deoxycholate-induced events (data not shown). This finding is also consistent with the modeling and FP results described above that show that the N-terminal domain is not required for the docking of deoxycholate onto IpaD in silico or for the binding of FITC-deoxycholate in vitro. Furthermore, a recently proposed model of the IpaD-MxiH interaction by Zhang et al. (31) suggested that the IpaD N terminus may be displaced by a MxiH unit upon needle tip complex formation, thus minimizing the role of the N-terminal domain of IpaD in the function of this protein at the needle tip in vivo. In contrast, mutation of residues predicted to be involved in deoxycholate binding based on docking simulations and that are part of the central coiled-coil of IpaD can have a significant effect on the ability of IpaD to direct the invasion of cultured epithelial cells (Table 1). In particular, conversion of Ser-158 to Ala (S158A) reduced invasiveness by 80% (Table 1). Combinations of mutations with S158A did not result in a further reduction of invasiveness, suggesting that Ser-158 alone among these particular residues has an important role in IpaD function.
TABLE 1.
The effect of directed IpaD mutations on Shigella invasiveness
| IpaD mutant | Relative invasiona |
|---|---|
| ipaD null strain (SF622) | 0 ± 0 |
| Wild-type | 100 ± 8 |
| K72A | 81 ± 10 |
| L75S/A79S | 90 ± 19 |
| K72A/L75S/A79S | 82 ± 15 |
| E154A/H155A | 104 ± 15 |
| S158A | 22 ± 4b |
| E154A/H155A/S158A | 21 ± 9b |
| S158A/T161A | 28 ± 5b |
| S158A/Q162A | 18 ± 1b |
| K151E | 421 ± 21b |
| K151E/E229K | 0 ± 0 |
| E229K | 58 ± 9 |
Invasion was measured using a standard gentamycin protection assay with S. flexneri SF622 making wild-type IpaD having 74 ± 6 colonies per well (n = 3). The normal centrifugation step was omitted to avoid the loss of subtle differences in invasion capacity.
Indicates a statistical difference in the presented values (p < 0.01) using a Student's t test.
It is noteworthy that mutation of Lys-151 to Glu (K151E) resulted in greater than a 4-fold increase in Shigella invasiveness (Table 1). When the crystal structure of IpaD was originally reported (10) it was suggested that within the context of the TTSA needle tip complex, IpaD could potentially form a pentamer with Lys-151 of one monomer forming a salt bridge with Glu-229 on the adjacent monomer. To test whether disruption of such a salt bridge could account for this change in IpaD function, the double mutant IpaDK151E/E229K was generated to restore any potential salt bridge and thereby restore the wild-type invasion phenotype to the ipaD null strain S. flexneri SF622. Instead, the double mutation completely eliminated the ability of IpaD to restore invasiveness to SF622 (Table 1). Oddly, the single mutant, IpaDE229K in SF622 led to a 40% reduction in invasiveness (Table 1). Although this is a significant reduction, it does not account for the phenotype of IpaDK151E/E229K. These results suggest that Lys-151 and Glu-229 work together in controlling IpaD function by a mechanism that will need further examination.
