Abstract
Zymogens of the chymotrypsin-like serine protease family are converted to the protease state following insertion of a newly formed, highly conserved N terminus. This transition is accompanied by active site formation and ordering of several surface loops in the catalytic domain. Here we show that disruption of this transition in factor X through mutagenesis (FXaI16L and FXaV17A) not only alters active site function, but also significantly impairs Na+ and factor Va binding. Active site binding was improved in the presence of high NaCl or with saturating amounts of factor Va membranes, suggesting that allosteric linkage exists between these sites. In line with this, irreversible stabilization of FXaI16L with Glu-Gly-Arg-chloromethyl ketone fully rescued FVa binding. Furthermore, the Km for prothrombin conversion with the factor Xa variants assembled into prothrombinase was unaltered, whereas the kcat was modestly reduced (3- to 4-fold). These findings show that intramolecular activation of factor X following the zymogen to protease transition not only drives catalytic site activation but also contributes to the formation of the Na+ and factor Va binding sites. This structural plasticity of the catalytic domain plays a key role in the regulation of exosite expression and prothrombinase assembly.
Proteolysis of zymogen proteins to their enzymatically active form is a central feature of many physiological processes, including blood coagulation (1). The paradigm for this type of activation mechanism is the zymogen to protease transition in the chymotrypsin-like serine protease family. Bond cleavage at a highly conserved site (Arg15–Ile16; the chymotrypsin numbering system is used throughout (2)) unmasks a new N terminus, which becomes an intramolecular ligand for Asp194 within a hydrophobic environment (3, 4). This transition is associated with a conformational change in the so-called “activation domain” (residues comprising 16–19, 142–152, 184–193, and 216–223) and is responsible for ordering the primary specificity pocket and oxyanion hole (4). These structural changes lead to the maturation of the active serine protease and impart function.
The zymogen and the mature protease exist in an equilibrium that typically favors the zymogen; however, this equilibrium can be shifted depending on various conditions. Bode and colleagues (5, 6) have elegantly demonstrated that trypsinogen can adopt an active trypsin-like structure upon strong ligand binding to the S1 specificity pocket2 and upon binding suitable ligands with high affinity for the Ile16 cleft. Additional examples of this induction to the protease state without cleavage of the Arg15–Ile16 bond include the binding of streptokinase to plasminogen, staphylocoagulase to prothrombin, and an autoantibody to prothrombin (7–11). Conversely, trypsin mutants harboring changes at Ile16 or Val17 or at loops flanking glycine hinge regions in the activation domain can unfavorably alter protease function (12–14). Collectively, these studies provide evidence that serine proteases, either in their zymogen or protease forms, can adopt alternate states with different functions.
Factor X (FX)3 is also conformationally activated following its conversion from the zymogen to the protease state (15, 16). Factor X is converted to factor Xa (FXa) by either the extrinsic (tissue factor/factor VIIa) or intrinsic (factor VIIIa/factor IXa) tenase enzymatic complexes following cleavage of the Arg15– Ile16 scissile bond, thereby liberating a 52-amino acid activation peptide (17). Factor Xa reversibly associates with its cofactor factor Va (FVa) on an anionic membrane surface in the presence of Ca2+ ions to form prothrombinase, the physiological activator of prothrombin (17). The interaction between membrane-bound FVa and FXa is mediated, at least in part, by a solvent-exposed α-helix (163–170) and residues within the heparin binding site (exosite II region) on the catalytic domain (18, 19). This helix is adjacent to the Na+ binding site (residues 183–189 and 221–225) and S1 specificity site; both of which are part of the activation domain.
In contrast to FXa, the zymogen FX is not known to bind FVa (20–22), indicating that an important step in the maturation of the protease is the exposure of the cofactor binding site. Although this could be due to removal of the large activation peptide, another possibility is that conformational activation of the protease domain following the zymogen to protease transition leads to the expression of the FVa binding site. Indirect evidence to support this comes from a recent study from our laboratory using a FXa Na+ binding site mutant (Y225P) (23). This mutant had markedly reduced Na+ binding and, surprisingly, had a destabilized Ile16–Asp194 salt bridge making the derivative “zymogen-like.” As a result, we and others have shown that the mutant has a lower affinity for both FVa and active site-directed probes (23–25). Similar results with respect to N-terminal stabilization and cofactor binding have also been found using factor IXaY225P (26).
In the current study, we have exploited the well defined mechanism of serine protease activation and prepared FXa variants with zymogen-like properties. These derivatives were employed to directly examine the relationship between the zymogen to protease transition and the expression of discrete structural determinants that govern the function of FXa.
EXPERIMENTAL PROCEDURES
Materials—The fluorophore 4-aminobenzamidine (pAB) was from Aldrich, and its concentration was determined in water using E293 = 15,000 m–1 cm–1 (27). Glutamylglycylarginyl chloromethyl ketone (EGR-CH2Cl) was obtained from EMD Chemicals, Inc. (San Diego, CA). The peptidyl substrate methoxycarbonylcyclohexylglycylglycylarginine-p-nitroanilide (SpecXa) was from American Diagnostica (Greenwich, CT). H-d-Phenylalanylpipecolylarginine-p-nitroanilide (S-2238) and N-α-benzyloxycarbonylarginylglycylarginine-p-nitroanilide (S-2765) were purchased from Diapharma Group, Inc. (West Chester, OH). Substrate solutions were prepared in water, and concentrations were verified using E342 = 8270 m–1 cm–1 (28). All tissue culture reagents were from Invitrogen (Carlsbad, CA) except insulin-transferrin-sodium selenite, which was from Roche Applied Science. Small unilamellar phospholipid vesicles (PCPS) composed of 75% (w/w) hen egg l-α-phosphatidylcholine and 25% (w/w) porcine brain l-α-phosphatidylserine (Avanti Polar Lipids, Alabaster, AL) were prepared and characterized as described previously (29).
Proteins—Human prothrombin, FX and factor V (FV) were isolated from plasma as described previously (30–32). Thrombin and prethrombin-1 were prepared and purified by established procedures (33, 34). The FX activator from Russell's viper venom (RVVX-CP) was purified as described previously (35). Human FVa was prepared by proteolytic activation of FV by thrombin and purified as described before (36). Oregon Green488-FXa (OG488-FXa) was prepared as described (30). Molecular weights and extinction coefficients (E0.1% 280 nm) of the various proteins used were taken as follows: RVVX-CP, 93,000 and 1.18 (37); prothrombin, 72,000 and 1.47 (33); prethrombin-1, 49,900 and 1.78 (33); thrombin, 37,500 and 1.94 (38); FVa, 173,000 and 1.78 (39); FX, 59,000 and 1.16 (40); and FXa, 46,000 and 1.16 (40).
Preparation of Recombinant FX—Recombinant FX (rFX) variants were generated with the QuikChange site-directed mutagenesis kit (Stratagene) using appropriate mutagenic complementary oligonucleotides. For each variant, the entire FX cDNA was sequenced to confirm the presence of the desired mutation and to ensure that there were no polymerase induced errors. Wild-type or mutant rFX in the mammalian expression plasmid pCMV4 were stably expressed in human embryonic kidney (HEK) 293 cells and purified as previously described (41, 42).
Preparation of FXa—Plasma-derived FX (PDFX) and rFX (wild-type or variants) were activated using RVVX-CP and subsequently purified using benzamidine-Sepharose or Sephacryl S-200 as described (34, 41). Following purification, the proteins were stored in 50% glycerol at –20 °C.
