Abstract
Cas is a multidomain signaling protein that resides in focal adhesions. Cas possesses a large central substrate domain containing 15 repeats of the sequence YXXP, which are phosphorylated by Src. The phosphorylation sites are essential for the roles of Cas in cell migration and in regulation of the actin cytoskeleton. We showed previously that Src catalyzes the multisite phosphorylation of Cas via a processive mechanism. In this study, we created mutant forms of Cas to identify the determinants for processive phosphorylation. Mutants containing single or multiple YXXP mutations were phosphorylated processively by Src, suggesting that individual sites are dispensable. The results also suggest that there is no defined order to the Cas phosphorylation events. We also studied the effects of these mutations by reintroducing Cas into Cas-deficient fibro-blasts. Mutants lacking some or all YXXP sites augment the ability of Src to promote anchorage-independent growth. On the other hand, deletion of YXXP sites compromises the ability of Cas to promote tumor cell migration.
p130cas (Crk-associated substrate, 130 kDa) was originally identified as a focal adhesion protein that is highly tyrosine-phosphorylated during transformation by v-Src and v-Crk and that also forms stable complexes with these oncoproteins (1–3). Activated Src does not induce a complete transformed phenotype when expressed in Cas-deficient fibroblasts, indicating a role for Cas in cellular transformation by Src (4). Recent studies have revealed multiple signaling roles for Cas in normal cells (5, 6). Mouse embryos lacking Cas die in utero because of the defects in cardiovascular development, disorganization of myofibrils, impaired actin stress fiber association, and growth retardation (4). Cas-deficient fibroblasts contain short, disorganized actin filaments, and actin stress fiber organization is restored after ectopic expression of Cas (4, 7). These results suggest a crucial role for Cas in the regulation of the actin cytoskeleton.
Cas possesses an N-terminal SH32 domain, a large central substrate domain containing 15 repeats of the motif YXXP, and a C-terminal Src binding region (Fig. 1A). The YXXP motifs are substrates for several tyrosine kinases and serve as binding sites for the adapter protein Crk and other SH2 domain-containing signaling proteins (5, 6). The YXXP motifs of Cas can be divided into YDXP and YQXP motifs (Fig. 1B). The YDXP motifs are important for actin cytoskeleton organization and cell migration (8). We and others have mapped the sites of Src phosphorylation within the Cas substrate domain (9, 10). These phosphorylation sites play an important role in Cas-associated functions such as cell migration and regulation of the actin cytoskeleton.
FIGURE 1. Schematic representation of wild-type and mutant Cas proteins.
A, domain structure of Cas showing the SH3 domain (SH3), proline-rich region (Pro), substrate region containing 15 repeats of YXXP, serine-rich region (Ser), C-terminal Src binding sequence (SBS), and helix-loop-helix region (HLH). B, diagram showing WT Cas and mutant forms of Cas used in this study with YDXP and YQXP motifs indicated above the diagram. In mutants CasF7, CasF12, and CasF17, 7, 12, and 17 tyrosine residues were mutated to Phe, respectively. Tyrosine residues in the substrate domain are numbered and shown in black, and mutated Tyr residues are shown in gray.
Multisite phosphorylation can be carried out by two basic mechanisms: (i) a processive mechanism in which the enzyme binds to the substrate and catalyzes all the phosphorylation events before dissociating from the fully phosphorylated product; or (ii) a distributive mechanism in which each phosphorylation is the result of a separate enzyme-substrate binding event. The dual phosphorylation of mitogen-activated protein kinases by mitogen-activated protein kinases takes place in a two-collision, distributive manner (11, 12). In contrast, recent kinetic studies on the spliceosomal SR proteins show that they are phosphorylated processively by SR protein-specific Ser/Thr kinases (13). We showed previously that Src phosphorylates Cas by a processive mechanism in which Src binds to the C-terminal polyproline region of Cas and catalyzes the multiple phosphorylation events (14).
In many systems, the biological effects of multisite phosphorylation are distinguishable from those of single-site phosphorylation. For example, multisite phosphorylation of the cyclin-dependent kinase inhibitor Sic1 is important in setting a threshold for the onset of DNA synthesis (15). The biological significance of processive phosphorylation of the YXXP motifs in Cas remains unclear. Cas mutants lacking the entire substrate domain or the YDXP motifs are defective in cell migration (8, 16). In cells transformed by oncogenic Src, the YXXP motifs are important for invasiveness and formation of podo-some structures (17, 18). Shin et al. (10) demonstrated that Cas variants containing single or double tyrosine mutations promote cell migration at levels comparable with wild-type Cas, whereas mutants containing as few as 6 tyrosines retain some activity. Previous work from our laboratory highlighted the importance of a particular tyrosine in the substrate domain (Tyr253) in migration of Src-transformed cells (9). It was not clear, however, whether this single site would be sufficient for migration.
In this study, we carried out mutational experiments to identify the determinants for processive phosphorylation of Cas by Src. We also studied the effects of these mutations on cell migration and anchorage-independent growth in Src-transformed cells. Results from these studies suggest that no single YXXP site is essential to subsequent phosphorylation of other sites. Mutant forms of Cas lacking multiple sites are also phosphorylated processively by Src. In addition, we show that Cas augments the ability of Src to promote anchorage-independent growth, regardless of whether Cas is phosphorylated. In contrast to nonanchored growth, Cas must be phosphorylated on at least 6 tyrosine residues by Src to promote tumor cell migration.
