Abstract
Background
The phosphatidylinositol 3-kinase (PI3K)-AKT signal transduction pathway is critical to cell growth and survival. In vitro functional studies indicate that the candidate schizophrenia susceptibility gene DTNBP1 influences AKT signaling to promote neuronal viability. The AKT1 gene has also been implicated in schizophrenia by association studies and decreased protein expression in the brains of schizophrenic patients.
Methods
The association of DTNBP1 in the Irish Study of High Density Schizophrenia Families (ISHDSF) prompted our investigation of AKT1 for association with disease in this sample. Eight SNPs spanning AKT1 were analyzed for association with schizophrenia across 4 definitions of affection, and according to Operational Criteria Checklist of Psychotic Illness (OPCRIT) symptom scales. We examined expression of AKT1 mRNA from post-mortem brain tissue of schizophrenic, bipolar and control individuals.
Results
No single marker showed significant association, but the risk haplotype previously found over-transmitted to Caucasian schizophrenic patients was significantly under-transmitted in the ISHDSF (0.01<p<0.05), across all OPCRIT symptom dimensions. Exploratory haplotype analysis confirmed association with schizophrenia toward the 5' end of AKT1 (0.008<p<0.049, uncorrected). Consistent with reduced AKT1 protein levels reported in schizophrenic brain, we found significantly decreased RNA levels in prefrontal cortex of schizophrenic individuals.
Conclusions
The replication of association of AKT1 gene variants in a further Caucasian family sample adds support for involvement of AKT signaling in schizophrenia, perhaps encompassing a broader clinical phenotype that includes mood dysregulation. We show that AKT1 signaling may be compromised in schizophrenic and bipolar patients via reduced RNA expression of specific AKT isoforms.
Keywords: AKT1, schizophrenia, association, gene expression, clinical features
Introduction
Schizophrenia (MIM 181500) is a severe psychiatric illness with complex etiology and lifetime prevalence of ~1% worldwide (1). Family, twin and adoption studies have consistently demonstrated a genetic component to schizophrenia, together with developmental and environmental influences (1, 2). Current pathophysiological models posit the disease to be the cognitive, perceptual and behavioural outcome of subtle alterations in brain development occurring long before clinical symptoms, and emphasize glutamatergic, dopaminergic and GABAergic systems (3).
In recent years, positional candidate genes have been widely studied for association with schizophrenia. A parallel approach toward identifying susceptibility variants is via ‘functional candidate’ genes or molecular pathways based on disease etiology. Alterations in intracellular signaling pathways are known to be important in treating neuropsychiatric disorders, e.g. two medications for bipolar disorder and schizophrenia, lithium and clozapine (respectively), enhance AKT signaling (4). Indeed, as a key signaling intermediate downstream of dopamine receptor D2 (DRD2), the best-established target of antipsychotic drugs, AKT is instrumental in normal dopaminergic transmission and expression of dopamine-associated behaviours (4). DNA sequence variants in AKT1 were associated with bipolar disorder following linkage to 14q32 (5), and more recently with schizophrenia (6).
AKT1 belongs to a serine/threonine kinase family, including AKT2 and AKT3, integral to a signal transduction pathway (with phosphatidylinositol-3 kinase (PI3K) and glycogen synthase kinase 3β (GSK3β)) that regulates cellular functions such as survival, growth, transcriptional regulation and nutrient metabolism (7). AKT1 activation can suppress apoptosis in a transcription-independent manner. AKT1, AKT2 and AKT3 are encoded by separate genes, each playing unique as well as common roles in cells: Akt1 knockout mice are growth retarded, Akt2 knockout mice develop diabetes-like symptoms through impaired insulin response, and Akt3 knockout mice show reduced brain size (reviewed in 8). AKT1, although ubiquitous, is highly expressed in brain, hence critically involved in neuronal survival, but also important for neuronal excitability and synaptic plasticity by directly regulating GABA-A receptor mediated synaptic inhibition (9). It appears to be involved in neurodevelopment and working memory formation, domains potentially impaired in schizophrenia (10, 11).
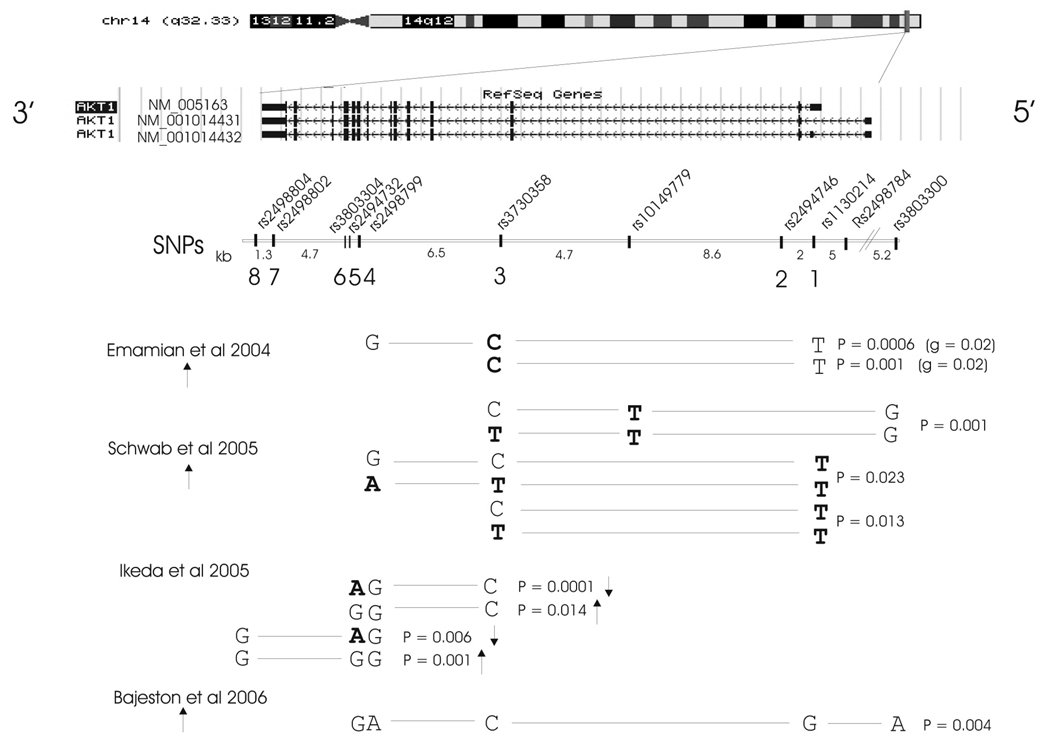
The study of Emamian et al (6) provided converging lines of evidence to link dysregulation of AKT signaling with schizophrenia. Reduced AKT1 protein levels and phosphorylation of its substrate GSK3β were found in brain tissue of schizophrenic patients, while C57Bl/6 mice treated with haloperidol displayed increased phosphorylation of total AKT and GSK3β. Furthermore, DNA variants within the AKT1 gene were associated with schizophrenia in 268 families of European origin. Most significantly associated was SNP rs3730358 in intron 3 (p=0.05), with one haplotype (rs1130214-rs3730358-rs2498799, alleles T-C-G) spanning intron 2-intron 7, over-transmitted to affected offspring (1.8:1, p=0.0006) (Fig.1). The AKT1 core risk haplotype (rs1130214-rs3730358, T-C, p=0.001) was associated with lower AKT1 protein levels in lymphocyte-derived cell lines of schizophrenia patients.