Preparation of EGR-FXa—Recombinant FXa (7 mg) or rFXaI16L (10 mg) was incubated with a 5- or 100-fold molar excess, respectively, of EGR-CH2Cl. Following modification, the proteins were applied to a Sephacryl S-200 column equilibrated in 20 mm Hepes/150 mm NaCl/5 mm EDTA, pH 7.4. Fractions were pooled, precipitated with ammonium sulfate, collected by centrifugation, dissolved in 50% glycerol, and stored at –20 °C. EGR-rFXa and EGR-rFXaI16L have <0.1% chromogenic activity toward SpecXa.
Characterization of FX and FXa—Protein purity was assessed by SDS-PAGE using 4–12% gels (Invitrogen) under reducing (50 mm dithiothreitol, final) and non-reducing conditions using the MES buffer system followed by staining with Coomassie Brilliant Blue R-250. N-terminal sequence analysis was performed in the laboratory of Dr. Alex Kurosky and Steven Smith at the University of Texas Medical Branch at Galveston. Chemical γ-carboxyglutamic acid (Gla) analysis was carried out in our laboratory as described (41, 42). This analysis indicates that rFX, rFXI16L, and rFXV17A have essentially the full complement of Gla residues (10.5–10.8 mol of Gla/mol of FX) compared with PDFX (10.7 mol of Gla/mol of FX; theoretical = 11 mol of Gla/mol of FX).
Determination of Kinetic Parameters for Peptidyl Substrate Hydrolysis—All kinetic measurements were performed in 20 mm Hepes, 0.15 m NaCl, 0.1% (w/v) polyethylene glycol-8000, 2 mm CaCl2, pH 7.5 (assay buffer), unless otherwise indicated. The kinetic parameters of peptidyl substrate hydrolysis (SpecXa) were measured using increasing concentrations of substrate (10–500 μm) and initiated with either free FXa (2.0 nm wild-type FXa; 6.0 nm mutant FXa) or FXa assembled into prothrombinase (5 nm FXa, 30 nm FVa, and 50 μm PCPS).
Carbamylation of Ile16 by Reaction with NaNCO—Mixtures containing wild-type or mutated FXa (2 μm) in assay buffer were reacted with 0.2 m NaNCO essentially as described (23). The final pH of the reaction mixture upon addition of NaNCO was pH 7.45. At selected time intervals (15–300 min) an aliquot of the reaction mixture was diluted in assay buffer, and the residual enzymatic activity was determined from initial steadystate rates of SpecXa hydrolysis.
Inhibition of FXa and Prothrombinase by Pefabloc tPA/Xa—The inhibitory constant (Ki) of Pefabloc tPA/Xa (Pefabloc, Pentapharm, Basel, Switzerland) for FXa or prothrombinase was assessed assuming classic competitive inhibition by initial velocity measurements of chromogenic substrate (S-2765) hydrolysis. Reaction mixtures (200 μl) contained: Pefabloc (0.02–5.0 μm), FXa (3 nm wild-type or 10 nm mutant FXa), or in the case of prothrombinase, PCPS (60 μm) and FVa (20 nm or 100 nm, for wild-type and mutant rFXa, respectively). Reactions were prepared in the wells of a 96-well plate and allowed to incubate for 2 min at room temperature. The reaction was initiated with S-2765 (100 μm or 500 μm, for wild-type and mutant rFXa, respectively).
Inhibition of FXa and Prothrombinase by pAB—The inhibitory constant (Ki) of pAB for FXa or prothrombinase was assessed assuming classic competitive inhibition. Initial velocity measurements of S-2765 hydrolysis for free FXa (1.0 or 3.0 nm, for wild-type or mutant rFXa) or prothrombinase (50 μm PCPS, 20 nm FVa, and 1.0 or 3.0 nm, for wild-type or mutant rFXa) were made in the presence of increasing concentrations of pAB (0–1.0 mm) in assay buffer.
Functional Binding Studies: The Effect of FVa on Peptidyl Substrate Cleavage by FXaI16L—Reaction mixtures (200 μl) containing SpecXa (10–800 μm), PCPS (50 μm), with different fixed concentrations of FVa (1, 2, 5, 8, 12, 18, and 25 nm) were prepared in the wells of a 96-well plate and allowed to incubate for 5 min at room temperature. Because the substrate stock solution was prepared in water, an appropriate volume of 10× assay buffer solution adjusted to pH 7.75 was added to each mixture to ensure that the final pH and concentration of buffer solutes was invariant. The reaction was initiated with rFXaI16L (10 nm, final).
Assessment of Na+ Binding—Cleavage of SpecXa (100 μm) was measured using either 2 nm wild-type or 50 nm mutant rFXa in the presence of increasing concentrations of Na+ (0–450 mm) in 50 mm Tris, 5 mm CaCl2, 0.1% polyethylene glycol 8000, pH 7.5, at 25 °C. To adjust the ionic strength of the reaction buffer, LiCl was used as the compensating chloride salt.
Fluorescence Intensity Measurements—Reaction mixtures containing 10 nm OG488-FXa, 50 μm PCPS, and 8 or 10 nm FVa in assay buffer were titrated with increasing concentrations of a non-fluorescent FXa competitor (i.e. rFXaS195A) at 25 °C in 1 × 1-cm2 stirred quartz cuvettes, and steady-state fluorescence intensity was measured using λex = 480 and λem = 520 nm with a long pass filter (KV500, Schott, Duryea, PA) in the emission beam essentially as described (30).
Kinetics of Protein Substrate Cleavage—The kinetic parameters of prothrombinase-catalyzed prothrombin activation (Km and Vmax) were determined in assay buffer by measuring the initial rate of thrombin formation at increasing concentrations of macromolecular substrate as described (23, 43, 44). Assay mixtures contained PCPS vesicles (20 μm), FVa (20 nm), and various concentrations of prothrombin (0–2.0 μm) or prethrombin-1 (0–12 μm). The reaction was initiated with FXa (0.1 nm and 0.4 nm for wild-type rFXa and mutant rFXa, respectively) for prothrombin or 0.5 nm for prethrombin-1. Prothrombin cleavage by FXa alone was measured using 1.4 μm prothrombin, 50 μm PCPS, and 10 nm rFXa or 50 nm FXaI16L. The initial rate of thrombin generation was measured as described (43).
Data Analysis—Data were analyzed according to the referenced equations by nonlinear least squares regression analysis using the Marquardt algorithm (45). The qualities of the fits were assessed by the criteria described (46). Reported estimates of error represent ± 2 S.D.
For competition experiments with the non-fluorescent derivatives of FXa, titration curves were analyzed as described previously (47, 48). Non-linear least square regression analysis, assuming both n and ncomp equal to 1, yielded fitted values of Kd, fixed describing the binding of OG488-FXa to FVa on the membrane surface and Kd, comp describing the equivalent interaction between the competitor FXa species and FVa membranes.
Initial velocity measurements of peptidyl substrate or macromolecular substrate hydrolysis by FXa or prothrombinase were analyzed by fitting the data to the Henri-Michaelis-Menten equation (49), to yield fitted values for Km and Vmax.
Initial velocity measurements of SpecXa hydrolysis by FXa or prothrombinase using increasing concentrations of pAB or Pefabloc were analyzed according to the rate expression for classical competitive inhibition (49), to yield the fitted value for Ki.
