Abstract
Bif-1 interacts with Bax and enhances its conformational rearrangement, resulting in apoptosis. However, the molecular mechanism governing the interaction between Bif-1 and Bax is poorly defined. Here we provide evidence that Bif-1 is phosphorylated, an event that can be repressed by apoptotic stimuli. The protein kinase c-Src binds to and directly phosphorylates Bif-1 on tyrosine 80. Moreover, Src phosphorylation of Bif-1 suppresses the interaction between Bif-1 and Bax, resulting in the inhibition of Bax activation during anoikis. Together, these results suggest that phosphorylation of Bif-1 impairs its binding to Bax and represses apoptosis, providing another mechanism by which Src oncogenic signaling can prevent cell death.
Apoptosis is a multistep process that culminates in the self-destruction of an individual cell. This self-killing mechanism proceeds through a highly regulated set of events that ultimately leads to the activation of cellular cysteine proteases known as caspases (1). Bcl-2 family members regulate caspase activation by determining the integrity of the outer mitochondrial membrane (OMM).4 The Bcl-2 family is composed of opposing subsets of proteins, such as anti-apoptotic members Bcl-2, Bcl-XL, and Mcl-1, that protect the integrity of the OMM through inhibition of the multidomain pro-apoptotic proteins Bax and Bak whose activation is associated with mitochondrial membrane dysfunction. BH3-only proteins, also part of the Bcl-2 family, can promote Bax and Bak activation through binding and inhibiting the ability of anti-apoptotic proteins to inhibit Bax and Bak (2). It has also been suggested that a subset of BH3-only proteins, including tBid, Bim, and Puma, can directly interact with Bax and Bak and cause their activation (3). The loss of OMM integrity leads to the release of apoptogenic proteins such as cytochrome c and SMAC, which activate caspases and inhibit caspase inhibitory molecules, respectively (4).
Bax activation preceding the loss of OMM integrity is a multistep process involving conformational rearrangement, recruitment to the OMM, membrane insertion, oligomerization, and ultimately pore formation. Although a subset of the BH3-only proteins, including tBid, Bim and Puma, are likely able to directly activate Bax (3, 5, 6), it has been speculated that another membrane protein, or proteins, is also required for Bax activation as pretreatment of purified mitochondrial membranes with protease K prevents tBid-induced Bax oligomerization (7). Similarly, tBid and Bax can efficiently release preloaded dextran from outer mitochondrial vesicles compared with chemically defined protein-free liposomes (8). However, controversy persists as to which mitochondrial proteins are required for Bax activation (9). It should also be considered that not all apoptotic stimuli are equal and different pathways may be activated independently.
Interestingly, Bax pore formation in large unilamellar vesicles is accompanied by structural changes in the lipid bilayer caused by monolayer curvature (10), and Bax has a tendency to accumulate at sites of fission and fusion on the OMM where lipids are likely to deviate from a bilayer structure (11). Together, these previous findings suggest that Bax pore formation can be regulated by mitochondrial proteins and those that alter membrane curvature are increasingly likely to affect Bax conformational activation.
Bif-1, also known as SH3GLB1 or endophilin B1, was initially identified by two independent groups as a Bax-interacting molecule (12, 13) even though it lacks homology with the Bcl-2 family. Members of the endophilin family are known to bind membranes through their N-BAR domain and promote membrane curvature (14-16). Bif-1, like other members of the endophilin family, also contains a carboxyl-terminal Src homology 3 domain. Bif-1 localizes on the membranes of intracellular organelles such as the Golgi and mitochondria (17-19). Importantly, it has been determined that Bif-1 directly interacts with Bax and enhances the kinetics of apoptosis induction by promoting conformational activation of Bax and Bak in response to intrinsic apoptotic signals (12, 18).
Anoikis, Bax-dependent detachment-induced apoptosis, is a natural response of a cell to loss of integrin engagement or improper integrin signaling (20). This process is responsible for the prevention of metastasis by clearing potential tumorigenic cells from circulation (21). Src, the classic oncogenic kinase, is known to repress anoikis and promote metastatic dissemination of cancers (22, 23). Src-mediated inhibition of anoikis is dependent on its ability to repress Bax activation (24). We have recently shown that Src signaling can prevent the initiation of anoikis by inhibiting Mcl-1 degradation and Bim induction (25). However, depletion of Mcl-1 along with induction of Bim by blocking the Akt and ERK1/2 signaling is unable to elicit a robust anoikis response in the presence of Src activity. Therefore, Src must be acting at multiple levels to prevent Bax activation during detachment.
In this study, we found that Src interacts with Bif-1 and phosphorylates Bif-1 on tyrosine. This phosphorylation event has a direct impact on the ability of Bif-1 to bind Bax, which correlates with a repression of Bax activation during anoikis.