Fig. 1.
Most significant AKT1 SNP haplotypes associated with schizophrenia to date. Exon locations of the 3 main alternative AKT1 transcripts are shown at the top (http://genome.ucsc.edu/, human genome build 35). SNPs are positioned underneath with intervening distances in kilobases. Numbers 1–8 refer to SNPs typed in our study. The p values of the most significant haplotypes are given for each report of association between AKT1 and schizophrenia. The arrows depict over-representation (up arrow) or under-representation (down arrow) in cases. SNP alleles highlighted in bold are significantly associated with schizophrenia as single SNPs. Marker order and haplotypes are given in the reverse order here to that in the text, in accordance with the orientation of the gene on the UCSC genome browser.
Association between AKT1 and schizophrenia has been replicated in 3 independent studies: 79 Caucasian sib-pair families (12), a Japanese 507 case −437 control sample (13) and an Iranian 321 case −383 control sample (14). Schwab et al (12) found association with rs3730358 (p=0.027), but more significantly with 5’UTR SNPs, rs1130214 (p=0.011) and rs10149779 (p=0.002). Most significant association was detected with haplotype rs3803300-rs10149779-rs3730358 (p=0.001), although others were also over-transmitted to affected offspring, including the T-C-G risk haplotype of Emamian et al (6; Fig.1). Ikeda et al (13) found most significant association with rs2494732, in intron 7 (p=0.004), and with haplotypes overlapping the Emamian et al ‘risk’ haplotype at the 3’ end (Fig.1). The most significant haplotype rs3730358-rs2498799-rs2494732 (C-G-A) was more frequent in controls than cases, indicative of a ‘protective’ effect. Bajestan et al (14) showed modest evidence of association with schizophrenia in the Iranian population with a different haplotype of the 5 original SNPs of Emamian et al (6). Norton et al (15) in a UK 673 case −716 control sample recently reported weak evidence for association (p=0.04, uncorrected) with a 4-marker haplotype encompassing the Emamian et al core haplotype. However, a second Japanese case-control sample (16), and Japanese and Taiwanese family-based studies (17, 18) detected no association between these SNPs and schizophrenia. A family-based study in the Finnish population also failed to demonstrate association between AKT1 or DTNBP1 and schizophrenia (19).
Functional studies suggest that defective PI3K-AKT signaling in schizophrenic brains may be due to decreased expression of dysbindin (DTNBP1), which may normally support neuronal viability through a neurotrophic effect via AKT signaling (20). Defective dysbindin-AKT signaling may therefore produce long-term vulnerability leading to psychiatric illness. The presence of a high-risk DTNBP1 haplotype in the ISHDSF previously implicating dysbindin in the etiology of schizophrenia (21), prompted our investigation of association between AKT1 and schizophrenia in this sample.
Materials and Methods
Subjects and phenotypes
The ISHDSF sample comprises 265 high-density schizophrenia families with 1408 individuals available for genotyping (22). All participating individuals gave appropriate informed consent to the study. The sample was divided into 4 concentric diagnostic categories for analysis purposes, ranging from narrow to very broad spectrum disease. Briefly, D2 (core schizophrenia) includes schizophrenia, poor-outcome schizoaffective disorder and simple schizophrenia, D5 (narrow psychosis spectrum) additionally includes schizotypal personality disorder and all other non-affective psychotic disorders, D8 (broad spectrum) additionally includes mood-congruent and incongruent psychotic affective illness, paranoid, avoidant and schizoid personality disorders, while D9 (very broad spectrum) includes family members with any psychiatric disorder. All patients with history of any psychotic episode were assessed by K.S.K. for lifetime psychotic symptoms using the Operational Criteria Checklist of Psychotic Illness (OPCRIT; 23), to reflect both symptom severity and chronicity.
The Stanley Foundation postmortem brain series (SBS) donated by the Stanley Medical Research Institute comprises DNA and total RNA extracted from dorsolateral prefrontal cortex (Brodmann’s area 46) of 35 individuals each with schizophrenia, bipolar disorder and controls (24).
SNPs and genotyping
We assessed 8 AKT1 SNPs, 4 common to prior positive studies (rs1130214, rs2494732, rs2498799, rs3730358) and 4 additional SNPs (http://www.hapmap.org/) to increase marker density and include the 3’UTR (rs2494746, rs3803304, rs2498802, rs2498804). SNP rs3803300 (6), failed to perform well by template-directed dye-terminator incorporation with fluorescence polarization detection (FP-TDI).
SNP genotyping was performed by FP-TDI using AcycloPrime SNP detection kits (PerkinElmer Life Sciences, Boston, MA), manufacturer’s instructions, and an automated allele scoring platform (25). PCR and single-base extension primers were designed manually and are available on request. Average genotyping completion rate was 93%. Genotypes were examined for incompatibilities within families by PEDCHECK v1.1 (26), unlikely recombinations by MERLIN (27), and were corrected as necessary. Where errors could not be unambiguously resolved, respective marker genotypes were deleted for the whole family.
Statistical methods
Genotype deviation from Hardy-Weinberg equilibrium (HWE) and pairwise intermarker linkage disequilibrium (LD) were assessed using HAPLOVIEW v3.2 from genotypes of unrelated founders (28). We performed association analysis by transmission disequilibrium test (TDT) using TRANSMIT v2.5.4 (29) and PDTPHASE v4.0, an implementation of the pedigree disequilibrium test (PDT) with extensions to deal with uncertain haplotypes and missing data (30, 31). P values were empirically derived from 10,000 bootstrap replicates in each TRANSMIT analysis to allow inclusion of multiple affected offspring per family, and multiple families per pedigree. PDTPHASE creates a measure of marker-disease associations for each pedigree, averaged from that defined for each constituent triad and discordant sibpair, thus has greater power over methods using only a subset of the data. PDTPHASE was also the analytic tool in the original AKT1 association with schizophrenia (6). Throughout, we analyzed only haplotypes of frequency >2% to avoid the application of statistics to small numbers of chromosomes and multiple rare alleles/haplotypes.
Association analysis was performed in stages across diagnostic categories: 1) Single SNPs 2) A replication study, specifically examining reported AKT1 schizophrenia risk haplotypes 3) Investigation into etiological heterogeneity, by testing the original risk haplotype (6) for association with OPCRIT symptom dimensions 4) False Discovery Rate control (32) following this directed testing phase, to minimize risk of Type I errors 5) ‘Sliding windows’ analysis of 2-4 marker haplotypes and 6) In the absence of ‘tag SNPs’, exploratory analysis of all 2-4 SNP haplotypes for association with disease. The large number of possible haplotypes precludes presenting all these exploratory data systematically. Therefore, the latter two stages remain uncorrected for multiple testing and are presented with that caveat. The statistical significance of these exploratory findings, though based on empirical p values obtained by simulation, should be viewed accordingly.
Quantitative real-time PCR
Expression of AKT1, AKT2, and AKT3 was analyzed by quantitative real-time PCR using SYBR Green I on a Bio-Rad (Hercules, CA) iCycler platform, normalized against two reference genes GAPDH and TBP by the 2−ΔΔCT algorithm (33; 34), implemented in the Relative Gene Expression Macro Software Package (Bio-Rad Inc). For details of the real-time PCR experiments, statistical analysis of gene expression levels, and AKT1 and DTNBP1 haplotype ascertainment see Supplementary Methods.