MATERIALS AND METHODS
Antibodies
Anti-Cas monoclonal antibody was obtained from BD Transduction Laboratories. Monoclonal antibody against avian pp60v-Src kinase was obtained from Upstate Biotechnology, and anti-β-actin monoclonal antibody was obtained from Santa Cruz Biotechnology.
Mutagenesis of Cas
Cas mutants were produced with the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol. A baculovirus transfer vector encoding wild-type Cas (14) was used as a template. Mutagenic primers contained 25–45 nucleotides, and the annealing temperature was 55°C. Multisite mutants were obtained by mutagenizing 1 tyrosine at a time in the template. Tyrosine to phenylalanine mutations were verified by DNA sequencing on an ABI 373 automated DNA sequencer. The following tyrosines were mutated to phenylalanine3,: for CasF7, Tyr119, Tyr196, Tyr253, Tyr291, Tyr310, Tyr366, and Tyr414; for CasF12, the 7 tyrosines of CasF7 plus Tyr132, Tyr169, Tyr183, Tyr657, and Tyr668; and for CasF17, the 12 tyrosines of CasF12 plus Tyr238, Tyr271, Tyr331, Tyr376, and Tyr391.
Cas and Src Production in Sf9 Cells
N-terminal polyhistidine-tagged v-Src and Cas were expressed using the Bac-to-Bac baculovirus expression system (Invitrogen) as described (14). Sf9 cells were grown at 27 °C in Excel 401 (JRH Biosciences) in the presence of 2.5% heat-inactivated fetal bovine serum (Sigma) and 1% penicillin/streptomycin/amphotericin B (Invitrogen). Approximately 500–600 ml of cells at a density of 1.5 × 106 cells/ml were infected with recombinant baculovirus at a multiplicity of infection of 5–10 plaque-forming units/cell for a period of 72 h. The cells were centrifuged and stored at −70 °C until needed. Wild-type and mutant forms of Cas were purified using nickel-nitrilotriacetic acid resin (Qiagen) as described previously (14). All purified proteins were stored at −20 °C.
In Vitro Kinase Assays
For experiments in which the total phosphorylation of Cas was monitored, Cas was incubated in the presence of 50 nm v-Src at 30 °C in kinase assay buffer (150 mm Tris-HCl, pH 7.5, 100 mm MgCl2, 5 mg/ml bovine serum albumin). Reactions were started by the addition of 0.1 mm unlabeled ATP plus [γ-32P]ATP (5 µCi), and aliquots were withdrawn at various time points. The reactions were stopped by mixing with Laemmli buffer and boiling and were analyzed by SDS-PAGE followed by autoradiography. To quantitate [γ-32P]ATP incorporation, the gels were dried and exposed to a PhosphorImager screen (GE Healthcare), and the band intensity was quantitated using ImageQuant software.
For pulse-chase assays in which the concentration of v-Src was varied, 2 µm Cas (wild-type or mutants) was incubated with 800 nm v-Src in kinase assay buffer in the presence of 0.1 µm [γ-32P]ATP for 3 min on ice. To analyze the pulse reaction, an aliquot was removed and stopped by adding Laemmli buffer and boiling. Additional aliquots of the reaction were removed and diluted into four separate tubes containing kinase buffer and 0.5 mm unlabeled ATP to give final enzyme concentrations of 100, 200, 400, and 600 nM. These chase reactions were carried out at 30 °C for an additional 25 min. The reactions were stopped by adding Laemmli buffer and boiling. Equal amounts of Cas from each reaction were analyzed by 8% SDS-PAGE and autoradiography.
To determine the stoichiometry of 32P incorporation, 1 µm Cas (wild-type or mutant) was incubated with 200 nm v-Src in kinase assay buffer at 30 °C. Reactions were started by the addition of 0.1 mm unlabeled ATP plus [γ-32P]ATP (5 µCi), and aliquots were withdrawn at various time points. The reactions were stopped by mixing with Laemmli buffer and boiling and analyzed by SDS-PAGE followed by staining with Coomassie Blue. Bands were excised from the gel and dissolved in H2O2, and 32P incorporation was measured using a liquid scintillation counter.
Stable Expression of Cas and Src in Mammalian Cells
The BamHI/NotI fragments encoding WT Cas or mutant Cas were excised from pFastBac HTb and subcloned into the BamHI/SnaBI sites of pBABEhygro (19). v-Src was expressed using the vector pBABEpuro (9). Fibroblasts from homozygous null Cas knock-out cells were used as described previously (4, 8). Cas knock-out cells were transfected with equal concentrations of both plasmids (1 µg/well, 6-well plate). For controls, cells were transfected with empty vectors (pBABEhygro and/or pBABE-puro) as appropriate, and all the transfectants were selected for hygromycin resistance and puromycin resistance conferred by the transfection vectors. Clones were not taken from the resultant stable cells to avoid the potential consequences of clonal variation.
