Abstract
Nucleostemin (NS) is expressed in the nucleoli of adult and embryonic stem cells and in many tumors and tumor-derived cell lines. In coimmunoprecipitation experiments, nucleostemin is recovered with the tumor suppressor p53, and more recently we have demonstrated that nucleostemin exerts its role in cell cycle progression via a p53-dependent pathway. Here, we report that in human osteosarcoma cells, nucleostemin interacts with nucleophosmin, a nucleolar protein believed to possess oncogenic potential. Nucleostemin (NS) and nucleophosmin (NPM) displayed an extremely high degree of colocalization in the granular component of the nucleolus during interphase, and both proteins associated with prenucleolar bodies in late mitosis before the reformation of nucleoli. Coimmunoprecipitation experiments revealed that NS and NPM co-reside in complexes, and yeast two-hybrid experiments confirmed that they are interactive proteins, revealing the NPM-interactive region to be the 46-amino acid N-terminal domain of NS. In bimolecular fluorescence complementation studies, bright nucleolar signals were observed, indicating that these two proteins directly interact in the nucleolus in vivo. These results support the notion that cell cycle regulatory proteins congress and interact in the nucleolus, adding to the emerging concept that this nuclear domain has functions beyond ribosome production.
INTRODUCTION
Although the most extensively studied and well-established function of the nucleolus is its role in ribosome biosynthesis, various nonribosomal proteins began to be observed in the nucleolus a decade ago (Pederson, 1998b). Subsequently, several cell cycle regulatory proteins have been observed in the nucleolus by proteomics analysis of purified nucleoli (Andersen et al., 2002; Scherl et al., 2002) and by in situ localization methods (Pederson, 1998a; Boisvert et al., 2007). Meanwhile, and possibly related, we have recently discovered that the granular component of the nucleolus, long assumed to be populated solely by nascent ribosomes, in fact consists of both RNA-containing complexes and proteinaceous particles that lack detectable RNA (Politz et al., 2005). We have therefore begun to consider the possibility that the nucleolus is a staging site for the assembly of cell cycle regulatory machinery, either to function within the nucleolus by regulating ribosome synthesis or the production of other ribonucleoprotein complexes, or to shuttle to the nucleoplasm to impact DNA replication or other key steps in cell cycle progression.
Nucleostemin (NS) is a nucleolar protein required for embryogenesis and cell cycle progression (Beekman et al., 2006; Zhu et al., 2006), but its mode of action is unclear. NS shuttles between the nucleolus and surrounding nucleoplasm based on its state of guanosine triphosphate (GTP) binding (Tsai and McKay, 2005). A likely key to understanding the action of NS would be to define the proteins with which it interacts. An interaction between N-terminal basic region of NS and the tumor suppressor p53 was initially observed by pull-down and coimmunoprecipitation experiments (Tsai and McKay, 2002), and subsequently we demonstrated a role of p53 in the arrest of cell cycle progression in NS-depleted cells (Ma and Pederson, 2007). Moreover, the regulatory subunit B of human protein phosphatase-2 (PPP2R5A) has been identified as an NS-interactive protein by yeast two-hybrid experiments (Yang et al., 2005). In addition, the protein RSL1D1, which contains a ribosomal protein-homologous element, was found to interact with both the N-terminal basic domain and the GTP binding domain of NS and also was found to be important for the nucleolar location of NS (Meng et al., 2006). Finally, NS has also been found to interact with telomeric repeat-binding factor 1 (TRF1) and to negatively regulate the stability of TRF1 via ubiquitination (Zhu et al., 2006).
Nucleophosmin (NPM, also known as B23 protein) is an abundant and multifunctional nucleolar phosphoprotein that has been implicated in rRNA processing, ribosome assembly, centrosome duplication, cell proliferation, and malignancy (Grisendi et al., 2006; Naoe et al., 2006). NPM has been variously reported to have either oncogenic or tumor suppressor-like activities, and these difference are thought to be attributable, at least in part, to the p53 expression status of the cell (e.g., Colombo et al., 2002). In the present investigation, we have found that NPM directly interacts with NS in the nucleoli of living human tumor cells.
