Abstract
In Saccharomyces cerevisiae, more than 180 assembly factors associate with preribosomes to enable folding of pre-rRNA, recruitment of ribosomal proteins, and processing of pre-rRNAs to produce mature ribosomes. To examine the molecular architecture of preribosomes and to connect this structure to functions of each assembly factor, assembly subcomplexes have been purified from preribosomal particles. The Nop7-subcomplex contains three assembly factors: Nop7, Erb1, and Ytm1, each of which is necessary for conversion of 27SA3 pre-rRNA to 27SBS pre-rRNA. However, interactions among these three proteins and mechanisms of their recruitment and function in pre-rRNPs are poorly understood. Here we show that Ytm1, Erb1, and Nop7 assemble into preribosomes in an interdependent manner. We identified which domains within Ytm1, Erb1, and Nop7 are necessary for their interaction with each other and are sufficient for recruitment of each protein into preribosomes. Dominant negative effects on growth and ribosome biogenesis caused by overexpressing truncated Ytm1, Erb1, or Nop7 constructs, and recessive phenotypes of the truncated proteins revealed not only interaction domains but also other domains potentially important for each protein to function in ribosome biogenesis. Our data suggest a model for the architecture of the Nop7-subcomplex and provide potential functions of domains of each protein.
INTRODUCTION
Ribosome assembly in eukaryotes requires the association of 79 different ribosomal proteins with ribosomal RNAs, concomitant with folding, modification, and processing of the rRNA primary transcript to form mature 5.8S, 18S, and 25–28S rRNAs. Genetic screens and new proteomic tools have identified more than 180 different assembly factors necessary for maturation of these preribosomes (pre-rRNPs) in the yeast Saccharomyces cerevisiae (reviewed in Fatica and Tollervey, 2002; Fromont-Racine et al., 2003; Raué, 2003; Granneman and Baserga, 2004). Homologues of most of these assembly factors are found in metazoans from Drosophila to humans (reviewed in Boisvert et al., 2007; Leung et al., 2003).
The major challenge now before us is to understand the precise functions of each of these assembly factors. Identification of networks of interactions among the assembly factors, ribosomal proteins, and ribosomal RNAs is an important first step to address this question. To begin to identify this internal structure of preribosomes and to connect this structural information to function of ribosomal molecules, we have used a combined genetic and biochemical approach to identify neighborhoods or subcomplexes of interacting proteins within preribosomes (Harnpicharnchai et al., 2001; Miles et al., 2005; Zhang et al., 2007).
One such neighborhood is the Nop7-subcomplex, which contains assembly factors Nop7, Ytm1, and Erb1 (Harnpicharnchai et al., 2001; Du and Stillman, 2002; Krogan et al., 2004, Miles et al., 2005). Two-hybrid assays and GST pulldown experiments show that Nop7 and Ytm1 interact directly with Erb1, but not with each other (Miles et al., 2005). Mammalian counterparts of these three proteins, Pes1, WDR12, and Bop1, also associate with each other to form the PeBoW complex (Hölzel et al., 2005). Each of these proteins is essential, conserved among eukaryotes, located primarily in the nucleolus, and required for the assembly of 60S ribosomal subunits (Strezoska et al., 2000; Pestov et al., 2001; Adams et al., 2002; Lerch-Gaggl et al., 2002; Oeffinger et al., 2002; Miles et al., 2005; Hölzel et al., 2005). The Nop7-subcomplex is also a member of a “functional cluster” in yeast; depletion of each of these three proteins causes identical defects in rRNA precursor (pre-rRNA) processing: altered conversion of 27SA3 pre-rRNA to 27SBS pre-rRNA (Adams et al., 2002; Miles et al., 2005; Oeffinger et al., 2002; Pestov et al., 2001). Knock-down of the mammalian proteins also affects processing of the corresponding pre-rRNAs. Nop7 and Erb1 assemble into 90S preribosomal particles containing 35S pre-rRNA, whereas Ytm1 is recruited later, into 66S A2 preribosomes (Miles et al., 2005). Thus, Nop7, Ytm1, and Erb1 function several steps after they associate with pre-ribosomes, suggesting that there may be separate means to recruit and to utilize these proteins in subunit assembly.
Nop7, Ytm1, and Erb1 each contain potential protein–protein interaction domains, which may enable them to form a microscaffold within nascent ribosomes, to organize a neighborhood necessary for processing of 27SA3 pre-rRNA. Here, we used several different methods to investigate how these three ribosome assembly factors interact with each other and are recruited into and function in preribosomes: 1) identifying ligands of these interaction domains both in vitro and in vivo, 2) testing whether truncated Ytm1, Erb1, or Nop7 proteins containing or lacking each potential interaction domain can associate with preribosomal particles, and 3) assaying effects of the truncations on cellular localization, ribosome assembly and pre-rRNA processing in both wild-type cells and cells lacking the corresponding full-length proteins. We found that Ytm1 binds to Erb1 via the C-terminal WD40 domain of Ytm1, the N-terminal conserved region of Erb1 interacts with Nop7 and Ytm1, and the central region of Nop7 binds to Erb1. Surprisingly, the conserved C-terminal WD40 domain of Erb1 is not essential for growth or ribosome biogenesis. We also show that recruitment of Nop7, Ytm1, and Erb1 into nascent ribosomes is mutually interdependent. Interactions between these specific domains in the three proteins are important for the assembly of each protein into preribosomes. Moreover, dominant negative effects on growth and ribosome biogenesis caused by overexpressing truncated Ytm1, Erb1, or Nop7 proteins reveal domains potentially important for each protein to function in preribosome maturation. The methods described here can be used as an experimental system to discover interactions among other assembly factors and ultimately contribute to understand the molecular architecture of preribosomes.
MATERIALS AND METHODS
Yeast Strains and Media
All yeast strains were derived from JWY3400 (MATa ura3-52 lys2-801 trp1-1 leu2-1 his3-Δ200 pep4::HIS3 prb1-Δ1.6R can). Yeast cells were grown at 30°C in YEPD medium (2% dextrose, 2% peptone, and 1% yeast extract), or else synthetic medium lacking leucine (C-leu) or uracil (C-ura) and supplemented with 1% raffinose (C-leu or C-ura + 1% raffinose) or with 1% raffinose + 1% galactose (C-leu or C-ura + 1% raffinose and 1% galactose). Cells were harvested at 5–107 cell/ml unless otherwise indicated. Yeast strains expressing C-terminal triple hemagglutinin (3HA)-tagged proteins, C-terminal 13 Myc-tagged proteins, or N-terminal 3HA-tagged proteins were created as described previously (Longtine et al., 1998). RPF2-TAP strains were generated by integrating the tandem affinity purification (TAP) tag at the 3′ end of the genomic RPF2 allele (Rigaut et al., 1999).
Cloning of Full-Length or Truncated YTM1, ERB1, or NOP7 Genes into Yeast Expression Plasmids
Full-length or truncated YTM1, ERB1, or NOP7 genes were PCR-amplified from yeast genomic DNA and cloned into the pGREG535 or pGREG536 CEN plasmids using the SalI restriction site (Jansen et al., 2005). Both plasmids have a galactose-inducible and glucose-respressible GAL1 promoter. To generate plasmids expressing TAP-tagged truncated proteins, plasmids containing different truncations of the YTM1, ERB1, or NOP7 genes in pGREG535 were linearized by XhoI, and the TAP tag was introduced into the 3′ end of each gene by PCR and gap-repair (Rigaut et al., 1999). Sequences of oligonucleotides used for PCR to clone or tag genes are available upon request.
Assays for Growth Defects
Growth of yeast cells was assayed by plating serial dilutions. Yeast transformed with plasmids containing full-length or truncated YTM1, ERB1, or NOP7 were grown in C-leu + 1% raffinose medium or C-ura + 1% raffinose medium to early log phase. Cells were diluted 10-, 100-, 1000-, and 10,000-fold; 10 μl of cultures was spotted onto C-leu+ glucose or C-leu + galactose medium where specified.
Assaying Ribosome Production
Ribosome assembly subcomplexes, preribosomes, ribosomes, and polyribosomes were resolved on 7–47% sucrose gradients as described previously (Harnpicharnchai et al., 2001).