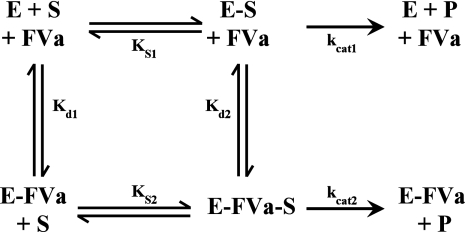
Global Analysis of Initial Velocity Data: Effect of FVa on Peptidyl Substrate Cleavage by rFXaI16L—The equilibrium dissociation constants for the binding of FVa to membrane-bound rFXaI16L in the absence (Kd1) and presence of peptidyl substrate (Kd2), as well as the substrate dissociation constant for rFXaI16L (Ks1) and membrane-bound FXa saturated with FVa (Ks2), were calculated from initial velocity measurements of SpecXa hydrolysis at different fixed concentrations of FVa, according to the system of ordinary differential equations that comprise SCHEME 1 and by using the rapid equilibrium assumption. The entire data set was globally fit using the program Dynafit (50) to extract Ks1, Kd1, kcat1, Ks2, Kd2, and kcat2.
SCHEME 1.
RESULTS
Generation of Zymogen-like FXa Variants—To investigate the role of N-terminal insertion on the expression of various structural determinants on FXa we modified positions 16 or 17 with the intent of creating zymogen-like variants. Ile16 was modified to Leu, Phe, Asp, or Gly, and position Val17 was changed to Leu, Ala, or Gly. Variants were initially transiently transfected in HEK 293 cells, and 48 h post-transfection conditioned media was collected. Subsequently, FX antigen levels were determined by enzyme-linked immunosorbent assay, and FXa activity levels were assessed by a chromogenic-based assay following activation with RVVX-CP. Whereas total antigen was comparable to wild-type FX, activity levels following processing with RVVX-CP varied among the mutants and ranged from <1% to 25% (data not shown). We speculate that these differences in activity reflect the extent to which the zymogen to protease transition was perturbed, with the lowest activity variants being more zymogen-like. Two mutants, rFXaI16L and rFXaV17A, whose activities were 5 and 3% of wild-type, respectively, were chosen for further study.
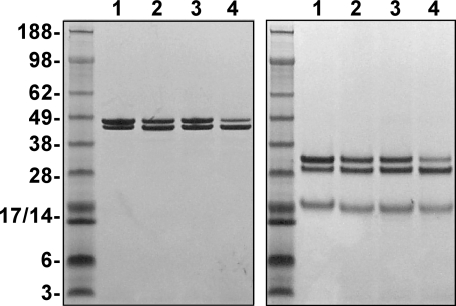
Expression and Purification of Recombinant Proteins—We established high producing stable clones expressing rFX, rFXI16L, or rFXV17A in HEK 293 cells. Each protein was purified to homogeneity and activated using established procedures (23, 42). SDS-PAGE analysis before and after disulfide bond reduction showed that the proteases were purified to homogeneity and consisted of the characteristic α- and β-forms of FXa (Fig. 1). N-terminal sequence analysis (data not shown) and chemical Gla analysis each yielded the expected results.
FIGURE 1.
SDS-PAGE analysis. Purified proteins (4 μg/lane) were subjected to SDS-PAGE under non-reducing (left) or reducing (right) conditions and visualized by staining with Coomassie Blue R-250. Lane 1, human PDFXa; lane 2, rFXa; lane 3, rFXaV17A; and lane 4, rFXaI16L. The apparent molecular weights of the standards are indicated on the gels.
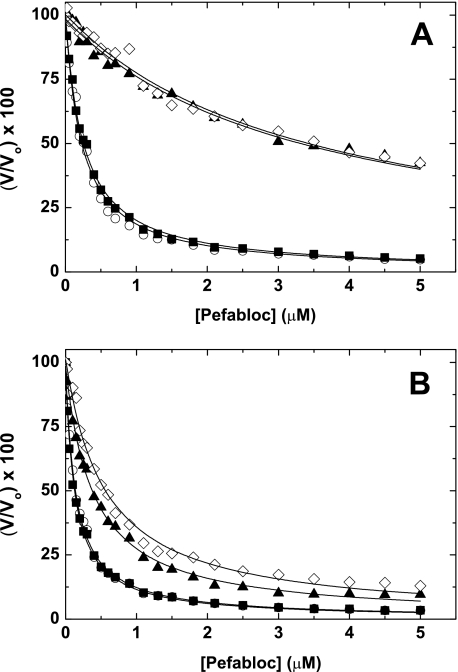
Assessment of Active Site Binding: Free FXa—To examine how changes at position 16 or 17 influence the active site, we performed kinetic experiments with several probes. Using either SpecXa, Pefabloc, or pAB, the results showed that both mutants exhibited reduced binding as the Km or Ki was increased ∼10–25-fold relative to wild type (Table 1 and Fig. 2A). We also found that the kcat for Spec Xa hydrolysis was reduced 2- to 4-fold (Table 1). These data indicate that changes to the native N terminus of FXa have a dramatic effect on catalytic function. These results are consistent with the idea that active site formation and N-terminal stabilization are allosterically linked, a finding well established in the trypsinogen/trypsin system (4).
TABLE 1.
Active site assessment for factor Xa and prothrombinase
The errors in the fitted constants represent ± 2 S.D. For each probe, data are representative of two to three similar experiments.
|
Enzyme species
|
SpecXaa
|
Pefabloc,bKi
|
pAB,bKi
|
|
|---|---|---|---|---|
| Km | kcat | |||
| μm | s-1 | nm | nm | |
| Free FXa | ||||
| PDFXa | 72 ± 7.4 | 140 ± 4.5 | 64 ± 3.0 | 80 ± 2.4 |
| rFXa | 89 ± 11 | 210 ± 13 | 70 ± 2.0 | 78 ± 1.5 |
| rFXa116L | 1150 ± 200 | 58 ± 8.2 | 1700 ± 65 | 730 ± 40 |
| rFXaV17A | 1380 ± 330 | 73 ± 14 | 1690 ± 72 | 990 ± 37 |
| Prothrombinase | ||||
| PDFXa | 170 ± 12 | 72 ± 2.2 | 53 ± 1.0 | 45 ± 0.6 |
| rFXa | 130 ± 11 | 140 ± 4.4 | 50 ± 2.0 | 49 ± 0.6 |
| rFXa116L | 290 ± 54 | 33 ± 3.4 | 210 ± 5.0 | 140 ± 9.0 |
| rFXaV17A | 490 ± 28 | 91 ± 2.7 | 290 ± 13 | 190 ± 6.4 |
For free FXa; reaction conditions: 2.0 nm wild-type or 6.0 nm mutant rFXa was incubated with increasing concentrations of SpecXa (10-500 μm). For prothrombinase; reaction conditions: 5.0 nm wild-type or mutant FXa was incubated with 30 nm FVa, 50 μm PCPS, and increasing concentrations of SpecXa (10-500 μm).
Inhibition kinetics of free FXa or prothrombinase were determined from initial velocity studies conducted with the peptidyl substrate S-2765 or SpecXa. Details of the experimental design and reactant concentrations can be found under “Experimental Procedures.”
FIGURE 2.