EXPERIMENTAL PROCEDURES
Metabolic Labeling and in Vitro Kinase Assays—For detection of Bif-1 phosphorylation, 293T cells were incubated for overnight in phosphate-free Dulbecco's modified Eagle's medium containing 5% dialyzed fetal bovine serum (FBS) and then labeled with 0.5-1.0 mCi of [32P]orthophosphate for 6 h in the presence of 10% dialyzed FBS. Plates were washed with ice-cold phosphate-buffered saline and lysed with radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors. Lysates were precleared with protein G-agarose prior to immunoprecipitation with anti-Bif-1 or anti-Myc monoclonal antibodies. For in vitro kinase assays, a recombinant c-Src was obtained from Upstate Biotechnology. The phosphorylation reaction was carried out at 30 °C for 15 min in 40 μl of reaction mixture (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 0.1 mm EGTA, 1 mm dithiothreitol, 0.015% Brij 35, 0.1 mg/ml bovine serum albumin, 10 μm ATP, 5 μCi of [γ-32P]ATP (3000 Ci/mmol), 5 units of Src kinase) containing 1 μg of indicated GST fusion protein. The reaction was stopped by addition of 14 μl of 4× SDS loading buffer and heat denaturation, resolved by SDS-PAGE, and analyzed by autoradiography.
Yeast Two-hybrid Assays—Two-hybrid assays were performed as described previously (12). Briefly, c-Src fused to the LexA DNA binding domain was transfected into Saccharomyces cerevisiae EGY48 cells with the full-length or deletion mutants of Bif-1 fused to the B42 transactivation domain. Five independent transformants were grown on either galactose- or glucose-containing agar plates to induce or repress β-galactosidase activity, respectively.
Expression and Purification of GST-Bif-1—The GST-Bif-1 wild type and deletion mutant proteins were expressed by pGEX-4T-1 plasmid in the DH5α strain of Escherichia coli. Briefly, transformed cells were grown in Luria Bertani medium containing ampicillin (100μg/ml) at 37 °C to an A600 nm of 0.8, and then 1 mm isopropyl-1-thio-β-d-galactopyranoside was added to induce protein expression at 37 °C for 3 h. Cells were lysed in PBS, pH 7.4, containing protease inhibitors by sonication and centrifuged at 14,000 × g for 30 min. The resulting supernatant was incubated with glutathione-Sepharose 4B (Amersham Biosciences) at 4 °C for 1 h and then washed three times with PBS. The protein was eluted with 10 mm reduced glutathione in 50 mm Tris-HCl, pH 8.0, and dialyzed against 50 mm Tris-HCl buffer, pH 7.5.
Immunoprecipitation and Immunoblot Assay—Cells were lysed using 1% Chaps lysis buffer (150 mm NaCl, 10 mm HEPES, pH 7.4, 1% Chaps) containing protease and phosphatase inhibitors. Immunocomplexes were pulled down with the indicated antibodies and washed three times in lysis buffer. Immunoprecipitates were resolved using SDS-PAGE and immunoblotted with the indicated antibodies. For detection of active Bax, anti-Bax 6A7 monoclonal antibody (Sigma) was used for immunoprecipitation and anti-Bax N20 polyclonal antibody (Santa Cruz Biotechnology) was used for subsequent immunoblot analysis. The Bax/Bif-1 interaction was determined by immunoprecipitation of Bax using polyclonal anti-mouse/rat Bax 1696 antiserum (26) followed by immunoblotting with anti-Bif-1 monoclonal antibody (Imgenex). The Src/Bif-1 interaction was determined in H1299 cell lysate in 1% Chaps lysis buffer by immunoprecipitation of Bif-1 with monoclonal antibody (Imgenex) and subsequent immunoblotting for Src (Cell Signaling catalogue number 2108). For detection of tyrosine phosphorylation of Bif-1, A431, 293T, and NIH3T3 cells were deprived of serum for 18 h in Dulbecco's modified Eagle's medium containing 0.1% bovine serum albumin. Cells were then incubated with Src inhibitors PD180970 or dasatinib for 2 h. Cells were then stimulated by the addition of epidermal growth factor to 50 ng/ml for 5 min, washed once with 5 ml of ice-cold PBS, and collected in 900 μl of Buffer A (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 25 mm NaF, 5 mm sodium pyrophosphate, 1 mm Na3VO4, 2 μg/ml aprotinin, 2 μg/ml leupeptin, 100 μg/ml phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, 20 mm p-nitrophenyl phosphate, 1% Triton X-100) as described previously (27). One mg of precleared cell lysate was incubated with Bif-1 rabbit polyclonal antibody for 1 h on ice, and 20 μl of protein-A agarose was added and rocked overnight at 4 °C. Beads were washed three times with Buffer A and then boiled in 20 μl of Laemmli sample buffer and resolved by SDS-PAGE. Proteins were transferred onto nitrocellulose membrane (Bio-Rad) and blocked in 3% chicken egg white albumin in Tris-buffered saline-Tween 20 for 2 h. The phosphorylated Bif-1 protein was detected with horseradish peroxidase-conjugated anti-phospho-tyrosine (PY20) mouse monoclonal antibody (BD Biosciences).