Tandem Affinity Purification, One-Step Purification, and Mass Spectrometry of Preribosomes
Tandem affinity purification of preribosomes or ribosome assembly subcomplexes from whole cell extracts was carried out by the methods of Miles et al. (2005). Preribosome purification after a high-speed spin was performed as described in Krogan et al. (2004). One-step purification of preribosomes from whole cell extracts was performed as described in Oeffinger et al. (2007). Proteins copurifying with Ytm1-C-TAP were identified by mass spectrometry as described in Horsey et al. (2004).
GST Pulldown and Yeast Two-Hybrid Assays
GST pulldown and yeast two-hybrid assays of protein–protein interactions were performed as described in Miles et al. (2005).
Indirect Immunofluorescence Microscopy
Yeast transformed with plasmids containing truncated YTM1, ERB1, or NOP7 were grown at 30°C in synthetic medium containing 1% raffinose to ∼3·107 cells/ml. Galactose was added to each culture to a final concentration of 1% to induce the expression of truncated proteins. Cells were harvested after 4-h induction and prepared for microscopy (Pringle et al., 1989). Cells were incubated with monoclonal anti-HA antibodies in phosphate-buffered saline containing 0.5% bovine serum albumin overnight at 4°C. After washing, cells were incubated with anti-mouse fluorescein-conjugated antibodies (Invitrogen, Carlsbad, CA). To detect nuclear DNA, cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI) for 3.5 min. Slides were viewed using a Zeiss Axioskope 2 plus microscope equipped with a 100× objective (Thornwood, NY). Photographs were taken on a Canon Powershot G5 camera. Digital images were processed using Adobe Photoshop (San Jose, CA).
Analysis of RNA
RNA isolated from whole cell extracts was assayed by Northern analysis or primer extension as described in Horsey et al. (2004).
Western Blotting and Antibodies
Proteins present in whole cell extracts, purified preribosomes, or assembly subcomplexes were assayed by Western blot analysis according to standard protocols (Ausubel et al., 1994). TAP-tagged proteins were detected using alkaline phosphatase conjugated to IgG (Pierce, Rockford, IL). HA-tagged proteins were identified with mouse mAb 12CA5. Rabbit polyclonal Nop7 antibody, rpL5 antibody, Sec61 antibody, and mouse monoclonal Myc antibody were used against Nop7, rpL5, Sec61, or Myc-tagged proteins, respectively.
RESULTS
Assembly of Erb1, Nop7, and Ytm1 into Preribosomes Is Interdependent
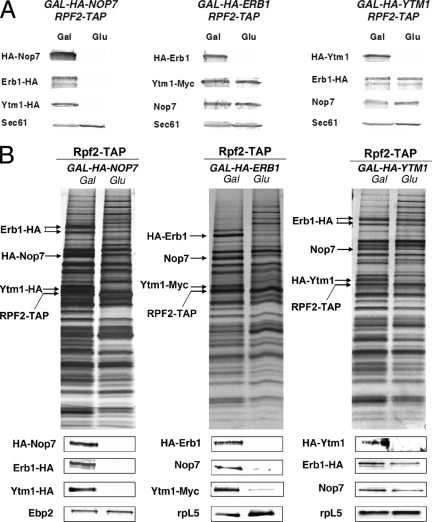
Do individual components of the heterotrimeric Nop7-subcomplex influence the association of the other two proteins with preribosomes? To address this question, we assayed the constituents of preribosomes when Nop7, Erb1, or Ytm1 was depleted. Strains that express each of the three proteins under the control of the galactose-inducible and glucose-repressible GAL promoter were constructed. Western blotting of proteins from whole cell extracts confirmed Nop7, Erb1, or Ytm1 were depleted to very low or undetectable levels in the respective strains upon shifting each strain from galactose- to glucose-containing medium (Figure 1A). Consequently, growth of the cells slowed significantly (Pestov et al., 2001; Adams et al., 2002; Miles et al., 2005). Preribosomal particles were affinity-purified from these strains using TAP-tagged Rpf2, which is present in 90S preribosomes and each of the consecutive 66S precursors to mature 60S ribosomal subunits (Zhang et al., 2007). Rpf2 is required for a different pre-rRNA processing step than the Nop7-subcomplex (Zhang et al., 2007); thus it is unlikely to directly interact with the Nop7-subcomplex. SDS-PAGE and silver staining of proteins from preribosomes purified from the undepleted and depleted strains revealed that although a specific subset of proteins was absent, pre-rRNPs were otherwise largely intact upon depletion of Nop7, Ytm1, or Erb1 (Figure 1B). When either Nop7 or Erb1 was depleted, the other two components of the Nop7-subcomplex were completely or almost completely undetectable in preribosomes. In contrast, when Ytm1 was depleted, amounts of Nop7 or Erb1 in preribosomes were reduced to a lesser extent (Figure 1B). These results are consistent with our previous findings that Ytm1 assembles into preribosomes after Nop7 and Erb1 (Miles et al., 2005). The decreased levels of Nop7 and Erb1 in preribosomes from Ytm1-depleted cells indicate that assembly of Ytm1 into preribosomal particles is important for the retention of these two earlier assembled proteins. Because preribosomes undergo multiple consecutive rearrangements during their maturation, it seems very likely that dynamic intermolecular interactions would play an important role to maintain association of factors with particles. To assess whether the inability of Nop7, Erb1, or Ytm1 to associate with preribosomes upon depletion of one is due to instability of those proteins in cells, Western blots from whole cell lysates were carried out. On depletion of either Erb1 or Ytm1, wild-type levels of the other two proteins were detected in whole cell extracts, indicating no effects on their stability (Figure 1A). Thus their absence from preribosomes was not simply caused by their absence from cells. In contrast, amounts of Erb1 and Ytm1 in whole cell extracts were substantially decreased upon depletion of Nop7 (Figure 1A), indicating that their inability to associate with preribosomes upon depletion of Nop7 arises due to their instability in the absence of Nop7. We conclude that association of Erb1, Nop7, and Ytm1 with pre-RNPs occurs in an interdependent manner, whereas association of most other proteins with preribosomes is not affected in the absence of these three proteins.
Figure 1.
Interdependence of assembly of Erb1, Nop7, and Ytm1 into preribosomes. Yeast strains containing GAL-NOP7, GAL-ERB1, or GAL-YTM1 were grown at 30°C in galactose medium to 3·107 cells/ml. A second culture of each strain was grown in galactose medium and shifted to glucose medium for 16 h to 3 × 107 cells/ml, to deplete the respective proteins. Preribosomes were isolated from whole cell extracts by affinity purification, using Rpf2-TAP. (A) Proteins in whole cell extracts were assayed by Western blotting. (B) Proteins in affinity-purified preribosomes were resolved by SDS-PAGE and stained with silver (top) or subjected to Western blot analysis (bottom). Positions of stained Erb1, Nop7, and Ytm1 are indicated.
Ytm1 Binds Erb1 via Its C-terminal WD40 Repeats
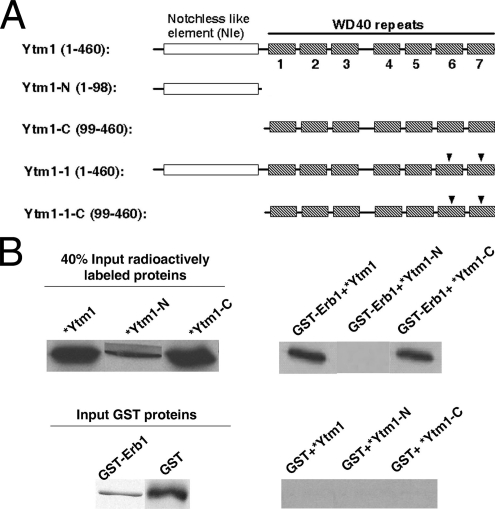
Previously, we showed that Ytm1 interacts directly with Erb1 (Miles et al., 2005). To determine which portion of Ytm1 is responsible for this interaction, we assayed binding in vitro to GST-Erb1 of the N-terminal 98 amino acids of Ytm1 (Ytm1-N) containing the Notchless-like element (Nal et al., 2002), and the C-terminal 362 amino acids of Ytm1 (Ytm1-C) containing seven WD40 repeats (Miles et al., 2005; Figure 2A). Although the function of the Notchless-like element is unknown (see Discussion), the WD40 repeats form a beta-propeller structure, a well-characterized protein–protein interaction domain (reviewed in Smith et al., 1999). [35S]methionine labeled Ytm1, Ytm1-N, and Ytm1-C were synthesized in vitro and assayed for binding to purified GST-Erb1. Ytm1-C bound to GST-Erb1 almost as well as full-length Ytm1, whereas Ytm1-N did not bind to GST-Erb1 (Figure 2B). Therefore, Ytm1 binds Erb1 via its C-terminal WD40 domain.