Bioinformatics
50bp of DNA sequence encompassing rs1130214 was analyzed for transcription-factor binding sites using Matinspector (http://www.genomatrix.de; 35) and putative splice sites using the Berkeley Drosophila Genome Project predictor (http://www.fruitfly.org/seq_tools/splice.html). VISTA was used to define conserved regions in AKT1 genomic sequence surrounding rs1130214 (36). Other features were ascertained from the University of California Santa Cruz genome browser (http://genome.ucsc.edu/).
Results
Table 1 lists the AKT1 SNP locations and allele frequencies in the ISHDSF, which are similar to those of Emamian et al (6). AKT1 has 14–15 known exons (depending on splice variant) spanning ~24.2kb (Fig.1). Genotype frequencies of all SNPs were in HWE (data not shown).
Table 1.
Position and allele frequencies of AKT1 SNPs used in this study
| SNP number | SNP name | Position (bp)* | AKT1 gene location # | Distance (bp) | Major allele | Minor allele | Minor allele frequency in ISHDSF |
|---|---|---|---|---|---|---|---|
| 1 | rs1130214 | 104,330,779 | 5’ UTR | 2015 | G | T | 0.31 |
| 2 | rs2494746 | 104,328,764 | Intron 2 | 11312 | G | C | 0.08 |
| 3 | rs3730358 | 104,317,452 | Intron 3 | 6513 | C | T | 0.17 |
| 4+ | rs2498799 | 104,310,939 | Exon 9 | 702 | G | A | 0.25 |
| 5 | rs2494732 | 104,310,237 | Intron 11 | 46 | A | G | 0.45 |
| 6 | rs3803304 | 104,310,191 | Intron 11 | 4704 | G | C | 0.28 |
| 7 | rs2498802 | 104,305,487 | 3’UTR | 1347 | C | G | 0.34 |
| 8 | rs2498804 | 104,304,140 | 3’UTR | G | T | 0.34 |
University of California at Santa Cruz (UCSC) Human Genome Browser Gateway assembly May 2004
Based on AKT1 gene transcript variant 1 (NM_005163) which has 14 exons
rs2498799 is a synonymous coding variant
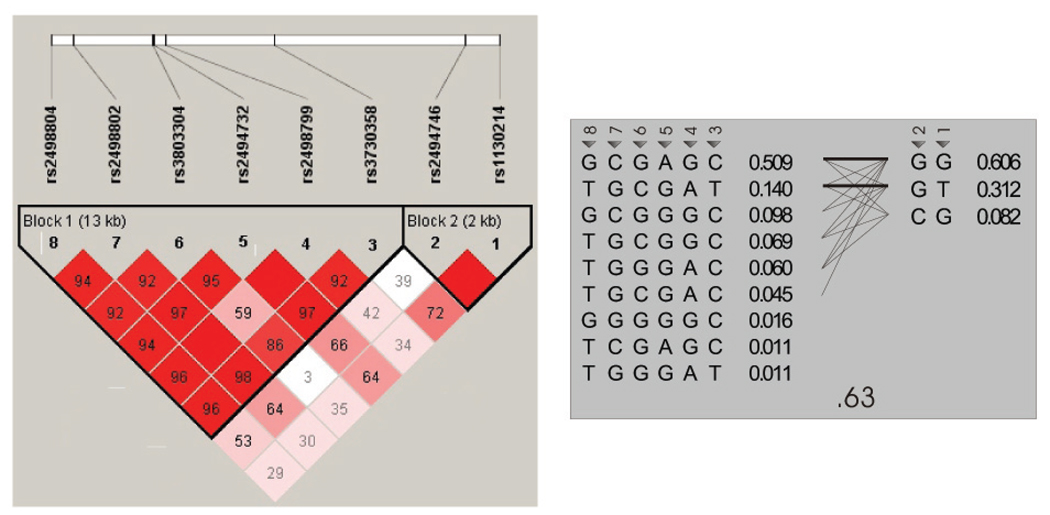
Linkage disequilibrium (LD) map
Pairwise intermarker LD in the ISHDSF revealed 2 haplotype ‘blocks’ across AKT1, with strong LD between 5’ SNPs rs1130214-rs2494746, then further 3’ between rs3730358-rs2498804 (Fig.2). We see a similarly complex LD structure to that reported in cases by Ikeda et al (13), with low LD between rs3803304-rs2498799 (SNPs 4-6 in Fig.1) in the midst of otherwise high intermarker LD. This may contribute to the haplotypic diversity of AKT1 and underlie the lack of tag SNPs for either block (Fig.2).
Fig. 2.
LD map of the AKT1 region in the ISHDSF. The map is viewed through Haploview v 3.2 (the solid spine rule for block structure is shown), with gene direction 3’–5’. Pairwise LD is indicated by the numbers (related to D’) and depth of red colour. SNPs and chromosomal positions are indicated along the top. The region spans ~26kb. Despite apparent ‘blocks’, the haplotypes on the right show no tag SNPs.
Single marker association analysis
AKT1 single markers showed no association with schizophrenia, although rs1130214 and rs2494732 show a modest trend (0.10<p<0.20, two-tailed) towards significance with TRANSMIT and PDTPHASE across several diagnostic categories (Supplemental Table 1). Interestingly, rs2494732 was the most significant SNP of Ikeda et al (13; p=0.0037), whereas Schwab et al (12) found rs1130214 to be associated with disease (p=0.011).
Haplotype replication
Several AKT1 haplotypes have been associated with schizophrenia: in Caucasians (6, 12), haplotype [rs1130214-rs3730358]-rs2498799 (T-C-G, SNPs [1-3]-4 in our study; Fig.1) and in a Japanese sample (13), association is more pronounced toward 3’ AKT1 (Fig.1). Given the ethnicity of our sample, we focused on the Caucasian over-transmitted risk haplotype T-C-G (rs1130214-rs3730358-rs2498799; 6). We see a contrasting trend towards under-transmission in the ISHDSF, with less striking over-transmission of the all-common-allele haplotype G-C-G (Table 2 and Table 3). This effect is most evident with TRANSMIT for haplotype rs1130214-rs2498799 (SNPs 1-4; global p=0.002 at D5; Table 2) and with PDTPHASE, for haplotype rs1130214-rs3730358 (SNPs 1-3, the ‘core’ haplotype of Emamian et al (6); global p=0.031 at D9; Table 3). Our results also contrast those of Schwab et al (12) who consistently saw over-transmission of the T allele of rs1130214 (SNP 1), in 2 versions of the ‘risk’ haplotype (T-C-G and T-T-A; Fig.1). We see the T-T-A haplotype at similar frequency in the ISHDSF (~9%), trending towards over-transmission to affected individuals (data not shown). The overall picture in the ISHDSF (over-transmission of SNPs 1-3-4 haplotype G-C-G, under-transmission of T-C-G) becomes stronger with broader diagnosis, D5-D9 (especially D5; Table 2 and Table 3).
Table 2.