Critical to invasion, and thus the virulence process, is the ability of IpaD to localize to the TTSA needle tip and recruit IpaB to the surface after an appropriate signal is detected. Therefore, immunofluorescence microscopy was used to correlate the impact of the IpaD mutations on invasion (Table 1) with proper Shigella surface localization of IpaD and IpaB. As previously demonstrated IpaB staining was not seen on the surface of Shigella prior to deoxycholate addition, whereas IpaD staining was seen on the surface (Table 2). After deoxycholate addition, increased IpaD staining was observed and all the cells had IpaB on their surfaces. In contrast, no IpaD or IpaB are found on the surface of the ipaD null strain SF622 under any conditions tested. IpaDS158A had IpaD on its surface in the absence or presence of deoxycholate, but did have a slight reduction in the ability to surface localize IpaB when compared with wild-type levels (Table 2). Although this decrease may not entirely account for the decrease in the invasion of IpaDS158A, it could significantly contribute to it if the strain is unable to mobilize the first pore translocator efficiently. Despite the greatly elevated invasiveness seen for Shigella making IpaDK151E, the numbers of bacteria possessing IpaD on the surface is similar to that seen for wild-type bacteria in the absence of deoxycholate (Table 2). There did, however, seem to be a greater than normal intensity of IpaD staining on the surface of these bacteria after deoxycholate was added while having at least wild-type levels of IpaB. For the double mutant IpaDK151E/E229K, <20% of the bacteria appeared to have IpaD on the bacterial surface in the absence or presence of deoxycholate (Table 2). Perhaps more importantly, only a very small number of bacteria have visible IpaB appear on their surfaces after deoxycholate addition (Table 2). This behavior is distantly similar to that of the ipaB null mutant strain S. flexneri SF620 where IpaD is lost from the Shigella surface with deoxycholate addition (11). The secretion profile for this mutant also resembles that seen for SF620 with greatly elevated levels of IpaB and IpaC in the overnight culture supernatants (data not shown). Little IpaD, however, is seen here despite the fact that these cells make normal levels of IpaD based on Western blot analysis (data not shown). On the other hand, IpaDE229K behaved essentially like wild-type bacteria with respect to IpaD surface localization (Table 2), whereas only a fraction of these bacteria had IpaB on their surfaces after the addition of deoxycholate. It is interesting that IpaDS158A was less invasive than IpaDE229K because the former appeared to only be somewhat attenuated in its response to deoxycholate (Table 2), whereas IpaDE229K appeared to be more dysfunctional in how it responded to the presence of deoxycholate.
TABLE 2.
Examination of the numbers of Shigella with IpaD or IpaB on the surface in the absence and presence of deoxycholate
|
ipaD mutant expressed in SF622
|
Deoxycholate added
|
Number of bacteria with surface
exposeda
|
|
|---|---|---|---|
| IpaDb | IpaBc | ||
| Wild-type | - | + | - |
| + | ++d | + (100%)e | |
| ipaD null | - | - | - |
| + | - | - | |
| S158A | - | + | - |
| + | + | +/- (70%) | |
| K151E | - | + | - |
| + | +++ | + (100%) | |
| K151E/E229K | - | -/+ | - |
| + | -/+ | -/+ (<20%) | |
| E229K | - | + | - |
| ± | + (80%) | -/+ (20%) | |
The number of bacteria having specific proteins on their surface was determined by immunofluorescence staining of the bacteria as described under “Experimental Procedures.” In each case more than 10 representative fields were examined with at least 20 bacteria per field.
IpaD staining was with monoclonal antibodies (16F8) prepared in the laboratory of Dr. E. V. Oaks with rabbit anti-mouse IgG labeled with Alexa 488 dye as the secondary antibody.
IpaB staining was with rabbit antisera generated against IpaB in the laboratory of Dr. E. V. Oaks with mouse anti-rabbit IgG labeled with Alexa 568 dye as the secondary antibody.
Changes in the intensity of perceived labeling (independent of the percentage of cells stained) are estimated relative to SF622 expressing wild-type ipaD grown in the absence of deoxycholate. Increased apparent labeling is shown with ++ or +++.
In samples where only a limited number of bacteria are stained for a given protein, the approximate percentage of cells showing fluorescence staining as indicated by the values given in parentheses.
Monitoring the Ability for FITC-deoxycholate to Bind to Different Recombinant IpaD Mutant Proteins—Because there are alterations in the function of IpaD when certain amino acids thought to be involved in deoxycholate binding are changed, we tested selected mutants to determine whether the binding of FITC-deoxycholate to the purified mutants could be detected. Because mutations at Lys-151, Glu-229, and Ser-158 resulted in the most significant changes in Shigella invasiveness, these mutant forms of IpaD were targeted for purification and analysis of FITC-deoxycholate binding by FP (Fig. 6). Although IpaDS158A gave rise to the largest decrease in Shigella invasiveness, it still bound FITC-deoxycholate although the apparent kd was increased slightly (Fig. 6). This was not further altered by simultaneously introducing mutations Q165A and Q162A, which is consistent with invasion data showing that these additional mutations do not further negatively affect the entry process (Fig. 6 and Table 1). IpaDK151E likewise had a slightly higher apparent kd value relative to wild-type protein, suggesting that its dramatic positive effect on invasiveness is not due to increased affinity for deoxycholate. Interestingly, IpaDE229K was greatly reduced in its ability to bind FITC-deoxycholate, which may correlate with the in vivo observation that it may have a major defect in recruiting IpaB to the bacterial surface (Table 2). Although this mutation is not in the putative deoxycholate binding pocket, because Glu-229 is proposed to be involved in a salt bridge with Lys-151, a conformational change in IpaD may have occurred resulting in a decreased affinity for deoxycholate and therefore preventing the recruitment of IpaB to the bacterial surface.