MATERIALS AND METHODS
Cell Culture, Transfection, and Establishment of Stable Cell Line
U2OS (human osteosarcoma cells) were cultured at 37°C in DMEM supplemented with 10% fetal bovine serum (FBS). The kDa.1 derivative of U2OS was kindly provided by Dawn E. Quelle (College of Medicine, University of Iowa; Meng et al., 2006), and it was maintained in DMEM containing 10% FBS and 500 μg/ml geneticin. A stably transformed, green fluorescent protein (GFP)-NS–inducible cell line, U2OSiNS-GFP, was constructed by cotransfection of U2OS cells at 40–60% confluence with pcDNA4-rNS-GFP-TO (described under Plasmids) and pcDNA6/TR (Invitrogen, Carlsbad, CA) (1:6 ratio) by using Lipofectamine 2000, followed by selection with 500 μg/ml hygromycin B and 5 μg/ml blasticidin S. After 2 wk in the selection medium, single cells were cultured in 96-well plates. After another 2 wk, each clone was subcultured and examined for expression of NS-GFP after induction by 0.5 μg/ml doxycycline for 24–48 h.
Plasmids
The plasmid encoding red fluorescent human nucleostemin (pmRFP-hNS-C1) was described previously (Ma and Pederson, 2007). pEGFP-B23-C1 was kindly provided by Sui Huang (Feinberg School of Medicine, Northwestern University; Chen and Huang, 2001). pEGFP-rNS was kindly provided by Robert Tsai (Texas A&M Health Science Center, Houston, TX; Tsai and McKay, 2002). The nucleostemin (rNS) coding region was cloned into pEGFP-N1, and the resulting rNS-GFP coding region was subcloned into the pcDNA4/TO vector to generate pcDNA4-rNS-GFP-TO for construction of the NS-GFP–inducible stable cell line (see above). The yeast two-hybrid plasmids were kindly provided by Peter Pryciak (University of Massachusetts Medical School). DNA fragments encoding human NPM, wild-type human NS, human NS mutants consisting of amino acids 1–46 (NSB), amino acids 1–267 (NSBG), amino acids 47-549 (NSdB), or amino acids 268–549 (NSdBG) were inserted into the vector containing the activation domain to obtain pAD-NPM, pAD-NS, pAD-NSB, pAD-NSBG, pAD-NSdB, and pAD-NSdBG. NPM was inserted into the vector containing the DNA binding domain and reporter genes to obtain pBD-NPM. Yeast transformation and filter β-galactosidase assays were performed as described previously (Winters and Pryciak, 2005). Parental BiFC plasmids were kindly provided by Tom Kerppola (Hu et al., 2002) and modified by insertion of cyan fluorescent protein (CFP) and monomeric red fluorescent protein (mRFP) into the YN and YC vectors, respectively, to obtain pBiFC-CFP-YN and pBiFC-mRFP-YC. DNA fragments encoding wild-type NS, the N-terminal 46 amino acids (NSB), or NS lacking the 46-amino acid N-terminal domain (i.e., amino acids 47-549) (NSdB) were inserted into pBiFC-CFP-YN to obtain pBiFC-NS-CFP-YN, pBiFC-NSB-CFP-YN, and pBiFC-NSdB-CFP-YN. The DNA fragment encoding NPM was inserted into pBiFC-mRFP-YC to obtain pBiFC-NPM-mRFP-YC.
Immunofluorescence
Cells grown on coverslips were fixed for 12 min in phosphate-buffered saline (PBS) containing 4% formaldehyde, followed by permeabilization with 0.5% Triton X-100 for 5 min. Coverslips were then incubated with primary antibodies in PBS, 1% bovine serum albumin for 1–2 h before washing and incubation with the appropriate secondary antibodies. All these steps were carried out at room temperature. Coverslips were mounted in Prolong Antifade (Invitrogen), and two- or three-dimensional images were captured and in some cases subjected to deconvolution as described previously (Ma and Pederson, 2007). The primary antibodies and dilutions were as follows: rabbit anti-human NS polyclonal antibody (1:200; Millipore Bioscience Research Reagents, Temecula, CA) and mouse anti-human NPM monoclonal antibody (1:500; Santa Cruz Biotechnology, Santa Cruz, CA).