Caucasian AKT1 haplotype replication by TRANSMIT. Significant p values and haplotypes are highlighted in bold.
| DIAG | HAPLO | * | Alleles | No.fams# | p val | Obs | Exp | Freq | Global P |
|---|---|---|---|---|---|---|---|---|---|
| D2 | 1-3 | ↑ | G-C | 326 | 0.079 | 693 | 678 | 0.64 | 0.039 |
| ↓ | T-C | 0.056 | 183 | 196 | 0.20 | ||||
| D5 | 1-3 | ↑ | G-C | 363 | 0.036 | 842 | 822 | 0.058 | |
| ↓ | T-C | 0.078 | 223 | 236 | |||||
| D8 | 1-3 | ↑ | G-C | 376 | 0.125 | 899 | 884 | 0.208 | |
| ↓ | T-C | 0.178 | 247 | 258 | |||||
| D9 | 1-3 | ↑ | G-C | 405 | 0.057 | 1111 | 1089 | 0.140 | |
| ↓ | T-C | 0.056 | 322 | 339 | |||||
| D2 | 1-4 | ↑ | G-G | 321 | 0.106 | 604 | 589 | 0.56 | 0.009 |
| ↓ | T-G | 0.010 | 171 | 190 | 0.19 | ||||
| D5 | 1-4 | ↑ | G-G | 359 | 0.045 | 731 | 712 | 0.002 | |
| ↓ | T-G | 0.003 | 205 | 228 | |||||
| D8 | 1-4 | ↑ | G-G | 372 | 0.068 | 783 | 765 | 0.009 | |
| ↓ | T-G | 0.009 | 228 | 248 | |||||
| D9 | 1-4 | ↑ | G-G | 403 | 0.071 | 964 | 945 | 0.006 | |
| ↓ | T-G | 0.003 | 299 | 324 | |||||
| D2 | 1-3-4 | ↑ | G-C-G | 326 | 0.130 | 607 | 593 | 0.55 | 0.095 |
| ↓ | T-C-G | 0.024 | 172 | 187 | 0.09 | ||||
| D5 | 1-3-4 | ↑ | G-C-G | 363 | 0.087 | 731 | 714 | 0.029 | |
| ↓ | T-C-G | 0.023 | 207 | 225 | |||||
| D8 | 1-3-4 | ↑ | G-C-G | 376 | 0.133 | 781 | 765 | 0.121 | |
| ↓ | T-C-G | 0.054 | 230 | 245 | |||||
| D9 | 1-3-4 | ↑ | G-C-G | 405 | 0.085 | 957 | 937 | 0.187 | |
| ↓ | T-C-G | 0.017 | 301 | 320 | |||||
up and down arrows indicate over- or under-transmission to affected individuals
number of families with transmissions to affected offspring
Table 3.
Caucasian AKT1 haplotype replication by PDTPHASE. Significant p values and haplotypes are highlighted in bold.
| DIAG | HAPLO | * | Alleles | p val | Trio T | Trio NT | Aff Sib | Unaff Sib | Freq | Global P |
|---|---|---|---|---|---|---|---|---|---|---|
| D2 | 1-3 | ↑ | G-C | 0.257 | 700 | 675 | 925 | 909 | 0.65 | 0.199 |
| ↓ | T-C | 0.114 | 181 | 204 | 302 | 301 | 0.19 | |||
| D5 | 1-3 | ↑ | G-C | 0.045 | 850 | 817 | 1001 | 959 | 0.051 | |
| ↓ | T-C | 0.070 | 219 | 246 | 305 | 315 | ||||
| D8 | 1-3 | ↑ | G-C | 0.062 | 907 | 879 | 973 | 949 | 0.087 | |
| ↓ | T-C | 0.102 | 244 | 265 | 310 | 311 | ||||
| D9 | 1-3 | ↑ | G-C | 0.047 | 1123 | 1091 | 801 | 770 | 0.031 | |
| ↓ | T-C | 0.014 | 319 | 343 | 245 | 265 | ||||
| D2 | 1-4 | ↑ | G-G | 0.380 | 614 | 590 | 836 | 814 | 0.57 | 0.429 |
| ↓ | T-G | 0.139 | 162 | 180 | 273 | 275 | 0.16 | |||
| D5 | 1-4 | ↑ | G-G | 0.121 | 744 | 709 | 915 | 874 | 0.190 | |
| ↓ | T-G | 0.049 | 194 | 219 | 280 | 299 | ||||
| D8 | 1-4 | ↑ | G-G | 0.133 | 797 | 762 | 894 | 863 | 0.263 | |
| ↓ | T-G | 0.086 | 217 | 237 | 287 | 295 | ||||
| D9 | 1-4 | ↑ | G-G | 0.158 | 985 | 947 | 736 | 695 | 0.086 | |
| ↓ | T-G | 0.010 | 281 | 308 | 227 | 247 | ||||
| D2 | 1-3-4 | ↑ | G-C-G | 0.274 | 620 | 593 | 850 | 829 | 0.56 | 0.337 |
| ↓ | T-C-G | 0.097 | 164 | 185 | 279 | 283 | 0.17 | |||
| D5 | 1-3-4 | ↑ | G-C-G | 0.080 | 747 | 712 | 917 | 882 | 0.149 | |
| ↓ | T-C-G | 0.050 | 198 | 224 | 285 | 303 | ||||
| D8 | 1-3-4 | ↑ | G-C-G | 0.090 | 798 | 764 | 896 | 872 | 0.250 | |
| ↓ | T-C-G | 0.083 | 220 | 242 | 292 | 299 | ||||
| D9 | 1-3-4 | ↑ | G-C-G | 0.092 | 982 | 947 | 737 | 700 | 0.087 | |
| ↓ | T-C-G | 0.010 | 289 | 314 | 230 | 255 | ||||
up and down arrows indicate over- or under-transmission to affected individuals
AKT1 ‘risk haplotype’ analysis with OPCRIT symptom scales
To clarify the putative association between AKT1 and schizophrenia in the ISHDSF, we examined transmission of the Caucasian risk haplotype (6) within OPCRIT symptom dimensions. From factor analysis of OPCRIT item lifetime ratings for ISHDSF subjects, five factors were extracted and used to derive scales; negative symptoms, delusions, hallucinations, mania and depression (37). For each scale, patients were divided into high and low-scoring categories to perform a family-based association test appropriate for our sample and design (38). Multiple thresholds (upper 20%, 30%, 40%) were used, as threshold delineation is somewhat arbitrary, above which patients were coded affected, and below, unaffected.
We tested, for each OPCRIT factor at each threshold, for TD to affected offspring of the AKT1 haplotype rs1130214-rs2498799 (SNPs 1-4; T-G) using TRANSMIT (29). This haplotype tagged the Emamian et al risk haplotype T-C-G (SNPs 1-3-4) in the ISHDSF and showed significant evidence for association (albeit under-transmission) in the whole sample (Table 2). As for the schizophrenia diagnostic categories, the T-G haplotype was significantly under-transmitted for all symptom dimensions at the 40% threshold (Table 4). Associations are less significant at more stringent thresholds, perhaps as power decreases with less “affected” individuals. However, at the 30% threshold, significant associations remain for hallucinations (p<0.003) and depression (p<0.023) (Table 4).
Table 4.