FIGURE 6.
The binding of FITC-deoxycholate to different IpaD site-specific mutants. Titration of 50 nm FITC-deoxycholate with increasing concentrations of protein was used to compare the binding properties of different IpaD site-specific mutants. The proteins used were wild-type IpaD (closed circles), IpaDE229K (open circles), IpaDS158A/Q162A/Q165A (closed triangles), IpaDS158A (open triangles), and IpaDK151E (closed squares). The average starting FP value for FITC-deoxycholate in these experiments was about 35 mP units (n = 3).
DISCUSSION
The TTSS is an essential determinant for Shigella virulence because it promotes invasion of the cells of the human intestine. It has been recently shown that the essential virulence protein IpaD works primarily as a TTSA needle tip protein (9) with bile salts acting as environmental signals for the stable recruitment of IpaB onto the Shigella needle tip complex (11). In this study, we show that the bile salt deoxycholate binds directly to IpaD and this interaction can be studied in detail using established in vitro analyses. Although it is not known whether other environmental small molecules, especially those within the colonic submucosa, can have a similar effect on the Shigella TTSA tip complex composition, it is almost certain that bile salts elicit an effect on the Shigella TTSA as this pathogen traverses the human gastrointestinal tract. Consistent with the prior observation that different bile salts can influence Shigella virulence functions (15) and participate in the recruitment of IpaB to the TTSA needle tip (11), other bile salts were competitive for the binding of FITC-deoxycholate by IpaD. One bile salt derivative (dehydrocholate), however, was not competitive. This bile salt also fails to induce the recruitment of IpaB to the Shigella surface (data not shown). This implicates the presence and position of hydroxyl groups on deoxycholate with having a role in interacting with specific amino acid side chains on IpaD.
It is intuitively attractive that the Salmonella homologue of IpaD (SipD) also binds FITC-deoxycholate in vitro, because both organisms are enteric pathogens, whereas BipD from the systemic or pulmonary pathogen B. pseudomallei does not. Yet, when expressed in S. flexneri SF622 (the ipaD null mutant), BipD localizes to the MxiH needle tip (10, 38) as would be predicted for SipD as well, because all three are nearly identical at their C termini at the sequence level, which is the region of IpaD implicated in anchoring it to the needle (9). When deoxycholate is added, however, BipD is lost from the SF622 needle tip as was seen for IpaD when deoxycholate was added to the S. flexneri ipaB null strain SF620 (10). Therefore, if IpaB cannot associate with the needle tip, the tip protein, which may have undergone a conformational change in response to the environmental signal (see IpaDE229K), is unable to maintain its association with the needle tip and is thus released. Thus, the inability to grasp IpaB probably stems from sequence differences at positions other than the C terminus of the tip proteins. More importantly, however, this suggests that BipD has the information within its structure needed to bind deoxycholate, but that the unusual stability and complexity of its N-terminal domain (10, 17) may block deoxycholate binding in vitro, whereas the BipD-MxiH needle tip complex is able to recognize the presence of deoxycholate. This once again implicates the central coiled-coil of these tip complex proteins in the recognition of environmental small molecules. It was recently reported by Zhang et al. (31) that the N terminus of IpaD may be displaced by MxiH when the former assumes a stable position at the Shigella TTSA needle tip. It is thus possible that the BipD N-terminal globular domain is similarly displaced, which allows it to take up the observed position at the MxiH needle tip. Based upon molecular modeling and docking simulations, it is possible that tip-associated BipD has a newly created deoxycholate binding site that does not exist for full-length BipD alone in solution.