Figure 2.
The C-terminal WD-40 domain of Ytm1 (i.e., Ytm1-C) interacts directly with Erb1. (A) Schematic representation of Ytm1 and truncation constructs. Shown are the notchless-like element (NLE, white rectangle), and WD40 repeats 1–7 (hatched boxes). Black triangles (▾) indicate two mutations: G398D and S442N. (B) Left, 40% of the input [35S] labeled proteins in each assay and 100% of the pulled down GST proteins (Coomassie blue staining) are shown. Right, synthetic 35S-methionine–labeled full-length Ytm1, Ytm1-N, or Ytm1-C proteins (*) were incubated with purified GST-Erb1 protein. Complexes were eluted from glutathione beads, subjected to SDS-PAGE, and detected by autoradiography.
Binding to Erb1 Enables Ytm1-C to Assemble Into 66S Preribosomes
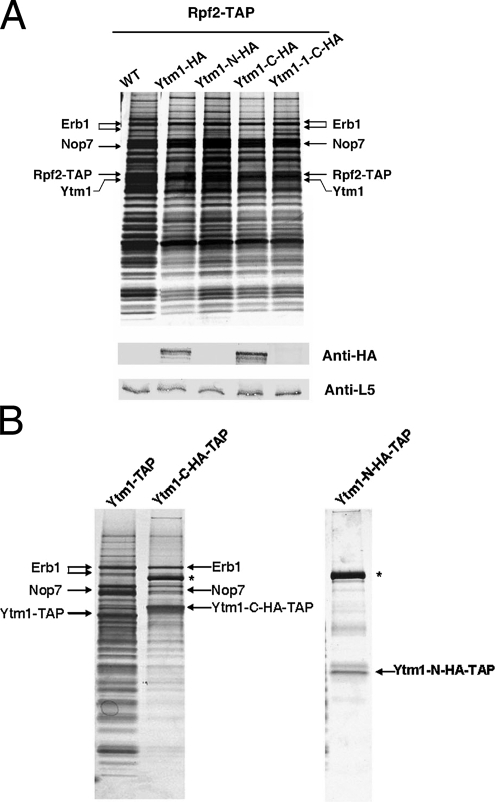
We then asked whether binding to Erb1 is required for association of Ytm1 with assembling ribosomes. If this is true, Ytm1-C should copurify with preribosomal particles, whereas Ytm1-N should not. To test this prediction, we expressed HA-tagged Ytm1, Ytm1-N, or Ytm1-C under control of the GAL promoter in cells also expressing endogenous Ytm1. Western blotting indicated that each protein was expressed at similar levels, 4 h after induction of the GAL constructs (Supplementary Figure S1A). Pre-rRNPs from cells expressing HA-tagged Ytm1, Ytm1-N, or Ytm1-C were purified using TAP-tagged Rpf2. Consistent with our prediction, Ytm1-C, but not Ytm1-N, associated with preribosomal particles to the same extent as full-length HA-tagged Ytm1 (Figure 3A). Indirect immunofluorescence microscopy showed that both truncated Ytm1 proteins localized to the nucleus (Supplementary Figure S2A). Thus, the absence of Ytm1-N from preribosomes is unlikely to result from its inability to enter the nucleus.
Figure 3.
Ytm1-C assembles into preribosomes and the Nop7-subcomplex in vivo. Yeast strains containing each of the GAL-inducible full-length or truncated YTM1 genes were grown at 30°C in synthetic medium containing 1% raffinose to ∼1.5·107 cells/ml. Galactose was added to each strain to a final concentration of 1% to induce the expression of Ytm1 proteins. Cells were harvested after 4-h induction (one doubling time). (A) Lysates were prepared from cells expressing endogenous Ytm1 plus HA-tagged full-length Ytm1, Ytm1-N, Ytm1-C, or Ytm1-1-C, or from cells expressing only endogenous Ytm1 (WT). Pre-rRNPs were purified using TAP-tagged Rpf2. Proteins present in the affinity-purified preribosomes were resolved by SDS-PAGE and stained with silver (top) or subjected to Western blot analysis (bottom) using anti-HA or anti-rpL5 antiserum. (B) Left, preribosomes purified from wild-type cells using TAP-tagged Ytm1 and purified from cells using TAP-tagged Ytm1-C. Right, preribosomes purified from cells expressing TAP-tagged Ytm1-N. Proteins in affinity-purified samples were resolved by SDS-PAGE and stained with silver. Protein bands in the Ytm1-C-TAP purified samples were excised and subjected to mass spectrometry to identify Erb1, Nop7, and Ytm1-C-TAP. The band indicated by an asterisk is Ste23, a protein not necessary for ribosome biogenesis (L. Tang, unpublished data).
To verify the interaction between Ytm1-C and Erb1 in vivo, TAP-tagged Ytm1-C was used to affinity-purify Ytm1-C and associated molecules from whole cell extracts. Significant amounts of both Erb1 and Nop7 copurified with Ytm1-C (Figure 3B), indicating that Ytm1-C forms a heterotrimeric subcomplex with Erb1 and Nop7 in vivo. In contrast, neither Erb1 nor Nop7 copurified with TAP-tagged Ytm1-N (Figure 3B). Western blotting using antibodies against other ribosome assembly factors showed that preribosomes copurified with Ytm1-C, but not Ytm1-N (Figure 3B, and Supplementary Figure S3), providing independent evidence that Ytm1-C can assemble into preribosomal particles.
Previously we showed that the Ytm1–1 mutant protein contains two mutations, G398D and S442N, in the sixth and seventh WD40 repeats, which disrupt the interaction between Ytm1 and Erb1 and weaken the association of Ytm1 with preribosomes (Miles et al., 2005). To confirm that binding to Erb1 enables Ytm1-C to assemble into preribosomes, we tested whether these mutations in Ytm1-1-C that decrease binding to Erb1 affect the association of Ytm1-1-C with pre-RNPs. We constructed strains expressing Ytm1–1 or Ytm1-1-C, each of which contains these two mutations (Figure 2A). Although Ytm1-1-C was expressed at levels similar to Ytm1-C (Supplementary Figure S1A) and was primarily localized to the nucleus (Supplementary Figure S2A), it did not copurify with preribosomes (Figure 3A).
Overexpression of Ytm1-C Has Dominant Negative Effects on Growth and Ribosome Biogenesis
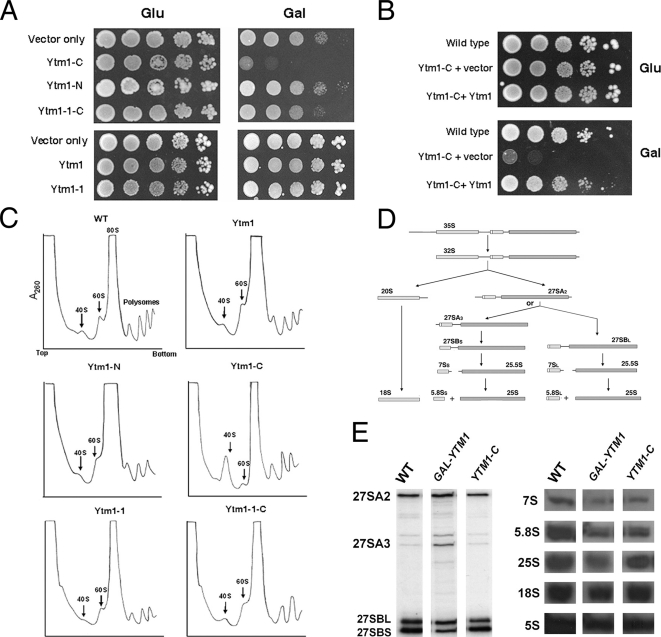
The above results indicate that Ytm1-C might compete with wild-type Ytm1 for binding to Erb1 and assembly into preribosomal particles. However, the absence of the N-terminal 98 amino acids may prevent the truncated Ytm1-C protein from carrying out all necessary functions of full-length Ytm1. Thus preribosomes containing Ytm1-C might not undergo proper maturation. Alternatively, the truncated Ytm1-C protein might fold into an aberrant structure (“poison subunit”) and interfere with ribosome biogenesis (Herskowitz, 1987). Consistent with these ideas, overexpression of Ytm1-C had a dominant negative effect on growth, whereas overexpression of either full-length Ytm1 or Ytm1-N did not affect growth rate (Figure 4A). To determine the effect of overexpression of Ytm1-C on ribosome biogenesis, we assayed levels of ribosomal subunits by sucrose gradient sedimentation. Cells overexpressing either full-length Ytm1 or Ytm1-N contained wild-type levels of 60S subunits (Figure 4C). In contrast, yeast overexpressing Ytm1-C contained significantly fewer free 60S subunits (Figure 4C), indicating a defect in 60S ribosomal subunit assembly almost as strong as observed upon depletion or inactivation of Ytm1 (Harnpicharnchai et al., 2001; Miles et al., 2005). To further investigate defects in 60S subunit biogenesis in strains overexpressing Ytm1-C, we assayed the relative amounts of pre-rRNA processing intermediates and mature rRNAs. Surprisingly, we observed no significant differences in the relative amounts of pre-rRNA or mature rRNA in these cells compared with wild-type cells (Figure 4E). These results suggest that overexpression of Ytm1-C must cause a defect in production of 60S ribosomal subunits that does not include perturbations of pre-rRNA processing.