Immunoprecipitation and Immunoblotting
U2OS cells were lysed on ice in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40, containing 1 mM phenylmethylsulfonyl fluoride and a protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) as specified by the manufacturer. Protein concentration was determined using the bicinchoninic acid assay (Pierce Chemical, Rockford, IL). Samples were incubated with antibody for human NPM (see below) or with nonimmune human immunoglobulin G (IgG) (2 μg/each) at 4°C for overnight, and then protein A- or protein G-agarose beads were added for an additional 2 h. The beads were washed and the eluted proteins were separated by SDS-polyacrylamide gel electrophoresis followed by transfer to Immobilon-P membranes (Millipore, Billerica, MA) which were then incubated with specific primary antibodies followed by their detection with appropriate horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence substrate (Pierce Chemical). The primary antibodies and dilutions used for immunoblotting were rabbit anti-human NS (1:5000; Millipore Bioscience Research Reagents) and mouse anti-human NPM (1:5000; Santa Cruz Biotechnology).
Time-Lapse Fluorescence Microscopy of Mitotic Cells
The U2OSiNS-GFP cell line was grown on Lab-Tek chambered coverglasses (Nalge Nunc Intlernational, Rochester, NY) and transfected with pBiFC-NPM-mRFP-YC by using standard procedures. The microscope and chamber were kept in 37°C and 5% CO2 during observation and imaging. A Leica DM-IRB microscope, equipped with a 100× objective (numerical aperture 1.4), a Quantix 57 charge-coupled device camera (Photometrics, Tucson, AZ), the appropriate filter sets, and MetaMorph acquisition software (Molecular Devices, Sunnyvale, CA) were used, as detailed previously (Jacobson and Pederson, 1997; Politz et al., 2007).
Bimolecular Fluorescence Complementation (BiFC) Imaging in Living Cells
U2OS cells grown on a Lab-Tek chambered coverglasses were cotransfected with plasmids encoding the desired fusion proteins. Twenty-four to 48 h after transfection, cells were washed with PBS, and then they were incubated at 30°C (5% CO2) for 2–6 h. Fluorescence was observed in living cells as described above using a Leica DM-IRB microscope equipped with the appropriate filter sets: CFP (436/20, BP480/40), mRFP (546/14, LP580), and yellow fluorescent protein (YFP0 (510/20, BP560/40). BiFC signal was evaluated only in cells exhibiting similar levels of CFP and mRFP expression. Controls established that there was no spectral cross-talk between channels at the exposure times used in these experiments.
RESULTS AND DISCUSSION
Nucleolar Colocalization of NS and NPM during Interphase
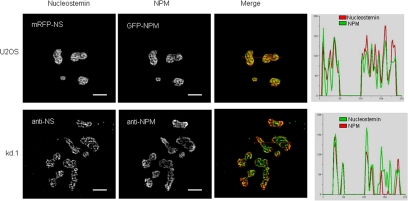
We previously demonstrated that NS resides in the granular component of the nucleolus at sites that lack 28S rRNA (Politz et al., 2005). NPM had been shown previously to be localized in the granular component of the nucleolus (Goessens, 1984), and we were therefore interested to know the degree to which these two proteins are colocalized. As shown in Figure 1, top, when mRFP-tagged human NS and GFP-tagged human NPM were coexpressed in U2OS cells, both mRFP-NS and GFP-NPM were concentrated in nucleoli. Using high-resolution digital imaging and deconvolution analysis, the intensity distribution of mRFP-nucleostemin was found to spatially overlap very extensively with GFP-NPM in subnucleolar regions, indicated by yellow in the merged image and the quantitative linescans (Figure 1). To assess whether these results might be due to overexpression of the two fluorescent proteins, we also determined the degree of intranucleolar colocalization of endogenous NS and NPM by immunostaining. Because resolving the intranucleolar distribution of NPM can be difficult due to its high concentration and issues of antibody accessibility, we used a U2OS-derived stably transformed cell line in which the level of NPM expression is depressed ∼50% due to small interfering RNA knockdown (Korgaonkar et al., 2005). The intranucleolar localizations of endogenous NS and NPM were again found to be extensively overlapping (Figure 1, bottom).
Figure 1.
Colocalization of NS and NPM within the nucleolus. Top row, mRFP-NS and GFP-NPM were cotransfected into U2OS cells and live imaging was performed 24 h after transfection. Bottom row, endogenous NS and NPM in nontransfected U2OS-derived kDa.1 cells determined by immunostaining. Far right, images were subjected to deconvolution and the intensity of the two colors at each pixel along red lines drawn in the merged panels was quantified and plotted. Bar, 5 μm.
