Abstract
The small guanosine triphosphate (GTP)-binding proteins of the Rho family are implicated in various cell functions, including establishment and maintenance of cell polarity. Activity of Rho guanosine triphosphatases (GTPases) is not only regulated by guanine nucleotide exchange factors and GTPase-activating proteins but also by guanine nucleotide dissociation inhibitors (GDIs). These proteins have the ability to extract Rho proteins from membranes and keep them in an inactive cytosolic complex. Here, we show that Rdi1, the sole Rho GDI of the yeast Saccharomyces cerevisiae, contributes to pseudohyphal growth and mitotic exit. Rdi1 interacts only with Cdc42, Rho1, and Rho4, and it regulates these Rho GTPases by distinct mechanisms. Binding between Rdi1 and Cdc42 as well as Rho1 is modulated by the Cdc42 effector and p21-activated kinase Cla4. After membrane extraction mediated by Rdi1, Rho4 is degraded by a novel mechanism, which includes the glycogen synthase kinase 3β homologue Ygk3, vacuolar proteases, and the proteasome. Together, these results indicate that Rdi1 uses distinct modes of regulation for different Rho GTPases.
INTRODUCTION
Small guanosine triphosphatases (GTPases) of the Rho family control fundamental processes of cell biology common to all eukaryotes, such as morphogenesis, polarity, movement, and cell division (Jaffe and Hall, 2005). In the budding yeast Saccharomyces cerevisiae, which encodes six Rho GTPases (Cdc42 and Rho1 to Rho5), these proteins play a pivotal role in the establishment of cell polarity (Park and Bi, 2007). Budding yeast cells undergo polarized growth during various phases of their life cycle, including budding during vegetative growth, mating between haploid cells of opposite mating types, and filamentous growth upon nutrient limitation. Although Cdc42 and Rho1 are well characterized, little is known about the other Rho proteins. Membrane association of Rho-type GTPases is essential for their function, and it depends on a C-terminal prenyl moiety and an adjacent polybasic region. Cdc42 localizes to internal membranes and the entire plasma membrane, where it clusters at sites of polarized growth, including the tips of mating projections, incipient bud sites, the tips and sides of growing buds, and the bud neck region of large-budded cells (Ziman et al., 1993; Richman et al., 2002). In addition to membrane-bound Cdc42, a cytoplasmic pool has been found (Ziman et al., 1993). Similar to Cdc42, Rho1 has been shown to localize at sites of polarized growth (Yamochi et al., 1994).
Like other regulatory GTPases, they act as molecular switches, cycling between an active guanosine triphosphate (GTP)-bound state and an inactive guanosine diphosphate (GDP)-bound state. This activity is highly regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). The exchange of GDP to GTP, and thus the activation of Cdc42, is catalyzed by GEFs. In the active GTP-bound state, Rho GTPases perform their regulatory function through a conformation-specific interaction with effector proteins. GAPs stimulate the intrinsic GTPase activity, leading to inactivation. However, the role of GAPs might be more complex. For Cdc42, it has been suggested that GTP hydrolysis, and thus a cycling between GDP and GTP states, is necessary for the establishment of polarity, septin assembly, and mating (Gladfelter et al., 2002; Caviston et al., 2003; Irazoqui et al., 2003; Barale et al., 2006). Apart from GEFs and GAPs, Rho GTPases are also regulated by guanine nucleotide dissociation inhibitors (GDIs) (DerMardirossian and Bokoch, 2005; Dovas and Couchman, 2005; Dransart et al., 2005). Three distinct biochemical activities have been attributed to Rho GDIs. First, they inhibit the dissociation of GDP from Rho GTPases. Second, they block intrinsic and GAP-stimulated GTPase activity. Third, Rho GDI extract Rho proteins from membranes and form high-affinity cytosolic complexes, in which the C-terminal prenyl group of the Rho protein inserts into the hydrophobic pocket formed by the immunoglobulin-like β sandwich of the GDI. The dissociation of the GTPase–GDI complex is regulated by various mechanisms. Several biologically relevant membrane lipids have been reported to decrease the affinity of Rho GDIs for GTPases (Chuang et al., 1993). Complex formation and membrane extraction also depend on the phosphorylation status of the GDI, the GTPase, or both (Bourmeyster and Vignais, 1996; Mehta et al., 2001; Forget et al., 2002; DerMardirossian et al., 2004). Furthermore, in higher eukaryotes some membrane-associated proteins, such as members of the ezrin/radixin/moesin family and the neurotrophin receptor p75NTR, have the ability to disrupt the GTPase–GDI complex (Takahashi et al., 1997; Yamashita and Tohyama, 2003).
In S. cerevisiae, the role of the sole Rho GDI Rdi1 is unclear. Conflicting results have been reported for the overexpression of RDI1. Masuda et al. (1994) show that high levels of RDI1 result in lethality, although the phenotype of these cells has not been characterized (Masuda et al., 1994). In contrast, others claim that RDI1 overexpression causes only a slightly rounder cell morphology (Tcheperegine et al., 2005). Deletion of RDI1 does not produce any detectable phenotypes in budding yeast (Masuda et al., 1994), but in the human fungal pathogen Candida albicans, cells lacking RDI1 exhibit reduced polarized growth (Court and Sudbery, 2007).
Rdi1 is found in the cytoplasm, but it also localizes to the tips of small buds and the bud neck region, where it interacts with Cdc42 (Koch et al., 1997; Richman et al., 2004; Cole et al., 2007). Rdi1 has the ability to extract Cdc42 and Rho1 from vacuolar membranes (Eitzen et al., 2001), and it can also extract Cdc42 from the plasma membrane (Richman et al., 2004; Tcheperegine et al., 2005). However, it has not been tested whether Rdi1 interacts with other Rho GTPases.
Here, we show that Rdi1 interacts only with a subset of Rho GTPases. Whereas Cdc42, Rho1 and Rho4 form a complex with Rdi1 and can be extracted from membranes, Rdi1 does not bind to Rho2, Rho3, and Rho5. Furthermore, we demonstrate that the p21-activated kinase (PAK) Cla4 disrupts Rdi1–Rho1 and Rdi1–Cdc42 complexes. Finally, we show that Rdi1 and the glycogen synthase kinase 3β (GSK-3β) homologue Ygk3 promote Rho4 degradation by vacuolar proteases and the proteasome.
MATERIALS AND METHODS
Growth Conditions, Yeast Strains, and Plasmids
All yeast strains are listed in Supplemental Table 1. The strains used in this study were in the YPH499 background, with the exception of strains used for filamentous growth. For these experiments strains in the Σ1278b background were used (CTY64, PC344, PPY966, THY697, THY705, and THY706). Yeast strains were grown in yeast extract, peptone, dextrose (YPD) or synthetic complete (SC) medium. For induction of the GAL1 promoter, yeast cells were grown in yeast extract, peptone (YP) or SC medium with 3% raffinose instead of glucose. Galactose (final concentration 2%) was added to induce the GAL1 promoter. SLAD medium contains 50 μM ammonium sulfate, 2% glucose, 2% Bacto-agar, and 0.67% YNB. Yeast strains were constructed using polymerase chain reaction (PCR)-amplified cassettes (Longtine et al., 1998; Knop et al., 1999; Janke et al., 2004).
All constructs used in this work are listed in Supplemental Table 2. For N-terminal tagging of Rho proteins, these genes were first amplified by PCR from genomic DNA and cloned into pRS315. A NotI site was introduced 3′ of the start codon by site directed mutagenesis. The plasmid GTEPI, which contains a DNA fragment encoding three adjacent copies of the hemagglutinin (HA) epitope cloned into the NotI site in the polylinker of pBluescript II, was cut with NotI, and the liberated fragment was ligated in frame into the NotI site of the pRS315 plasmids carrying the RHO genes. 3HA-tagged RHO genes were transferred into pRS305. These genes were inserted at the genomic LEU2 by transformation with EcoRV or KasI digested pRS305 carrying the 3HA-tagged RHO genes.
Immunoprecipitations and Membrane Extraction Assays
For immunoprecipitations and precipitations with glutathione-Sepharose (GE Healthcare, Chalfont St. Giles, United Kingdom) cells were disrupted with glass beads in lysis buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 10 mM EDTA, 1 mM EGTA, 5% glycerol, 1% Triton X-100, and protease inhibitors) and clarified by centrifugation at 13,000 rpm for 5 min. Protein concentration was determined using Bradford protein assay solution (Roth, Karlsruhe, Germany). Epitope-tagged proteins were immunoprecipitated by adding the respective antibody and protein G-Sepharose (GE Healthcare). Resin was washed three times with lysis buffer, resuspended in 2× SDS sample buffer, and analyzed by immunoblotting. For membrane extraction assays, cells were treated as for immunoprecipitations but without Triton X-100. Cleared lysates were centrifuged for 1 h at 100,000 × g. For dephosphorylation of 3HA-Rho4, the protein was precipitated with anti-HA antibodies, beads were washed three times with phosphatase buffer (20 mM Tris, pH 7.4, 10 mM NaCl, and 5% glycerol). Beads were then split into three and either no, 100 U of λ-phosphatase (New England Biolabs, Ispwich, MA), or 100 U of λ-phosphatase, which was heat-inactivated by treating it 30 min at 95°C, was added. Samples were incubated 30 min at 30°C, and proteins were eluted with SDS sample buffer.