Retrovirus Production—Bif-1 wild-type and Y80F mutant cDNAs were cloned into the BglII-XhoI sites of the pKI retrovirus plasmid. For Src expression, active c-Src531 mutant was cloned into the EcoRI-XhoI sites of pBMN-IRES-GFP vector. Retrovirus production was carried out in Amphotropic 293T cells. Briefly, cells were transfected with the indicated retroviral vector using the calcium phosphate method overnight in the presence of 25 mm chloroquine. Medium was replaced the next morning, and 36-48 h later the viral-containing medium was collected and used to infect the target Bif-1-/- mouse embryonic fibroblasts (MEFs) in the presence of 8 μg/ml polybrene.
Cell Culture and Transfection—293T, NIH3T3, 3Y1, and Bif-1-/- MEFs were maintained in Dulbecco's modified Eagle's medium containing 10% FBS supplemented with 100 μg/ml streptomycin and 100 units/ml penicillin. Transfection was performed using the calcium phosphate precipitation method. For anoikis assays, the indicated MEFs were grown to ∼80-90% confluency and then forcibly detached on poly(2-hydroxyethyl methacrylate)-coated culture plates for 24 h in 1% FBS as described previously (25). Control attached cells were also cultured for 24 h in 1% FBS before collection.
Caspase-3 Assay—Caspase-3 activation was assayed as DEV-Dase activity with the caspase-3 fluorescence assay kit according to the manufacturer's recommendations (Sigma).
RESULTS
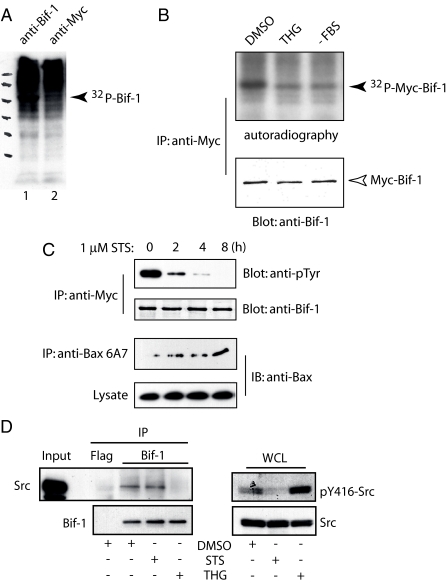
Bif-1 Phosphorylation in Vivo Is Repressed upon Apoptotic Stimuli—Because Bif-1 binding to Bax is induced by apoptotic stimuli (12, 18, 28), we reasoned that the ability of Bif-1 to activate Bax might be regulated by post-translational modifications. To determine whether Bif-1 could be phosphorylated in intact cells, 293T cells were metabolically labeled with 32PO4 and subjected to immunoprecipitation with anti-Bif-1 or control anti-Myc antibodies (Fig. 1A). A specific 32P-labeled band corresponding to Bif-1 was pulled down with anti-Bif-1, but not control anti-Myc, antibody. To determine the phosphorylation status of Bif-1 in stressed versus unstressed cells, 293T cells expressing Myc-tagged Bif-1 were metabolically labeled with 32PO4 and treated with DMSO, thapsigargin (THG), or serum starvation (Fig. 1B). The levels of phosphorylated Bif-1 were found to be highest in unstressed cells. Similarly, the level of tyrosine phosphorylation of Bif-1 was repressed in a time-dependent manner upon treatment with staurosporine (Fig. 1C, STS). The kinetics of Bif-1 dephosphorylation correlated with the activation of Bax as determined by immunoprecipitation of conformationally active Bax with the 6A7 monoclonal antibody (Fig. 1C). Together, these results clearly indicate that Bif-1 is phosphorylated in whole cells and apoptotic stresses promote the dephosphorylation of Bif-1.
FIGURE 1.
Bif-1 phosphorylation in whole cells is repressed after apoptotic stimuli. A, 293T cells were metabolically labeled with 32PO4 for 6 h, and lysates were collected and subjected to immunoprecipitation with anti-Bif-1 or anti-Myc antibodies followed by SDS-PAGE and autoradiography. B, 293T cells were transfected with Myc-Bif-1 and labeled with 32PO4 for 6 h after serum deprivation (-FBS) for 24 h or exposure to 2 μm thapsigargin (THG) or DMSO for 8 h. Myc-Bif-1 protein was immunoprecipitated with anti-Myc antibody and subjected to SDS-PAGE/immunoblot and autoradiography. C, 293T cells were transfected with Myc-Bif-1 and treated with 1 μm staurosporine (STS) for various times prior to immunoprecipitation/immunoblot analysis with the indicated antibodies. D, H1299 cells were treated with 1 μm STS, 2 μm THG, or DMSO for 8 h. Left panel, lysates were immunoprecipitated (IP) with anti-Bif-1 antibody or control FLAG antibody and then probed with anti-Src and anti-Bif-1 antibodies. Right panel, whole cell lysates (WCL) were probed for Src activity with anti-p416 antibody.