Within-family vertical transmission of AKT1 haplotype rs1130214-rs2498799 (SNPs 1-4, alleles T-G) for the 5 dimensions of the OPCRIT scale. All individuals affected with psychosis and with available OPCRIT data (n=749) were analyzed. Number of families with transmissions to affected offspring is shown in parentheses. p values (χ2, 1df) are highlighted using *<0.05, **<0.01.
| Threshold | Negative Symptoms | Delusions | Hallucinations | Mania | Depression |
|---|---|---|---|---|---|
| 20% | 0.641 (140) | 0.15 (115) | 0.003**a (184) | 0.089 (108) | 0.12 (101) |
| 30% | 0.226 (179) | 0.081 (169) | 0.003**a (184) | 0.345 (168) | 0.023* (140) |
| 40% | 0.003** (225) | 0.024* (201) | 0.008** (255) | 0.002** (221) | 0.004** (186) |
N.B. “a” denotes that the two thresholds resulted in the same cutoff score on the Hallucinations scale, which is comprised of only seven items. The data are identical at both levels, and the two separate entries represent a single test.
Control of false discoveries
The analysis of single markers, haplotype replication and OPCRIT analysis, generated in total 150 significance tests. To control for risk of false discoveries we used the adaptive (39) and more conservative step-up (32) FDR control methods. The FDR is robust to effects of correlated tests, hence especially applicable to this study (32, 40, 41). An FDR=0.1 provides a reasonable balance between control of false positives and power to detect true association (40). For FDR<0.1 the necessary sample size required to detect true effects increases dramatically. Using the adaptive method, we estimated the proportion of true null hypotheses to be as low as p0=0.6. Allowing an FDR=0.1 gave a p value threshold =0.017, whereby 17 of 150 tests were significant. The step-up method, with p0 assumed =1, gave a p value threshold =0.004, whereby 7 tests were significant. All are with the rs1130214-rs2498799 (T-G) haplotype, D5 and D9 diagnostic categories as before (Table 3), and with negative, hallucinations, mania and depression OPCRIT symptom factors (Table 4).
Exploratory analysis – ‘sliding window’ haplotypes
This analysis revealed a trend towards association at the 5’ of AKT1, with increasing significance moving from 2 to 4 SNP windows (Table 5 and Table 6). With PDTPHASE, strongest associations were with broadest diagnostic category D9 (haplotypic p=0.008, global p=0.017), then D5, with no association at D2, core schizophrenia (Table 5). Similar to the replication study, a pattern emerged of over-transmission of all-common-allele haplotypes, and under-transmission of these haplotypes with the rs1130214 rare allele (T) (SNP 1; Table 5). For TRANSMIT, this pattern held but with strongest associations at D5 (haplotypic p=0.025, global p=0.030; Table 6). No consistent evidence for association was observed for 3’ marker haplotypes (data not shown).
Table 5.
Significant ‘sliding windows’ AKT1 SNP haplotypes (in bold) by PDTPHASE.
| DIAG | HAPLO | * | Alleles | p val | Trio T | Trio NT | Aff Sib | Unaff Sib | Freq | Global P |
|---|---|---|---|---|---|---|---|---|---|---|
| D2 | 1-2 | ↑ | G-G | 0.217 | 650 | 626 | 863 | 849 | 0.60 | 0.188 |
| ↓ | T-G | 0.240 | 76 | 85 | 90 | 111 | 0.31 | |||
| D5 | 1-2 | ↑ | G-G | 0.098 | 788 | 759 | 930 | 900 | 0.264 | |
| ↓ | T-G | 0.125 | 371 | 393 | 473 | 490 | ||||
| D8 | 1-2 | ↑ | G-G | 0.116 | 841 | 816 | 905 | 897 | 0.304 | |
| ↓ | T-G | 0.400 | 405 | 423 | 483 | 484 | ||||
| D9 | 1-2 | ↑ | G-G | 0.028 | 1043 | 1014 | 752 | 716 | 0.078 | |
| ↓ | T-G | 0.072 | 517 | 540 | 388 | 406 | ||||
| D2 | 1-2-3 | ↑ | G-G-C | 0.133 | 638 | 607 | 857 | 833 | 0.58 | 0.107 |
| ↓ | T-G-C | 0.140 | 179 | 201 | 302 | 301 | 0.18 | |||
| D5 | 1-2-3 | ↑ | G-G-C | 0.037 | 769 | 735 | 913 | 870 | 0.078 | |
| ↓ | T-G-C | 0.049 | 215 | 243 | 301 | 314 | ||||
| D8 | 1-2-3 | ↑ | G-G-C | 0.054 | 818 | 789 | 888 | 866 | 0.133 | |
| ↓ | T-G-C | 0.075 | 239 | 262 | 306 | 310 | ||||
| D9 | 1-2-3 | ↑ | G-G-C | 0.022 | 1010 | 977 | 734 | 693 | 0.017 | |
| ↓ | T-G-C | 0.009 | 314 | 339 | 241 | 265 | ||||
| D2 | 1-2-3-4 | ↑ | G-G-C-G | 0.154 | 580 | 550 | 806 | 771 | 0.52 | 0.176 |
| ↓ | T-G-C-G | 0.136 | 163 | 183 | 282 | 283 | 0.16 | |||
| D5 | 1-2-3-4 | ↑ | G-G-C-G | 0.045 | 697 | 664 | 857 | 812 | 0.078 | |
| ↓ | T-G-C-G | 0.043 | 197 | 223 | 283 | 301 | ||||
| D8 | 1-2-3-4 | ↑ | G-G-C-G | 0.057 | 742 | 712 | 841 | 808 | 0.216 | |
| ↓ | T-G-C-G | 0.075 | 219 | 240 | 290 | 298 | ||||
| D9 | 1-2-3-4 | ↑ | G-G-C-G | 0.035 | 912 | 881 | 691 | 641 | 0.046 | |
| ↓ | T-G-C-G | 0.008 | 287 | 312 | 228 | 253 | ||||
up and down arrows indicate over- or under-transmission to affected individuals.
Table 6.
Significant ‘sliding windows’ AKT1 SNP haplotypes (in bold) by TRANSMIT.
| DIAG | HAPLO | * | Alleles | p val | Obs | Exp | Freq | Global P |
|---|---|---|---|---|---|---|---|---|
| D2 | 1-2 | ↑ | G-G | 0.083 | 648 | 632 | 0.59 | 0.078 |
| ↓ | T-G | 0.494 | 313 | 324 | 0.32 | |||
| D5 | 1-2 | ↑ | G-G | 0.055 | 783 | 765 | 0.079 | |
| ↓ | T-G | 0.116 | 374 | 388 | 0.32 | |||
| D8 | 1-2 | ↑ | G-G | 0.165 | 837 | 823 | 0.191 | |
| ↓ | T-G | 0.239 | 407 | 418 | ||||
| D9 | 1-2 | ↑ | G-G | 0.047 | 1039 | 1018 | 0.128 | |
| ↓ | T-G | 0.178 | 518 | 532 | ||||
| D2 | 1-2-3 | ↑ | G-G-C | 0.037 | 628 | 610 | 0.56 | 0.037 |
| ↓ | T-G-C | 0.059 | 181 | 193 | 0.19 | |||
| D5 | 1-2-3 | ↑ | G-G-C | 0.025 | 756 | 734 | 0.030 | |
| ↓ | T-G-C | 0.059 | 219 | 234 | ||||
| D8 | 1-2-3 | ↑ | G-G-C | 0.114 | 806 | 789 | 0.162 | |
| ↓ | T-G-C | 0.145 | 243 | 255 | ||||
| D9 | 1-2-3 | ↑ | G-G-C | 0.028 | 993 | 969 | 0.142 | |
| ↓ | T-G-C | 0.079 | 317 | 332 | ||||
| D2 | 1-2-3-4 | ↑ | G-G-C-G | 0.097 | 566 | 551 | 0.51 | 0.111 |
| ↓ | T-G-C-G | 0.046 | 170 | 183 | 0.18 | |||
| D5 | 1-2-3-4 | ↑ | G-G-C-G | 0.105 | 675 | 659 | 0.067 | |
| ↓ | T-G-C-G | 0.031 | 206 | 222 | ||||
| D8 | 1-2-3-4 | ↑ | G-G-C-G | 0.194 | 719 | 706 | 0.278 | |
| ↓ | T-G-C-G | 0.066 | 228 | 242 | ||||
| D9 | 1-2-3-4 | ↑ | G-G-C-G | 0.138 | 878 | 862 | 0.296 | |
| ↓ | T-G-C-G | 0.045 | 297 | 314 | ||||
up and down arrows indicate over- or under-transmission to affected individuals.