Figure 4.
Expression of Ytm1-C causes dominant negative effects on growth and ribosome biogenesis. (A) Wild-type cells or yeast expressing endogenous Ytm1 plus full-length or truncated Ytm1 proteins were grown to early log phase, serially diluted (10-, 100-, 1000 and 10,000-fold) onto Glu (glucose) or Gal (galactose) medium, and incubated at 30°C. (B) Wild-type cells or yeast overexpressing Ytm1-C or co-overexpressing Ytm1-C and full-length Ytm1 were grown to early log phase and serially diluted as above. (C) Whole cell extracts were subjected to centrifugation on sucrose gradients to separate 40S and 60S ribosomal subunits, 80S ribosomes, and polyribosomes. (D) Pre-rRNA processing pathway in the yeast S. cerevisiae. (E) RNA was extracted from wild-type yeast (WT) or from strains in which Ytm1 was depleted (GAL-YTM1) or Ytm1-C was overexpressed (YTM1-C). Pre-rRNAs were assayed by primer extension (left) or by Northern blotting (right), using specific oligonucleotide primers or probes, respectively.
If Ytm1-C binds to Erb1 and competes with full-length Ytm1 to assemble into preribosomes, then disrupting the interaction between Ytm1-C and Erb1 should suppress the dominant negative phenotypes caused by Ytm1-C. Consistent with this idea, overexpression of the mutant Ytm1-1-C protein did not affect either growth or ribosome biogenesis (Figure 4, A and C). In addition, when full-length Ytm1 was co-overexpressed together with Ytm1-C, the growth defect caused by Ytm1-C was suppressed (Figure 4B).
All of these findings are consistent with our previous results that Ytm1 assembles into preribosomal particles later than Erb1 (Miles et al., 2005) and that Erb1 is required for Ytm1 recruitment (Figure 1). Additionally, our findings help explain why overexpression of WDR12 ΔNle, a similarly truncated version of WDR12, the mammalian homologue of Ytm1, causes dominant negative effects on cell proliferation and ribosome biogenesis in cultured mammalian cell lines (Hölzel et al., 2005).
The N-terminal Conserved Region of Erb1 Interacts with Nop7 and Ytm1
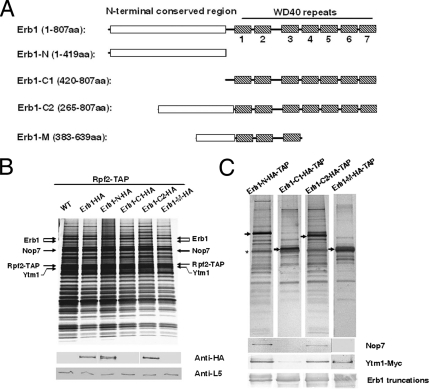
Like Ytm1, Erb1 also contains seven WD40 repeats (Supplementary Figure S4; D. Wilson, personal communication). By binding directly to both Nop7 and Ytm1, Erb1 plays a central role in the formation of the Nop7-subcomplex (Miles et al., 2005). However, which domain of Erb1 (or its mammalian homologue Bop1) is required for these interactions has not been determined. To systematically investigate domains involved in the interaction between Erb1 and other components in the Nop7-subcomplex and domains required for the recruitment of Erb1 into preribosomes, full-length Erb1 and a panel of four different truncated Erb1 constructs (Figure 5A) were expressed from the GAL promoter in cells also expressing endogenous full-length Erb1. Erb1-N contains the N-terminal 419 amino acids of Erb1, which includes a highly conserved region of unknown function (Pestov et al., 2001). Erb1-C1 contains the C-terminal 388 amino acids of Erb1, which includes the seven WD40 motifs. Erb1-C2 contains amino acids 265–807, including half of the N-terminal conserved region and all of the WD40 repeats. Erb1-M, containing amino acids 383–639 (Figure 5A), was identified as a ligand of Ytm1 in a genome-wide two-hybrid screen (data not shown). Western blotting indicated that all four truncated constructs were stably expressed (Supplementary Figure S1B). Indirect immunofluorescence microscopy showed that all four truncated proteins were present in both the cytoplasm and the nucleus. Among them, Erb1-C2 accumulated most strongly in the nucleolus (Supplementary Figure S2B).
Figure 5.
The N-terminal conserved region of Erb1 interacts with both Nop7 and Ytm1 and assembles into preribosomes. (A) Schematic representation of full-length and truncated Erb1 constructs. Shown are the N-terminal conserved region of Erb1 (white rectangle) and the WD40 repeats 1–7 (hatched boxes). (B) Lysates were prepared from cells expressing endogenous Erb1 plus HA-tagged full-length Erb1 or each Erb1 truncation or from wild-type cells only expressing endogenous Erb1 (WT), as in Figure 3. Pre-rRNPs were purified using TAP-tagged Rpf2. Proteins present in the affinity-purified preribosomes were resolved by SDS-PAGE and stained with silver (top) or subjected to Western blot analysis (bottom) using antibodies against the HA tag, or ribosomal protein L5. (C) TAP-tagged Erb1-N, Erb1-C1, Erb1-C2, or Erb1-M was expressed using the GAL promoter by the addition of galactose to a final concentration of 1% for 4 h. Proteins associated with each tagged, truncated protein were affinity-purified, resolved by SDS-PAGE, and stained with silver (top) or subjected to Western blot analysis using antibodies against Nop7, the Myc tag on Ytm1, or the HA tag on the Erb1 proteins (bottom). The black arrow (→) in each panel indicates the corresponding TAP-tagged Erb1 truncation.
We first tested which of these Erb1 fragments is present in preribosomal particles. Pre-rRNPs were purified from cells expressing each of the truncated Erb1 constructs, using TAP-tagged Rpf2. Western blotting showed that Erb1-N and Erb1-C2 assemble into preribosomes at levels similar to full-length Erb1, whereas Erb1-C1 and Erb1-M do not (Figure 5B). Thus, amino acids 265–383, which are present in both Erb1-N and Erb1-C2, but not in Erb1-C1 and Erb1-M (Figure 5A), are important for recruitment of Erb1 into pre-rRNPs.
To identify whether Nop7, Ytm1, or any other molecules associate with each domain of Erb1 in vivo, we carried out affinity purification with each TAP-tagged truncated Erb1 protein. Nop7 and Ytm1, as well as small amounts of other preribosomal proteins (Supplementary Figure S3), copurified with Erb1-N-TAP and with Erb1-C2-TAP (Figure 5C). This confirmed our previous findings that both Erb1-N and Erb1-C2 assemble into preribosomes. No Nop7 and very little Ytm1 copurified with Erb1-C1-TAP (Figure 5C). Consistent with the two-hybrid interaction observed between Ytm1 and Erb1-M (data not shown), Ytm1, but not Nop7, copurified with TAP-tagged Erb1-M (Figure 5C). Erb1-M contains only three WD40-repeats (amino acids 420–639), which may not be able to fold into a β-propeller structure and serve as a stable functional interaction domain. Thus we conclude that the N-terminal region of Erb1-M lacking the WD40 domains (amino acids 383–419) most likely interacts with Ytm1. Other protein(s) might contribute to the interaction between each truncated Erb1 and these ligand(s), but this seems unlikely because both Nop7 and Ytm1 bind to Erb1 directly in vitro (Miles et al., 2005). Additionally, interactions between Erb1-N and Ytm1 or Nop7 were confirmed by yeast two-hybrid assays (data not shown).