Western Blot Analysis of Mammalian Cells
Western blot analysis was performed on Cas-deficient cells grown for 72 h after plating. Briefly, cells were quickly aspirated, washed with phosphate-buffered saline, and lysed in modified radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5mm EDTA, 1% Nonidet P-40, 1% sodium deoxycholate, 5 mg/liter aprotinin, 5 mg/liter leupeptin, 0.1 mm phenylmethylsufonyl fluoride, 1 mm Na3VO4) at 4 °C for 45 min. The cell lysates were centrifuged at 14,000×g for 10 min at 4 °C. Protein concentrations were determined using Bio-Rad protein assay. Lysates (30 µg) were separated on 8% SDS-PAGE and transferred onto polyvinylidene difluoride membrane. Membranes were incubated with anti-Cas primary antibody overnight followed by incubation with horseradish peroxidase-conjugated secondary antibody and detected by ECL plus (Amersham Biosciences).
Cell Growth Assays
Cas-deficient fibroblasts were analyzed by methods described previously (9, 20, 21). Cells were grown in Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum at 37 °C in 100% humidity. All comparisons were done in parallel to avoid any changes in culture conditions between experiments. To examine anchored growth, 20,000 cells were seeded at 1 ml/well in tissue culture-treated 12-well cluster plates (Falcon). To measure nonanchored growth, 20,000 cells were seeded in ultra low attachment 24-well cluster plates (Corning). Cell numbers were obtained by Coulter counter.
To measure cell migration, 200,000 cells were plated in 6-well cluster plates on cell culture inserts with a 3- or 8-µm pore size (Transwell-Clear, Costar) and grown for 72 or 24 h, respectively. Cells were then released separately from the top of the membrane, the bottom of the membrane, and the well beneath the membrane. Migration was then quantitated as the percent of cells found in the well over the total cell number.
RESULTS
Single-site Mutants of Cas Are Phosphorylated by Src
We first tested whether selected substrate motifs in Cas play a critical role in directing Src phosphorylation. Based on our previous synthetic peptide studies, we selected 4 tyrosine residues in the substrate domain of Cas for analysis: Tyr253, Tyr310, Tyr366, and Tyr414. Synthetic peptide substrates modeled on these four sites showed the highest values of kcat/Km for Src phosphorylation of the peptides tested (9). We expressed and purified wild-type Cas as well as Cas mutants containing Tyr to Phe substitutions at these four sites. To study the effect of these single-site mutations on Cas phosphorylation by Src, we carried out in vitro kinase assays using purified v-Src and [ γ-32P]ATP. The phosphorylation rates of WT Cas and the single-site mutants were comparable as determined by densitometry (Fig. 2). These results show that of the residues tested, no single site is crucial for in vitro phosphorylation of Cas by Src.
FIGURE 2. Single-site mutants of Cas are phosphorylated by Src.
In vitro kinase assays were carried out at 30 °C using purified Cas proteins (WT or mutant), purified v-Src, and [γ-32P]ATP. Aliquots were withdrawn at the indicated times and analyzed by SDS-PAGE and autoradiography. To quantify the rates of phosphorylation, densitometry was carried out on a Bio-Rad GS-710 imaging densitometer with QuantityOne software. Rates are presented on the right.
Src Phosphorylates Multisite Mutants of Cas in a Processive Manner
We generated three multisite mutants of Cas with multiple tyrosine to phenylalanine substitutions: CasF7, CasF12, and CasF17 (Fig. 1B). In CasF7, we mutated 7 tyrosine residues that were phosphorylated efficiently by Src in the context of synthetic peptides (9). In CasF12, we mutated 7 tyrosines of CasF7, an additional 3 tyrosine residues in the substrate domain of Cas that were mapped by mass spectrometry (9), and the two potential tyrosine phosphorylation sites in the C terminus of Cas. In CasF17, we mutated 12 tyrosines of CasF12 plus an additional 5 tyrosines in the substrate domain of Cas. Thus, in CasF17, all the YXXP tyrosines of the substrate domain were eliminated.
We carried out in vitro kinase reactions with purified Src and Cas proteins. We removed aliquots of the reactions at different time points and analyzed them by SDS-PAGE and silver staining. As we reported previously (14), multisite phosphorylation of wild-type Cas produces a large shift in its electrophoretic mobility (Fig. 3). The single discrete band is consistent with a processive mechanism because no partially phosphorylated forms are observed. Src phosphorylation of the multisite mutants CasF7 and CasF12 also produced a shift in electro-phoretic mobility (Fig. 3). The shift was not as large as for WT Cas, presumably because of the lower number of phosphorylation sites present in these mutants. The shifted bands for CasF7 and CasF12 remained sharp. We did not observe significant phosphorylation of CasF17 in this experiment (Fig. 3A) or in experiments with [γ-32P]ATP (data not shown).
FIGURE 3. Phosphorylation of Cas multisite mutants.
A, in vitro reactions were carried out using Cas proteins and 100 µm unlabeled ATP for 20 min in the presence or absence of v-Src. The reactions were analyzed by SDS-PAGE and silver staining. B, time courses for Cas phosphorylation were carried out using methods similar to those described in A. Aliquots were withdrawn at the indicated times and analyzed by SDS-PAGE and silver staining.