Monoclonal mouse anti-HA (12CA5) was obtained from Roche Diagnostics (Mannheim, Germany), and goat anti-glutathione transferase (GST) was from GE Healthcare. Mouse monoclonal anti-Myc (9E10), anti-Cdc42, anti-Cdc11, and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Secondary antibodies were from Jackson ImmnuoResearch Laboratories (West Grove, PA).
Microscopy
F-actin of yeast cells was stained with rhodamine-phalloidin (Invitrogen, Carlsbad, CA). One milliliter of cells was harvested and resuspended in phosphate-buffered saline (PBS), 0.1% Triton X-100, and 3.7% formaldehyde. After 30-min incubation at room temperature, cells were washed once with PBS and fixed for 2 h at 30°C with PBS, 3.7% formaldehyde. Cells were washed two times with PBS and resuspended in 90 μl of PBS. Three units of rhodamine-phalloidin was added, cells were incubated for 2 h in the dark, and then they were washed three times with PBS. Cells were examined with an Axiovert 200M fluorescence microscope (Carl Zeiss, Jena, Germany) equipped with a 100× Plan oil-immersion objective, and images were captured using an AxioCam MRm charge-coupled device camera (Carl Zeiss).
Filamentous Growth Assays
For agar invasion assays, 105 cells of an overnight culture were spotted on YPD and grown for 2 d at 30°C. Plates were photographed before and after being rinsed under a gentle stream of deionized water. For pseudohyphal growth assays, cells were grown overnight, and 100 cells were spread on solid SLAD medium. Plates were incubated for 4 d at 30°C, and images were taken using a 10× objective.
RESULTS
Rdi1 Has a Role in Pseudohyphal Growth and Mitotic Exit
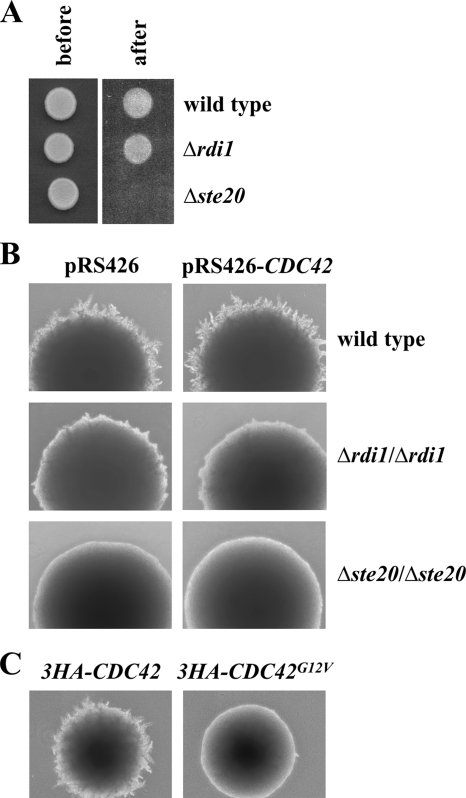
To assess the function of Rdi1 in S. cerevisiae, we analyzed the phenotype of cells deleted for and overexpressing RDI1, respectively. The RDI1 deletion strain was indistinguishable from wild-type cells in terms of growth, cell morphology, and mating (data not shown). It has been reported that in diploid C. albicans loss of the homologous gene RDI1 greatly reduces filamentous growth (Court and Sudbery, 2007). In addition, RDI1 deletion abrogates the enhanced filamentation of cells deleted for the Cdc42 GAPs BEM3 and RGA2 (Court and Sudbery, 2007). Therefore, we tested whether similar phenotypes can be observed for S. cerevisiae, where filamentous growth occurs in both, haploid (invasive growth) and diploid (pseudohyphal growth) cells (Palecek et al., 2002; Park and Bi, 2007). Haploid cells penetrate agar in response to nutritional limitation. To analyze the role of RDI1 in haploid invasive growth, we deleted this gene in cells of the Σ1278b background, which is necessary for tests of filamentous growth. The haploid Δrdi1 strain invaded agar like wild-type cells (Figure 1A). On nitrogen starvation, diploid cells assume an elongated morphology and generate chains of filamentous-form cells projecting from the main colony of yeast-form cells (Figure 1B). The homozygous Δrdi1/Δrdi1 diploid strain exhibited a defect in pseudohyphal differentiation when grown on low nitrogen medium (Figure 1B). Notably, overexpression of CDC42 from a multicopy plasmid did not rescue this defect (Figure 1B). Thus, it seems unlikely that Rdi1 regulates Cdc42 activity like the GEF Cdc24. It is conceivable that the phenotype of the Δrdi1/Δrdi1 strain reflects elevated, deregulated Cdc42 activity. Possibly Cdc42 removal from membranes by Rdi1 is necessary to restrict active Cdc42 to the growing tip. In line with this model is our observation that the constitutively active CDC42G12V mutant (Ziman et al., 1991) displayed a severe pseudohyphal growth defect (Figure 1C).
Figure 1.
Rdi1 and filamentous growth. (A) RDI1 deletion does not affect haploid invasive growth. Cells (105) of the indicated strains were spotted on a YPD plate and incubated for 2 d at 30°C. Pictures were taken before and after gentle rinsing with water. Δste20 cells, which fail to invade the agar, served as control. (B) Δrdi1/Δrdi1 cells exhibit reduced diploid pseudohyphal growth. The indicated strains were grown on low nitrogen SLAD medium for 4 d at 30°C. Δste20/Δste20 cells, which are defective in filamentous growth, were used as control. (C) Constitutively active Cdc42 impairs pseudohyphal growth. 3HA-CDC42 and 3HA-CDC42G12V under control of the CDC42 promoter were integrated at the URA3 locus by using an integrative plasmid. The indicated strains were treated as described in B. The results shown in this figure are representative of two independent experiments.
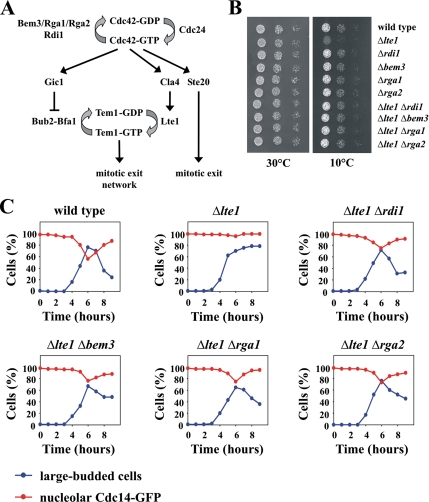
Cdc42 and its effectors Ste20, Cla4, and Gic1 promote exit from mitosis via at least three independent mechanisms (Höfken and Schiebel, 2002; Jensen et al., 2002; Seshan et al., 2002; Höfken and Schiebel, 2004; Bosl and Li 2005) (Figure 2A). The small GTPase Tem1 and its putative GEF Lte1 are the most upstream components of the mitotic exit network, the signaling cascade that controls exit from mitosis (Bosl and Li, 2005). Cells in which LTE1 is deleted grow normally at 30°C, but arrest in anaphase with large buds, two separated 4,6-diamidino-2-phenylindole staining regions, and an elongated mitotic spindle when grown at low temperatures (Bosl and Li, 2005). We used this mitotic exit defect of Δlte1 cells to assay the activity of Cdc42 regulators. Previously, it has been demonstrated that the deletion of LTE1 in a temperature-sensitive mutant of CDC24, the sole GEF of Cdc42, is lethal at all temperatures (Höfken and Schiebel, 2002). Here, we show that the individual deletion of the Cdc42 GAPs BEM3, RGA1, and RGA2 suppressed the cold-sensitive growth defect of Δlte1 (Figure 2B). Interestingly, the double mutant Δlte1Δrdi1 also grew like the wild-type strain at 10°C (Figure 2B). These results are consistent with a role for Cdc24 as a positive regulator and for Rdi1, Bem3, Rga1, and Rga2 as negative regulators of Cdc42. We then asked whether deletion of RDI1 and the GAPs rescued the mitotic exit defect of the Δlte1 strain. Cells were arrested in G1 with α-factor and released at 10°C. In addition to the percentage of large-budded cells, the release of Cdc14-green fluorescent protein (GFP) from the nucleolus was monitored through the cell cycle. Cdc14 is released from the nucleolus during anaphase and thus serves as a reliable marker for mitotic exit (Bosl and Li, 2005). At 10°C, wild-type cells released Cdc14 and underwent cytokinesis at ∼6 h (Figure 2C). In contrast, most Δlte1 cells arrested in anaphase with Cdc14-GFP trapped in the nucleolus (Figure 2C). Δlte1 cells in which either RDI1 or the Cdc42 GAPs were deleted did not arrest in anaphase and released Cdc14 from the nucleolus with the same kinetics as wild-type cells (Figure 2C).
Figure 2.