Therefore, these results indicate the following: 1) amino acids 265–383, which are present in both Erb1-N and Erb1-C2, but not in Erb1-M, are required for interaction with Nop7, and are important for Erb1 to assemble into preribosomes, 2) the central region of Erb1 containing amino acids 383–419 is involved in interactions between Erb1 and Ytm1 (see Figure 9), and 3) interactions with both Nop7 and Ytm1 enable Erb1-N and Erb1-C2 to stably associate with preribosomal particles. Conversely, the inability to bind to Nop7 most likely prevents Erb1-C1 and Erb1-M from assembling into or remaining associated with pre-rRNPs. These results are consistent with the findings that efficient association of Erb1 with preribosomes is dependent upon both Nop7 and Ytm1 (Figure 1B).
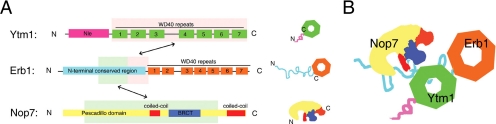
Figure 9.
(A) Interaction domains within the Nop7-subcomplex. Light pink or light green boxes in each protein indicate the smallest region shown to interact with their full-length ligands. (B) Architecture of the Nop7-subcomplex.
Effects of Overexpression of Erb1 Truncations on Growth and Ribosome Biogenesis
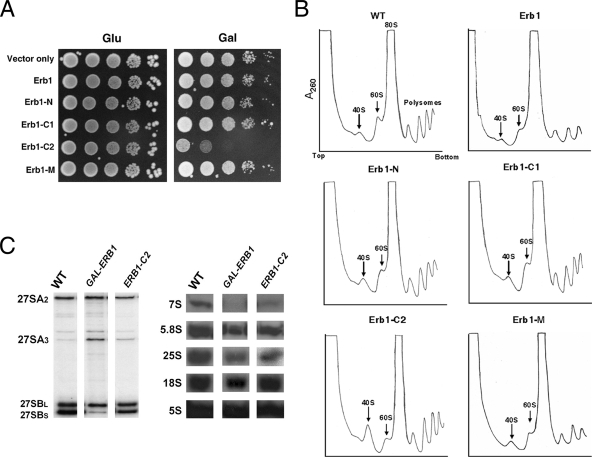
We also investigated effects of overexpression of Erb1 truncations on growth rate and ribosome biogenesis. As predicted, overexpression of Erb1-C1 and Erb1-M, two Erb1 truncations that cannot associate with preribosomes, did not cause any defects in growth (Figure 6A) or ribosome biogenesis (Figure 6B). Similarly, overexpression of full-length Erb1 did not affect cell growth or ribosome assembly (Figure 6, A and B). This result is different from two previous observations, which showed that overexpressing Bop1 in cultured mouse or human cells has mild or strong dominant negative effects on cell proliferation and pre-rRNA processing (Strezoska et al., 2000; Rohrmoser et al., 2007). The differences between these results in mouse and human cells and yeast may indicate subtle structural, functional, and regulatory changes during evolution.
Figure 6.
Expression of Erb1-C2 causes dominant negative effects on growth and ribosome biogenesis. (A) Wild-type cells or yeast expressing endogenous Erb1 plus full-length or truncated Erb1 proteins were grown to early log phase and serially diluted as in Figure 4A. (B) Extracts from wild-type cells (WT) or from cells overexpressing the indicated forms of Erb1 were subjected to centrifugation on sucrose gradients to separate 40S and 60S ribosomal subunits, 80S ribosomes, and polyribosomes. (C) RNA was extracted from wild-type yeast (WT) or from strains in which Erb1 was depleted (GAL-ERB1) or Erb1-C2 was overexpressed (ERB1-C2). Pre-rRNAs were assayed by primer extension (left) or Northern blotting (right), using specific oligonucleotide primers or probes, respectively.
In contrast, overexpression of Erb1-C2 had a dominant negative effect on growth (Figure 6A) and on levels of 60S ribosomal subunits (Figure 6B), indicating a defect in maturation of 66S preribosomes. Primer extension and Northern blotting experiments showed that in the strains overexpressing Erb1-C2, the relative amounts of the 27SA3 pre-rRNA intermediate increased, whereas 27SB and 7S pre-rRNAs, as well as 25S and 5.8S rRNAs, decreased (Figure 6C). These pre-rRNA processing defects are similar to those in the Erb1 depletion strain, but are not as strong (Pestov et al., 2001; Figure 6C). These results with Erb1-C2 suggest that the N-terminal 264 amino acids of Erb1, missing from Erb1-C2, are important for ribosome biogenesis and pre-rRNA processing, or else that Erb1-C2 might function as a “poison subunit” to interfere with assembly of 60S ribosomal subunits. Consistent with these results, overexpression of Bop1Δ, a mammalian Erb1 deletion construct containing the same domains as Erb1-C2, causes dominant negative effects on cell proliferation and 60S ribosomal subunit assembly (Strezoska et al., 2000, 2002).
Surprisingly, although Erb1-N also binds to Nop7 and Ytm1 and assembles into pre-rRNPs, overexpression of Erb1-N does not cause dominant negative effects on growth, 60S ribosomal subunit assembly, or pre-rRNA processing (Figure 6, A–C).
The WD40 Motifs of Erb1 Are Not Essential
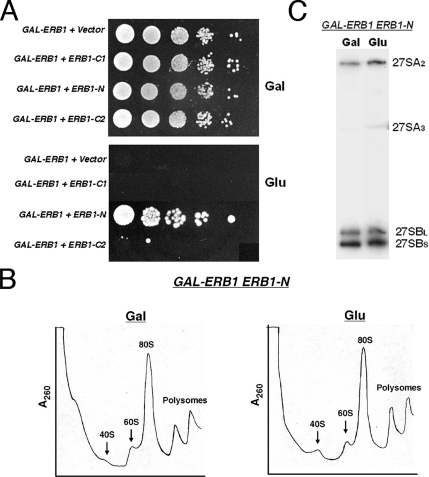
The absence of effects on ribosome biogenesis caused by overexpression of Erb1-N, despite the significant association of this truncated protein with preribosomes, suggests that Erb1-N is sufficient to perform all essential functions of Erb1. To test this hypothesis, Erb1-N was expressed from its own promoter in a strain that also contains a GAL-ERB1gene. Thus, when cells grown in galactose are shifted to glucose, Erb1-N but not full-length Erb1 will be expressed. Consistent with our hypothesis, expression of ERB1-N rescued the lethal phenotype of shutting off GAL-ERB1 (Figure 7A). In contrast, neither Erb1-C1 nor Erb1-C2 could complement the lethality caused by depletion of wild-type Erb1 (Figure 7A), consistent with our observations that Erb1-C1 cannot assemble into preribosomes and that Erb1-C2 can assemble into preribosomal particles, but causes dominant negative effects. Compared with wild-type cells expressing full-length Erb1, cells expressing only the truncated Erb1-N protein showed no obvious defects in 60S ribosomal subunit assembly and polysome formation (Figure 7B). Consistent with this lack of noticeable effects on ribosome production or function, the relative levels of 27SA3 or 27SB pre-rRNAs in cells expressing Erb1-N were only slightly different from those in wild-type cells (Figure 7C). Mild defects in ribosome assembly in vivo are sometimes exacerbated by growing cells under less than ideal conditions, such as high or low temperatures. However, cells expressing only Erb1-N exhibited no growth defects at 37 or 13°C, compared with 30°C (data not shown). Nevertheless, the reproducible mild defects in pre-rRNA processing in cells expressing Erb1-N leave open the possibility that the conserved C-terminal WD40 domain of Erb1 may be involved in some function of Erb1 in ribosome biogenesis.
Figure 7.