Inhibition of FXa by Pefabloc. The initial velocity (V) of peptidyl substrate hydrolysis (S-2765; 100 μm for wild-type rFXa or 500 μm for mutant rFXa) catalyzed by free FXa (A) (3 nm wild-type rFXa; 10 nm mutant rFXa) or prothrombinase (B) (60 μm PCPS and 20 or 100 nm FVa for wild-type rFXa and mutant rFXa, respectively) was determined at increasing concentrations of Pefabloc. For visual purposes, the data are normalized to V/Vo × 100, where V is the velocity at a given concentration of Pefabloc and Vo is the starting initial velocity. ▪, PDFXa; ○, rFXa; ▴, rFXaI16L; and ⋄, rFXaV17A. The data are representative of two to three similar experiments.
Based on the nature of the mutations, we hypothesized that N-terminal insertion during the zymogen to protease transition would be suboptimal. This could be due to the new N terminus not fully inserting into the Ile16 hydrophobic pocket, or rather the new N terminus could be misaligned or not properly oriented to efficiently facilitate the necessary conformational changes. To discriminate between these possibilities, we subjected each of the proteases to chemical modification using NaNCO, which preferentially modifies the N terminus of proteins (51). The data did not show any significant difference in the rate of carbamylation between wild-type, rFXaI16L, and rFXaV17A (data not shown). This indicates that the N terminus may be inserted into the protein and be protected from chemical modification. Perhaps the N terminus is suboptimally positioned and cannot properly facilitate the zymogen to protease transition in the usual way.
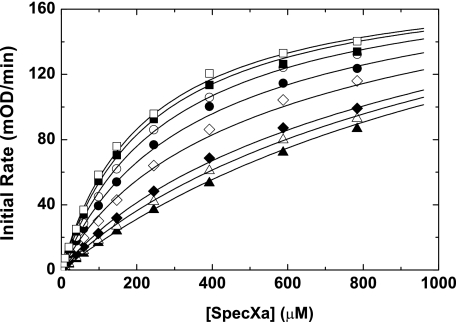
Assessment of Active Site Binding: Prothrombinase—In contrast to the results with free FXa, full saturation of the variants with FVa membranes to form prothrombinase resulted in almost complete rescue of active site binding (Table 1 and Fig. 2B). This was generally consistent with each of the three active site probes, including pAB, which targets the S1 specificity pocket. These data indicate that FVa somehow stabilizes the active site of each variant, thereby improving the affinity for active site-directed probes. Based on these data and our prior observations (23), one interpretation is that N-terminal insertion is allosterically linked to expression of the FVa and S1 binding sites. To provide evidence for this, initial velocity measurements of Spec Xa cleavage by membrane-bound rFXaI16L at different fixed concentrations of FVa were made (Fig. 3), followed by global analysis of all relevant equations describing the binding interactions shown in SCHEME 1. The data indicate that membrane-bound rFXaI16L binds with a reduced affinity to FVa (Kd1 = 3.1 ± 0.6 nm) and peptidyl substrates (KS1 = 1392 ± 131 μm); however, occupation of the active site with SpecXa restored FVa binding (Kd2 ∼ 0.42 nm) and saturating membrane-bound FXaI16L with FVa restored peptidyl substrate binding (KS2 = 192 ± 8.5 μm). The rates of catalysis of rFXaI16L in the absence of FVa (kcat1 = 70 ± 4.6 s–1) or in the presence of saturating concentrations of FVa (kcat2 = 51 ± 0.5 s–1) are essentially the same, indicating that the binding of FVa to FXaI16L does not substantially influence the rate constant for peptidyl substrate hydrolysis. Similar results were obtained with rFXaV17A (data not shown). These data provide evidence that N-terminal insertion is allosterically linked to the formation of the catalytic site as well as to expression of a productive FVa binding site.
FIGURE 3.
Effect of FVa on peptidyl substrate cleavage by membrane bound rFXaI16L. The initial rate of chromogenic substrate cleavage by membrane-bound FXaI16L (10 nm; 50 μm PCPS) was determined using increasing concentrations of SpecXa in the presence of different fixed concentrations of FVa (0 nm (▴), 1 nm (▵), 2 nm (♦), 5 nm (⋄), 8 nm (•), 12 nm (○), 18 nm (▪), and 20 nm (□)) in assay buffer. The lines are drawn following global analysis of the initial velocity data as described under “Data Analysis.” The data are representative of two similar experiments.
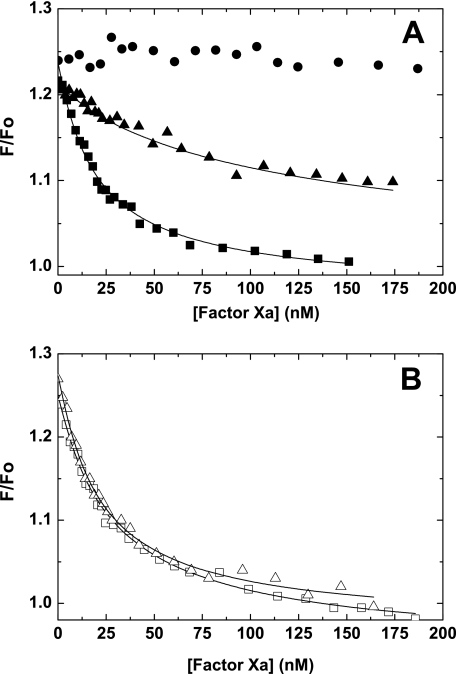
Competitive Binding Experiments—To directly assess FVa binding and provide further evidence for allosteric linkage between N-terminal stabilization, active site, and the FVa binding site, competitive binding experiments were established. We took advantage of the change in fluorescence intensity, which accompanies the binding of FVa to OG488-FXa membranes to monitor the FXa-FVa interaction (30, 39). Titration of reaction mixtures containing fixed amounts of OG488-FXa, PCPS, and FVa with increasing concentrations of inactive but non-fluorescent rFXaS195A (active site open) decreased the fluorescence intensity to near baseline levels (Fig. 4A). Analysis of the displacement curves indicates that rFXaS195A bound FVa membranes with Kd ∼ 1 nm (Table 2). In contrast to the protease, the zymogen rFXS195A had no appreciable effect on the fluorescence signal, suggesting that it does not bind, or binds very weakly to FVa membranes (Fig. 4A). Similar results were obtained with wild-type rFX (data not shown). Consistent with the idea that rFXaI16L and rFXaV17A are zymogen-like, each variant had a ∼10-fold reduced affinity for FVa membranes (Table 2 and Fig. 4A). These data provide evidence that full expression of the FVa binding site is dependent upon proper stabilization of the new N terminus in the Ile16 cleft. Consistent with the notion that the zymogen to protease transition is linked to the expression of the FVa binding site and active site, we were able to fully rescue FVa binding following covalent modification of the active sites of rFXaI16L and rFXaV17A with a peptidyl chloromethyl ketone (EGR-CH2Cl) (Fig. 4B and Table 2). These data show that stabilization of the active site can shift the variants from a zymogen-like state back to the protease-like state thus restoring FVa binding. These data provide evidence that N-terminal insertion is allosterically linked to both catalytic site expression and the formation of the FVa binding site.
FIGURE 4.