Deletion of RDI1 suppresses the mitotic exit defect of Δlte1 cells. (A) The role of Cdc42 in mitotic exit. Cdc42 is activated by its GEF Cdc24, whereas the GAPs Bem3, Rga1, and Rga2 stimulate GTP hydrolysis. Activated Cdc42 triggers mitotic exit by three independent mechanisms. Gic1 disrupts the interaction between the small GTPase Tem1 and its two-component GAP Bub2-Bfa1, which results in the activation of Tem1. Cla4 stimulates Lte1, the putative GEF of Tem1, which causes Tem1 activation. Ste20 promotes mitotic exit independently of Gic1 and Cla4 by an unknown mechanism. (B) Suppression of the Δlte1 growth defect by deletion of either RDI1 or Cdc42 GAPs. Serial dilutions (1:10) of the indicated strains grown on YPD at 10 and 30°C, respectively. (C) Deletion of either RDI1 or Cdc42 GAPs suppresses the mitotic exit defect of Δlte1 cells. The indicated strains with CDC14-GFP were arrested in G1 with α-factor. Cells progressed synchronously through the cell cycle at 10°C upon removal of α-factor by washing with YPD medium. The number of cells with large buds and nucleolar Cdc14-GFP (n > 100 at each time point) was determined over time. Because RDI1 and Cdc42 GAP single deletion strains grew like wild-type cells at 10°C (Figure 1B), these strains were not included for this analysis. The results shown in this figure are representative of two independent experiments.
RDI1 overexpression has been reported to either result in lethality (Masuda et al., 1994) or produce slightly rounder cells (Tcheperegine et al., 2005). Therefore, we tested the consequences of higher RDI1 levels in our background. RDI1 under control of its own promoter on a multicopy plasmid had no effect on cell growth and morphology (Supplemental Figure S1A; data not shown). The same result was obtained for a strain in which a single copy of genomic RDI1 was under control of the strong GAL1 promoter (Supplemental Figure S1B). In contrast, RDI1 under control of the GAL1 promoter expressed from a 2 μm-based multicopy plasmid (Masuda et al., 1994) resulted in lethality (Supplemental Figure S1C). To examine the cause of this lethality, we monitored cell morphology over time after RDI1 overexpression. Four hours after induction of GAL1-RDI1 from a multicopy plasmid, ∼90% of cells arrested as slightly bigger round, unbudded cells (Supplemental Figure S1, D and E). The actin cytoskeleton was polarized in only 6% of these round cells, whereas 70% of unbudded cells with normal Rdi1 levels had an polarized actin cytoskeleton (Supplemental Figure S1E).
Rdi1 Specifically Interacts with Cdc42, Rho1, and Rho4
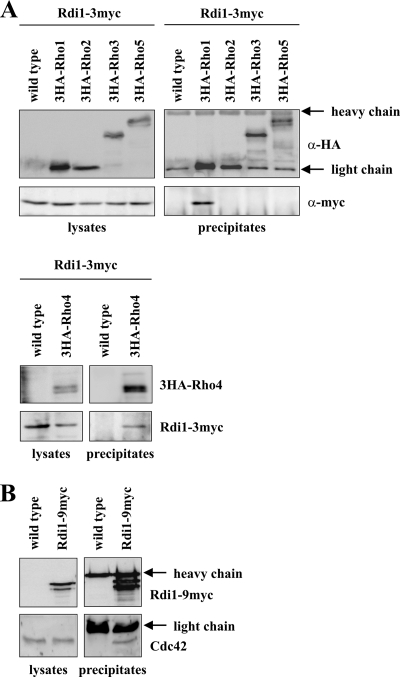
The most likely reason for the failure of cells to polarize after strong RDI1 overexpression is the removal of Rho GTPases from sites of polarized growth by Rdi1. It has been shown that Rdi1 is able to extract Cdc42 and Rho1 from membranes (Koch et al., 1997; Eitzen et al., 2001; Richman et al., 2004; Tcheperegine et al., 2005), but it has not been tested whether Rdi1 interacts with other Rho GTPases as well. Therefore, we performed immunoprecipitation experiments with strains in which Rho GTPases were N-terminally tagged with the HA and Rdi1 C-terminally with the myc epitope. Coimmunoprecipitation of Rho1 and Rho4, but not of Rho2, Rho3, and Rho5 with Rdi1 was observed (Figure 3A). The multiple bands observed for 3HA-Rho4 are due to phosphorylation of this protein (see below). In a similar immunoprecipitation experiment using antibodies against Cdc42, it was demonstrated that Cdc42 binds to Rdi1 (Figure 3B). Because all proteins were expressed from their own promoters for this experiment, Rho protein levels might not be sufficiently high for detection of a Rdi1–Rho complex. To circumvent this problem, RHO genes were fused with GST and placed under the control of the strong GAL1 promoter. Although all Rho GTPases were expressed at high levels upon induction, again only Cdc42, Rho1 and Rho4, but not the other Rho proteins, formed a complex with Rdi1 (Supplemental Figure S2).
Figure 3.
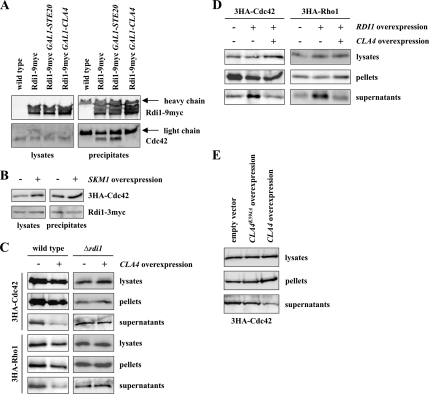
Rdi1 interacts with Cdc42, Rho1, and Rho4, but not with other Rho proteins. (A) Rdi1 coimmunoprecipitates with Rho1 and Rho4. Cells expressing RDI1–3myc, and RDI1–3myc with 3HA-tagged RHO1, RHO2, RHO3, RHO4, and RHO5 were lysed, and equal amounts of protein extract were precipitated with anti-HA antibodies. Immunoprecipitates were analyzed by immunoblotting with antibodies against the myc and HA epitopes. 3HA-Rho1 and 3HA-Rho2 run at the same height as the light chain. However, a stronger signal indicates that these proteins were precipitated, compared with the heavy chain, which occurs with the same intensity in all samples. (B) Coimmunoprecipitation of Rdi1 with Cdc42. Cells expressing RDI1–9myc and wild-type cells were lysed, and equal amounts of protein were precipitated with anti-myc antibodies. Precipitates were examined with antibodies raised against Cdc42 and the myc epitope, respectively. The results shown in this figure are representative of two independent experiments.
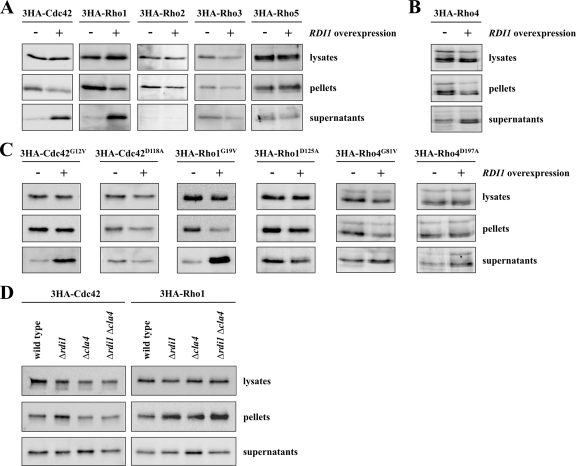
To examine the amount of membrane-associated Rho GTPases, cell lysates were submitted to cell fractionation by high-speed centrifugation. Interestingly, the ratio between protein in soluble supernatant and the membrane pellet was quite different for the various Rho GTPases (Supplemental Figure S3). To characterize which Rho proteins could be extracted from membranes by Rdi1, lysates of cells with and without RDI1 overexpression, respectively, were fractionated and analyzed by immunoblotting. After overexpression of RDI1, the amount of cytosolic protein increased for Cdc42 and Rho1 (Figure 4A). Consistently, less Rho1 and Cdc42 protein was found in the membrane fraction of cells overexpressing RDI1 (Figure 4A). However, because Rho proteins predominantly associate with membranes, these changes can be most easily observed in the cytosolic fraction. Higher levels of RDI1 had no effect on the distribution of Rho2, Rho3, and Rho5 (Figure 4A). Interestingly, the total amount of Rho4 dropped following RDI1 overexpression (Figure 5A). Because Rho4 protein level were only moderately reduced in cells lacking PEP4 after RDI1 overexpression (see below), the membrane extraction assay for Rho4 was performed in the Δpep4 background. Higher RDI1 levels caused an increase of Rho4 in the supernatant and a reduction of membrane-associated Rho4 (Figure 4B). The ability of Rdi1 to extract Rho4 from membranes also was examined by fluorescence microscopy. GFP-Rho4 under control of its own promoter could be detected by immunoblotting, but it was too weak to be visualized by fluorescence microscopy. Therefore, we expressed GFP-RHO4 from the inducible GAL1 promoter. GFP-Rho4 was associated with internal membranes and the entire plasma membrane (Figure 6A). The protein clustered at the presumptive bud site and the bud neck of large buds. Unlike most other proteins that have a role in polarity, Rho4 was not or only weakly enriched at the tip of small buds and absent from mating projections (Figure 6, A and B). As expected, RDI1 overexpression resulted in the loss of membrane-bound Rho4 (Figure 6, C and D).
Figure 4.