The C-terminal WD40 domain of Erb1 is not essential for growth or ribosome assembly. (A) Empty vector or a plasmid containing each ERB1 truncation driven by the ERB1 promoter was introduced into the GAL-ERB1 strain. Cells were grown to early log phase in galactose medium, serially diluted (10-, 100-, 1000-, and 10,000-fold) onto Glu (glucose) medium, to turn off the wild-type GAL-ERB1 gene, or Gal (galactose) medium, to express GAL-ERB1, and incubated at 30°C. (B) Erb1-N was expressed from its own promoter in a GAL-ERB1 strain. This GAL-ERB1 ERB1-N strain was grown at 30°C in galactose medium to 3 ×107 cells/ml. A second culture was grown in galactose medium and shifted to glucose medium for 16 h to 3 ×107 cells/ml, to deplete wild-type Erb1 protein. Whole cell extracts were subjected to centrifugation on sucrose gradients to separate 40S and 60S ribosomal subunits, 80S ribosomes, and polyribosomes. (C) RNA was extracted from the GAL-ERB1 ERB1-N strain grown in galactose, or shifted to glucose for 16 h. The 27SA2, 27SA3, 27SBL, and 27SBS pre-rRNAs present in each strain were identified by primer extension.
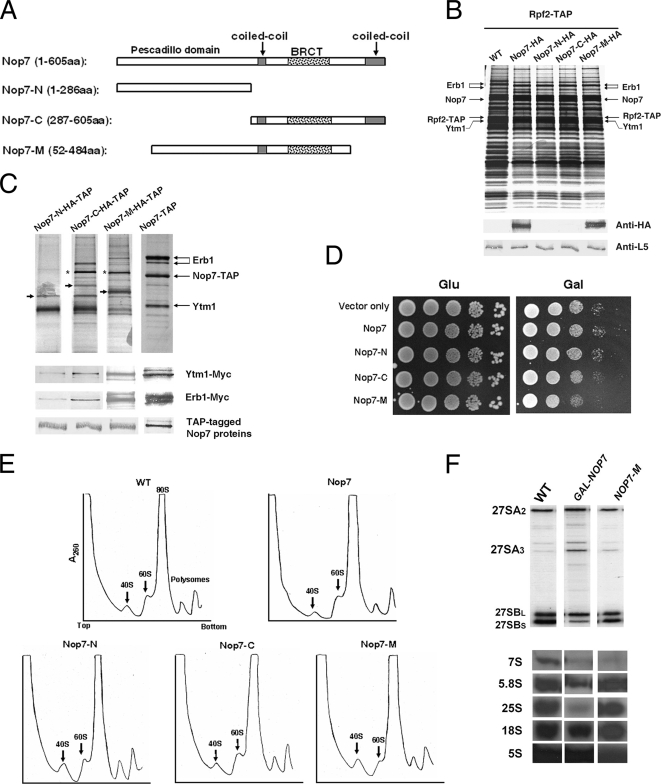
The Central Region of Nop7 (Nop7-M) Binds to Erb1, Assembles into 66S Preribosomal Particles, and Causes Mild Dominant Negative Effects
Previously, we showed that Nop7 interacts directly with Erb1 but not Ytm1 in vitro (Miles et al., 2005). Wild-type Nop7 and a panel of Nop7 truncations were generated to investigate domains involved in interaction between Nop7 and Erb1 and in recruitment of Nop7 into preribosomes (Figure 8A). The N-terminal half of Nop7 (Nop7-N) contains a highly conserved pescadillo-like sequence of unknown function, and the C-terminal half (Nop7-C) contains three predicted protein–protein interaction domains: a BRCT motif (Clapperton et al., 2004) and two coiled-coil domains (Figure 8A). Nop7-M lacks the amino-terminal 51 amino acids and the carboxyl-terminal 120 amino acids of wild-type Nop7, but contains most of the amino-terminal pescadillo-like region, the BRCT domain, and one coiled-coil domain (Figure 8A). HA-tagged full-length Nop7 and these three HA-tagged Nop7 truncations were expressed from the GAL promoter in cells also expressing endogenous full-length Nop7 (Supplementary Figure S1C). To identify the domains of Nop7 required for its recruitment into preribosomes, pre-rRNPs were purified from cells expressing each truncated Nop7 construct, using TAP-tagged Rpf2. Western blot analysis demonstrated that Nop7-M, but not Nop7-N or Nop7-C, was present in purified preribosomal particles at levels similar to full-length Nop7 (Figure 8B). Indirect immunofluorescence microscopy showed that each of the truncated Nop7 proteins was localized to the nucleus (Supplementary Figure S2C). Thus, the absence of Nop7-N and Nop7-C from preribosomes is unlikely to result from their inability to enter the nucleus. When each truncated Nop7 construct was TAP-tagged and used for affinity purification, more Erb1 and Ytm1 copurified with Nop7-M than with Nop7-N or Nop7-C (Figure 8C), indicating domains present only in Nop7-N or Nop7-C are not sufficient for Nop7 to bind Erb1 stably. Although the interaction between Erb1 and each Nop7 truncation could be indirect, we believe that these interactions are direct, because Nop7 and Erb1 bind to each other directly in vitro (Miles et al., 2005). We also observed that overexpression of Nop7-M caused mild defects in growth (Figure 8D) and ribosome biogenesis (Figure 8E). Consistent with these mild defects in cells overexpressing Nop7-M, only small amounts of 27SA3 pre-rRNA accumulated relative to 27SB pre-rRNAs (Figure 8F). In contrast, overexpression of full-length Nop7, Nop7-N, or Nop7-C did not affect growth or ribosome assembly (Figure 8, D and E). Taken together, these results indicate that Nop7 binds to Erb1 via amino acids 52–484 containing multiple potential protein–protein interaction motifs. The mild dominant phenotype of Nop7-M suggests that either the amino- or carboxyl-terminal portion of Nop7, or both, missing from Nop7-M, might be important for ribosome biogenesis.
Figure 8.
Interaction domains of Nop7. (A) Schematic representation of Nop7 and its truncation constructs. Shown are the N-terminal pescadillo domain of Nop7 (white rectangle), BRCT domain (dotted box), and two coil-coiled domains (gray boxes). (B) Pre-rRNPs were purified from cells expressing endogenous Nop7 plus HA-tagged full-length Nop7 or each Nop7 truncation or from wild-type cells only expressing endogenous Nop7 (WT) as in Figure 3. Proteins present in the affinity-purified preribosomes were resolved by SDS-PAGE and stained with silver (top), or subjected to Western blot analysis (bottom) using antibodies against the HA epitope, or ribosomal protein rpL5. (C) TAP-tagged Nop7-N, Nop7-C, Nop7-M, or full-length Nop7 was expressed from the GAL promoter by the addition of galactose to a final concentration of 1% for 4 h. Cell extracts from these strains were prepared and subjected to a high-speed spin (42K) for 3 h. Proteins associated with each tagged truncation were affinity-purified from the supernatants, resolved by SDS-PAGE, and stained with silver (top), or subjected to Western blot analysis using antibodies against the Myc tag on Ytm1 or Erb1, or the protein A tag on Nop7-TAP (bottom). The black arrow in each panel indicates the corresponding TAP-tagged Nop7 truncation. The asterisk (*) indicates Ste23, a protein not necessary for ribosome biogenesis (L. Tang, unpublished data). (D) Wild-type cells or yeast expressing full-length or truncated Nop7 proteins were grown to early log phase and serially diluted as in Figure 4A. (E) Cell extracts were subjected to centrifugation on sucrose gradients to separate 40S and 60S ribosomal subunits, 80S ribosomes, and polyribosomes. (F) RNA was extracted from wild-type yeast (WT) or from strains in which Nop7 was depleted (GAL-NOP7) or Nop7-M was overexpressed (NOP7-M). Pre-rRNAs were assayed by primer extension (top) or Northern blotting (bottom), using specific oligonucleotide primers or probes, respectively.
Previous studies on Pes1, the mammalian homologue of Nop7, are consistent with and support our results. Transposon-based insertion mutagenesis of Pes1 suggested that a stretch of seven amino acids located in the pescadillo-like domain is involved in interaction of Pes1 with Bop1 and that this interaction is important for Pes1 to function in pre-rRNA processing and cell proliferation (Lapik et al., 2004). Systematic deletions of Pes1 and mutation of the BRCT domain in Pes1 indicated that the BRCT domain, a coiled-coil domain, and most of the N-terminal pescadillo-like protein domains are important for incorporation of Pes1 into the PeBoW-complex (Grimm et al., 2006; Hölzel et al., 2006).