Mutational analyses focused on a region between Lys-151 and Ser-158 and the corresponding putative salt bridge partner at Glu-229. The effects of some of the mutations were subtle, whereas others were quite substantial, both positively and negatively for the pathogen. Based on molecular modeling, IpaDS158A was made and invasion was reduced by 80% but IpaB surface localization was only reduced by 20%. If, however, the efficiency or rate of IpaB mobilization is severely restricted, this could cause the significant reduction in invasion that is observed. Another mutant near the putative deoxycholate binding site, IpaDK151E, gave rise to more than a 4-fold increase in Shigella invasiveness. Based on FP IpaDK151E does not greatly alter deoxycholate binding, which gives indirect evidence (along with the other two mutants) that the salt bridge may in fact be real. Because previous reports have proposed that Lys-151 could be involved in salt bridge formation with Glu-229 on the adjacent IpaD monomer within the IpaD needle tip complex (10), the double mutant IpaDK151E/E229K was made but it rendered Shigella entirely noninvasive. This could not be explained in part by IpaDE229K because this mutant was still invasive, but at reduced levels relative to Shigella expressing wild-type ipaD. This does suggest that Glu-229 and Lys-151 work cooperatively, but there is not an obvious mechanism to account for the observed effects of the double mutation. Interestingly, IpaDE229K was more compromised in FITC-deoxycholate binding in vitro than any of the mutations directed at the identified deoxycholate pocket. Again, it is possible that this residue is critical for dynamic structural changes in IpaD that are part of the deoxycholate-signaling process.
In light of the data concerning the K151E and E229K mutations, it must be acknowledged that introducing mutations that reverse local charges can influence more than a protein-ligand interaction. It is certainly possible that such mutations could induce localized or even distant changes in protein conformation that could drastically alter protein function. Such an event could account for the positive and negative effects, respectively, that the K151E and E229K mutations have on the invasion function of IpaD. In fact, the observation that recombinant IpaDE229K appears to have a slightly reduced affinity for deoxycholate could be explained by a mutation-induced change in the conformation of the deoxycholate binding pocket. This would also be consistent with the data presented in Table 2 for this mutant. In the case of IpaDK151E/E229K the complete loss of the invasion function of IpaD, the reduced maintenance of IpaD on the Shigella surface, and the altered ability for deoxycholate to recruit IpaB to the bacterial surface are most easily explained by proposing that IpaD has undergone a conformational change in response to the double mutation.
Taken together, the biochemical, mutagenesis, and computational data support a model in which deoxycholate interacts with IpaD, the Shigella TTSA needle tip protein, to promote the stable recruitment and retention of IpaB to form a ternary complex with IpaD and MxiH atop the Shigella TTSA needle tip. Although unequivocal evidence of the residues required for deoxycholate binding will necessitate detailed structural analyses, mutational studies clearly show that amino acid residues within the putative deoxycholate binding pocket appear to have roles in: 1) IpaB recruitment to the TTSA needle tip; 2) the strength of the IpaD-deoxycholate interaction; and 3) Shigella invasion efficiency for cultured cells. The biochemical, structural, and docking data presented here and in preceding works (10, 11, 31) provide a first step toward understanding how the Shigella TTSS is involved in responding to its environment. Furthermore, because the molecular structures of IpaD and MxiH have been solved, the use of molecular modeling and docking simulations can now provide a basis for the rational design of drugs targeting the Shigella TTSA. The finding that BipD at the MxiH needle tip is affected by deoxycholate addition whereas BipD does not interact with deoxycholate in vitro suggests that anti-infective agents that block Shigella TTSA function may have broad spectrum effects that could also influence other type III secretion systems.
Supplementary Material
Acknowledgments
Shyam Mehta is acknowledged for technical contributions. Microscopy was performed in the Microscopy and Analytical Imaging Laboratory at the University of Kansas. Critical reading of the manuscript by members of the Picking group is gratefully acknowledged.
This work was supported, in whole or in part, by National Institutes of Health Grants P20 RR016475 awarded to the Kansas IDeA Network of Biomedical Research Excellence (K-INBRE) (to P. R. A.) and R25GM62232 awarded to the Initiative for Maximizing Student Diversity (IMSD) (to C. D. L.). This work was also supported by United States Public Health Service Grants AI034428 (to W. D. P.), AI067858 and AI072510 (to W. L. P.), and R01 GM72910 (to R. S. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Scheme 1, Materials, Figs. S1 and S2, and Table S1.