Rdi1 specifically extracts Rho1 and Cdc42 from membranes. (A) Rdi1 extracts Cdc42 and Rho1 from membranes. Cells expressing 3HA-tagged Rho proteins and carrying either GAL1-RDI1 on a plasmid or the empty vector were grown in selective medium with 3% raffinose. RDI1 was overexpressed for 3 h by addition of galactose. Cells were lysed and equal amounts of protein extract were separated by centrifugation at 100,000 × g to obtain the soluble supernatant and membrane pellet fractions. Rho proteins in different fractions were detected by Western blotting with antibodies against the HA epitope. (B) RDI1 extracts Rho4 from membranes. The experiment was performed as described in A but in a Δpep4 background to counteract Rho4 degradation caused by RDI1 overexpression (C), Rdi1 preferentially extracts GTP-bound forms of Rho GTPases from membranes. The experiment was carried out as described in A. For Rho4 proteins the assay was performed in cells lacking PEP4. (D) Effect of RDI1 and CLA4 deletion on cytosolic levels of Cdc42 and Rho1. Cells of the indicated strains were lysed and protein extracts were fractionated by centrifugation at 100,000 × g. Cdc42 and Rho1 were detected by immunoblotting using anti-HA antibodies. The results shown in this figure are representative of two independent experiments.
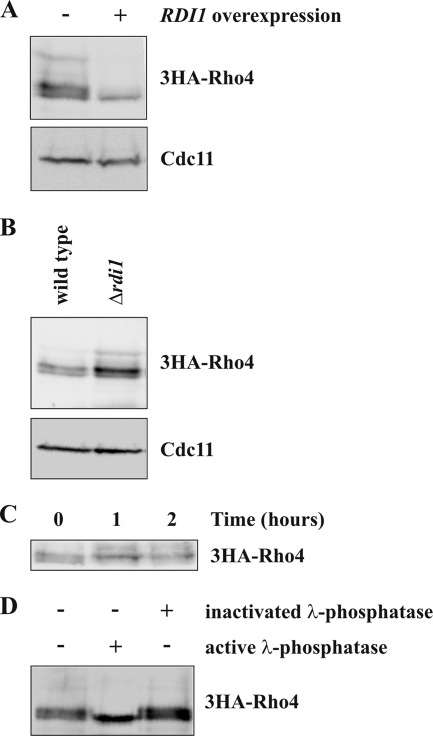
Figure 5.
Rdi1 targets Rho4 for degradation. (A) RDI1 overexpression results in lower Rho4 protein levels. 3HA-RHO4 cells carrying either GAL1-RDI1 on a vector or the empty plasmid were grown in YP medium with 3% raffinose. RDI1 was overexpressed for 2 h by addition of galactose. Subsequently, cells were lysed, and equal amounts of protein extract were examined by immunoblotting using antibodies against the HA epitope and Cdc11 (loading control). (B) Δrdi1 cells have higher Rho4 protein levels. Cells expressing 3HA-RHO4 in a wild-type and Δrdi1 background were analyzed by immunoblotting with the indicated antibodies. (C) Rho4 is a relatively stable protein. Exponentially growing 3HA-Rho4 cells were incubated with 50 μg/ml cycloheximide. At the indicated times, samples were taken and analyzed by immunoblotting. (D) Rho4 is a phosphoprotein. 3HA-tagged Rho4 was immunoprecipitated with anti-HA antibodies and treated with buffer, active, or heat-inactivated λ-phosphatase. Samples were analyzed by immunoblotting. The results shown in this figure are representative of two independent experiments.
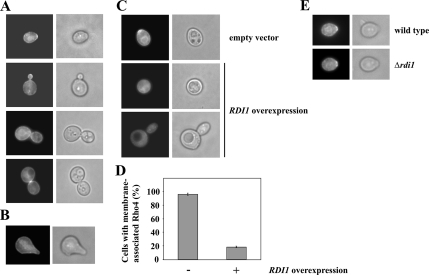
Figure 6.
Localization of Rho4. (A) Rho4 localization during budding. GAL1-GFP-RHO4 of exponentially growing cells was induced for 120 min by the addition of galactose. Cells were fixed with formaldehyde and analyzed by fluorescence microscopy. (B) Rho4 is not enriched at the tip of mating projections. Logarithmically growing GAL1-GFP-RHO4 cells were incubated with 1 μg/ml α-factor and 2% galactose for 150 min. Fixed cells were examined by fluorescence microscopy. (C) Rdi1 extracts Rho4 from plasma membranes. Logarithmically growing GAL1-GFP-RHO4 cells carrying either GAL1-RDI1 on a plasmid (pKT10-GAL-RDI1) or the empty vector (pKT10-GAL) were induced with galactose for 120 min. (D) Quantification of C. The percentage of cells with membrane-associated Rho4 is given as the mean of three independent experiments with SD bars (n > 100 for each experiment). (E) RDI1 deletion does not affect polarized Rho4 localization. GFP-Rho4 of wild type and Δrdi1 cells was induced for 120 min.
We next asked whether Rdi1 preferentially extracts the GDP- or GTP-bound form of Cdc42, Rho1, and Rho4 from membranes. The mutations CDC42G12V and CDC42D118A lock Cdc42 in the GTP- and GDP-bound state, respectively (Ziman et al., 1991). Therefore, we compared the intracellular distribution of these mutated proteins and also of the paradigmatic mutant proteins Rho1G19V, Rho1D125A, Rho4G81V, and Rho4D197A after RDI1 overexpression. Unexpectedly, Rdi1 extracted the GTP-bound Cdc42G12V and Rho1G19V like wild-type proteins from membranes, whereas the cytosolic levels of GDP-bound Cdc42D118A and Rho1D125A did not increase in cells overexpressing RDI1 (Figure 4, A and C). In contrast, wild type Rho4, Rho4G81V, and Rho4D197A seemed to be equally well extracted from membranes by Rdi1 (Figure 4, B and C). However, the effect of Rdi1 on Rho4 was less pronounced; therefore, it was more difficult to assess.
Because Rdi1 extracts Rho1 and Cdc42 from membranes, we examined whether these Rho proteins were present in the cytosol of cells deleted for RDI1. Unexpectedly, the cytoplasmic pool of Rho1 and Cdc42 was the same in wild type and Δrdi1 cells (Figure 4D).
In summary, our data indicate that Rdi1 only interacts with Cdc42, Rho1, and Rho4, but not with other Rho proteins.
PAK-Family Kinase Cla4 Regulates Association of Rdi1 with Cdc42 and Rho1
The association of Rho GTPases with Rho GDIs can be modulated by phosphorylation of these proteins (Bourmeyster and Vignais, 1996; Mehta et al., 2001; Forget et al., 2002; DerMardirossian et al., 2004). Because the Cdc42 effectors Cla4 and Ste20 play a key role in cell polarity, we asked whether elevated levels of these kinases would reduce the efficiency of the coimmunoprecipitation of Cdc42 and Rdi1. Anti-myc epitope antibodies not only efficiently precipitated Rdi1–9myc but also Cdc42 (Figures 3B and 7A). Overexpression of CLA4 abolished binding between Rdi1 and Cdc42 (Figure 7A). Importantly, higher expression of either STE20 or SKM1, which are both Cdc42 effectors and PAK-family kinases as well, had no effect on the Cdc42–Rdi1 interaction (Figure 7, A and B). Thus, Cla4, but not related proteins, can specifically disrupt binding between Cdc42 and Rdi1. We also tested, whether CLA4 overexpresssion has the same effect on the Rho1–Rdi1 complex. However, although the coimmunoprecipitation of Rho1 and Rdi1 was highly reproducible, we came to varying results for the Rho1–Rdi1 complex after CLA4 overexpression (data not shown). Therefore, using coimmunoprecipitations, it cannot conclusively be demonstrated whether Cla4 has an effect on the Rho1–Rdi1 complex. To circumvent this problem, we tested whether high levels of CLA4 change the intracellular distribution of Rho1 and Cdc42. Overexpression of CLA4 decreased the amount of cytosolic Rho1 and Cdc42 (Figure 7C). Importantly, this effect was not observed in the absence of RDI1 (Figure 7C). These results suggest that Cla4 disrupts the binding between Rdi1 and Cdc42 as well as between Rdi1 and Rho1. Consistently, in Δcla4 cells a slightly higher portion of Rho1 was found in the cytosol compared with the wild-type strain, whereas the cytosolic amount of Rho1 in the Δcla4 Δrdi1 double mutant was similar to the wild-type and Δrdi1 (Figure 4D). For Cdc42, there was only a weak or no effect for Cdc42 in the CLA4 deletion strain (Figure 4D). Finally, using the membrane extraction assay it was tested whether Cla4 antagonizes Rdi1. Whereas higher Rdi1 levels increased the cytosolic pool of Cdc42 and Rho1 (Figures 4A and 7D), simultaneous overexpression of CLA4 and RDI1 reversed this effect (Figure 7D). Under these conditions, cytosolic amounts of Cdc42 and Rho1 are comparable with the wild-type situation. Next, we tested whether the kinase activity of Cla4 is required for its effect on Rdi1 complexes. To this end, we examined the localization of 3HA-Cdc42 after overexpression of wild-type CLA4 and the kinase-dead CLA4K594A. As described above, overexpression of wild-type CLA4 results in a reduced cytosolic pool of 3HA-Cdc42 compared with the wild-type strain (Figure 7, C and E). In contrast, this effect was much weaker in cells overexpressing the kinase-dead CLA4 (Figure 7E).
Figure 7.