Colocalization of NS and NPM in Anaphase and during Nucleolar Reformation
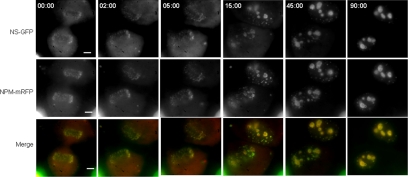
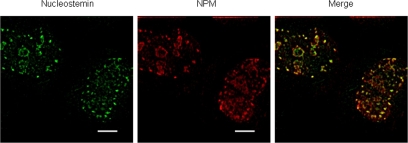
Given the high degree of colocalization of NS and NPM in the nucleoli of interphase cells, we examined the temporal and spatial features of their association as nucleoli reform during the late stages of mitosis. We constructed a stably transformed U2OS cell line that conditionally expresses NS-GFP (Supplemental Figure S1) and tracked this protein along with transiently expressed NPM-mRFP in live mitotic cells. NPM is known to be released as nucleoli disassemble during prophase and to then associate with prenucleolar bodies (PNBs) during telophase, followed by the coalescence of PNBs into nucleoli once active rRNA polymerase I and early rRNA processing factors have been recruited to the nucleolar organizer regions (Dundr et al., 2000; Savino et al., 2001; Angelier et al., 2005). We found NS and NPM to be extensively colocalized in the immediate periphery of the chromosomes in anaphase (0.00 min; Figure 2) and to remain colocalized because PNBs occurred in early telophase (15 min) and after complete nucleolar reformation had occurred in late telophase (45 min) and early G1 (90 min). To verify that the mitotic association of NS and NPM is not due to the GFP tag on the NS, we carried out double antibody staining experiments with the parental U2OS cells of the stable cell line (Figure 3), which confirmed that endogenous, non-GFP–tagged NS displays a high degree of colocalization with NPM during telophase. These results (Figures 2 and 3) parallel the previous description of NPM during anaphase and telophase (Dundr et al., 2000; Savino et al., 2001; Angelier et al., 2005), and they demonstrate that the colocalization of NPM with NS seen in interphase is reflected in the earliest stages of nucleolar reformation during late mitosis.
Figure 2.
Localization of NS and NPM during anaphase and telophase. NS-GFP was induced in U2OSiNS-GFP cells (see Supplemental Figure S1) by addition of doxycycline together with transient transfection with NPM-mRFP. Twenty-four hours later NS-GFP and NPM-mRFP were imaged in the same focal plane in dividing cells. Top row, NS-GFP in a cell entering anaphase (0.00 min) and proceeding through telophase and into G1. Middle row, NPM-mRFP in the same cell. Bottom row, merged NS-GFP and NPM-mRRF. Bar, 10 μm.
Figure 3.
Double immunostaining of NS and NPM in telophase. Parental U2OS cells were immunostained for NS (left) and NPM (middle). Shown is a typical telophase cell. The images were subjected to deconvolution and then merged (right). Bar, 5 μm.
A 46-Amino Acid N-Terminal Domain of NS Is Essential for Interaction with NPM
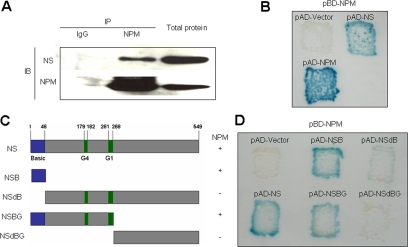
Notwithstanding the very high degree of colocalization of NS and NPM in nucleoli and during mitosis, these results do not address actual molecular complexing. As shown in Figure 4A, NS was detected in complexes precipitated by NPM antibody but not by nonimmune IgG. Moreover, an interaction between NS and NPM was observed in yeast-two hybrid experiments (Figure 4B, top right), as was a homodimerization of NPM (Figure 4B, bottom left) in confirmation of the well-known self-association of this protein as determined in biochemical studies (Yung and Chan, 1987; Chan and Chan, 1995; Herrera et al., 1996). To identify the region(s) within NS required for interaction with NPM, mutants were constructed containing only a 46-amino acid N-terminal basic region (Figure 4C, NSB) as well as mutants lacking this N-terminal basic region (Figure 4C, NSdB), containing the N-terminal basic and GTP binding region domains (Figure 4C, NSBG), or containing only the C-terminal half of NS (Figure 4C, NSdBG). Yeast two-hybrid analyses with these mutants demonstrated that the 46-amino acid N-terminal domain of NS is both necessary (Figure 4D, top right) and indeed sufficient (Figure 4D, top center) for NPM interaction.