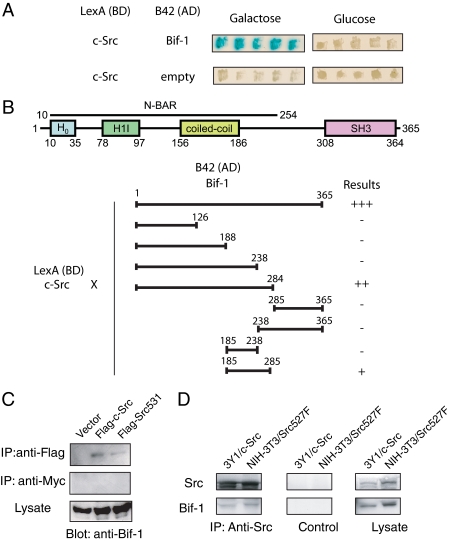
Src Interacts with Bif-1 in Both Yeast and Mammalian Cells—A search of Bif-1-interacting proteins through the use of a yeast two-hybrid assay resulted in the identification of c-Src kinase. The LexA-c-Src fusion protein specifically interacted with the wild type Bif-1 fused to the B42 transactivation domain in yeast (Fig. 2A). To map the binding site of Bif-1 to c-Src, a series of deletion mutants of Bif-1 were generated. It was found that full-length Bif-1 was the most effective at binding c-Src but appeared to require the presence of the region between amino acid residues 238-285 (Fig. 2B).
FIGURE 2.
Bif-1 interacts with Src in both yeast and mammalian cells. A, LexA-c-Src and B42-Bif-1 fusion proteins interact in yeast. B, schematic representation of the domain structure of Bif-1 and mapping of the Src binding domain of Bif-1. LexA-c-Src was transfected into yeast with the indicated deletion mutants of Bif-1 fused to B42. Interactions of pairs of fusion proteins were determined by visual examination of β-galactosidase activity and are represented graphically. SH3, Src homology 3. AD, transactivation domain; BD, DNA binding domain; H0, helix zero; H1I, helix one insert, N-BAR, N-terminal bin/amphiphysin/rvs. C, 293T cells were transiently transfected with pFLAG-tagged c-Src or c-Src531 or parental pFLAG-CMV2 vector for 2 days. Immunoprecipitation was performed using anti-FLAG antibody or anti-Myc (control) followed by SDS-PAGE/immunoblot analysis with anti-Bif-1 polyclonal antiserum. D, 3Y1 rat fibroblasts stably expressing c-Src or NIH3T3 cells stably expressing a constitutively active c-Src (Y527F) were subjected to immunoprecipitation with anti-c-Src or control antibody followed by SDS-PAGE/immunoblot analysis.
To determine whether Bif-1 was a binding partner for c-Src in mammalian cells, 293T cells were transfected with empty vector, FLAG-tagged c-Src, or the naturally occurring Src mutant Src531, which results in a truncated protein that has been shown to be activating, transforming, and tumorigenic (29). Immunoprecipitation with anti-FLAG antibody revealed that endogenous Bif-1 could indeed interact with both c-Src and mutant Src531 (Fig. 2C). Similarly, Bif-1 was found to interact with wild type c-Src and mutant c-Src (Y527F) in 3Y1 rat fibroblasts and NIH3T3 cells, respectively (Fig. 2D). Src binding to Bif-1 is therefore a common event with implications in Bif-1 regulation.
Because both STS and THG prevented the phosphorylation of Bif-1 (Fig. 1, B and C), we sought to determine whether apoptotic stimulation affects the binding of Src to Bif-1. As shown in Fig. 1D, H1299 cells treated with STS did not prevent the association of Bif-1 with Src at endogenous levels as compared with DMSO control but instead prevented the kinase activity of Src as determined by the activation-associated Tyr(P)-416 Western blot. Alternatively, THG treatment could effectively prevent the association between Bif-1 and Src. Notably, treating cells with THG did not suppress, but rather enhanced, the activation of Src, which is consistent with previous observations (30). Together, these results suggest that apoptotic stresses can prevent the phosphorylation of Bif-1 either through inhibition of Src interaction with Bif-1 or by preventing Src activity.
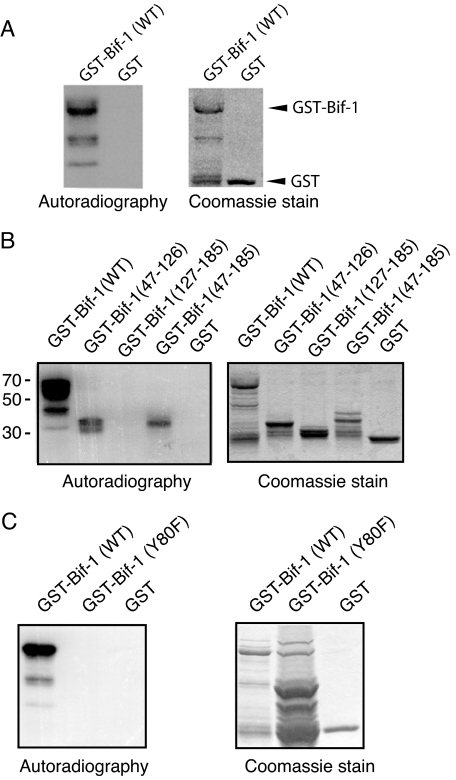
Src Phosphorylation of Bif-1 at Tyr-80—We next sought to determine whether Src could directly phosphorylate Bif-1 using an in vitro kinase assay. Incubation of recombinant c-Src with GST-Bif-1 fusion protein, but not GST alone, resulted in detectable phosphorylation determined by autoradiography (Fig. 3A). The subsequent Src phosphorylation site on Bif-1 was narrowed down through the use of deletion mutants (Fig. 3B). Src-mediated phosphorylation of Bif-1 occurred in the amino-terminal region of Bif-1 encompassing amino acids 47-126. A closer evaluation of this region revealed a potential Src phosphorylation consensus motif (EEFVY80EKLD), and substitution of Tyr-80 to Phe abrogated Src-mediated Bif-1 phosphorylation (Fig. 3C).