Exploratory analysis – all 2-4 SNP haplotypes
The pattern of over-transmission of all-common-allele SNP haplotypes including SNP 1 (rs1130214;G) and under-transmission with the rare T allele was observed, D5 and D9 maintaining strongest associations (Supplementary Table S2). Interestingly, haplotype 1-5 (rs1130214-rs2494732) whose 2 SNPs singly showed trends toward significance (Supplementary Table S1), is most significantly associated with TRANSMIT/PDTPHASE at broadest diagnostic category D9 (0.0002<global p<0.045; Supplementary Table S2). We saw no association with ‘risk’ haplotypes of Ikeda et al (13), or the tested portion of the Bajestan et al (14) risk haplotype.
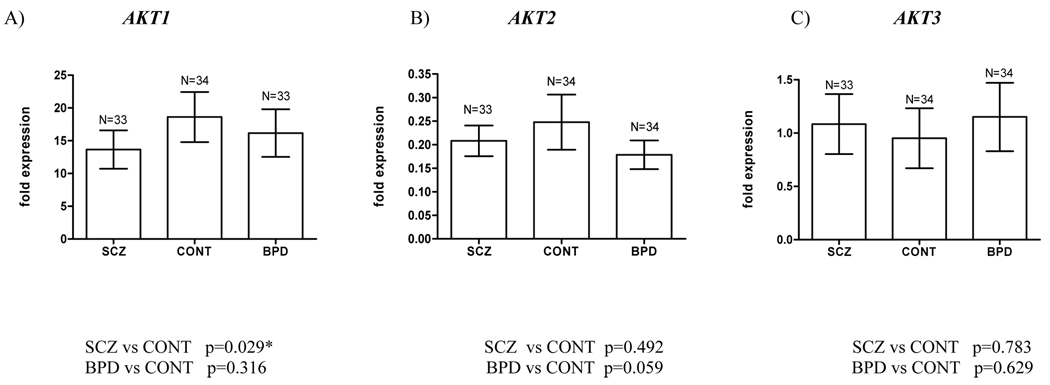
AKT gene expression in the Stanley brain series
We investigated whether the reduced AKT1 protein in schizophrenic brain (6) was reflected in lower levels of AKT1 mRNA, and analyzed transcriptional levels of AKT2 and AKT3, no different between cases and controls at the protein level, yet which have functional redundancy with AKT1 as well as specific roles (6, 8). Decreased AKT1 expression was evident in dorsolateral prefrontal cortex of both schizophrenic and bipolar patients compared to controls, significantly for schizophrenics, and of moderate effect size (Fig.3A; p=0.029, Cohen’s d=0.44). A non-significant reduction in AKT2 expression was observed in schizophrenic and bipolar brain compared to controls (Fig.3B), whilst no difference between diagnostic groups in AKT3 expression level was detected (Fig.3C). Analysis of potential confounder variables, AKT1 and DTNBP1 haplotypic effects are described in the Supplementary Results.
Fig. 3.
AKT gene expression relative to reference genes GAPDH and TBP analyzed by real-time qPCR of the Stanley Consortium brain series. Expression differences between diagnostic groups were evaluated by one-tailed Mann-Whitney U test for AKT1 (SCZ vs CONT) and two-tailed tests for all other comparisons.
In silico functional analysis of rs1130214
Putative function for this SNP was investigated owing to its complex role in haplotype association both here and in prior studies (6, 12). VISTA analysis revealed that it lies upstream of the AKT1 5’UTR in a region conserved in mammals and rodents and of high regulatory potential (Supplementary Fig S1; 42). Although within an alternative 5’ exon (Supplementary Fig S1), it is not predicted to affect splicing (data not shown). However, rs1130214 lies within a cluster of functional STAT3 binding sites and the T allele abolishes a putative E2F transcription factor binding site (Supplementary Fig. S1), so it potentially influences regulation of AKT1 expression (8).
Discussion
As is increasingly observed in complex disease association studies, reports on AKT1 and schizophrenia reveal inconsistent replication of a particular allele or allelic combination, yet replication at a broader level, i.e. the SNP or haplotype and ultimately the gene itself (43, 44). Potential reasons for such diverse findings are sample size, heterogenous phenotypes, marker sets, allele frequencies or LD structure, genetic admixture, multiple testing issues, and indeed true heterogeneity in variants that influence disease.
In this study we sought to replicate association of AKT1 with schizophrenia in the ISHDSF, prompted by the susceptibility haplotype in this sample within the dysbindin (DTNBP1) gene (21). Although no single AKT1 marker showed association with disease, the data were consistent with global association of common alleles for selected SNP haplotypes including rs1130214, and a striking under-transmission of these haplotypes with the rare allele (T) at rs1130214. This contrasts with Emamian et al (6) and Schwab et al (12) who found this allele in over-transmitted haplotypes, but is similar to Ikeda et al (13) in the most significant association being a ‘protective’ (albeit different) haplotype. Our study is not unique – an increasing number of publications are replicating a previously reported disease-marker association with the risk allele(s) reversed. One explanation may be the ‘Flip-Flop Phenomenon’ of Lin et al (45), whereby such associations may indeed be confirmations, with the ‘flip-flop’ due to interactive effects, haplotypic heterogeneity or linkage disequilibrium with a causal variant at another locus.
There is still debate over the ‘level of replication’ required to demonstrate true replication of association. Sullivan (46) has explored the capacity of association studies to produce false positive findings and impact of various definitions of replication. He defines 4 levels of replication, A-D, being highly precise (same SNP, statistical test/phenotype, direction of association) and D very loose (any SNP, test, direction). Our study corresponds to definition B (same SNPs, test, different direction), predicted to give 0–10% false replications. We therefore consider it unlikely that our results represent false positive findings.
We note in our study the slight difference in results between TRANSMIT and PDTPHASE (e.g. Table 2 and Table 3). This is due to differences in how several key factors are treated in their algorithms (family structure, frequency of missing parental genotypes, number of informative families and assumed genetic model) creating differences in power. Nicodemus et al (47) have evaluated various algorithms commonly used to implement TD-based tests, employing the joint null hypothesis of no linkage and no association, hence their results are applicable to our study. Although overall, PDTPHASE may be considered most rigorous for extended pedigrees, we included TRANSMIT as it was required for the FDR approach to correct for multiple testing, recovers missing parental data efficiently from unaffected sibling data, and with no linkage to 14q32, whereby using multiple families within pedigrees was permitted, TRANSMIT can use the maximal amount of data present in the sample. Both algorithms have strengths for use with the ISHDSF, both broadly concur, thus results of both are presented accordingly.