Cla4 regulates binding of Rdi1 to Cdc42 and Rho1. (A) CLA4 overexpression inhibits complex formation between Cdc42 and Rdi1. The indicated strains were grown in YP medium with raffinose. We added 2% galactose to induce expression of STE20 and CLA4, respectively. Cells were then lysed and equal amounts of protein were precipitated with anti-myc antibodies. Immunoblots were probed with antibodies raised against Cdc42 and the myc epitope. (B) SKM1 overexpression has no effect on the Cdc42–Rdi1 complex. 3HA-CDC42 RDI1–3myc cells carrying either GAL1-GST-SKM1 on a plasmid or the empty plasmid were induced with galactose for 120 min. Cells were subjected to immunoprecipitation with anti-HA antibodies. Immunoblots were analyzed with anti-HA and anti-myc antibodies. (C) CLA4 overexpression decreases the cytoplasmic pool of Rho1 and Cdc42. Cells expressing the indicated 3HA-tagged Rho GTPase and carrying either GAL1-myc-CLA4 on a plasmid or the empty plasmid, respectively, were induced for 120 min. Cells were lysed, and equal amounts of protein extract were separated by centrifugation at 100,000 × g. Rho proteins were detected by immunoblotting using antibodies against the HA epitope. (D) CLA4 overexpression reverses RDI1-dependent membrane extraction of Cdc42 and Rho1. 3HA-CDC42, 3HA-CDC42 GAL1–3HA-RDI1, 3HA-RHO1, and 3HA-RHO1 GAL1–3HA-RDI1 cells carrying either an empty plasmid or a plasmid encoding GAL1-myc-CLA4 were induced with galactose for 2 h. Subsequently, lysed cells were fractionated by centrifugation at 100,000 × g and analyzed by immunoblotting. (E) Cla4 kinase activity is required for its effect on Cdc42-Rdi1. 3HA-CDC42 cells carrying either an empty plasmid, GAL1-myc-CLA4, or GAL1-myc-CLA4K594A were treated as described in C. The results shown in this figure are representative of two independent experiments.
Interestingly, Cla4 did not affect the amount of Rho4 in the supernatant and pellet after high-speed centrifugation (Supplemental Figure S4, A and B). Thus, the regulatory role of Cla4 is specific for Cdc42 and Rho1.
Rdi1 and the GSK-3β Homologue Ygk3 Promote Degradation of Rho4
As mentioned above, RDI1 overexpression reduces the total amount of Rho4 protein (Figure 5A). Conversely, higher protein levels of Rho4 were observed in Δrdi1 cells compared with the wild-type strain (Figure 5B). This could be achieved by two different mechanisms. Either Rdi1 targets Rho4 for degradation or, alternatively, Rdi1 regulates Rho4 protein levels through gene expression. Because Rho4 levels dropped markedly after only 2 h of RDI1 overexpression, a regulation at the transcriptional or translational level would only have an effect if Rho4 were a short-lived protein. To test this, we blocked protein synthesis with cycloheximide. Even after 2-h incubation with cycloheximide, Rho4 protein levels remained unchanged (Figure 5C), suggesting that Rho4 is a relatively stable protein. Thus, Rdi1 seems to regulate Rho4 levels through proteolysis. Because usually multiple bands were observed for 3HA-Rho4 in immunoblots, it was examined whether these are phospho-forms of 3HA-Rho4. In fact, slower migrating bands disappeared following phosphatase treatment (Figure 5D).
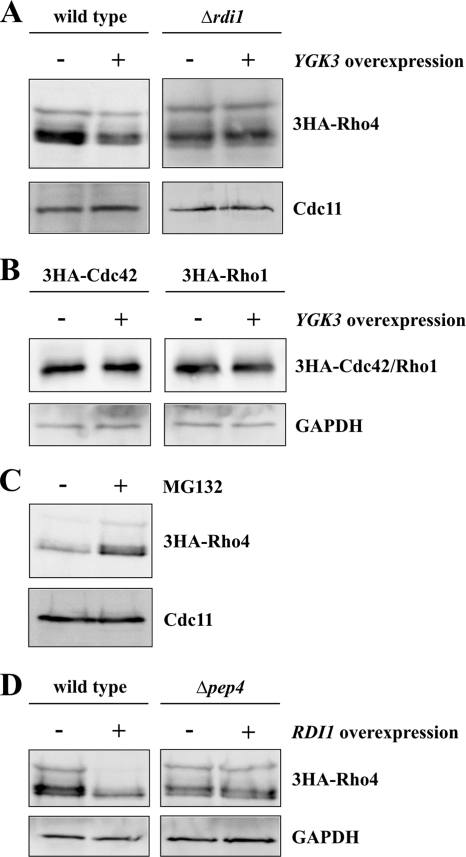
In a proteomic approach, phosphorylation of Rho4 by Ygk3, a homologue of GSK-3β kinase, has been demonstrated previously (Ptacek et al., 2005). Furthermore, Ygk3 has a role in ubiquitin-dependent proteolysis (Andoh et al., 2000). Therefore, we tested whether Ygk3 is involved in the degradation of Rho4. In fact, YGK3 overexpression resulted in reduced Rho4 levels (Figure 8A), indicating that this protein, like Rdi1, promotes Rho4 proteolysis. Importantly, in the absence of RDI1 Ygk3 fails to degrade Rho4 (Figure 8A), suggesting that Rdi1 and Ygk3 act in the same pathway. In contrast YGK3 overexpression had no effect on protein levels of Cdc42 and Rho1 (Figure 8B), which are also extracted from membranes by Rdi1. Because the ubiquitin ligase Rsp5 is involved in GSK-3β–mediated proteolysis (Andoh et al., 2000), we examined the role of Rsp5 in Rho4 degradation using a temperature-sensitive mutant. At both, the permissive (23°C) and restrictive (37°) temperature, Rho4 degradation after RDI1 overexpression in rsp5-1 cells was indistinguishable from the wild type (Supplemental Figure S5). Thus, the ubiquitin ligase Rsp5 is not required for Rho4 degradation. Eukaryotic cells contain two distinct systems for protein degradation: the vacuolar system and the proteasome. To find out whether Rho4 is degraded in a proteasome-dependent manner, Rho4 protein levels after RDI1 overexpression were examined in the presence and absence of the proteasome-specific inhibitor MG132 (Lee and Goldberg, 1996). Treatment with MG132 stabilized Rho4 (Figure 8C), indicating an involvement of the proteasome in Rho4 degradation. We also examined whether the vacuolar system plays a role in Rho4 degradation. To this end, RDI1 was overexpressed in Δpep4 cells, which lack most vacuolar proteases (Ammerer et al., 1986). Unexpectedly, Rho4 was not degraded in the absence of vacuolar proteases (Figure 8D), suggesting that Rho4 proteolysis depends on the vacuolar system as well as the proteasome.
Figure 8.
Degradation of Rho4. (A) YGK3 overexpression leads to Rho4 degradation. Cells in the wild-type and Δrdi1 background, respectively, expressing 3HA-RHO4 and carrying either an empty plasmid or a plasmid encoding GAL1-GST-YGK3 were induced for 2 h. Equal amounts of proteins were separated by SDS-PAGE, and Rho4 was detected by antibodies against the HA epitope. Cdc11 was used as loading control. (B) Overexpression of YGK3 has no effect on protein levels of Cdc42 and Rho1. The experiment was performed as described in A. (C) Rho4 degradation depends on the proteasome. GAL1-RDI1 was overexpressed from a 2-μm plasmid (pKT10-GAL-RDI1) in 3HA-RHO4 cells in the presence of the proteasome-specific inhibitor MG132 (50 μM) or with dimethyl sulfoxide alone. The experiment was carried out in a Δerg6 background, because these cells have enhanced permeability for MG132 (Lee and Goldberg, 1996). (D) Rho4 is degraded by vacuolar proteases. Wild-type and Δpep4 cells with HA-tagged Rho4 carrying an empty plasmid and a plasmid with GAL1-RDI1, respectively, were incubated for 2 h with galactose to overexpress RDI1. Rho4 was detected by immunoblotting using anti-HA antibodies. GAPDH served as loading control. The results shown in this figure are representative of two independent experiments.
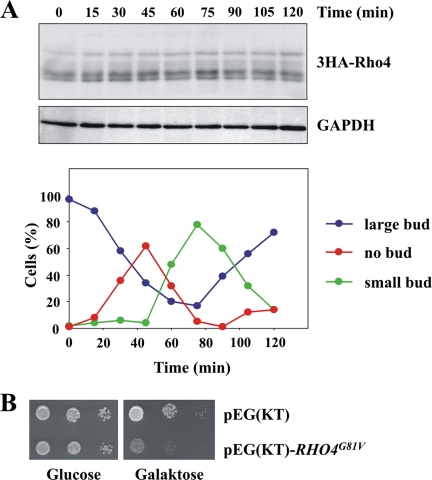
Next, we examined whether Rho4 was degraded in a cell cycle-dependent manner. However, no major changes were observed at different stages of the cell cycle (Figure 9A). It is conceivable, that Rho4 degradation is necessary to restrict the protein to a certain membrane domain. Nevertheless, GFP-Rho4 is enriched at the presumptive bud site in wild-type cells and in the absence of RDI1 (Figure 6E). Therefore, it seems unlikely, that the polarized localization of Rho4 is achieved by its proteolysis. It is also possible, that only active Rho4 is extracted from the membrane and subsequently degraded to terminate its function. To test whether hyperactive Rho4 causes any damage to cells, we overexpressed RHO4G81V. As described above, the paradigmatic rho4G81V mutant is locked in the active GTP-bound state due to a defect in GTP hydrolysis. This constitutively active form of Rho4 reduces cell growth markedly (Figure 9B). Unfortunately, no morphological or other changes were observed in these cells (data not shown). Together, our data show that Rdi1 and Ygk3 promote Rho4 degradation, a process which depends on both, vacuolar proteases and the proteasome. Further, the degradation of Rho4 may be important to terminate its activity.