Figure 4.
Interaction of NS and NPM by both coimmunoprecipitation and yeast two-hybrid analysis. (A) Coimmunoprecipitation of NS and NPM. Proteins captured from U2OS cell extracts by NPM antibody (middle lane) or nonimmune IgG (left lane) were subjected to immunoblotting for NS or NPM. Right lane, immunoblot of total cell protein. The two regions of the blot containing the NS and NPM antibody-reactive bands are juxtaposed in this composite figure. (B) Yeast two-hybrid analysis of NS-NPM interaction. Shown are the β-galactosidase signals for strains carrying pBD-NPM and pAD (activation domain) vector alone (top left), pBD-NPM and pAD-NS (top right), and pBD-NPM and pAD-NPM (bottom left). (C) Mutants of NS. “Basic” denotes the N-terminal domain previously implicated in nucleolar localization, and “G1” and “G4” indicate GTP binding domains. The + and − signs at the right indicate the interaction of each mutant with NPM, as determined by two-hybrid analysis (D). (D) Yeast two-hybrid analysis of NPM interaction with mutant forms of NS.
Molecular Interaction of NS and NPM in Living U2OS Cells
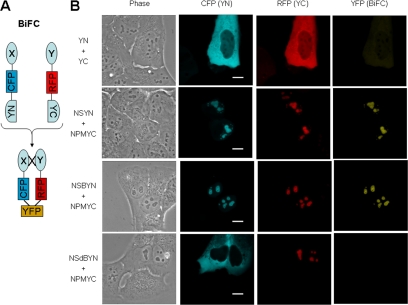
We next investigated the occurrence and subcellular localization of NS and NPM interaction in living U2OS cells by using BiFC. BiFC is based on the generation of a fluorescent signal when the two halves of YFP combine and refold, only when in sufficient proximity to one another by virtue of their attachment to two other dimerizing proteins (Hu et al., 2002). For our experiments, we used a modified system initially reported by Wolff et al. (2006) that enabled the levels of the two expressed proteins to be assessed in a given cell by also inserting cyan or red fluorescent protein coding sequences into each plasmid, as diagrammed in Figure 5A. When cells were transfected with the two plasmids lacking NS and NPM, the expressed proteins were predominantly cytoplasmic (Figure 5B, top row, CFP and RFP) and little interaction was observed, reflected by the very low intensity of yellow signal (Figure 5B, top row, YFP). In contrast, when NS and NPM were present on the plasmids, the NS and NPM fusion proteins were directed to nucleoli (Figure 5, second row, CFP and RFP, respectively) showing that the presence of the cyan or red fluorescent protein elements and the hemi-YFP did not interfere with the ability of NS and NPM to properly localize in nucleoli. Significantly, bright yellow BIFC signals were observed in the nucleoli (Figure 5B, second row, YFP) indicating a direct molecular complexing of the two proteins. When the NS BiFC plasmid carried only the 46-amino acid N-terminal region of NS, this protein displayed strong nucleolar localization (Figure 5B, third row, CFP), and as shown in the YFP panel (Figure 5B, third row, YFP), a BiFC signal was observed comparable with the intensity seen with wild-type NS. These results strongly confirm the yeast two-hybrid results and indicate that the N-terminal domain of NS is sufficient for the heterodimerization of NS and NPM in the nucleoli of living human cells. Finally, as shown in the bottom row, a NS mutant lacking this N-terminal domain failed to localize in nucleoli, or even the nucleus, and no BiFC was observed despite the demonstrable nucleolar presence of NPM (Figure 5B, bottom row).
Figure 5.
BiFC of NS and NPM in U2OS cells. (A) Schematic representation of modified BiFC. X, NS or mutant NS; Y, NPM; CFP and RFP, cyan and red fluorescent protein, respectively; YN, N-terminal domain of YFP; YC, C-terminal domain of YFP; YFP, reconstituted yellow fluorescent protein. (B) Expression and BiFC in U2OS cells expressing the pairs of plasmids indicated on the left. Images were acquired 36 h after transfection. The images in each column were scaled the same. Bar, 5 μm.