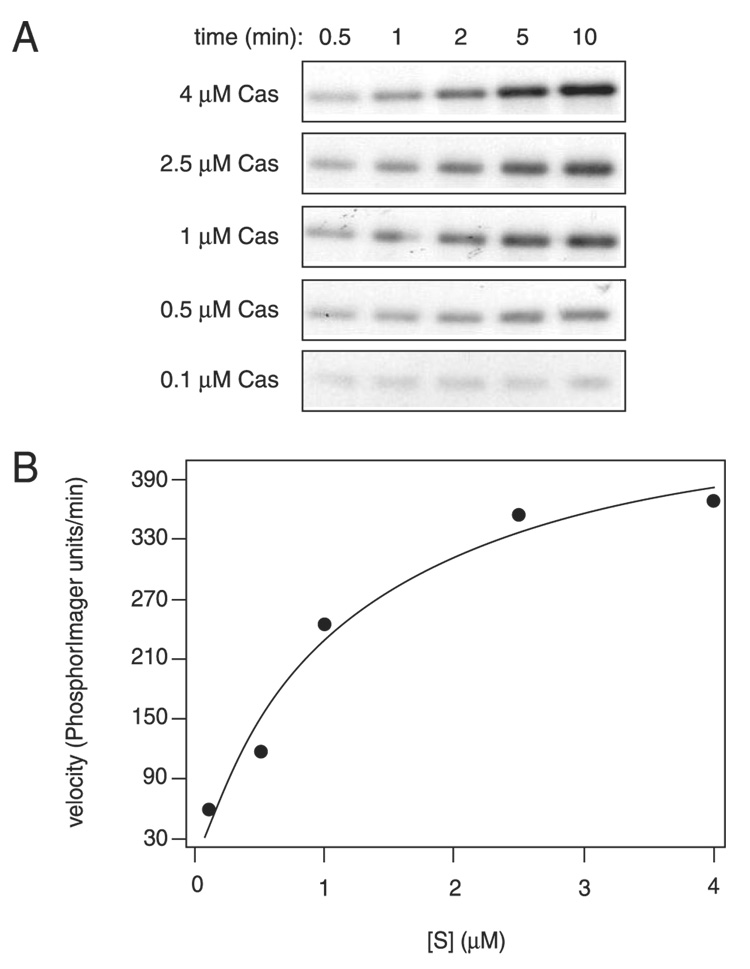
We analyzed the overall phosphorylation rates of WT Cas, CasF7, and CasF12 by in vitro kinase assays using [γ-32P]ATP. A characteristic of processive phosphorylation is that the production of fully phosphorylated product follows classical Michaelian kinetics. In contrast, for a nonprocessive mechanism, increasing the concentration of substrate results in decreased accumulation of fully phosphorylated product (11, 12). This characteristic of nonprocessive reactions is because of the competition between unphosphorylated substrate and partially phosphorylated substrates each time the substrate and enzyme dissociate. We carried out the reactions at five different concentrations of Cas (0.1, 0.5, 1, 2.5, and 4 µm). We removed aliquots of the reaction at different time points and analyzed them by SDS-PAGE and autoradiography. To quantitate 32P incorporation, the gels were dried and exposed to a Phos-phorImager screen (GE Healthcare). As we reported previously (14), higher concentrations of WT Cas result in increased accumulation of phosphorylated Cas, consistent with a processive mechanism (Fig. 4). To ensure that the reaction had gone to completion, we carried out stoichiometry measurements using 1 µm Cas. 32P-Labeled Cas was excised from the gel, solubilized, and analyzed in a liquid scintillation counter. These experiments gave a stoichiometry of 15.1±1.9 mol of phosphate/mol of protein for wild-type Cas, indicating that most of the 17 sites were phosphorylated in this experiment. We also observed an increase in the accumulation of phosphorylated CasF7 and CasF12 with increasing substrate concentrations (Fig. 5). Stoi-chiometry measurements for CasF7 and CasF12 were 6.1 ± 1.9 and 4.7 ± 0.9 mol of phosphate/mol of protein, respectively. The dependence of product formation on Cas concentration in these experiments suggests that the multisite mutants CasF7 and CasF12 are phosphorylated processively by Src.
FIGURE 4. Kinetics of wild-type Cas phosphorylation at different substrate concentrations.
A, WT Cas was incubated in the presence of 50 nm v-Src at 30 °C in kinase assay buffer. Reactions were started by the addition of 100 µm unlabeled ATP plus [γ-32P]ATP (5 µCi). Aliquots were withdrawn at the indicated times and analyzed by 8% SDS-PAGE and autoradiography. B, incorporation of 32P into Cas was measured by exposing the dried gels to a PhosphorImager screen (GE Healthcare) and quantifying the band intensity using ImageQuant software. A graph of velocity (PhosphorImager units/min) versus substrate concentration is shown. The data were fit to the Michaelis-Menten equation using nonlinear regression analysis.
FIGURE 5. Kinetics of mutant Cas phosphorylation at different substrate concentrations.
A, CasF7 was incubated in the presence of 50 nm v-Src at 30 °C in kinase assay buffer. Reactions were started by the addition of 100 µm unlabeled ATP plus [γ-32P]ATP (5 µCi). Aliquots were withdrawn at the indicated times and analyzed by 8% SDS-PAGE and autoradiography. B, incorporation of 32P into Cas was measured by exposing the dried gels to a PhosphorImager screen and quantifying the band intensity using ImageQuant software. A graph of velocity (PhosphorImager units/min) versus substrate concentration is shown. The data were fit to the Michaelis-Menten equation using nonlinear regression analysis. C, CasF12 phosphorylation reactions were analyzed by SDS-PAGE as described above in A. D, CasF12 phosphorylation was quantified by PhosphorImager as shown in B.