Footnotes
The abbreviations used are: TTSS, type III secretion system; TTSA, type III secretion apparatus; Ipa, invasion plasmid antigen; ipg, invasion plasmid gene; mxi, genes encoding the major exporters of Ipa proteins; spa, genes encoding proteins for the surface presentation of Ipa antigens; MxiH, the Shigella flexneri TTSA needle protein; FITC, fluorescein isothiocyanate; FP, fluorescence polarization; mP, millipolarization units; FRET, Förster resonance energy transfer; CPM, 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin; EDC, 1-ethyl-3-(3-dimethyl aminopropyl) carbodiimide hydrochloride; CAPS, N-cyclohexyl-3-aminopropanesulfonic acid; PBS, phosphate-buffered saline.
References
- 1.Mohle-Boetani, J. C., Stapleton, M., Finger, R., Bean, N. H., Poundstone, J., Blake, P. A., and Griffin, P. M. (1995) Am. J. Public Health 85 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buttner, D., and Bonas, U. (2002) Trends Microbiol. 10 186–192 [DOI] [PubMed] [Google Scholar]
- 3.Galan, J. E., and Wolf-Watz, H. (2006) Nature 444 567–573 [DOI] [PubMed] [Google Scholar]
- 4.Schroeder, G. N., and Hilbi, H. (2008) Clin. Microbiol. Rev. 21 134–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blocker, A., Jouihri, N., Larquet, E., Gounon, P., Ebel, F., Parsot, C., Sansonetti, P., and Allaoui, A. (2001) Mol. Microbiol. 39 652–663 [DOI] [PubMed] [Google Scholar]
- 6.Kubori, T., Matsushima, Y., Nakamura, D., Uralil, J., Lara-Tejero, M., Sukhan, A., Galan, J. E., and Aizawa, S. I. (1998) Science 280 602–605 [DOI] [PubMed] [Google Scholar]
- 7.Cordes, F. S., Komoriya, K., Larquet, E., Yang, S., Egelman, E. H., Blocker, A., and Lea, S. M. (2003) J. Biol. Chem. 278 17103–17107 [DOI] [PubMed] [Google Scholar]
- 8.Menard, R., Sansonetti, P. J., and Parsot, C. (1993) J. Bacteriol. 175 5899–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espina, M., Olive, A. J., Kenjale, R., Moore, D. S., Ausar, S. F., Kaminski, R. W., Oaks, E. V., Middaugh, C. R., Picking, W. D., and Picking, W. L. (2006) Infect. Immun. 74 4391–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, S., Roversi, P., Espina, M., Olive, A., Deane, J. E., Birket, S., Field, T., Picking, W. D., Blocker, A. J., Galyov, E. E., Picking, W. L., and Lea, S. M. (2007) J. Biol. Chem. 282 4035–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olive, A. J., Kenjale, R., Espina, M., Moore, D. S., Picking, W. L., and Picking, W. D. (2007) Infect. Immun. 75 2626–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayward, R. D., Cain, R. J., McGhie, E. J., Phillips, N., Garner, M. J., and Koronakis, V. (2005) Mol. Microbiol. 56 590–603 [DOI] [PubMed] [Google Scholar]
- 13.Lafont, F., Tran Van Nhieu, G., Hanada, K., Sansonetti, P., and van der Goot, F. G. (2002) EMBO J. 21 4449–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skoudy, A., Mounier, J., Aruffo, A., Ohayon, H., Gounon, P., Sansonetti, P., and Tran Van Nhieu, G. (2000) Cell Microbiol. 2 19–33 [DOI] [PubMed] [Google Scholar]
- 15.Pope, L. M., Reed, K. E., and Payne, S. M. (1995) Infect. Immun. 63 3642–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquart, M. E., Picking, W. L., and Picking, W. D. (1995) Biochem. Biophys. Res. Commun. 214 963–970 [DOI] [PubMed] [Google Scholar]