FIGURE 3.
Src phosphorylates Bif-1 at Tyr-80 in vitro. Recombinant c-Src kinase was incubated with GST, GST-Bif-1, or various GST-Bif-1 mutants and [γ-32P]ATP at 30 °C for 15 min before analysis by SDS-PAGE and autoradiography. Coomassie staining shows the input of GST and GST-Bif-1 proteins.
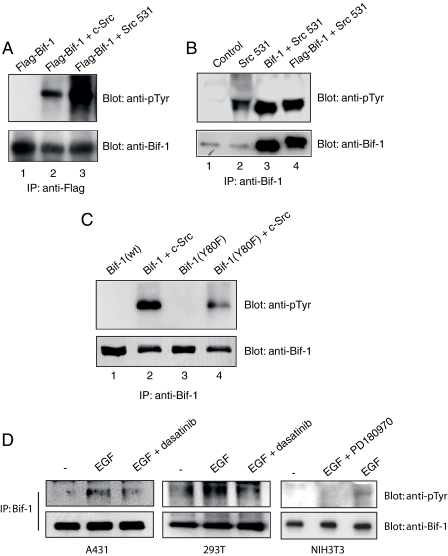
Src Phosphorylates Bif-1 at Tyr-80 in Intact Cells—Although the in vitro kinase assay illustrates that Src directly targets Tyr-80 in Bif-1, it remained to be determined whether this event could be recapitulated in whole cells. Therefore, 293T cells were co-transfected with FLAG-Bif-1 along with empty, c-Src, or Src531 constructs. Bif-1 was then immunoprecipitated from lysates using anti-FLAG antibody and subjected to immunoblotting with antibodies specific for phosphotyrosine or Bif-1 (Fig. 4A). Co-expression of Src with Bif-1 led to a marked increase in tyrosine phosphorylation of Bif-1. It was also found that expression of Src531 could induce the phosphorylation of both endogenous and ectopic Bif-1 regardless of tag presence in 293T cells (Fig. 4B).
FIGURE 4.
Src phosphorylates Bif-1 in intact cells. A, 293T cells were transiently co-transfected with FLAG-Bif-1 and either c-Src, c-Src531 mutant, or empty vector. After 2 days, FLAG-Bif-1 was immunoprecipitated with anti-FLAG antibody, followed by SDS-PAGE/immunoblot analysis with anti-Bif-1 rabbit antiserum or anti-phosphotyrosine PY20 antibody. B, 293T cells were transiently co-transfected with Src531 and plasmids encoding Bif-1 or FLAG-Bif-1 or parental vector DNA. Cells were lysed 2 days later, and Bif-1 was immunoprecipitated with anti-Bif-1 monoclonal antibody and subjected to SDS-PAGE/immunoblot analysis with antibodies specific for Bif-1 or phosphotyrosine. C, 293T cells were transfected with Bif-1 or Bif-1(Y80F) alone or together with c-Src expression plasmids. Bif-1 proteins were immunoprecipitated and analyzed 2 days later. D, A431, 293T, and NIH3T3 cells were treated with or without 500 nm PD180970 or 50 nm dasatinib for 2 h and then stimulated with 50 ng/ml recombinant epidermal growth factor (EGF) for 5 min. Cell lysates were prepared and subjected to immunoprecipitation with anti-Bif-1 antibody, followed by SDS-PAGE/immunoblot with anti-phosphotyrosine or anti-Bif-1 antibody.
By performing the same co-transfection experiments, we confirmed that Tyr-80 was the major phosphorylation target of Src as the Bif-1Y80F mutant showed a significant reduction in the level of tyrosine phosphorylation of Bif-1 (Fig. 4C). However, it appears as if there is some level of phosphorylation still present in the Bif-1Y80F mutant. This is likely due to phosphorylation at other tyrosine residues regulated by Src or the immunoprecipitation of endogenous Bif-1. Similarly, A431, 293T, and NIH3T3 cells were assayed for the ability of epidermal growth factor to stimulate endogenous Bif-1 phosphorylation (Fig. 4D). Epidermal growth factor was found to promote an increase in Bif-1 phosphorylation, which could be inhibited by co-treatment with the Src-specific inhibitor PD180970 in NIH3T3 cells as well as dasatinib in A431 and 293T cells, indicating that this phosphorylation is associated with intracellular signals that are associated with oncogenic suppression of apoptosis.