Figure 9.
Rho4 protein levels do not change during the cell cycle. (A) Rho4 protein levels remain constant throughout the cell cycle. GAL1-CDC20 3HA-RHO4 cells were arrested in metaphase by incubating cells in YP raffinose medium (no expression of CDC20). Galactose was added to the synchronized cells to induce CDC20 expression and to trigger anaphase onset. At the indicated times, samples were taken, cells were lysed, and equal amounts of protein were analyzed by immunoblotting with antibodies against the HA epitope and GAPDH (loading control). Cell cycle progression was determined by following the number of cells without, with a small and with a large bud by microscopy (n > 100 at each time point). (B) Overexpression of constitutively active RHO4 inhibits growth. Cells carrying either GAL1-GST-RHO4G81V or the corresponding empty plasmid pEG(KT) were spotted on SC-Ura plates with glucose and galactose, respectively, and they were grown for 2 d at 30°C. The results shown in this figure are representative of two independent experiments.
DISCUSSION
In this work, we show that the regulation of Rho proteins by the sole budding yeast Rho GDI Rdi1 is rather complex. Rdi1 interacts only with Cdc42, Rho1, and Rho4, but not with the other Rho proteins (summarized in Figure 10). We demonstrate that Rdi1 extracts Cdc42 and Rho1 from membranes and forms a soluble complex with these proteins, a process that is regulated by the PAK-family kinase Cla4. In contrast, membrane extraction of Rho4 by Rdi1 results in the degradation of this Rho protein. We found that this proteolytic pathway includes the proteasome, vacuolar proteases, and the GSK-3β homologue Ygk3.
Figure 10.
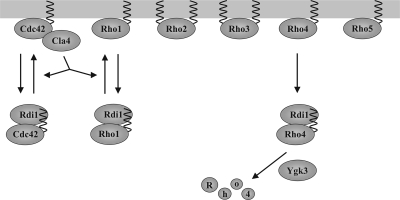
Model for the regulation of Rho GTPases by Rdi1. Rdi1 selectively extracts Cdc42, Rho1, and Rho4 from membranes and forms a complex with these proteins. Rho2 and Rho3 not only attach to the membrane through a prenyl group but also are palmitoylated. Cla4 disrupts binding between Rdi1 and Cdc42 and between Rdi1 and Rho1. Because Cla4 acts as a downstream effector of Cdc42, both proteins may constitute a positive feedback loop involved in the establishment of cell polarity. Furthermore, Cdc42 may regulate Rho1 activity via Cla4. The GSK-3β homologue Ygk3 and Rdi1 target Rho4 for degradation by vacuolar proteases and the proteasome after membrane extraction.
Functions of Rdi1
Very few phenotypes for the overexpression or deletion of RDI1 are known. Conflicting results have been reported for RDI1 overexpression. Tcheperegine et al. (2005) show that high levels of RDI1 cause only slightly rounder cell morphology, whereas others claim that RDI1 overexpression results in lethality (Masuda et al., 1994). In our hands, effects of RDI1 overexpression were clearly dose dependent. Only RDI1 expressed from the GAL1 promoter of a 2-μm plasmid was lethal. The actin cytoskeleton was depolarized in these cells and consequently buds were no longer present. This lack of polarization can be explained by the extraction of the essential Rho proteins Cdc42 and Rho1 from the plasma membrane by Rdi1.
To date, no phenotype has been described for Δrdi1 cells. Here, we show that deletion of RDI1 suppresses the mitotic exit defect of Δlte1 cells at low temperatures, possibly due to Cdc42 activation. Cdc42 regulates exit from mitosis via at least three independent pathways involving the Cdc42 effectors Cla4, Ste20, and Gic1 (Figure 2A) (Höfken and Schiebel, 2002; Jensen et al., 2002; Seshan et al., 2002; Höfken and Schiebel, 2004). Previously, we could demonstrate that a temperature-sensitive allele of the Cdc42 GEF CDC24 in a Δlte1 background is lethal at all temperatures (Höfken and Schiebel, 2002). Here, we observed that deletion of either RDI1 or of one of the Cdc42 GAPs results in suppression of the mitotic exit defect of Δlte1 cells. Consistently, overexpression of either STE20, GIC1, or GIC2 has the same effect on cells deleted for LTE1 (Höfken and Schiebel, 2002; Höfken and Schiebel, 2004). This suggests that deletion of the GAP genes or RDI1 leads to a slight activation of Cdc42 and its effectors, which trigger exit from mitosis independently of Lte1 (Figure 2A).
Interestingly, we observed a pseudohyphal growth defect for the diploid homozygous RDI1 deletion strain. Because CDC42 overexpression does not rescue this phenotype, it is unlikely that Rdi1 activates Cdc42. Because the deregulated CDC42G12V mutant exhibited a similar filamentation defect, Rdi1 may rather be necessary to restrict active Cdc42 to the growing tip. At first sight removal of Rho GTPases from the plasma membrane seems to suggest that Rdi1 acts as an inhibitor. However, Rho proteins could be targeted back to the membrane, where they are required, e.g., to sites of polarized growth. Furthermore, the extraction of Rho GTPases by Rdi1 from the membrane could play an important role in the maintenance of polarized distribution of these molecules. Cdc42 and other polarity proteins are not immobilized via scaffolds, but they are instead maintained in dynamically polarized states (Wedlich-Soldner et al., 2004). Endocytosis is required for the maintenance of cell polarization, e.g., by counteracting lateral diffusion of polarized proteins within the membrane (Valdez-Taubas and Pelham, 2003; Marco et al., 2007). Therefore, Rdi1-dependent extraction of Rho GTPases from membranes and return transport could be equally important for their polarized localization.
It has been reported that the expression of the constitutively active CDC42G12V or CDC42Q61L results in enhanced pseudohyphal growth (Mösch et al., 1996; Roberts et al., 1997). For these experiments, the CDC42 alleles were overexpressed from the inducible GAL1 promoter, whereas in our analysis CDC42 was placed under the control of its own promoter. In both cases, the endogenous copies of CDC42 were also present in the cell. Therefore, the protein levels seem to be critical. Possibly, physiological levels of Cdc42G12V may interfere with Cdc42 signaling pathways, causing a defect in pseudohyphal growth, whereas high amounts due to overexpression of constitutively active forms of Cdc42 cause enhanced pseudohyphal growth. Similarly, dose dependent polarity phenotypes were observed for other Cdc42 mutants which were attributed to differences in the capacity for GTP hydrolysis by Cdc42 of yeast cells after expression of physiological or supraphysiological levels (Irazoqui et al., 2004). Further work is needed to analyze whether molecular mechanisms along this line play a role for Cdc42G12V effects on pseudohyphal growth.
In contrast to the diploid homozygous Δrdi1/Δrdi1 strain, which exhibited a marked filamentation defect, haploid Δrdi1 cells invaded agar like the wild-type strain. The reason for this difference is unclear. Notably, RDI1 deletion in the diploid fungal pathogen C. albicans shows a similar defect in filamentation, when grown on solid medium (Court and Sudbery, 2007). Therefore, the role of Rdi1 might be well conserved.
Complex formation between Rdi1 and Rho GTPases
Although the association of Rdi1 with Cdc42 and Rho1 has been demonstrated (Koch et al., 1997; Eitzen et al., 2001; Richman et al., 2004; Tcheperegine et al., 2005), it remained unclear whether Rdi1 also interacts with other Rho GTPases. Therefore, we systematically examined the binding specificity of Rdi1. By using coimmunoprecipitations and membrane extraction assays, we could demonstrate that Rdi1 only binds to Cdc42, Rho1, and Rho4. Even after strong overexpression of RHO2, RHO3, and RHO5, the corresponding proteins did not form a complex with Rdi1. In line with these data, Rdi1 failed to extract the Rho GTPases Rho2, -3, and -5 from membranes. It seems likely that the specific association between Rdi1 and Rho GTPases depends on protein–protein interactions. X-ray crystallography revealed the binding of Rho GDIs to the switch I and II regions of Rho proteins (Hoffman et al., 2000; Scheffzek et al., 2000). Specific residues in these regions might determine whether Rdi1 binds to a Rho protein. Furthermore, Rho2 and Rho3 not only carry a prenyl moiety but also were identified as being palmitoylated (Roth et al., 2006). This additional lipid group could possibly prohibit the release of Rho2 and Rho3 from membranes by Rdi1.
Unexpectedly, we found that Rdi1 preferentially extracts the GTP-bound state of Cdc42 and Rho1 from membranes. In contrast, Rdi1 extracted comparable amounts of wild-type, GDP-, and GTP-associated Rho4 from membranes. However, it cannot be excluded that the point mutations, that were introduced to lock the Rho proteins either in the GDP- or GTP-bound state, also affect the interaction with Rdi1. Whether Rho GDIs form a complex with the GDP- or GTP-bound form of Rho GTPases is still controversial. Using fluorescence spectroscopy, it has been demonstrated that RhoGDI binds both forms of human Cdc42 equally well (Nomanbhoy and Cerione, 1996). In contrast, budding yeast Rho1 exclusively binds in the GDP-bound state to Rdi1 (Koch et al., 1997). The reason for these discrepancies is unclear, and further experiments are needed to reconcile these conflicting results.