DISCUSSION
Rearrangements of pre-rRNP structure and processing of pre-rRNAs during ribosome biogenesis occur in multiple consecutive steps. Each step is likely to require the concerted action of discrete subcomplexes within the large preribosomal particles. Therefore, dissecting the mechanisms of pre-rRNA processing and ribosome assembly could be greatly facilitated by resolving large preribosomal particles into individual, functional subcomplexes and determining the dynamics of interactions within and between subcomplexes. For example, understanding the functions of proteins within the kinetochore (De Wulf et al., 2003; Shang et al., 2003) or the nuclear pore complex (Alber et al., 2007a,b) has benefited greatly from identifying subcomplexes and interactions within or among them.
At least fourteen different subcomplexes have been identified from preribosomal particles in yeast, revealing the modular nature of ribosome assembly (Harnpicharnchai et al., 2001; Du and Stillman, 2002; Granneman et al., 2003; Dosil and Bustelo, 2004; Galani et al., 2004; Krogan et al., 2004; Miles et al., 2005; Karbstein and Doudna, 2006; Rashid et al., 2006; Schäfer et al., 2006; Rosado et al., 2007; Rudra et al., 2007; Zhang et al., 2007). Interactions among components of a subcomplex, such as the Nop7- or PeBoW-subcomplex (Lapik et al., 2004; Miles et al., 2005), the Rpf2-subcomplex (Morita et al., 2002; Miyoshi et al., 2004; Nariai et al., 2005; Zhang et al., 2007), or the Arx1-subcomplex (Lebreton et al., 2006), have been inferred from indirect yeast two-hybrid experiments in vivo, GST pulldown assays in vitro, or by assays for genetic interactions.
Interaction Domains Are Necessary for the Recruitment of Ytm1, Erb1, and Nop7
In this study, we have shown that assembly of Nop7, Erb1, and Ytm1 into preribosomes is interdependent. By purifying TAP-tagged truncated Ytm1, Erb1, and Nop7, and associated molecules and by purifying preribosomes in cells expressing truncated versions of these proteins, we have identified domains in each protein responsible for interactions among these three proteins in vivo and sufficient for their assembly into preribosomes. Examination of dominant and recessive phenotypes upon expression of each truncated protein revealed not only interaction domains, but also which sequences are important or not for other functions of these proteins.
Our results indicate that amino acids 265–419 within the N-terminal conserved region of Erb1 bind to the central region of Nop7 containing most of the N-terminal pescadillo-like protein domain, a coiled-coil domain, and the BRCT domain (Figure 9). This interaction is important for association of both proteins with preribosomal particles. However, it is not clear whether Erb1 and Nop7 form a heterodimeric complex before entering into pre-RNPs. Once in preribosomes, Erb1 recruits Ytm1 by the interaction between amino acids 383 and 419 within the amino-terminal conserved region of Erb1 and the carboxyl-terminal seven WD40 domains of Ytm1 (Figure 9). Assembly of Ytm1 into pre-rRNPs stabilizes association of previously incorporated Nop7 and Erb1. These associations might otherwise be weakened as a result of the structural rearrangements that occur during maturation of preribosomes.
Hölzel et al. (2006) found that mammalian counterparts of these three proteins, Pes1, Bop1, and WDR12, form the PeBoW complex. Our results are consistent with previous findings with Pes1, Bop1, and WDR12 and provide additional mechanisms for their assembly with each other and into preribosomes. Pes1 and Bop1 require each other to localize in the nucleolus and to cosediment with preribosomes (Lapik et al., 2004; Rohrmoser et al., 2007). Coimmunoprecipitation and cosedimentation experiments with mutant Pes1 or Bop1 proteins suggested domains required for their incorporation into the PeBoW complex or into preribosomes (Strezoska et al., 2000, 2002; Lapik et al., 2004; Grimm et al., 2006). Studies on WDR12 inferred that the C-terminal WD40 domain of this protein is important for its nucleolar localization (Hölzel et al., 2005).
Functions of WD40 Motifs in Ytm1 and Erb1
One interesting feature about the Nop7-subcomplex is that it contains two proteins with WD40 motifs. There are 111 WD40 repeat-containing proteins in yeast; 22 are ribosome assembly factors (Saveanu et al., 2007). However, the roles of WD40 domains in ribosome biogenesis have only been explored in a few cases (Miles et al., 2005). Here, our data provide in vivo and in vitro evidence that the carboxyl-terminal WD40-repeats of Ytm1 interact with Erb1 and are essential for Ytm1 to associate with pre-rRNPs. In contrast, the seven conserved WD40 motifs of Erb1 are not necessary for interaction of Erb1 with Nop7 or Ytm1, nor for the assembly of Erb1 into preribosomes, and surprisingly, are not required for ribosome biogenesis or any other essential functions of Erb1. Results with mammalian Bop1, the mammalian homologue of Erb1, are consistent with these findings. Overexpression of the Bop1 N4 truncation, which is homologous to Erb1-N, but extends 80 additional amino acids C-terminal into Bop1, causes no defects in cell cycle or pre-rRNA processing. However, overexpression of Bop1 N2, which is ∼40 amino acids shorter than Erb1-N, does cause defects in cell proliferation and ribosome biogenesis. Together, these results with Erb1 and Bop1 truncations suggest that the amino-terminal conserved region of Erb1 (419aa) or Bop1 (391aa), lacking the WD40 motifs, are largely sufficient for their function. Conserved amino acids 346–419 in Erb1-N or 324–391 in Bop1 N4, which are missing from Bop1 N2, likely are important for ribosome biogenesis but not necessary for assembly of Erb1 or Bop1 into preribosomes. The WD40 motifs of Erb1 may bridge interactions in preribosomal particles that are not essential for 60S subunit assembly or might participate in a different nonessential function of Erb1. Genome-wide two-hybrid assays using Erb1-C1 identified several ER binding proteins as potential ligands of the WD40 domain of Erb1, suggesting a potential “moonlighting” function of the WD40 repeats of Erb1 (data not shown).
Roles for Domains Other Than Interactions within the Nop7-Subcomplex
The dominant negative phenotype caused by Ytm1-C demonstrates the functional importance of the amino-terminal 98 amino acids of Ytm1, containing the Notchless-like element. This sequence was originally identified in the N-terminal part of Notchless, a protein implicated in the modulation of Notch signaling in Drosophila (Royet et al., 1998). Previously, Nal et al. (2002) found a direct interaction between GST-WDR12 and the intracytoplasmic domain of the Notch1 protein in vitro, presumably via the Nle of WDR12. Because there are no known Notch homologues in yeast, the function of the Nle in Ytm1 remains unclear. The Nle of Ytm1 might interact with other preribosomal molecules to recruit them into assembling ribosomes, or to initiate conformational changes in preribosomes essential for their further maturation.
To assemble into and function in preribosomal particles, assembly factors are likely to interact with multiple protein or RNA ligands. Domains not required for interactions among Nop7, Erb1, and Ytm1, such as the WD40 domain of Erb1, or the Nle element of Ytm1, may bind to other assembly factors. Genetic assays could provide valuable information about potential candidates that interact with the Nop7-subcomplex proteins. For example, the nop7-1 mutation is synthetically lethal with specific mutant alleles of DRS1, a putative DEAD box helicase necessary for 60S subunit assembly (Adams et al., 2002), and overexpression of DRS1 suppresses certain nop7 mutants (L. Tang, unpublished data). Overexpression of NOP7 suppresses the thermosensitive growth defects of certain mutations in ribosome assembly factor Nog1 (Honma et al., 2006). More experiments need to be done to discover proteins that physically associate with the Nop7-subcomplex within nascent ribosomes. Moreover, identification of these molecules will enable us to walk out of the Nop7-neighborhood and expand our knowledge of the architecture of preribosomes.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Mike Snyder (Yale University) for yeast strains expressing GST-Erb1 fusion protein, Jeffrey Brodsky (University of Pittsburgh) for anti-Sec61 antibody, Michael McAlear (Wesleyan University) for anti-Ebp2 antibody, Dieter H. Wolf (University of Stuttgart) for anti-Cic1 antibody, and Adam Linstedt (Carnegie Mellon University) for anti-Myc antibody. We thank Jingyu Zhang and Dimitri Pestov for useful discussions and critical evaluation of the manuscript and Marissa Vignali for information about proteins that interact with the Nle element of Ytm1 and the WD40 domain of Erb1. We are grateful to Susan Dowd and Mark Bier for assistance with mass spectrometry. This work was supported by National Institutes of Health Grant GM28301 to J.L.W.