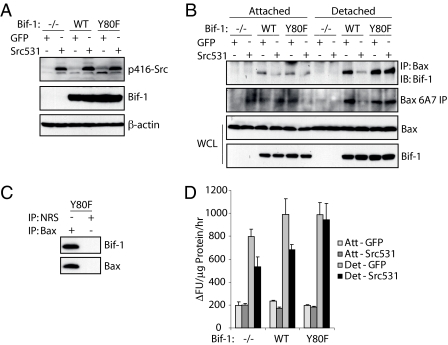
Bif-1Y80 Phosphorylation by Src Prevents Bif-1-mediated Bax Activation during Anoikis—Based on the previous knowledge of Bif-1 activation of Bax upon apoptotic stimuli and our current findings of Src-mediated phosphorylation of Bif-1, we sought to determine the role of Src-mediated Bif-1 phosphorylation in the context of anoikis initiation. Bif-1-/- MEFs expressing empty control vector (pKI) or re-established wild type or Y80F mutant Bif-1 in the presence of Src531 or control green fluorescence protein expression were created (Fig. 5A) and detached on poly(2-hydroxyethyl methacrylate)-coated plates for 24 h or left attached on normal culture dishes. Lysates were then collected and the interaction between Bif-1 and Bax was determined (Fig. 5B). Detachment caused an increase in Bif-1/Bax binding, consistent with our previous findings that apoptotic stimuli increase this interaction (12, 18). The Bif-1Y80F mutant was able to bind Bax with the same affinity as wild type. Importantly, co-expression of Src531 with wild type Bif-1 repressed the ability of Bif-1 to interact with Bax in response to detachment. However, the non-phosphorylatable Bif-1Y80F mutant retained Bax binding potential even in the presence of Src expression (Fig. 5B). This interaction was specific, as immunoprecipitation with normal rabbit serum (NRS) was unable to pull down either Bax or Bif-1 (Fig. 5C).
FIGURE 5.
Src phosphorylation of Bif-1 inhibits Bax conformational activation during anoikis. A, Bif-1-/- MEFs were infected with retrovirus encoding empty control, Bif-1 wild type (WT), or Bif-1 mutant (Y80F) and selected on 1.0 μg/ml puromycin for 10 days. These cells were then re-infected with retrovirus encoding green fluorescence proteins (GFP) or Src531 proteins. Expression of the transgenes was confirmed by immunoblot. B, the cells created in A were left attached or detached on poly(2-hydroxyethyl methacrylate)-coated plates for 24 h. Lysates were prepared and subjected to immunoprecipitation with anti-Bax polyclonal antiserum or monoclonal 6A7 antibody. The resulting immunocomplexes and whole cell lysates were analyzed by immunoblotting with anti-Bax or anti-Bif-1 antibody. C, lysates prepared from Bif-1(Y80F) cells detached for 24 h were subjected to immunoprecipitation with anti-Bax polyclonal antibodies or normal rabbit serum. D, caspase-3 assay was performed on the samples from B. The data shown are the means ± S.D. (n = 3). Det, detached; GFP, green fluorescence protein.
Detachment-induced Bax activation was increased in cells expressing Bif-1. Importantly, the enhanced Bax conformational change was inhibited by co-expressing Src531 in wild type Bif-1, but not Bif-1Y80F, cells (Fig. 5B). Similarly, co-expression of Src531 could partially rescue cells expressing empty or wild type Bif-1 from caspase-3 activation, but overexpression of Src531 had no inhibitory effect on cells expressing the non-phosphorylatable Bif-1Y80F mutant (Fig. 5D). These results were confirmed by a cell death assay measuring lactate dehydrogenase release (data not shown). These findings clearly indicate that Src phosphorylation of Bif-1 has direct biological impacts that affect the ability of a cell to undergo apoptosis.
DISCUSSION
Bax activation is known to control the initiation of apoptosis. However, the exact mechanism by which the conformational rearrangement and insertion of Bax into the OMM is controlled remains elusive. Although it is generally believed that activating BH3-only molecules such as tBid, Puma, and Bim can directly facilitate the activation of Bax, there remains debate as to whether Bax can directly interact with these molecules (3, 31). Furthermore, it appears that activating molecules, other than BH3-only proteins, are crucial to the ability of Bax to form pores in membranes (7, 9, 32). These observations have led to the pursuit and discovery of novel Bax-interacting proteins such as Bif-1.