Many AKT1 haplotypes including rs1130214 appeared to be associated with schizophrenia in the ISHDSF to varying degrees. However, results from our exploratory haplotype analysis may have emerged through multiple testing and reflect false positives. It is unlikely that rs1130214 is an appreciable risk variant itself as single marker association does not reach significance. That it has functional relevance is suggested by bioinformatic analysis, which indicates a transcriptional regulatory role. Our data is consistent overall with prior Caucasian studies in highlighting AKT1 5’ in schizophrenia susceptibility.
The C allele of rs3730358 (in the T-C-G haplotype) was associated with schizophrenia (6, 12) and in the core risk haplotype, rs1130214-rs3730358 (T-C), with decreased AKT1 protein levels in lymphocyte cell lines (6). However, lymphocytes from normal Caucasians homozygous for the C-G portion of the haplotype T-C-G have higher levels of AKT1 protein, and lowered reponse to apoptotic stimuli, than those harbouring the T-A haplotype (48). As we found no clear relationship between haplotype and AKT1 transcription in the SBS, further studies are required to elucidate any differential effect in schizophrenic patients (associated with AKT1 or not) versus controls. We did observe reduced expression of total AKT1 RNA from schizophrenic and bipolar brain compared to controls, statistically significant for the schizophrenic cases (p=0.029), and consistent with the reduced AKT1 protein in schizophrenic brain (6). We found no significant differences between cases and controls in total RNA expression of AKT2 and AKT3, reflecting again the protein level (6), and pinpointing AKT1 especially for a role in schizophrenia.
Familial factors are thought not only to influence liability to psychiatric illness but also specific clinical features (49). Along with shared environment, a major phenotype modifier is genetic, specific susceptibility alleles/haplotypes conferring liability to particular clinical subtypes (37, 50). The significant under-transmission in our study of the Emamian et al risk haplotype across all OPCRIT symptom dimensions may indicate an effect on risk for psychosis unspecific to symptom factors. This may be consistent with the strongest associations being at broader diagnostic categories, which include individuals with mood disorders. Using this approach at other schizophrenia risk loci, associations were identified with 1–2 symptom factors (37, 51). Prior AKT1/schizophrenia associations in Caucasian samples (6, 12) are with core schizophrenia or schizoaffective disorder (our D2 category). However, our results suggest the AKT1 effect to be general in nature, and underscore the possibility that AKT1 plays a role in multiple psychiatric syndromes. Although clear differences exist between schizophrenia and bipolar disorder (52), neuropharmacological studies implicate dopamine dysregulation in both, and molecular studies suggest the conditions may share predisposing genes (53).
In summary, our data is consistent with global association for AKTI haplotypes including SNP rs1130214 with schizophrenia in the ISHDSF. We note an equally striking under-transmission of haplotypes comprising the same markers but with the rare allele at rs1130214, in contrast to prior reports. We highlight the 5’UTR of AKT1 in disease association, and emphasise the role of genes, not specific alleles or haplotypes, as ‘units or replication’ in association studies (43). We conclude that whilst AKT1 may play a minor role in susceptibility to schizophrenia in the ISHDSF, it appears to impact risk for a class of psychiatric syndrome with symptoms of schizophrenia and mood dysregulation.
Supplementary Material
Acknowledgements
Special thanks go to all the patients and family members who participated in this study. We are grateful also to Dr Frank Dudbridge for helpful suggestions implementing PDTPHASE for our sample, and to some very useful comments from the reviewers. This work was supported by NIH Grant No. MH041953.
Footnotes
The authors declare no financial conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
‘Supplementary information is available at Biological Psychiatry’s website’
References
- 1.Mäki P, Veijola J, Jones PB, Murray GK, Koponen H, Tienari P, et al. Predictors of schizophrenia – a review. Brit Med Bull. 2005;73–74:1–15. doi: 10.1093/bmb/ldh046. [DOI] [PubMed] [Google Scholar]
- 2.Kendler KS, Diehl SR. The genetics of schizophrenia: a current, genetic-epidemiologic perspective. Schizophr Bull. 1993;19:261–285. doi: 10.1093/schbul/19.2.261. [DOI] [PubMed] [Google Scholar]
- 3.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 4.Beaulieu JM, Sotnikova TD, Yao WD, Kockeritz L, Woodgett JR, Gainetdinov RR, et al. Lithium antagonizes dopamine-dependent behaviors mediated by an AKT/glycogen synthase kinase 3 ignaling cascade. Proc Natl Acad Sci USA. 2004;101(14):5099–5104. doi: 10.1073/pnas.0307921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyota T, Yamada K, Detera-Wadleigh SD, Yoshikawa T. Analysis of a cluster of polymorphisms in the AKT1 gene in bipolar pedigrees. Neurosci Lett. 2003;339:5–8. doi: 10.1016/s0304-3940(02)01428-3. [DOI] [PubMed] [Google Scholar]
- 6.Emamian ES, Happ D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3B signaling in schizophrenia. Nat Genet. 2004;36(2):131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- 7.Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med. 2005;9(1):59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S, Kim D, Kaneko S, Szewczyk KM, Nicosia SV, Yu H, et al. Molecular cloning and characterization of the human AKT1 promoter uncovers its up-regulation by the Src/Stat3 pathway. J Biol Chem. 2005;280(47):38932–38941. doi: 10.1074/jbc.M504011200. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Liu L, Pei L, Ju W, Ahmadian G, Lu J, et al. Control of synaptic strength, a novel function of Akt. Neuron. 2003;38:915–928. doi: 10.1016/s0896-6273(03)00356-8. [DOI] [PubMed] [Google Scholar]
- 10.Lin CH, Yeh SH, Lin CH, Lu KT, Leu TH, Chang WC, et al. A role for the PI3 kinase signaling pathway in fear conditioning and synaptic plasticity in the amygdyla. Neuron. 2001;31:841–851. doi: 10.1016/s0896-6273(01)00433-0. [DOI] [PubMed] [Google Scholar]
- 11.Lai W-S, Xu B, Westphal KGC, Paterlini M, Olivier B, Pavlidis P, et al. Akt1 deficiency affects neuronal morphology and predisposes to abnormalities in prefrontal cortex functioning. Proc Natl Acad Sci USA. 2006;103(45):16906–16911. doi: 10.1073/pnas.0604994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwab SG, Hoefgen B, Hanses C, Hassenbach MB, Albus M, Lerer B, et al. Further evidence for association of variants in the AKT1 gene with schizophrenia in a sample of European sib-pair families. Biol Psychiatry. 2005;58(6):446–450. doi: 10.1016/j.biopsych.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda M, Iwata N, Suzuki T, Kitajima T, Yamanouchi Y, Kinshita Y, et al. Association of AKT1 with schizophrenia confirmed in a Japanese population. Biol Psychiatry. 2004;56:698–700. doi: 10.1016/j.biopsych.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 14.Bajestan SN, Sabouri AH, Nakamura M, Takashima H, Keikhaee MR, Behdani F, et al. Association of AKT1 haplotype with the risk of schizophrenia in Iranian population. Am J Med Genet B Neuropsychiatr Genet. 2006;141(4):383–386. doi: 10.1002/ajmg.b.30291. [DOI] [PubMed] [Google Scholar]