These results establish that NPM is a NS-interactive protein within the nucleoli of human osteosarcoma cells and identify the NPM-interactive region of NS as its 46-amino acid N-terminal domain, both by yeast two-hybrid and BiFC experiments. Although NPM has previously been implicated as a RNA-binding protein involved in rRNA processing (Wang et al., 1994; Savkur and Olson, 1998), not all of the NPM in the nucleolus seems to be RNA-associated based on the observation that a fraction of NPM is resistant to release by extensive RNase digestion (unpublished data). The fact that NPM and NS are demonstrably heterodimerized suggests that they may contribute to the RNA-deficient proteinaceous particles we previously observed in the granular component at the electron microscopic level (Politz et al., 2005). This idea is also compatible with the known homo-oligomerization (Yung and Chan, 1987; Chan and Chan, 1995; Herrera et al., 1996) and protein chaperone (Szebeni and Olson, 1999) activities of NPM, which might be expected to result in large complexes. Indeed, NS and NPM have recently been reported to co-reside in large (∼670-kDa) complexes with the Myb-binding protein 1a in HeLa cells (Yamauchi et al., 2008). Further studies are now in order to potentially extend the list of NS- and NPM-interactive proteins in the nucleolus, by using the same approaches as taken in the present investigation.
The present study does not address whether NS and NPM function in a dimeric/multimeric form to control the cell cycle, nor whether the initially reported interactivity of NS with p53, in coimmunoprecipitation and pull-down assays (Tsai and McKay, 2005), means there are heterotrimeric complexes of NS, NPM, and p53. Beyond the obvious need for further studies on these protein–protein interactions, there is the vexing problem of the conflicting reports on the role of NPM in the cell cycle, with some studies suggesting an oncoprotein-like function and others a tumor suppressor-like property. It has been suggested that these disparate findings on NPM reflect differences in the levels of p53 expression in the cell types investigated (Colombo et al., 2002), but it is also plausible that variations in the expression levels of other NPM-interactive proteins, including proteins yet to be discovered, may modulate its cell cycle progression activity, and, of course, the same point applies to NS-interactive proteins. The conceptual landscape should now envision these two nucleolar proteins as potential binding partners in any cell cycle control scenarios investigated, with the possibility of a multitude of additional proteins co-residing with them in complex machines.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joan Ritland Politz and Fan Zhang for constructive advice and comments on the manuscript, and we gratefully acknowledge the investigators, identified in the text, for kindly providing materials. This investigation was supported by National Science Foundation grant MCB-0445841 (to T.P.).
Abbreviations used:
- BiFC
bimolecular fluorescence complementation
- NPM
nucleophosmin
- NS
nucleostemin
- PNB
prenucleolar body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0128) on April 30, 2008.
REFERENCES
- Andersen J. S., Lyon C. E., Fox A. H., Leung A.K.L., Lam Y. W., Steen H., Mann M., Lamond A. I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Angelier N., Tramier M., Louvet E., Coppey-Moisan M., Savino T. M., De Mey J. R., Hernandez-Verdun D. Tracking the interactions of rRNA processing proteins during nucleolar assembly in living cells. Mol. Biol. Cell. 2005;16:2862–2871. doi: 10.1091/mbc.E05-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman C., Nichane M., De Clercq S., Maetens M., Floss T., Wurst W., Bellefroid E., Marine J. C. Evolutionarily conserved role of nucleostemin: controlling proliferation of stem/progenitor cells during early vertebrate development. Mol. Cell. Biol. 2006;26:9291–9301. doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F.-M., van Koningsbruggen S., Navascues J., Lamond A. I. The multifunctional nucleolus. Nature. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Chan P. K., Chan F. Y. Nucleophosmin/B23 (NPM) oligomer is a major and stable entity in HeLa cells. Biochim. Biophys. Acta. 1995;1262:37–42. doi: 10.1016/0167-4781(95)00044-h. [DOI] [PubMed] [Google Scholar]
- Chen D., Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E., Marine J. C., Danovi D., Falini B., Pelicci P. G. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 2002;4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- Dundr M., Misteli T., Olson M.O.J. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessens G. Nucleolar structure. Int. Rev. Cytol. 1984;87:107–158. doi: 10.1016/s0074-7696(08)62441-9. [DOI] [PubMed] [Google Scholar]