Identification of Bif-1 as a novel Bax-interacting factor has led to the finding that Bif-1 is able to potentiate the activation of Bax as well as Bak (12, 18). Reasons why Bif-1 is able to enhance the activation of Bax are not entirely understood. One possible explanation is that Bif-1 can alter the mitochondrial membrane structure such that it becomes suitable for conformational rearrangement and/or insertion of Bax into the mitochondria. As a member of the endophilin family of proteins, Bif-1 contains the N-BAR domain, which is known to promote membrane curvature (14, 16). Given that Bax accumulates at fission and fusion sites on the mitochondrial membrane and that Bif-1 is known to regulate mitochondrial morphology (11, 19), it is likely that the Bif-1/Bax interaction observed is the consequence of changes in membrane structure mediated by Bif-1. Interestingly, the use of deletion mutants has identified the first eleven amino acids in Bif-1 as essential for its ability to bind Bax (13). This region of Bif-1 is a part of the amino-terminal amphipathic helix known as helix zero, which is essential for binding and tubulating membranes (16, 33). The overlapping Bax and membrane binding functions of this region in Bif-1 may indicate that the interaction between Bif-1 and Bax is dependent on membrane dynamics.
Given the aforementioned role of Bif-1 in the activation of Bax and apoptosis (12, 18), as well as its newly defined role in autophagy (34), the mechanisms that regulate Bif-1 function are of increasing importance. Yeast two-hybrid analysis has identified Src as a Bif-1-interacting protein, an association that is also evident in both exogenous and endogenous expression systems. Furthermore, we have shown that Src can directly phosphorylate Bif-1 at Tyr-80. This residue is part of the internal amphipathic helix common to members of the endophilin family but weakly conserved therein. Previous deletion mutation analysis revealed that this domain is required for membrane binding and tubulating activities of endophilin A1 (16). Similarly, mutating positively charged residues to negative ones in this region of endophilin A1 inhibits membrane binding and tubulation (16). Phosphorylation of Tyr-80 on Bif-1 is likely to alter membrane binding by changing the charge distribution in the internal amphipathic helix, as well as altering the helix organization with the addition of a bulky phosphate group. Mutant Bif-1Y80F would lack the ability of Src to alter the charge structure of the helix and allow binding to membranes, which could explain why this mutant maintained Bax binding potential in cells with active Src. Future studies will be needed to determine specifically how Src-mediated phosphorylation at Tyr-80 of Bif-1 affects helix organization, membrane binding, and tubulation potential of the protein.
This work was supported, in whole or in part, by the National Institutes of Health. This work was also supported by the American Cancer Society and the Flight Attendant Medical Research Institute. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: OMM, outer mitochondrial membrane; ERK1/2, extracellular signal-regulated kinase 1/2; FBS, fetal bovine serum; THG, thapsigargin; STS, staurosporine; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; tBid, truncated Bid; MEF, mouse embryonic fibroblast; GST, glutathione S-transferase.
References
- 1.Wolf, B. B., and Green, D. R. (1999) J. Biol. Chem. 274 20049-20052 [DOI] [PubMed] [Google Scholar]
- 2.Green, D. R. (2006) Cancer Cell 9 328-330 [DOI] [PubMed] [Google Scholar]
- 3.Kim, H., Rafiuddin-Shah, M., Tu, H. C., Jeffers, J. R., Zambetti, G. P., Hsieh, J. J., and Cheng, E. H. (2006) Nat. Cell Biol. 8 1348-1358 [DOI] [PubMed] [Google Scholar]
- 4.Green, D. R. (2005) Cell 121 671-674 [DOI] [PubMed] [Google Scholar]
- 5.Kuwana, T., Bouchier-Hayes, L., Chipuk, J. E., Bonzon, C., Sullivan, B. A., Green, D. R., and Newmeyer, D. D. (2005) Mol. Cell 17 525-535 [DOI] [PubMed] [Google Scholar]
- 6.Letai, A., Bassik, M. C., Walensky, L. D., Sorcinelli, M. D., Weiler, S., and Korsmeyer, S. J. (2002) Cancer Cell 2 183-192 [DOI] [PubMed] [Google Scholar]
- 7.Roucou, X., Montessuit, S., Antonsson, B., and Martinou, J. C. (2002) Biochem. J. 368 Pt. 3, 915-921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuwana, T., Mackey, M. R., Perkins, G., Ellisman, M. H., Latterich, M., Schneiter, R., Green, D. R., and Newmeyer, D. D. (2002) Cell 111 331-342 [DOI] [PubMed] [Google Scholar]