Fluorescence (F) measurements of prothrombinase assembly. Reaction mixtures containing 10 nm OG488-FXa, 10 nm FVa, and 50 μm PCPS in assay buffer were titrated with increasing concentrations of rFX (•), rFXaS195A (▪), or rFXaI16L (▴) for panel A, or EGR-rFXa (□) or EGR-rFXaI16L (▵) for panel B. Fluorescence intensity was measured at 25 °C. The lines are drawn following analysis to independent, non-interacting sites, and the fitted values are given in Table 2. The data are representative of two to three similar experiments.
TABLE 2.
Equilibrium binding constants for prothrombinase assembly
The errors in the fitted constants represent ± 2 S.D. Data are representative of two to three similar experiments.
| Competitor speciesa | Kd, compb | Kd, fixedc | |
|---|---|---|---|
| nm | |||
| rFXaS195A | 1.34 ± 0.17 | 1.04 ± 0.17 | |
| rFXaI16L | 13.8 ± 1.07 | 1.22 ± 0.07 | |
| rFXaV17A | 7.25 ± 0.65 | 1.04 ± 0.23 | |
| Active site blockedd | |||
| EGR-rFXa | 1.80 ± 0.42 | 1.05 ± 0.31 | |
| EGR-rFXaI16L | 1.92 ± 0.20 | 1.05 ± 0.14 | |
Reaction mixtures containing 10 nm OG488-FXa, 50 μm PCPS and either 8 or 10 nm FVa in assay buffer were titrated with increasing concentrations of the competitor species. Fluorescence intensity was recorded as described under “Experimental Procedures.” For simplicity, the primary data in Fig. 4 is shown with only one fixed concentration of FVa.
Represents the equilibrium dissociation constant of the competitor species.
Represents the equilibrium dissociation constant of the fixed species (i.e. OG488-FXa).
Wild-type rFXa and rFXaI16L were covalently modified at the active site with EGR-CH2Cl, as described under “Experimental Procedures.”
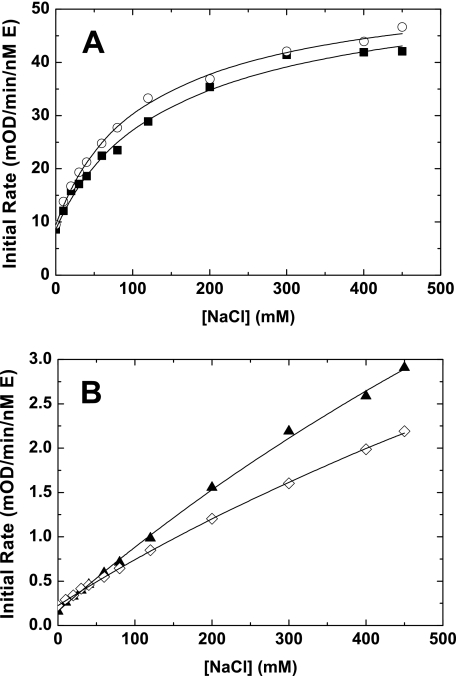
Interaction with Na+—We have previously shown that mutation of the Na+ binding site in FXa (rFXaY225P) alters FVa binding and N-terminal insertion (23). Based on those results we speculated that rFXaI16L and rFXaV17A should have reduced Na+ binding. To test this we used a chromogenic-based assay to infer binding of Na+ to FXa. To estimate the affinity of FXa and the FXa mutants for Na+, we measured the initial rate of peptidyl substrate hydrolysis at different concentrations of Na+ in the presence of saturating amounts of CaCl2. Using this method, we found that rFXaI16L and rFXaV17A bound Na+ with a much lower affinity (estimated values: rFXaI16L Kd ≥ 1673 ± 283 mm and rFXaV17A Kd ≥ 1624 ± 152 mm) relative to wild-type FXa (rFXa Kd = 114 ± 6.6 mm and PDFXa Kd = 138 ± 12.6 mm). This indicates that alteration of the zymogen to protease transition has a major influence on the structural integrity of the Na+ binding site (Fig. 5). Because the Na+ and S1 sites of FXa are known to be thermodynamically linked, proper analysis of binding data needs to take into account all relevant binding interactions (25). However, because the individual binding constants for SpecXa (in the absence of NaCl, data not shown) and Na+ are substantially altered for the variants, we were not able to perform a comprehensive linkage analysis as described by Underwood et al. (25). Thus, the binding constants derived from our experiments should be considered as estimates of the actual values. Additional studies with rFXaI16L indicated that high concentrations of Na+ (500 mm) alone (Km = 430 ± 16 μm) or together with saturating concentration of FVa membranes (Km = 156 ± 14 μm) significantly improved binding for SpecXa.
FIGURE 5.
Influence of Na+ on the chromogenic activity of FXa. The initial velocity of peptidyl substrate hydrolysis (SpecXa, 100 μm) catalyzed by free FXa as a function of the NaCl concentration (0–450 mm) as described under “Experimental Procedures.” A, PDFXa (▪), rFXa (○), 2 nm final. B, rFXaI16L (▴), rFXaV17A (⋄), 50 nm final. The solid lines were drawn following analysis of all data sets to a rectangular hyperbola; the fitted parameters are given in the text. The data are representative of two to three similar experiments.
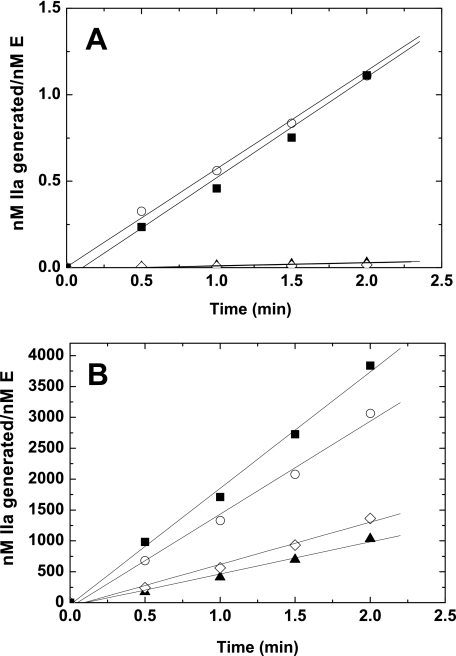
Kinetics of Macromolecular Substrate Cleavage—To further characterize these zymogen-like variants we pursued initial velocity measurements using the macromolecular substrates prothrombin and prethrombin-1. Consistent with results using active site-directed probes, both rFXaI16L and rFXaV17A exhibited a marked reduction (40-fold) in prothrombin activation in the absence of FVa relative to wild-type FXa (Fig. 6A). However, saturation of the variants with FVa membranes almost completely rescued (3-fold) the reduced rate of prothrombin activation (Fig. 6B). Further experimentation with the variants assembled in prothrombinase revealed that the variants exhibited a modest reduction in the maximal catalytic rate (kcat reduced ∼2–3-fold) with no obvious change in the Km for prothrombin (Table 3). Overall, these findings are consistent with the chromogenic substrate data as the kcat for Spec Xa hydrolysis was reduced ∼2- to 4-fold, whereas the Km was unchanged when the variants were assembled in prothrombinase. Essentially equivalent results were obtained using the alternative macromolecular substrate, prethrombin-1 (Table 3). These data indicate that the zymogen-like variants are poor enzymes in the absence of FVa membranes, but can efficiently cleave the physiological substrate prothrombin when assembled into prothrombinase. Additional experiments were performed to evaluate the pathway of prothrombin activation by SDS-PAGE using established procedures (52). Consistent with the wild-type enzyme, cleavage of prothrombin by rFXaI16L or rFXaV17A incorporated into prothrombinase gave bands consistent with sequential cleavage at Arg320 giving rise to meizothrombin as the intermediate, followed by cleavage at Arg271 to yield thrombin (data not shown). These data show that the FXa variants in prothrombinase did not detectably alter the pathway of prothrombin activation.