We carried out similar experiments with varying concentrations of Src. Because the enzyme and substrate remain bound during the multiple phosphorylation events in a processive mechanism, the rate of progression from an intermediate phosphorylated form to the fully phosphorylated product is independent of the enzyme concentration. In contrast, for a nonprocessive mechanism in which each phosphorylation depends on enzyme-substrate binding, the rate of progression depends on enzyme concentration. We performed pulse-chase assays in which the chase reactions contained different concentrations of Src. We first incubated Src (800 nm) and WT Cas, CasF7, or CasF12 (2 µm) with 0.1 µm [γ-32P]ATP for 3 min (pulse) and then diluted the reaction to yield final Src concentrations ranging from 100 to 600 nm. The dilutions contained an excess of unlabeled ATP (0.5 mm final) to reduce the specific activity of [γ-32P]ATP to levels that were undetectable in our assay. Our results showed comparable shifts in electrophoretic mobility at different concentrations of Src for WT Cas, CasF7, and CasF12 (Fig. 6). Time courses at 100 and 600 nm Src showed similar rates of phosphorylation (Fig. 6). This lack of dependence on Src concentration was consistent with a processive mechanism. In addition, phosphorylated Cas migrated as a single band in each case. Nonprocessive phosphorylation produces diffuse bands because of the formation of partially phosphorylated products, which migrate at various molecular weights (14). Our results suggest that even in the case of a mutant lacking 12 phosphorylation sites (CasF12), Src bound Cas once and completely phosphorylated the remaining sites before dissociating.
FIGURE 6. Phosphorylation of WT and mutant forms of Cas at different Src concentrations.
A, left, pulse-chase experiments were carried out on WT Cas at different Src concentrations. In the pulse reaction, Cas and Src were incubated on ice in the presence of 0.1 µm[γ-32P]ATP. After 3 min, aliquots were removed from the reaction and diluted into four separate tubes containing kinase assay buffer and excess unlabeled ATP (0.5 mm final) to give final Src concentrations of 100, 200, 400, and 600 nm. The chase reaction was carried out at 30 °C for 25 min. Reactions were stopped by adding SDS-PAGE sample buffer and boiling. Equal amounts of Cas from each reaction were analyzed by SDS-PAGE and autoradiography. Aliquots removed after 3-min pulse reaction were also analyzed on the gels. Right, in separate pulse-chase reactions, time courses were followed at Src concentrations of 100 and 600 nm. B, similar experiments were performed for CasF7. C, similar experiments were performed for CasF12.
Add-back Mutants of Cas Are Phosphorylated
To test whether a threshold number of tyrosines are required for Src to phosphorylate the substrate domain of Cas, we generated two “add-back” mutants. As noted above, the YXXP motifs in the substrate domain of Cas can be divided into YDXP and YQXP motifs. Our previous synthetic peptide data show that a substrate with the Tyr253 sequence has the highest value of kcat/Km for Src phosphorylation among the YDXP motifs, whereas a Tyr196 peptide has the highest kcat/Km among the YQXP motifs (9). We also reported that Tyr253 plays a critical role in the migration of Src transformed cells (9). Therefore, we reintroduced Tyr253 alone or together with Tyr196 into CasF17 to generate the CasF17-Tyr253 and CasF17-Tyr253/Tyr196 mutants. We carried out in vitro kinase assays using purified Src, Cas (WT and mutants), and [γ-32P]ATP. The results show that Src is able to phosphorylate the add-back mutants (Fig. 7). The phosphorylation levels are clearly lower than for WT Cas, and there is no shift in electrophoretic mobility for the mutants. Nonetheless, the results show that Src can gain access to individual tyrosines in the middle of the substrate region of Cas, even in the absence of surrounding sites.
FIGURE 7. Add-back mutants of Cas are phosphorylated by Src.
In vitro kinase assays were carried out at 30 °C using Cas proteins (WT or add-back mutants), v-Src, and [γ-32P]ATP. Aliquots were withdrawn at the indicated times and analyzed by SDS-PAGE and autoradiography.A, phosphorylation of add-back mutant CasF17 + Tyr253. B, phosphorylation of add-back mutant CasF17 + Tyr253/Tyr196.