- 9.Ott, M., Norberg, E., Walter, K. M., Schreiner, P., Kemper, C., Rapaport, D., Zhivotovsky, B., and Orrenius, S. (2007) J. Biol. Chem. 282 27633-27639 [DOI] [PubMed] [Google Scholar]
- 10.Terrones, O., Antonsson, B., Yamaguchi, H., Wang, H. G., Liu, J., Lee, R. M., Herrmann, A., and Basanez, G. (2004) J. Biol. Chem. 279 30081-30091 [DOI] [PubMed] [Google Scholar]
- 11.Karbowski, M., Lee, Y. J., Gaume, B., Jeong, S. Y., Frank, S., Nechushtan, A., Santel, A., Fuller, M., Smith, C. L., and Youle, R. J. (2002) J. Cell Biol. 159 931-938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuddeback, S. M., Yamaguchi, H., Komatsu, K., Miyashita, T., Yamada, M., Wu, C., Singh, S., and Wang, H. G. (2001) J. Biol. Chem. 276 20559-20565 [DOI] [PubMed] [Google Scholar]
- 13.Pierrat, B., Simonen, M., Cueto, M., Mestan, J., Ferrigno, P., and Heim, J. (2001) Genomics 71 222-234 [DOI] [PubMed] [Google Scholar]
- 14.Peter, B. J., Kent, H. M., Mills, I. G., Vallis, Y., Butler, P. J., Evans, P. R., and McMahon, H. T. (2004) Science 303 495-499 [DOI] [PubMed] [Google Scholar]
- 15.Masuda, M., Takeda, S., Sone, M., Ohki, T., Mori, H., Kamioka, Y., and Mochizuki, N. (2006) EMBO J. 25 2889-2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallop, J. L., Jao, C. C., Kent, H. M., Butler, P. J., Evans, P. R., Langen, R., and McMahon, H. T. (2006) EMBO J. 25 2898-2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, J. S., Zhang, L., Lee, S. Y., Gad, H., Luini, A., and Hsu, V. W. (2006) Nat. Cell Biol. 8 1376-1382 [DOI] [PubMed] [Google Scholar]
- 18.Takahashi, Y., Karbowski, M., Yamaguchi, H., Kazi, A., Wu, J., Sebti, S. M., Youle, R. J., and Wang, H. G. (2005) Mol. Cell. Biol. 25 9369-9382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karbowski, M., Jeong, S. Y., and Youle, R. J. (2004) J. Cell Biol. 166 1027-1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frisch, S. M., and Francis, H. (1994) J. Cell Biol. 124 619-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douma, S., Van Laar, T., Zevenhoven, J., Meuwissen, R., Van Garderen, E., and Peeper, D. S. (2004) Nature 430 1034-1039 [DOI] [PubMed] [Google Scholar]
- 22.Windham, T. C., Parikh, N. U., Siwak, D. R., Summy, J. M., McConkey, D. J., Kraker, A. J., and Gallick, G. E. (2002) Oncogene 21 7797-7807 [DOI] [PubMed] [Google Scholar]
- 23.Tan, M., Li, P., Klos, K. S., Lu, J., Lan, K. H., Nagata, Y., Fang, D., Jing, T., and Yu, D. (2005) Cancer Res. 65 1858-1867 [DOI] [PubMed] [Google Scholar]
- 24.Gilmore, A. P., Metcalfe, A. D., Romer, L. H., and Streuli, C. H. (2000) J. Cell Biol. 149 431-446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woods, N. T., Yamaguchi, H., Lee, F. Y., Bhalla, K. N., and Wang, H. G. (2007) Cancer Res. 67 10744-10752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krajewski, S., Krajewska, M., Shabaik, A., Miyashita, T., Wang, H.-G., and Reed, J. C. (1994) Am. J. Pathol. 145 1323-1333 [PMC free article] [PubMed] [Google Scholar]
- 27.Ren, Y., Meng, S., Mei, L., Zhao, Z. J., Jove, R., and Wu, J. (2004) J. Biol. Chem. 279 8497-8505 [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi, H., Paranawithana, S. R., Lee, M. W., Huang, Z., Bhalla, K. N., and Wang, H.-G. (2002) Cancer Res. 62 466-471 [PubMed] [Google Scholar]
- 29.Irby, R. B., Mao, W., Coppola, D., Kang, J., Loubeau, J. M., Trudeau, W., Karl, R., Fujita, D. J., Jove, R., and Yeatman, T. J. (1999) Nat. Genet. 21 187-190 [DOI] [PubMed] [Google Scholar]
- 30.Chung, K. C., Sung, J. Y., Ahn, W., Rhim, H., Oh, T. H., Lee, M. G., and Ahn, Y. S. (2001) J. Biol. Chem. 276 2132-2138 [DOI] [PubMed] [Google Scholar]
- 31.Willis, S. N., Fletcher, J. I., Kaufmann, T., van Delft, M. F., Chen, L., Czabotar, P. E., Ierino, H., Lee, E. F., Fairlie, W. D., Bouillet, P., Strasser, A., Kluck, R. M., Adams, J. M., and Huang, D. C. (2007) Science 315 856-859 [DOI] [PubMed] [Google Scholar]
- 32.Lutter, M., Fang, M., Luo, X., Nishijima, M., Xie, X., and Wang, X. (2000) Nat. Cell Biol. 2 754-761 [DOI] [PubMed] [Google Scholar]
- 33.Farsad, K., Ringstad, N., Takei, K., Floyd, S. R., Rose, K., and De Camilli, P. (2001) J. Cell Biol. 155 193-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi, Y., Coppola, D., Matsushita, N., Cualing, H. D., Sun, M., Sato, Y., Liang, C., Jung, J. U., Cheng, J. Q., Mul, J. J., Pledger, W. J., and Wang, H. G. (2007) Nat. Cell Biol. 9 1142-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]