FIGURE 6.
Activation of prothrombin. Reaction mixtures containing 1.4 μm prothrombin and 50 μm PCPS were incubated at 25 °C in the absence (A) or in the presence (B) of 20 nm FVa and initiated with FXa 0.1 nm (wild-type rFXa) or 0.4 nm (mutant rFXa). Aliquots of the reaction mixture were quenched during the initial rate of the reaction (0, 0.5, 1, 1.5, and 2.0 min), and thrombin generation was determined by using the chromogenic substrate S-2238. ▪, PDFXa; ○, rFXa; ▴, rFXaI16L; and ⋄, rFXaV17A. The data are representative of two to three similar experiments.
TABLE 3.
Kinetic constants for macromolecular substrate cleavage by prothrombinase
Steady-state kinetic constants were derived from initial velocity studies conducted with increasing concentrations of substrate. Concentrations of reaction components can be found under “Experimental Procedures.” The errors in the fitted constants represent ± 2 S.D. The data are representative of two to three independent measurements.
|
Prothrombin
|
Prethrombin-1
|
|||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| μm | min-1 | μm-1·s-1 | μm | min-1 | μm-1·s-1 | |
| PDFXa | 0.42 ± 0.02 | 2424 ± 54 | 96 | 5.4 ± 0.3 | 1088 ± 30 | 3.3 |
| rFXa | 0.35 ± 0.01 | 1937 ± 26 | 92 | 5.2 ± 0.4 | 1050 ± 35 | 3.4 |
| rFXaI16L | 0.31 ± 0.02 | 619 ± 14 | 33 | 5.7 ± 1.8 | 399 ± 10 | 1.2 |
| rFXaV17A | 0.47 ± 0.03 | 887 ± 26 | 31 | 6.9 ± 0.3 | 388 ± 8.9 | 0.9 |
DISCUSSION
For serine proteases, the zymogen and protease conformational states exist in equilibrium (4). This equilibrium lies far to the left when the proteins are unactivated and typically lies far to the right following activation. Notable exceptions are tissue type plasminogen activator, which has significant activity as a zymogen (low zymogenicity), and factor VIIa, which fails to attain its catalytically competent conformation after cleavage of the Arg15–Ile16 bond (high zymogenicity) (53, 54). For trypsin and FXa, the zymogen-like conformation is negligibly populated. In the current study, we directly examined this transition pathway by generating FXa variants with suboptimal N-terminal insertion and zymogen-like properties. We speculate that destabilization of internal salt bridge formation by mutagenesis at positions 16 or 17 has altered the equilibrium position between the zymogen and the protease. Consistent with this proposal, both FXa derivatives had altered active site pockets as evidenced by impaired binding of small peptidyl substrates and inhibitors. Based on the nature of the mutations it is likely that the liberated N terminus is not positioned or oriented in the usual way. Replacing Ile16 with Leu, and Val17 with Ala, could have altered the electrostatic interaction between residues 16 and 194 or disrupted optimal hydrophobic contacts within the N-terminal binding cleft, because both interactions contribute to the transition from the inactive to the active state (12). For the FXa variants, the reduction in their perceived activity likely represents a change in the ratio between the zymogen-like and protease conformations.
Structural differences between the zymogen and the protease states are primarily found within the activation domain (4). As a result, binding sites allosterically linked to this region should be affected by mutations that induce a zymogen-like conformation. Our experimental findings with rFXaI16L and rFXaV17A support this, because active site binding as well as binding at the Na+ and FVa binding sites were all substantially altered. The catalytic site is a well known element of the activation domain, and the Na+ binding site has been identified by structural studies and involves two loop segments (183–189 and 221–225) within this region (55). Although the FVa binding site remains to be completely defined, it is clear that elements of the activation domain are also involved. Several groups have shown that the exosite II region (heparin binding site) and also an α-helix (163–170) on the catalytic domain contribute to FVa binding (18, 19, 56). Analysis of the FXa structure indicates that the putative FVa binding helix is connected to the Na+ binding site through van der Waals contacts (25). More recent studies have shown that parts of this helical region appear linked to the S1- and Na+ binding sites (56), lending further support to the idea that changes in the activation domain will likely impact FVa binding. Furthermore, mutagenesis studies have shown that the Na+ binding site and the 185–189 loop are important for FVa binding (19, 23, 24, 57). It is unlikely, however, that all features of the activation domain contribute to FVa binding in a direct fashion. A clear example of this is the binding of tick anticoagulant protein to FXa. Tick anticoagulant protein interacts with the Na+ binding site and autolysis loop, yet positively influences FVa binding (58, 59). A more likely explanation is that some regions contribute directly while other regions are energetically linked; thus changes at one site can impact binding at a related site in an indirect fashion. The data of the current study provide evidence for this and indicate that interpretation of mutagenesis data, attempting to define the FVa binding site or other binding sites involving the activation domain, needs to consider these allosteric effects.
Because the equilibrium position between the zymogen and the protease states determines activity, probes that stabilize the protease conformation should realign the equilibrium position and restore activity. We were able to document this in two different ways. First, saturation of the FXa variants with FVa membranes almost completely restored binding at the active site. Second, covalent modification of rFXaI16L with a peptidyl chloromethyl ketone in the active site completely rescued FVa binding. These findings indicate that the FVa binding site is thermodynamically linked to the active site and the zymogen to protease transition. This conclusion is in line with the observation that the zymogen FX does not bind, or binds very weakly to FVa membranes as documented here and in previous work (20–22). For expression of the FVa binding site, we cannot discern whether this conformational change represents all necessary facets required for FVa binding or whether the large activation peptide present on the zymogen could also alter cofactor binding in some fashion. Nevertheless, the zymogen to protease transition clearly plays an important role in the maturation or stabilization of the FXa activation domain and contributes to the ability of the protease to bind FVa membranes.
A question that emerges from our findings is why linkage between the primary specificity site, FVa binding site, and N-terminal stabilization is not evident using wild-type FXa. We speculate that this is because wild-type FXa is already stabilized in the protease configuration while the zymogen-like FXa variants are not. Thus strong probes such as FVa will only have a minor effect on the active site of wild-type FXa; an established observation in the field (60). The interplay between these binding sites can only be revealed if the equilibrium position between the zymogen and the protease has been altered, as with FXaI16L and FVaV17A.
In addition to changes at the catalytic site and FVa binding site, both zymogen-like variants also exhibited reduced Na+ binding. Altered Na+ binding is consistent with prior observations with FXaY225P and FIXaY225P, which have destabilized N-terminal regions, altered active sites, and reduced cofactor binding (23, 26). Thus it appears that the rFXaI16L, rFXaV17A, and the Y225P variants may operate in comparable ways by destabilizing the activation domain and inducing a zymogen-like conformation. This would imply that Na+ binding to FXa reinforces the activation domain and thus stabilizes the protease conformation. Collectively these findings support the conclusion that the FX zymogen to protease transition drives the formation of the Na+ binding site, implying that the zymogen does not bind Na+ or does so very weakly. Structural support for this comes from the prethrombin-2-hirugen complex, and the zymogen structure of FVII, which has a Na+ site that is not properly formed (55, 61). Additionally, molecular modeling studies comparing FX and FXa suggest that there are major structural differences in the Na+ binding site (62).