Cas Phosphorylation Sites Are Dispensable for Src-mediated Anchorage-independent Growth
We tested the ability of wild-type and mutant forms of Cas to promote growth of fibroblasts derived from Cas-deficient mice. We produced cell lines stably expressing wild-type Cas and the following Cas mutants in the presence or absence of v-Src: CasF7, CasF12, CasF17, and CasF17-Tyr253 (Fig. 8). For comparison, we included a single-site mutant (Cas Y253F) in our study. As we observed previously (9), Cas did not affect the anchored growth rates of Src-transformed or -non-transformed cells (data not shown). Anchorage-independent growth is a reliable hallmark of most tumor cells (22, 23). As reported previously (4, 7, 9), the ability of Src to promote anchor-age-independent growth was augmented by Cas (Fig. 9A). Cells transfected with control vectors or with wild-type Cas alone did not grow in suspension, whereas cells expressing Src alone doubled in number in 11 days (Fig. 9A). In contrast, Src transfectants expressing either wild-type Cas or Y253F Cas increased their numbers by over 7- or 14-fold, respectively, in the same time period (Fig. 9A). This level of anchorage-independent growth was significantly higher than for the cells transfected with Src alone (p < 0.0001). Src transfectants expressing mutant forms of Cas (CasF7, CasF12, CasF17, or the add-back mutant CasF17-Tyr253) grew at least as well as cells expressing wild-type Cas (Fig. 9,A and B). This anchorage-independent growth of Src transfectants expressing mutant forms of Cas lacking multiple phosphorylation sites (in particular, CasF17) suggests that Cas augments the ability of Src to override anchorage dependence, even in the absence of substrate domain phosphorylation.
FIGURE 8. Expression of WT and mutant Cas in Cas−/− fibroblasts.
Cas-deficient fibroblasts were transfected with wild-type or mutant forms of Cas (in pBABEhygro) and v-Src (in pBABEpuro) and selected for hygromycin and puromycin resistance. Cells marked minus were transfected with the appropriate vector controls. Cells were lysed in modified radioimmune precipitation assay buffer, and lysates (30 µg) were analyzed by immunoblotting using Cas, v-Src, or β-actin antibodies. A, Cas Y253F, CasF17, and CasF17 + Tyr253 mutants. B, CasF7 and CasF12 mutants.
FIGURE 9. Effects of Src and Cas on Src-mediated anchorage-independent growth and migration.
A and B, anchorage-independent growth. 20,000 cells were plated in 12-well tissue culture plates. Cell numbers were obtained using a Coulter counter after 11 days. Open bars, cells without Src; closed bars, cells expressing Src. C and D, cell migration assays. 200,000 cells were plated on cell culture inserts (3-µm pore size) in 6-well plates. Cell migration was measured after 72 h as the percent of cells found in the bottom well/total cell number. E, cell migration assays were carried out using 8-µm pore size membranes, and cell migration was examined after 24 h.
Cas Phosphorylation Sites Are Necessary for Src to Promote Cell Migration
Data described above indicate that Src-mediated phosphorylation of Cas is not necessary to promote non-anchored growth. On the other hand, phosphorylation of Cas by Src has been shown to play a critical role in cell migration (8–10, 16, 24). To test whether the Cas mutants are able to promote cell migration, we examined the ability of Cas-deficient cells expressing Cas and/or Src to migrate through pores in a migration chamber. As shown in Fig. 9C, there was a significant increase in migration for cells expressing wild-type Cas together with Src as compared with cells expressing Src or Cas alone (p <0.0002). Cells expressing Src together with CasF7, CasF12, or CasF17 showed levels of migration that were comparable with vector controls (Fig. 9D). We confirmed our results using 8 µm membranes employed by Honda et al. (7) (Fig. 9E).
We found previously that Tyr253 in the substrate domain of Cas significantly increases migration of Src-transformed cells (9). To test whether Tyr253 alone is sufficient to promote cell migration, we carried out migration assays for cells transfected with the add-back mutant CasF17-Tyr253. Fewer than 0.5% of these cells migrated, indicating that Tyr253 is not sufficient to promote cell migration (Fig. 9C). In agreement with previous reports (8, 10), our results show that phosphorylation of multiple sites in the substrate domain of Cas is required for cell migration.
DISCUSSION
Cas is representative of a class of efficient Src substrates that contain SH3 and SH2 ligands and are phosphorylated on multiple sites. Multi-site phosphorylation of Cas is important for the various physiological roles of Cas in normal and transformed cells (5, 7, 8, 10, 16, 18). Several lines of evidence indicate that Src is directly responsible for phosphorylating Cas (3, 25, 26). Here, we have investigated the mechanism by which Src recognizes Cas phosphorylation sites. Our previous results show that (at least in vitro) Src phosphorylates Cas by a processive mechanism (14). The biological importance of processive phosphorylation is not clear at present. In the case of the Cdk inhibitor Sic1, phosphorylation occurs by a nonprocessive mechanism. The necessity for multisite, nonprocessive phosphorylation introduces a highly cooperative step and creates a switch-like response (15). The processive phosphorylation of Cas (and lack of dependence on enzyme concentration) suggests that Cas phosphorylation is unlikely to display a switch-like response. Instead, the biological importance of processive phosphorylation may be to greatly increase the rate of multisite phosphorylation, enabling faster assembly of the Cas-SH2 docking interactions and a faster response time. In a distributive mechanism, partially phosphorylated intermediates of Cas would act effectively as competitive inhibitors. Src would need to phosphorylate a significant portion of the entire pool of Cas molecules to produce highly phosphorylated Cas and achieve a biological effect. Thus, the processive mechanism allows for a more potent effect of Src than would be realized by a distributive mechanism.