- 17.Espina, M., Ausar, S. F., Middaugh, C. R., Baxter, M. A., Picking, W. D., and Picking, W. L. (2007) Protein Sci. 16 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington, A. T., Hearn, P. D., Picking, W. L., Barker, J. R., Wessel, A., and Picking, W. D. (2003) Infect. Immun. 71 1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odom, O. W., Picking, W. D., and Hardesty, B. (1990) Biochemistry 29 10734–10744 [DOI] [PubMed] [Google Scholar]
- 20.Lakowicz, J. R. (1983) Principles of Fluorescence Spectroscopy, Plenum Press, New York
- 21.van der Meer, B. W. (2002) J. Biotechnol. 82 181–196 [DOI] [PubMed] [Google Scholar]
- 22.Ermolenko, D. N., Majumdar, Z. K., Hickerson, R. P., Spiegel, P. C., Clegg, R. M., and Noller, H. F. (2007) J. Mol. Biol. 370 530–540 [DOI] [PubMed] [Google Scholar]
- 23.SYBYL, 7.1 Ed., The Tripos Associates, St. Louis, MO
- 24.Clark, M., Cramer, R. D., III, and Van Opdenbosch, N. (1989) J. Comput. Chem. 10 982–1012 [Google Scholar]
- 25.Gasteiger, J., and Marsili, M. (1978) Tetrahedron Lett. 34 2181–3184 [Google Scholar]
- 26.Chantalat, L., Jones, N. D., Korber, F., Navaza, J., and Pavlovsky, A. G. (1995) Protein Pept. Lett. 2 333–340 [Google Scholar]
- 27.Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., Hart, W. E., Belew, R. K., and Olson, A. J. (1998) J. Comput. Chem. 19 1639–1662 [Google Scholar]
- 28.Picking, W. L., Nishioka, H., Hearn, P. D., Baxter, M. A., Harrington, A. T., Blocker, A., and Picking, W. D. (2005) Infect. Immun. 73 1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Espina, M., Ausar, S. F., Middaugh, C. R., Picking, W. D., and Picking, W. L. (2006) Biochemistry 45 9219–9227 [DOI] [PubMed] [Google Scholar]
- 30.Harrington, A., Darboe, N., Kenjale, R., Picking, W. L., Middaugh, C. R., Birket, S., and Picking, W. D. (2006) Biochemistry 45 626–636 [DOI] [PubMed] [Google Scholar]
- 31.Zhang, L., Wang, Y., Olive, A. J., Smith, N. D., Picking, W. D., De Guzman, R. N., and Picking, W. L. (2007) J. Biol. Chem. 282 32144–32151 [DOI] [PubMed] [Google Scholar]
- 32.Mills, C. O., Rahman, K., Coleman, R., and Elias, E. (1991) Biochim. Biophys. Acta 1115 151–156 [DOI] [PubMed] [Google Scholar]
- 33.Mills, C. O., Milkiewicz, P., Molloy, D. P., Baxter, D. J., and Elias, E. (1997) Biochim. Biophys. Acta 1336 485–496 [DOI] [PubMed] [Google Scholar]
- 34.Mills, C. O., Milkiewicz, P., Saraswat, V., and Elias, E. (1997) Yale J. Biol. Med. 70 447–457 [PMC free article] [PubMed] [Google Scholar]
- 35.Baxter, D. J., Rahman, K., Bushell, A. J., Mills, C. O., Elias, E., and Billington, D. (1995) Biochim. Biophys. Acta 1256 374–380 [DOI] [PubMed] [Google Scholar]
- 36.El-Seaidy, A. Z., Mills, C. O., Elias, E., and Crawford, J. M. (1997) Am. J. Physiol. 272 G298–G309 [DOI] [PubMed] [Google Scholar]
- 37.Van Der Meer, B. W., Coker, G., III, and Simon Chen, S.-Y. (1994) Resonance Energy Transer: Theory, Wiley, New York
- 38.Druar, C., Yu, F., Barnes, J. L., Okinaka, R. T., Chantratita, N., Beg, S., Stratilo, C. W., Olive, A. J., Soltes, G., Russell, M. L., Limmathurotsakul, D., Norton, R. E., Ni, S. X., Picking, W. D., Jackson, P. J., Stewart, D. I., Tsvetnitsky, V., Picking, W. L., Cherwonogrodzky, J. W., Ketheesan, N., Peacock, S. J., and Wiersma, E. J. (2008) FEMS Immunol. Med. Microbiol. 52 78–87 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.