The notion that the zymogen to protease transition can influence different aspects of protein function in the serine protease family is in line with several previous observations. For example, the zymogen to protease transition has been shown to influence the expression of a receptor tyrosine kinase binding site on hepatocyte growth factor (63), the expression of proexosite I on prothrombin (64), directing the prothrombin activation pathway (52), and the formation or stabilization of a Na+ binding site in enzymes such as thrombin (55). These findings together with results of our work indicate that the functional switch between the zymogen and protease states likely plays an important role in protease specificity and function in addition to producing a functional active site.
Although the zymogen-like FXa variants had poor catalytic activity in the absence of FVa, they could efficiently activate prothrombin once assembled in prothrombinase. Based on these differential functional states, we would expect that in a plasma environment these derivatives as free FXa would be refractory to inhibition by protease inhibitors like anti-thrombin III. Furthermore, the variants should not interfere with the initiation of coagulation following vascular damage, because they are not expected to bind tissue factor pathway inhibitor very well. However, whereas the zymogen-like variants of FXa bind FVa more weakly, they are almost completely rescued at sufficiently high cofactor concentrations and catalyze thrombin formation efficiently. Thus, zymogen-like forms of FXa with these properties may act as long-lived proteases in circulation that are otherwise inert but retain the ability to catalyze thrombin formation upon binding to FVa on the activated platelet surface. We speculate that these types of FXa variants may have the potential to serve as therapeutic procoagulants that bypass deficiencies in other clotting factors in the cascade.
In summary, altering the equilibrium position between the FX zymogen and FXa protease states has provided unique insight into macromolecular binding site expression. The allosterically linked structural changes that accompany this conformational transition play a major role in the formation of the primary specificity site as well as the Na+ and FVa binding sites. Because of the uniformity of the transition mechanism, our findings have broad implications for better understanding structure/function relationships related to exosite expression and protein complex assembly for coagulation factor proteases.
Acknowledgments
We are grateful to Drs. Sriram Krishnaswamy and Mettine H. A. Bos for useful suggestions and critical review of the manuscript. We also acknowledge Dr. Alex Kurosky and Steven Smith at the University of Texas Medical Branch at Galveston for N-terminal sequence analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 HL-74124-01, Project 2 (to R. M. C.). This work was also supported by National Research Service Award T32 HL-07439-26 (to R. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Nomenclature of Schechter and Berger (65).
The abbreviations used are: FX, factor X; FXa, activated FX; PDFX, plasma-derived FX; rFX, recombinant FX; FV, factor V; FVa, activated FV; RVVX-CP, FX activator from Russell's viper venom; pAB, 4-aminobenazmidine; EGR-CH2Cl, glutamylglycylarginyl chloromethyl ketone; SpecXa, methoxycarbonylcyclohexylglycylglycylarginine-p-nitroanilide; S-2238, H-d-phenylalanyl-l-pipecolyl-l-arginine-p-nitroanilide; S-2765, N-α-benzyloxycarbonyl-d-arginyl-l-glycyl-l-arginine-p-nitroanilide; rTAP, recombinant tick anticoagulant peptide; PCPS, small unilamellar vesicles composed of 75% (w/w) phosphatidylcholine and 25% (w/w) phosphatidylserine; OG488-FXa, FXa modified with Oregon Green488; HEK293, human embryonic kidney cells; Gla, γ-carboxyglutamic acid; MES, 4-morpholineethanesulfonic acid.
References
- 1.Khan, A. M., and James, M. N. G. (1998) Prot. Sci. 7 815–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bode, W., Mayr, I., Bauman, Y., Huber, R., Stone, S. R., and Hofsteenge, J. (1989) EMBO J. 8 3467–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freer, S. T., Kraut, J., Roberts, J. D., Wright, H. T., and Xuong, N. H. (1970) Biochemistry 9 1997–2009 [DOI] [PubMed] [Google Scholar]
- 4.Huber, R., and Bode, W. (1978) Acc. Chem. Res. 11 114–122 [Google Scholar]
- 5.Bode, W., Schwager, P., and Huber, R. (1978) J. Mol. Biol. 118 99–112 [DOI] [PubMed] [Google Scholar]
- 6.Bode, W. (1979) J. Mol. Biol. 127 357–374 [DOI] [PubMed] [Google Scholar]
- 7.Reddy, K., and Markus, G. (1972) J. Biol. Chem. 247 1683–1691 [PubMed] [Google Scholar]
- 8.Davidson, D., Higgins, D. L., and Castellino, F. J. (1990) Biochemistry 29 3585–3590 [DOI] [PubMed] [Google Scholar]
- 9.Hemker, H., Bas, B., and Muller, A. (1975) Biochim. Biophys. Acta 379 180–188 [DOI] [PubMed] [Google Scholar]
- 10.Kawabata, S., Morita, T., Iwanaga, S., and Igarashi, H. (1985) J. Biochem. 98 1603–1614 [DOI] [PubMed] [Google Scholar]
- 11.Madoiwa, S., Nakamura, Y., Mimuro, J., Furusawa, S., Koyama, T., Sugo, T., Matsuda, M., and Sakata, Y. (2001) Blood 97 3783–3789 [DOI] [PubMed] [Google Scholar]
- 12.Hedstrom, L., Lin, T., and Fast, W. (1996) Biochemistry 35 4515–4523 [DOI] [PubMed] [Google Scholar]
- 13.Pasternak, A., Liu, X., Lin, T., and Hedstrom, L. (1998) Biochemistry 37 16201–16210 [DOI] [PubMed] [Google Scholar]
- 14.Gombos, L., Kardos, J., Patthy, A., Medveczky, P., Szilagyi, L., Malnasi-Csizmadia, A., and Graf, L. (2008) Biochemistry 47 1675–1684 [DOI] [PubMed] [Google Scholar]
- 15.Robison, D., Furie, B., Furie, B. C., and Bing, D. H. (1980) J. Biol. Chem. 255 2014–2021 [PubMed] [Google Scholar]
- 16.Keyt, B., Furie, B. C., and Furie, B. (1982) J. Biol. Chem. 257 8687–8695 [PubMed] [Google Scholar]
- 17.Mann, K. G., Nesheim, M. E., Church, W. R., Haley, P. E., and Krishnaswamy, S. (1990) Blood 76 1–16 [PubMed] [Google Scholar]