One question investigated in this study is whether phosphorylation of the sites within the Cas substrate domain follows a defined sequence. Several examples are known in which phosphorylation at one site on a protein “primes” another site for subsequent phosphorylation. As an initial test of this possibility, we generated four single-site mutants of Cas in which the most favored Src phosphorylation sites (as defined by synthetic peptide substrates) were altered. Our results show that these mutants are phosphorylated processively by Src (Fig. 2 and data not shown). Thus, of the sites tested, no single site acted as a “linchpin” that permitted phosphorylation of subsequent sites, suggesting that Cas phosphorylation by Src does not follow an obligatory sequence.
The lack of a specific sequence of phosphorylations was reinforced by experiments on the multisite mutants CasF7 and CasF12. These mutants, which lack the “best” phosphorylation sites, were phosphorylated processively (Fig. 3 and Fig.5). Presumably, if there were an obligatory order of phosphorylation, these mutants would not have been phosphorylated at multiple sites. The data suggest that tyrosines in the Cas substrate domain are not kinetically distinguishable and that any group of tyrosines present in the substrate region would be sufficient for Src phosphorylation. Furthermore, Src phosphorylated mutant forms of Cas in which all but one or two sites had been eliminated (CasF17-Tyr253 and CasF17-Tyr253/Tyr196; Fig. 7). It is noteworthy that these sites (Tyr196 and Tyr253) were distant in primary sequence from the C-terminal Src-binding site of Cas. These data stand in contrast to our previous results using a simple synthetic peptide model of Cas. The peptide contained two YDXP motifs from Cas (modeled on the sequence of Tyr253) and a C-terminal Src SH2 binding sequence. The Src family kinase Hck phosphorylates the sites in a defined order, proceeding from C terminus to N terminus (27). Presumably, in the intact Cas protein, a large number of potential phosphorylation sites are available to Src. The structure of the Cas substrate domain appears to be flexible enough so that Src (which is presumably bound at the C terminus) can gain access to sites far from the C-terminal region, in the middle of Cas.
Our data also address the biological importance of the Cas phosphorylation sites. We reintroduced wild-type or mutant forms of Cas into fibroblasts derived from Cas knock-out cells, in the presence or absence of Src. In agreement with others (4, 7, 8) and with our previous results (9), we found that the ability of Src to promote anchorage-independent growth is augmented by Cas (Fig. 9A). In contrast to these results, Brabek et al. (17) recently reported that Cas does not alter the anchor-age-independent growth characteristics of Src-transformed cells. (Brabek et al. (17) did note an increase in the number of colonies formed in soft agar for cells expressing Cas and Src together, but the increase is not statistically significant.) The reason for this discrepancy is not clear at present; one possibility is that the expression levels of Src and Cas differ among the cell lines tested by various investigators. In any case, our data (Fig. 9A) show that the difference in nonanchored growth between Cas-deficient and Cas-expressing cells is a matter of degree rather than an all-or-none phenomenon.
It has been reported previously that Cas variants lacking the SH3 domain or the YDXP sites from the substrate domain can promote anchorage-independent growth of Src-transformed cells (8). Here, we show that Cas mutants lacking the major phosphorylation sites (CasF7 and CasF12) or even all of the phosphorylation sites (CasF17) can promote anchorage-independent growth of the Cas knock-out cells in the presence of Src. Thus, Cas has the capacity to promote anchorage-independent growth in the absence of phosphorylation. This may be because of a scaffolding function of Cas in which Cas (via its C-terminal region containing SH3 and SH2 ligands) recruits Src to phosphorylate focal adhesion kinase, paxillin, and other integrin-associated substrates (17). Cas lacking a substrate domain has been shown to act as a dominant-negative mutant in COS-7 cells, consistent with its ability to bind these components but not downstream effectors (28). An additional mechanism may involve the ability of the C-terminal polyproline region of Cas to bind to the SH3 domain, relieving Src autoinhibition and enhancing kinase activity (5, 29).
Phosphorylation of the YXXP motifs in the Cas substrate domain has been shown previously to be necessary for cell migration (8, 17, 18). Within these motifs, the YDXP sequences are shown to be of particular importance, as a Cas construct lacking the YDXP motifs is deficient in cell migration and actin stress fiber organization (8). In wound healing assays, a Cas mutant possessing as few as 6 tyrosines gives some activity (10), indicating that the full complement of sites is not required. Here, in our study, we used Cas mutants lacking multiple phosphorylation motifs (CasF7, CasF12, and CasF17) to determine whether there were a threshold number of tyrosine motifs for cell migration. As shown previously (18), CasF17 is incapable of supporting cell migration in Src-transformed cells (Fig. 9, C and D). Reintroduction of a single site (Tyr253) that was shown previously to be important in cell migration did not rescue this defect (Fig. 9C). Furthermore, the CasF7 and CasF12 variants did not promote migration (Fig. 9D) despite the fact that these mutants were phosphorylated processively (Fig. 3 and Fig.5). Thus, it is clear that multisite phosphorylation of Cas, although necessary for cell migration activity, is not sufficient. In contrast, multisite phosphorylation is dispensable for anchorage-independent growth in Src-transformed cells.
Acknowledgment
We thank Chris Gordon for assistance with baculovirus expression.
Footnotes
This work was supported by National Institutes of Health Grants CA58530 (to W. T. M.) and CA88805 and EY014479 (to G. S. G.)
The abbreviations used are: SH, Src homology; WT, wild type
CasF7, CasF12, and CasF17 refer to mutants lacking 7, 12, and 17 Tyr residues, respectively.