- Grisendi S., Mecucci C., Falini B., Pandolfi P. P. Nucleophosmin and cancer. Nat. Rev. Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Herrera J. E., Correia J. J., Jones A. E., Olson M.O.J. Sedimentation analyses of the salt- and divalent metal ion-induced oligomerization of nucleolar protein B23. Biochem. 1996;35:2668–2673. doi: 10.1021/bi9523320. [DOI] [PubMed] [Google Scholar]
- Hu C. D., Chinenov Y., Kerppola T. K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Jacobson M. R., Pederson T. RNA traffic and localization reported by fluorescent molecular cytochemistry in living cells. In: Richter J. D., editor. mRNA Formation and Function. New York: Academic Press; 1997. pp. 341–360. [Google Scholar]
- Korgaonkar C., Hagen J., Tompkins V., Frazier A. A., Allamargot C., Quelle F. W., Quelle D. E. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol. Cell. Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol. Biol. Cell. 2007;18:2630–2635. doi: 10.1091/mbc.E07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Yasumoto H., Tsai R.Y.L. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J. Cell Sci. 2006;119:5124–5136. doi: 10.1242/jcs.03292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoe T., Suzuki T., Kiyoi H., Urano T. Nucleophosmin: a versatile molecule associated with hematological malignancies. Cancer Sci. 2006;97:963–969. doi: 10.1111/j.1349-7006.2006.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. Growth factors in the nucleolus? J. Cell Biol. 1998a;143:279–281. doi: 10.1083/jcb.143.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998b;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz J. C., Polena I., Trask I., Bazett-Jones D. P., Pederson T. A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol. Biol. Cell. 2005;16:3401–3410. doi: 10.1091/mbc.E05-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz J.C.R., Zhang F., Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. USA. 2007;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino T. M., Gébrane-Younès J., De Mey J., Sibarita J. B., Hernandez-Verdun D. Nucleolar assembly of the rRNA processing machinery in living cells. J. Cell Biol. 2001;153:1097–1110. doi: 10.1083/jcb.153.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur R. S., Olson M.O.J. Preferential cleavage in pre-ribosomal RNA by protein B23 endoribonuclease. Nucleic Acids Res. 1998;26:4508–4515. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl A., Couté Y., Déon C., Callé A., Kindbeiter K., Sanchez J.-C., Greco A., Hochstrasser D., Diaz J.-J. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebeni A., Olson M. O. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 1999;8:905–912. doi: 10.1110/ps.8.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y.L., McKay R.D.G. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y.L., McKay R.D.G. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J. Cell Biol. 2005;168:179–184. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Baumann A., Szebeni A., Olson M.O.J. The nucleic acid binding activity of nucleolar protein B23.1 resides in its carboxyl-terminal end. J. Biol. Chem. 1994;269:30994–30998. [PubMed] [Google Scholar]
- Winters M. J., Pryciak P. M. Interaction with the SH3 domain protein Bem1 regulates signaling by the Saccharomyces cerevisiae p21-activated kinase Ste20. Mol. Cell. Biol. 2005;25:2177–2190. doi: 10.1128/MCB.25.6.2177-2190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff H., Hartl A., Eilken H. M., Hadian K., Ziegler M., Brack-Werner R. Live-cell assay for simultaneous monitoring of expression and interaction of proteins. BioTechniques. 2006;41:688–692. doi: 10.2144/000112291. [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Keough R. A., Gonda T. J., Ishii S. Ribosomal stress induces processing of Mybbp1a and its translocation from the nucleolus to the nucleoplasm. Genes Cells. 2008;13:27–39. doi: 10.1111/j.1365-2443.2007.01148.x. [DOI] [PubMed] [Google Scholar]
- Yang H.-X., Jin G.-L., Meng L., Zhang J.-Z., Liu W.-B., Shou C.-C. Screening and identification of proteins interacting with nucleostemin. World J. Gastroenterol. 2005;11:4812–4814. doi: 10.3748/wjg.v11.i31.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung B.Y.-M., Chan P. K. Identification and characterization of a hexameric form of nucleolar phosphoprotein B23. Biochim. Biophys. Acta. 1987;925:74–82. doi: 10.1016/0304-4165(87)90149-8. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Yasumoto H., Tsai R. Y. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol. Cell. Biol. 2006;26:9279–9290. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.