We and Koch et al. (1997) found Cdc42 and Rho1 in the cytosol of Δrdi1 cells to a similar extent as in the wild-type strain. Possibly another protein can bind to the prenyl group of these Rho GTPases and forms a soluble complex. In addition, the observed cytosolic protein might be newly synthesized, which is not yet prenylated and therefore remains in the cytosol.
We also presented data, which suggest that the PAK-family kinase and Cdc42 effector Cla4 negatively regulates binding between Rdi1 and Cdc42 as well as Rho1 (Figure 10). Several lines argue against the idea that Cla4 simply competes with Rdi1 for binding with Rho proteins. First, this effect is specific for Cla4. Skm1 and Ste20 belong to the same kinase family as Cla4, and they act as direct downstream effectors of Cdc42 as well (Park and Bi, 2007). However, these related proteins did not interfere with Rdi1. Second, Cla4 is an effector of Cdc42 but not of Rho1 (Park and Bi, 2007). Nevertheless, we could show that Cla4 efficiently regulates binding between Rdi1 and Rho1 and between Rdi1 and Cdc42. Third, a kinase-dead mutant of Cla4 had almost no effect on Rdi1 complexes. Because Cla4 kinase activity is required for its effect on Rdi1, it is tempting to speculate that complex formation is regulated through phosphorylation of either Rdi1 or the Rho GTPases by Cla4. However, we could not demonstrate such a phosphorylation event (data not shown). Thus, it is conceivable that the regulation of Rdi1 by Cla4 is mediated via an unknown protein. It is unclear how Cla4 interferes with Rdi1 function. Cla4 could either inhibit the formation of a Rho GTPase–Rdi1 complex or alternatively disrupt an already existing complex.
There is some evidence that kinases modulate the interaction between RhoGDIs and Rho GTPases (Bourmeyster and Vignais, 1996; Mehta et al., 2001; Forget et al., 2002; DerMardirossian et al., 2004). In mammalian cells, Pak1 phosphorylates RhoGDI at Ser101 and Ser174, which results in the disruption of Rac1–RhoGDI complex (DerMardirossian et al., 2004). Notably, these serine residues are not conserved in budding yeast Rdi1.
Because Cla4 acts downstream of Cdc42, but also has a function in the disruption of the Rdi1–Cdc42 complex, Cla4 and Cdc42 may constitute a positive feedback loop involved in the establishment of cell polarity. According to this model, GTP-bound Cdc42 might recruit and activate Cla4, which in turn disrupts binding between Rdi1 and Cdc42. As a consequence, more Cdc42 is targeted to the plasma membrane at sites of polarized growth, where it is activated by its GEF Cdc24. Interestingly, Cla4 also phosphorylates Cdc24 (Gulli et al., 2000; Bose et al., 2001), and it was suggested that both proteins restrict Cdc42 activation via a negative feedback loop (Gulli et al., 2000). Together, Cla4 might have a dual role in the regulation of Cdc42 activity. In the early stages Cla4 may be involved in the plasma membrane recruitment and subsequent activation of Cdc42, whereas at later stages Cla4 terminates polarity via Cdc24. Importantly, Cla4 also has an effect on Rho1. Thus, Rho1 activity is indirectly regulated by Cdc42. A similar cross talk between Rho GTPases has been proposed for Pxl1. This paxillin-like protein may coordinate the function of Rho1 and Cdc42 during bud formation (Gao et al., 2004; Mackin et al., 2004). We observed that Cla4 has no effect on the interaction between Rho4 and Rdi1. Thus, Cla4 is highly specific for Cdc42 and Rho1.
Degradation of Rho4
Here, we observed that Rho4 protein levels depend on Rdi1. Lower amounts of Rho4 were found in cells with high levels of Rdi1 and vice versa. Because Rho4 is a relatively stable protein and a marked reduction of Rho4 levels took place after only 2 h of RDI1 overexpression, it is unlikely that this reduction is due to diminished protein synthesis. Thus, Rdi1 promotes Rho4 proteolysis. Further analysis revealed that Rho4 degradation was almost completely blocked in the absence of vacuolar proteases. In addition, using the proteasome inhibitor MG132, we could demonstrate that Rho4 proteolysis, at least to some extent, also depends on the proteasome. Finally, we observed that Ygk3, which phosphorylates Rho4 (Ptacek et al., 2005), promotes Rho4 degradation. Very little is known about Ygk3. It is one of four yeast homologues of GSK-3β, and it probably has a role in ubiquitin-dependent protein degradation (Andoh et al., 2000; Kassir et al., 2006). GSK-3β has a wide range of functions in higher eukaryotes, including the establishment of cell polarity (Kim and Kimmel, 2006). Notably, in the Hedgehog and the canonical Wnt pathway, GSK-3β mediates degradation of a transcription factor through phosphorylation of these proteins (Kim and Kimmel, 2006). Together, we propose a model, in which Rdi1 and Ygk3 coordinately regulate Rho4 degradation by vacuolar proteases and the proteasome (Figure 10).
Very few examples for the regulation of small GTPases through degradation have been reported. In fibroblasts, the HECT domain E3 ubiquitin ligase Smurf1 promotes the local degradation of RhoA and regulates cell polarity and protrusion (Wang et al., 2003). Similarly, the related Smurf2 triggers Rap1B proteolysis during neuronal differentiation (Schwamborn et al., 2007). There are several important differences between these examples and our observations. RhoA and Rap1B degradation depends on HECT domain E3 ubiquitin ligases. In contrast, Rsp5, the sole yeast ubiquitin ligase with a HECT domain (Rotin et al., 2000), is not involved in Rho4 degradation. Furthermore, Rho4 is degraded by vacuolar proteases, which has not been reported for RhoA and Rap1B. Finally, we demonstrate the involvement of the Rho GDI Rdi1 and the GSK-3β homologue Ygk3 in Rho4 degradation. Thus, Rho4 is likely to be degraded by a distinct mechanism.
It seems unusual that Rho4 degradation depends on the proteasome and the vacuolar system, but this has been reported for other proteins as well. For example, a mutated version of the plasma membrane H+-ATPase Pma1 misfolds, and it is subsequently degraded by the proteasome and the autophagy pathway (Mazón et al., 2007). However, it remains unclear why some proteins are degraded by both systems.
The physiological relevance of Rho4 proteolysis is not clear. RhoA and Rap1B are both degraded to restrict these proteins to a defined membrane region (Wang et al., 2003; Schwamborn et al., 2007). This does not seem to be the case for Rho4, which is localized to the entire plasma membrane and enriched at the presumptive bud site. If this polarized localization was achieved by Rho4 degradation, major changes in Rho4 protein levels during the cell cycle would be expected, but they were not observed in this study. Alternatively, it is conceivable that Rdi1 removes only active Rho4 from the membrane and targets it for destruction to terminate Rho4 activity. Thus, only a relatively small portion of Rho4 would be degraded; consequently, cell cycle-dependent changes may not be detectable by immunoblotting. In support of this model, we could also demonstrate that overexpression of constitutively active RHO4 is deleterious for cells.
Importantly, Rho GTPases, Rho GDIs and GSK-3β can be found in higher eukaryotes, where they also play a pivotal role in cell polarity. Therefore, the proposed regulatory link may be conserved from yeast to mammalian cells.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Paul Cullen (State University of New York, Buffalo, Buffalo, NY), Linda Hicke (Northwestern University, Evanston, IL), Peter Pryciak (University of Massachusetts Medical School, Worcester, MA), Yoshimi Takai (Osaka University Graduate School of Medicine, Osaka, Japan), Kazuma Tanaka (Hokkaido University, Sapporo, Japan), and Jeremy Thorner (University of California, Berkeley, Berkeley, CA) for providing constructs and strains. We thank Melanie Boß for excellent technical support and Gunnar Dittmar for strains and helpful discussions. This work is part of the doctoral thesis of C.T. and the diploma thesis of I.S. The project was supported by the Deutsche Forschungsgemeinschaft HO 2098/2-1.
Abbreviations used:
- GAP
GTPase-activating protein
- GDI
guanine nucleotide dissociation inhibitor
- GEF
guanine nucleotide exchange factor
- PAK
p21-activated kinase
- GSK-3β
glycogen synthase kinase 3β.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1152) on April 16, 2008.