References
- 1.Reynolds AB, Kanner SB, Wang HC, Parsons JT. Mol. Cell. Biol. 1989;9:3951–3958. doi: 10.1128/mcb.9.9.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuda M, Mayer BJ, Fukui Y, Hanafusa H. Science. 1990;248:1537–1539. doi: 10.1126/science.1694307. [DOI] [PubMed] [Google Scholar]
- 3.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. EMBO J. 1994;13:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda H, Oda H, Nakamoto T, Honda Z, Sakai R, Suzuki T, Saito T, Nakamura K, Nakao K, Ishikawa T, Katsuki M, Yazaki Y, Hirai H. Nat. Genet. 1998;19:361–365. doi: 10.1038/1246. [DOI] [PubMed] [Google Scholar]
- 5.Bouton AH, Riggins RB, Bruce-Staskal PJ. Oncogene. 2001;20:6448–6458. doi: 10.1038/sj.onc.1204785. [DOI] [PubMed] [Google Scholar]
- 6.Chodniewicz D, Klemke RL. Biochim. Biophys. Acta. 2004;1692:63–76. doi: 10.1016/j.bbamcr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Honda H, Nakamoto T, Sakai R, Hirai H. Biochem. Biophys. Res. Commun. 1999;262:25–30. doi: 10.1006/bbrc.1999.1162. [DOI] [PubMed] [Google Scholar]
- 8.Huang J, Hamasaki H, Nakamoto T, Honda H, Hirai H, Saito M, Takato T, Sakai R. J. Biol. Chem. 2002;277:27265–27272. doi: 10.1074/jbc.M203063200. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg GS, Alexander DB, Pellicena P, Zhang ZY, Tsuda H, Miller WT. J. Biol. Chem. 2003;278:46533–46540. doi: 10.1074/jbc.M307526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin NY, Dise RS, Schneider-Mergener J, Ritchie MD, Kilkenny DM, Hanks SK. J. Biol. Chem. 2004;279:38331–38337. doi: 10.1074/jbc.M404675200. [DOI] [PubMed] [Google Scholar]
- 11.Burack WR, Sturgill TW. Biochemistry. 1997;36:5929–5933. doi: 10.1021/bi970535d. [DOI] [PubMed] [Google Scholar]
- 12.Ferrell JE, Jr, Bhatt RR. J. Biol. Chem. 1997;272:19008–19016. doi: 10.1074/jbc.272.30.19008. [DOI] [PubMed] [Google Scholar]
- 13.Aubol BE, Chakrabarti S, Ngo J, Shaffer J, Nolen B, Fu XD, Ghosh G, Adams JA. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellicena P, Miller WT. J. Biol. Chem. 2001;276:28190–28196. doi: 10.1074/jbc.M100055200. [DOI] [PubMed] [Google Scholar]
- 15.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 16.Klemke RL, Leng J, Molander R, Brooks PC, Vuori K, Cheresh DA. J. Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brabek J, Constancio SS, Shin NY, Pozzi A, Weaver AM, Hanks SK. Oncogene. 2004;23:7406–7415. doi: 10.1038/sj.onc.1207965. [DOI] [PubMed] [Google Scholar]
- 18.Brabek J, Constancio SS, Siesser PF, Shin NY, Pozzi A, Hanks SK. Mol. Cancer Res. 2005;3:307–315. doi: 10.1158/1541-7786.MCR-05-0015. [DOI] [PubMed] [Google Scholar]
- 19.Morgenstern JP, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander DB, Ichikawa H, Bechberger JF, Valiunas V, Ohki M, Naus CC, Kunimoto T, Tsuda H, Miller WT, Goldberg GS. Cancer Res. 2004;64:1347–1358. doi: 10.1158/0008-5472.can-03-2558. [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, Jia Z, Nagele RG, Ichikawa H, Goldberg GS. Cancer Res. 2006;66:1543–1552. doi: 10.1158/0008-5472.CAN-05-3152. [DOI] [PubMed] [Google Scholar]
- 22.Giancotti FG, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg GS, Jin Z, Ichikawa H, Naito A, Ohki M, El-Deiry WS, Tsuda H. Cancer Res. 2001;61:1334–1337. [PubMed] [Google Scholar]
- 24.Cho SY, Klemke RL. J. Cell Biol. 2000;149:223–236. doi: 10.1083/jcb.149.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlaepfer DD, Broome MA, Hunter T. Mol. Cell. Biol. 1997;17:1702–1713. doi: 10.1128/mcb.17.3.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Nishida J, Yazaki Y, Hirai H. J. Biol. Chem. 1994;269:32740–32746. [PubMed] [Google Scholar]
- 27.Scott MP, Miller WT. Biochemistry. 2000;39:14531–14537. doi: 10.1021/bi001850u. [DOI] [PubMed] [Google Scholar]
- 28.Cheresh DA, Leng J, Klemke RL. J. Cell Biol. 1999;146:1107–1116. doi: 10.1083/jcb.146.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burnham MR, Bruce-Staskal PJ, Harte MT, Weidow CL, Ma A, Weed SA, Bouton AH. Mol. Cell. Biol. 2000;20:5865–5878. doi: 10.1128/mcb.20.16.5865-5878.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]