- 18.Rezaie, A. R. (2000) J. Biol. Chem. 275 3320–3327 [DOI] [PubMed] [Google Scholar]
- 19.Rudolph, A. E., Porche-Sorbet, R., and Miletich, J. P. (2001) J. Biol. Chem. 276 5123–5128 [DOI] [PubMed] [Google Scholar]
- 20.Persson, E., Hogg, P. J., and Stenflo, J. (1993) J. Biol. Chem. 268 22531–22539 [PubMed] [Google Scholar]
- 21.Dahlbäck, B., and Stenflo, J. (1978) Biochemistry 17 4938–4945 [DOI] [PubMed] [Google Scholar]
- 22.Miletich, J. P., Jackson, C. M., and Majerus, P. W. (1978) J. Biol. Chem. 253 6908–6916 [PubMed] [Google Scholar]
- 23.Camire, R. M. (2002) J. Biol. Chem. 277 37863–37870 [DOI] [PubMed] [Google Scholar]
- 24.Rezaie, A. R., and He, X. (2000) Biochemistry 39 1817–1825 [DOI] [PubMed] [Google Scholar]
- 25.Underwood, M. C., Zhong, D., Mathur, A., Heyduk, T., and Bajaj, S. P. (2000) J. Biol. Chem. 275 36876–36884 [DOI] [PubMed] [Google Scholar]
- 26.Schmidt, A. E., Stewart, J. E., Mathur, A., Krishnaswamy, S., and Bajaj, S. P. (2005) J. Mol. Biol. 350 78–91 [DOI] [PubMed] [Google Scholar]
- 27.Evans, S. A., Olson, S. T., and Shore, J. D. (1982) J. Biol. Chem. 257 3014–3017 [PubMed] [Google Scholar]
- 28.Lottenberg, R., and Jackson, C. M. (1983) Biochim. Biophys. Acta 742 558–564 [DOI] [PubMed] [Google Scholar]
- 29.Higgins, D. L., and Mann, K. G. (1983) J. Biol. Chem. 258 6503–6508 [PubMed] [Google Scholar]
- 30.Buddai, S. K., Toulokhonova, L., Bergum, P. W., Vlasuk, G. P., and Krishnaswamy, S. (2002) J. Biol. Chem. 277 26689–26698 [DOI] [PubMed] [Google Scholar]
- 31.Katzmann, J. A., Nesheim, M. E., Hibbard, L. S., and Mann, K. G. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baugh, R., and Krishnaswamy, S. (1996) J. Biol. Chem. 271 16126–16134 [DOI] [PubMed] [Google Scholar]
- 33.Mann, K. G. (1976) Methods Enzymol. 45 123–156 [DOI] [PubMed] [Google Scholar]
- 34.Krishnaswamy, S., Church, W. R., Nesheim, M. E., and Mann, K. G. (1987) J. Biol. Chem. 262 3291–3299 [PubMed] [Google Scholar]
- 35.Kisiel, W., Hermodson, M. A., and Davie, E. W. (1976) Biochemistry 15 4901–4906 [DOI] [PubMed] [Google Scholar]
- 36.Kalafatis, M., Krishnaswamy, S., Rand, M. D., and Mann, K. G. (1993) Methods Enzymol. 222 224–236 [DOI] [PubMed] [Google Scholar]
- 37.Gowda, D. C., Jackson, C. M., and Davidson, E. A. (1994) J. Biol. Chem. 269 10644–10650 [PubMed] [Google Scholar]
- 38.Lundblad, R. L., Kingdon, H. S., and Mann, K. G. (1976) Methods Enzymol. 45 156–176 [DOI] [PubMed] [Google Scholar]
- 39.Toso, R., and Camire, R. M. (2004) J. Biol. Chem. 279 21643–21650 [DOI] [PubMed] [Google Scholar]
- 40.Di Scipio, R. G., Hermodson, M. A., Yates, S. G., and Davie, E. W. (1977) Biochemistry 16 698–706 [DOI] [PubMed] [Google Scholar]
- 41.Larson, P. J., Camire, R. M., Wong, D., Fasano, N. C., Monroe, D. M., Tracy, P. B., and High, K. A. (1998) Biochemistry 37 5029–5038 [DOI] [PubMed] [Google Scholar]
- 42.Camire, R. M., Larson, P. J., Stafford, D. W., and High, K. A. (2000) Biochemistry 39 14322–14329 [DOI] [PubMed] [Google Scholar]
- 43.Krishnaswamy, S., and Walker, R. K. (1997) Biochemistry 36 3319–3330 [DOI] [PubMed] [Google Scholar]
- 44.Toso, R., and Camire, R. M. (2006) J. Biol. Chem. 281 8773–8779 [DOI] [PubMed] [Google Scholar]
- 45.Bevington, P. R., and Robinson, K. D. (1992) Data Reduction and Error Analysis for the Physical Sciences, McGraw-Hill, New York
- 46.Straume, M., and Johnson, M. L. (1992) Methods Enzymol. 210 87–105 [DOI] [PubMed] [Google Scholar]
- 47.Olson, S. T., Bock, P. E., and Sheffer, R. (1991) Arch. Biochem. Biophys. 286 533–545 [DOI] [PubMed] [Google Scholar]
- 48.Betz, A., and Krishnaswamy, S. (1998) J. Biol. Chem. 273 10709–10718 [DOI] [PubMed] [Google Scholar]
- 49.Segal, I. H. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme Systems, John Wiley & Sons, Inc., New York
- 50.Kuzmic, P. (1996) Anal. Biochem. 237 260–273 [DOI] [PubMed] [Google Scholar]
- 51.Plapp, B. V., Moore, S., and Stein, W. H. (1971) J. Biol. Chem. 246 939–945 [PubMed] [Google Scholar]
- 52.Bianchini, E. P., Orcutt, S. J., Panizzi, P., Bock, P. E., and Krishnaswamy, S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10099–10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tachias, K., and Madison, E. (1996) J. Biol. Chem. 271 28749–28752 [DOI] [PubMed] [Google Scholar]
- 54.Higashi, S., Matsumoto, N., and Iwanaga, S. (1996) J. Biol. Chem. 271 26569–26574 [DOI] [PubMed] [Google Scholar]
- 55.Zhang, E., and Tulinsky, A. (1997) Biophys. Chem. 63 185–200 [DOI] [PubMed] [Google Scholar]
- 56.Levigne, S., Thiec, F., Cherel, G., Irving, J. A., Fribourg, C., and Christophe, O. D. (2007) J. Biol. Chem. 282 31569–31579 [DOI] [PubMed] [Google Scholar]
- 57.Rezaie, A. R., and Kittur, F. S. (2004) J. Biol. Chem. 279 48262–48269 [DOI] [PubMed] [Google Scholar]
- 58.Wei, A., Alexander, R. S., Duke, J., Ross, H., Rosenfeld, S. A., and Chang, C. (1998) J. Mol. Biol. 283 147–154 [DOI] [PubMed] [Google Scholar]
- 59.Krishnaswamy, S., Vlasuk, G. P., and Bergum, P. W. (1994) Biochemistry 33 7897–7907 [DOI] [PubMed] [Google Scholar]
- 60.Krishnaswamy, S. (2005) J. Thromb. Haemost. 3 54–67 [DOI] [PubMed] [Google Scholar]
- 61.Eigenbrot, C., Kirchhofer, D., Dennis, M. S., Santell, L., Lazarus, R. A., Stamos, J., and Ultsch, M. H. (2001) Structure 9 627–636 [DOI] [PubMed] [Google Scholar]
- 62.Venkateswarlu, D., Perera, L., Darden, T., and Pedersen, L. G. (2002) Biophys. J. 82 1190–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirchhofer, D., Lipari, M. T., Santell, L., Billeci, K. L., Maun, H. R., Sandoval, W. N., Moran, P., Ridgway, J., Eigenbrot, C., and Lazarus, R. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5306–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bock, P. E., Panizzi, P., and Verhamme, I. M. (2007) J. Thromb. Haemost. 5 Suppl. 1, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schechter, I., and Berger, A. (1967) Biochem. Biophys. Res. Commun. 27 157–162 [DOI] [PubMed] [Google Scholar]