REFERENCES
- Ammerer G., Hunter C. P., Rothman J. H., Saari G. C., Valls L. A., Stevens T. H. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol. Cell Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh T., Hirata Y., Kikuchi A. Yeast glycogen synthase kinase 3 is involved in protein degradation in cooperation with Bul1, Bul2, and Rsp5. Mol. Cell Biol. 2000;20:6712–6720. doi: 10.1128/mcb.20.18.6712-6720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barale S., McCusker D., Arkowitz R. A. Cdc42p GDP/GTP cycling is necessary for efficient cell fusion during yeast mating. Mol. Biol. Cell. 2006;17:2824–2838. doi: 10.1091/mbc.E05-11-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose I., Irazoqui J. E., Moskow J. J., Bardes E. S., Zyla T. R., Lew D. J. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J. Biol. Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- Bosl W. J., Li R. Mitotic-exit control as an evolved complex system. Cell. 2005;121:325–333. doi: 10.1016/j.cell.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bourmeyster N., Vignais P. V. Phosphorylation of Rho GDI stabilizes the Rho A-Rho GDI complex in neutrophil cytosol. Biochem. Biophys. Res. Commun. 1996;218:54–60. doi: 10.1006/bbrc.1996.0011. [DOI] [PubMed] [Google Scholar]
- Caviston J. P., Longtine M., Pringle J. R., Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang T. H., Bohl B. P., Bokoch G. M. Biologically active lipids are regulators of Rac.GDI complexation. J. Biol. Chem. 1993;268:26206–26211. [PubMed] [Google Scholar]
- Cole K. C., McLaughlin H. W., Johnson D. I. Use of bimolecular fluorescence complementation to study in vivo interactions between Cdc42p and Rdi1p of Saccharomyces cerevisiae. Eukaryot. Cell. 2007;6:378–387. doi: 10.1128/EC.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court H., Sudbery P. Regulation of Cdc42 GTPase activity in the formation of hyphae in Candida albicans. Mol. Biol. Cell. 2007;18:265–281. doi: 10.1091/mbc.E06-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerMardirossian C., Bokoch G. M. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- DerMardirossian C., Schnelzer A., Bokoch G. M. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol. Cell. 2004;15:117–127. doi: 10.1016/j.molcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Dovas A., Couchman J. R. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005;390:1–9. doi: 10.1042/BJ20050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dransart E., Olofsson B., Cherfils J. RhoGDIs revisited: novel roles in Rho regulation. Traffic. 2005;6:957–966. doi: 10.1111/j.1600-0854.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- Eitzen G., Thorngren N., Wickner W. Rho1p and Cdc42p act after Ypt7p to regulate vacuole docking. EMBO J. 2001;20:5650–5656. doi: 10.1093/emboj/20.20.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget M. A., Desrosiers R. R., Gingras D., Beliveau R. Phosphorylation states of Cdc42 and RhoA regulate their interactions with Rho GDP dissociation inhibitor and their extraction from biological membranes. Biochem. J. 2002;361:243–254. doi: 10.1042/0264-6021:3610243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. D., Caviston J. P., Tcheperegine S. E., Bi E. Pxl1p, a paxillin-like protein in Saccharomyces cerevisiae, may coordinate Cdc42p and Rho1p functions during polarized growth. Mol. Biol. Cell. 2004;15:3977–3985. doi: 10.1091/mbc.E04-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S., Bose I., Zyla T. R., Bardes E. S., Lew D. J. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 2002;156:315–326. doi: 10.1083/jcb.200109062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulli M. P., Jaquenoud M., Shimada Y., Niederhauser G., Wiget P., Peter M. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- Hoffman G. R., Nassar N., Cerione R. A. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- Höfken T., Schiebel E. A role for cell polarity proteins in mitotic exit. EMBO J. 2002;21:4851–4862. doi: 10.1093/emboj/cdf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfken T., Schiebel E. Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. J. Cell Biol. 2004;164:219–231. doi: 10.1083/jcb.200309080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J. E., Gladfelter A. S., Lew D. J. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- Irazoqui J. E., Gladfelter A. S., Lew D. J. Cdc42p, GTP hydrolysis, and the cell's sense of direction. Cell Cycle. 2004;3:861–864. [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Janke C., et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jensen S., Geymonat M., Johnson A. L., Segal M., Johnston L. H. Spatial regulation of the guanine nucleotide exchange factor Lte1 in Saccharomyces cerevisiae. J. Cell Sci. 2002;115:4977–4991. doi: 10.1242/jcs.00189. [DOI] [PubMed] [Google Scholar]
- Kassir Y., Rubin-Bejerano I., Mandel-Gutfreund Y. The Saccharomyces cerevisiae GSK-3 beta homologs. Curr. Drug Targets. 2006;7:1455–1465. doi: 10.2174/1389450110607011455. [DOI] [PubMed] [Google Scholar]
- Kim L., Kimmel A. R. GSK3 at the edge: regulation of developmental specification and cell polarization. Curr. Drug Targets. 2006;7:1411–1419. doi: 10.2174/1389450110607011411. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Koch G., Tanaka K., Masuda T., Yamochi W., Nonaka H., Takai Y. Association of the Rho family small GTP-binding proteins with Rho GDP dissociation inhibitor (Rho GDI) in Saccharomyces cerevisiae. Oncogene. 1997;15:417–422. doi: 10.1038/sj.onc.1201194. [DOI] [PubMed] [Google Scholar]
- Lee D. H., Goldberg A. L. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mackin N. A., Sousou T. J., Erdman S. E. The PXL1 gene of Saccharomyces cerevisiae encodes a paxillin-like protein functioning in polarized cell growth. Mol. Biol. Cell. 2004;15:1904–1917. doi: 10.1091/mbc.E04-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E., Wedlich-Soldner R., Li R., Altschuler S. J., Wu L. F. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007;129:411–422. doi: 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T., Tanaka K., Nonaka H., Yamochi W., Maeda A., Takai Y. Molecular cloning and characterization of yeast rho GDP dissociation inhibitor. J. Biol. Chem. 1994;269:19713–19718. [PubMed] [Google Scholar]
- Mazón M. J., Eraso P., Portillo F. Efficient degradation of misfolded mutant Pma1 by endoplasmic reticulum-associated degradation requires Atg19 and the Cvt/autophagy pathway. Mol. Microbiol. 2007;63:1069–1077. doi: 10.1111/j.1365-2958.2006.05580.x. [DOI] [PubMed] [Google Scholar]
- Mehta D., Rahman A., Malik A. B. Protein kinase C-alpha signals rho-guanine nucleotide dissociation inhibitor phosphorylation and rho activation and regulates the endothelial cell barrier function. J. Biol. Chem. 2001;276:22614–22620. doi: 10.1074/jbc.M101927200. [DOI] [PubMed] [Google Scholar]
- Mösch H. U., Roberts R. L., Fink G. R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomanbhoy T. K., Cerione R. Characterization of the interaction between RhoGDI and Cdc42Hs using fluorescence spectroscopy. J. Biol. Chem. 1996;271:10004–10009. doi: 10.1074/jbc.271.17.10004. [DOI] [PubMed] [Google Scholar]
- Palecek S. P., Parikh A. S., Kron S. J. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology. 2002;148:893–907. doi: 10.1099/00221287-148-4-893. [DOI] [PubMed] [Google Scholar]
- Park H. O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J., et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Richman T. J., Sawyer M. M., Johnson D. I. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot. Cell. 2002;1:458–468. doi: 10.1128/EC.1.3.458-468.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman T. J., Toenjes K. A., Morales S. E., Cole K. C., Wasserman B. T., Taylor C. M., Koster J. A., Whelihan M. F., Johnson D. I. Analysis of cell-cycle specific localization of the Rdi1p RhoGDI and the structural determinants required for Cdc42p membrane localization and clustering at sites of polarized growth. Curr. Genet. 2004;45:339–349. doi: 10.1007/s00294-004-0505-9. [DOI] [PubMed] [Google Scholar]
- Roberts R. L., Mösch H. U., Fink G. R. 14–3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D., Staub O., Haguenauer-Tsapis R. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 2000;176:1–17. doi: 10.1007/s00232001079. [DOI] [PubMed] [Google Scholar]
- Scheffzek K., Stephan I., Jensen O. N., Illenberger D., Gierschik P. The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat. Struct. Biol. 2000;7:122–126. doi: 10.1038/72392. [DOI] [PubMed] [Google Scholar]
- Schwamborn J. C., Muller M., Becker A. H., Puschel A. W. Ubiquitination of the GTPase Rap1B by the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. EMBO J. 2007;26:1410–1422. doi: 10.1038/sj.emboj.7601580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Seshan A., Bardin A. J., Amon A. Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 2002;12:2098–2110. doi: 10.1016/s0960-9822(02)01388-x. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Sasaki T., Mammoto A., Takaishi K., Kameyama T., Tsukita S., Takai Y. Direct interaction of the Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J. Biol. Chem. 1997;272:23371–23375. doi: 10.1074/jbc.272.37.23371. [DOI] [PubMed] [Google Scholar]
- Tcheperegine S. E., Gao X. D., Bi E. Regulation of cell polarity by interactions of Msb3 and Msb4 with Cdc42 and polarisome components. Mol. Cell Biol. 2005;25:8567–8580. doi: 10.1128/MCB.25.19.8567-8580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Taubas J., Pelham H. R. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Wang H. R., Zhang Y., Ozdamar B., Ogunjimi A. A., Alexandrova E., Thomsen G. H., Wrana J. L. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Wai S. C., Schmidt T., Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T., Tohyama M. The p75 receptor acts as a displacement factor that releases Rho from Rho-GDI. Nat. Neurosci. 2003;6:461–467. doi: 10.1038/nn1045. [DOI] [PubMed] [Google Scholar]
- Yamochi W., Tanaka K., Nonaka H, Maeda A., Musha T., Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J. Cell Biol. 1994;125:1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., O'Brien J. M., Ouellette L. A., Church W. R., Johnson D. I. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiaes gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol. 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., Preuss D., Mulholland J., O'Brien J. M., Botstein D., Johnson D. I. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol. Biol. Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.