- 15.Norton N, Williams HJ, Dwyer S, Carroll L, Peirce T, Moskvina V, Segurado R, Nikolov I, Williams NM, Ikeda M, Iwata N, Owen MJ, O’Donovan MC. Schizophrenia Res. 2007 doi: 10.1016/j.schres.2007.02.006. [e-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.Ohtsuki T, Inda T, Arinami T. Failure to confirm association between AKT1 haplotype and schizophrenia in a Japanese case-control population. Mol Psychiatry. 2004;6:981–983. doi: 10.1038/sj.mp.4001559. [DOI] [PubMed] [Google Scholar]
- 17.Ide M, Ohnishi T, Murayama M, Matsumoto I, Yamada K, Iwayama Y, Dedova I, Toyota T, Asada T, Takashima A, Yoshikawa T. Failure to support a genetic contribution of AKT1 polymorphisms and altered AKT signaling in schizophrenia. J Neurochem. 2006;99(1):277–287. doi: 10.1111/j.1471-4159.2006.04033.x. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y-L, Fann CS-J, Liu C-M, Wu J-Y, Hung S-I, Chan H-Y, et al. Absence of significant associations between four AKT1 SNP markers and schizophrenia in the Taiwanese population. Psychiat Genet. 2006;16(1):39–41. doi: 10.1097/01.ypg.0000180681.80546.f3. [DOI] [PubMed] [Google Scholar]
- 19.Turunen JA, Peltonen JO, Pietilainen OPH, Hennah W, Loukola A, Paunio T, Silander K, Ekelund J, Varilo T, Partonen T, Lonnqvist J, Peltonen L. The role of DTNBP1, NRG1 and AKT1 in the genetics of schizophrenia in Finland. Schizophrenia Res. 2007;91:27–36. doi: 10.1016/j.schres.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 20.Numakawa T, Yagasaki Y, Ishimoto T, Okada T, Suzuki T, Iwata N, et al. Evidence for novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum Mol Gen. 2004;13(21):2699–2708. doi: 10.1093/hmg/ddh280. [DOI] [PubMed] [Google Scholar]
- 21.Van den Oord EJCG, Sullivan PF, Jiang Y, Walsh D, O’Neill FA, Kendler KS, et al. Identification of a high-risk haplotype for the dystrobrevin binding protein 1 (DTNBP1) gene in the Irish study of high-density schizophrenia families. Mol Psychiatry. 2003;8:499–510. doi: 10.1038/sj.mp.4001263. [DOI] [PubMed] [Google Scholar]
- 22.Kendler KS, O’Neill FA, Burke J, Murphy B, Duke F, Straub RE, et al. Irish Study of High- Density Schizophrenia Families: Field methods and power to detect linkage. Am J Med Genet (Neuropsych Genet) 1996;67:179–190. doi: 10.1002/(SICI)1096-8628(19960409)67:2<179::AID-AJMG8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 23.McGuffin P, Farmer A, Harvey I. A polydiagnostic application of operational criteria in studies of psychotic illness. Development and reliability of the OPCRIT system. Arch Gen Psychiatry. 1991;48:764–770. doi: 10.1001/archpsyc.1991.01810320088015. [DOI] [PubMed] [Google Scholar]
- 24.Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The stanley foundation brain collection and neuropathology consortium. Schiz Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- 25.Van den Oord EJCG, Jiang Y, Riley BP, Kendler KS, Chen X. FP-TDI SNP genotype scoring by manual and statistical procedures: a study of error rates and types. Biotechniques. 2003a;34:610–624. doi: 10.2144/03343dd04. [DOI] [PubMed] [Google Scholar]
- 26.O’Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Clayton D. A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet. 1999;65:1170–1177. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 36.Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, et al. Strategies and Tools for Whole-Genome Alignments. Genome Res. 2003;13(1):73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fanous A, van den Oord E, Riley BP, Aggen SH, Neale MC, O’Neill FA, et al. Relationship Between a High-Risk Haplotype in the DTNBP1 (Dysbindin) Gene and Clinical Features of Schizophrenia. Am J Psychiatry. 2005;162(10):1824–1832. doi: 10.1176/appi.ajp.162.10.1824. [DOI] [PubMed] [Google Scholar]
- 38.Van den Oord EJCG. A comparison between different designs and tests to detect QTLs in association studies. Behav Genet. 1999;29:245–256. [Google Scholar]
- 39.Benjamini Y, Hochberg Y. The adaptive control of the false discovery rate in multiple hypotheses testing. J Behav Educ Statist. 2000;25:60–83. [Google Scholar]
- 40.Van den Oord EJCG, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003;19:537–542. doi: 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Storey JD. The false discovery rate: A Bayesian interpretation and the q-value. Annals Statist. 2003;31:2013–2035. [Google Scholar]
- 42.King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15(8):1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riley B, Kendler KS. Molecular genetic studies of schizophrenia. Eur J Hum Genet. 2006;14(6):669–680. doi: 10.1038/sj.ejhg.5201571. [DOI] [PubMed] [Google Scholar]
- 45.Lin P-I, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan PF. Spurious genetic associations. Biol Psychiatry. 2007;61(10):1121–1126. doi: 10.1016/j.biopsych.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 47.Nicodemus KK, Luna A, Shugart YY. An evaluation of power and type I error of single-nucleotide polymorphism transmission/disequilibrium-based statistical methods under different family structures, missing parental data, and population stratification. Am J Hum Genet. 2007;80:178–185. doi: 10.1086/510498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris SL, Gil G, Robins H, Hu W, Hirshfield K, Bond E, et al. Detection of functional single-nucleotide polymorphisms that affect apoptosis. Proc Natl Acad Sci USA. 2005;45(102):16297–16302. doi: 10.1073/pnas.0508390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kendler KS, Karkowski-Shuman L, O’Neill FA, Straub RE, MacLean CJ, Walsh D. Resemblance of psychotic symptoms and syndromes in affected sibling pairs from the Irish Study of high-Density Schizophrenia Families: Evidence for possible etiologic heterogeneity. Am J Psychiatry. 1997;154:191–198. doi: 10.1176/ajp.154.2.191. [DOI] [PubMed] [Google Scholar]
- 50.Cardno AG, Sham PC, Murray RM, McGuffin P. Twin study of symptom dimensions in psychosis. Br J Psychiatry. 2001;179:39–45. doi: 10.1192/bjp.179.1.39. [DOI] [PubMed] [Google Scholar]
- 51.McClay JL, Fanous A, van den Oord EJCG, Webb BT, Walsh D, O’Neill FA, et al. Catechol-O-methyltransferase and the clinical features of psychosis. Am J Med Genet B Neuropsychiatr Genet. 2006;141(8):935–938. doi: 10.1002/ajmg.b.30401. [DOI] [PubMed] [Google Scholar]
- 52.Murray RM, Sham P, Van Os J, Zanelli J, Cannon M, McDonald C. A developmental model for similarities and dissimilarities between schizophrenia and bipolar disorder. Schizophrenia Res. 2004;71(2–3):405–416. doi: 10.1016/j.schres.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Craddock N, O’Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32(1):9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen J. A power primer. Psychological Bulletin. 1992;112(1):155–159. doi: 10.1037//0033-2909.112.1.155. [DOI] [PubMed] [Google Scholar]
- 55.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.