Abbreviations used:
- rRNA
ribosomal RNA
- pre-rRNA
rRNA precursor
- RNP
ribonucleoprotein
- pre-rRNP
preribosomes.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1281) on April 30, 2008.
REFERENCES
- Adams C. C., Jakovljevic J., Roman J., Harnpicharnchai P., Woolford J. L., Jr. Saccharomyces cerevisiae nucleolar protein Nop7 is necessary for biogenesis of 60S ribosomal subunits. RNA. 2002;8:150–165. doi: 10.1017/s1355838202010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F., et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F., et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- Boisvert F. M., van Koningsbruggen S., Navascues J., Lamond A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Clapperton J. A., Manke I. A., Lowery D. M., Ho T., Haire L. F., Yaffe M. B., Smerdon S. J. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat. Struct. Mol. Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A. D., Sorger P. K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosil M., Bustelo X. R. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90S pre-ribosomal particle. J. Biol. Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- Du Y. C., Stillman B. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- Fatica A., Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Galani K., Nissan T. A., Petfalski E., Tollervey D., Hurt E. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J. Biol. Chem. 2004;279:55411–55418. doi: 10.1074/jbc.M406876200. [DOI] [PubMed] [Google Scholar]
- Granneman S., Baserga S. J. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell. Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Granneman S., Gallagher J. E., Vogelzangs J., Horstman W., van Venrooij W. J., Baserga S. J., Pruijn G. J. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60–80S ribonucleoprotein complexes. Nucleic Acids Res. 2003;31:1877–1887. doi: 10.1093/nar/gkg300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm T., Hölzel M., Rohrmoser M., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Eick D. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 2006;34:3030–3040. doi: 10.1093/nar/gkl378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnpicharnchai P., et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- Herskowitz I., et al. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Hölzel M., et al. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 2005;170:367–378. doi: 10.1083/jcb.200501141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzel M., Grimm T., Rohrmoser M., Malamoussi A., Harasim T., Gruber-Eber A., Kremmer E., Eick D. The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Res. 2006;35:789–800. doi: 10.1093/nar/gkl1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma Y., et al. TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients. EMBO J. 2006;25:3832–3842. doi: 10.1038/sj.emboj.7601262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsey E., Jakovljevic J., Miles T. D., Harnpicharnchai P., Woolford J. L., Jr. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 2004;10:813–827. doi: 10.1261/rna.5255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Wu C., Schade B., Thomas D. Y., Whiteway M. Drag & drop cloning in yeast. Gene. 2005;344:43–51. doi: 10.1016/j.gene.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Karbstein K., Doudna J. A. GTP-dependent formation of a ribonucleoprotein subcomplex required for ribosome biogenesis. J. Mol. Biol. 2006;356:432–443. doi: 10.1016/j.jmb.2005.11.052. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Lapik Y. R., Fernandes C. J., Lau L. F., Pestov D. G. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol. Cell. 2004;15:17–29. doi: 10.1016/j.molcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Lebreton A., Saveanu C., Decourty L., Rain J. C., Jacquier A., Fromont-Racine M. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch-Gaggl A., Haque J., Li J., Ning G., Traktman P., Duncan S. A. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. J. Biol. Chem. 2002;277:45347–45355. doi: 10.1074/jbc.M208338200. [DOI] [PubMed] [Google Scholar]
- Leung A. K., Andersen J. S., Mann M., Lamond A. I. Bioinformatic analysis of the nucleolus. Biochem. J. 2003;376:553–569. doi: 10.1042/BJ20031169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Miles T. D., Jakovljevic J., Horsey E. W., Harnpicharnchai P., Tang L., Woolford J. L., Jr. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K., Shirai C., Horigome C., Takenami K., Kawasaki J., Mizuta K. Rrs1p, a ribosomal protein L11-binding protein, is required for nuclear export of the 60S pre-ribosomal subunit in Saccharomyces cerevisiae. FEBS Lett. 2004;565:106–110. doi: 10.1016/j.febslet.2004.03.087. [DOI] [PubMed] [Google Scholar]
- Morita D., Miyoshi K., Matsui Y., Toh-E A., Shinkawa H., Miyakawa T., Mizuta K. Rpf2p, an evolutionarily conserved protein, interacts with ribosomal protein L11 and is essential for the processing of 27SB pre-rRNA to 25S rRNA and the 60S ribosomal subunit assembly in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:28780–28786. doi: 10.1074/jbc.M203399200. [DOI] [PubMed] [Google Scholar]
- Nal B., Mohr E., Silva M. I., Raggett R., Navarro C., Carrel P., Depetris D., Verthuy C., Jordan B. R., Ferrier R. Wdr12, a mouse gene encoding a novel WD-repeat protein with a notchless-like amino-terminal domain. Genomics. 2002;79:77–86. doi: 10.1006/geno.2001.6682. [DOI] [PubMed] [Google Scholar]
- Nariai M., Tanaka T., Okada T., Shirai C., Horigome C., Mizuta K. Synergistic defect in 60S ribosomal subunit assembly caused by a mutation of Rrs1p, a ribosomal protein L11-binding protein, and 3′-extension of 5S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:4553–4562. doi: 10.1093/nar/gki772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M., Leung A., Lamond A., Tollervey D. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA. 2002;8:626–636. doi: 10.1017/s1355838202020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M., Wei K. E., Rogers R., DeGrasse J. A., Chait B. T., Aitchison J. D., Rout M. P. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- Pestov D. G., Stockelman M. G., Strezoska Z., Lau L. F. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 2001;29:3621–3630. doi: 10.1093/nar/29.17.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J., Preston R. A., Adams A. E., Stearns T., Drubin D. G., Haarer B. K., Jones E. W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Rashid R., Liang B., Baker D. L., Youssef O. A., He Y., Phipps K., Terns R. M., Terns M. P., Li H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell. 2006;21:249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Raué H. A. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine. In: Olson M.O.J., editor. The Nucleolus. New York: Kluwer Academic/Plenum Publishers; 2003. pp. 1–24. [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rohrmoser M., Hölzel M., Grimm T., Malamoussi A., Harasim T., Orban M., Pfisterer I., Gruber-Eber A., Kremmer E., Eick D. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol. Cell. Biol. 2007;27:3682–3694. doi: 10.1128/MCB.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado I. V., Dez C., Lebaron S., Caizergues-Ferrer M., Henry Y., de la Cruz J. Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 2007;27:1207–1221. doi: 10.1128/MCB.01523-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royet J., Bouwmeester T., Cohen S. M. Notchless encodes a novel WD40-repeat-containing protein that modulates Notch signaling activity. EMBO J. 1998;17:7351–7360. doi: 10.1093/emboj/17.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D., Mallick J., Zhao Y., Warner J. R. Potential interface between ribosomal protein production and pre-rRNA processing. Mol. Cell. Biol. 2007;27:4815–4824. doi: 10.1128/MCB.02062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C., Rousselle J. C., Lenormand P., Namane A., Jacquier A., Fromont-Racine M. The p21-activated protein kinase inhibitor Skb15 and its budding yeast homologue are 60S ribosome assembly factors. Mol. Cell. Biol. 2007;27:2897–2909. doi: 10.1128/MCB.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- Shang C., Hazbun T. R., Cheeseman I. M., Aranda J., Fields S., Drubin D. G., Barnes G. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Gaitatzes C., Sacena K., Neer E. J. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Strezoska Ž., Pestov D. G., Lau L. F. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 2000;20:5516–5528. doi: 10.1128/mcb.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strezoska Ž., Pestov D. G., Lau L. F. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 2002;277:29617–29625. doi: 10.1074/jbc.M204381200. [DOI] [PubMed] [Google Scholar]
- Zhang J., Harnpicharnchai P., Jakovljevic J., Tang L., Guo Y., Oeffinger M., Rout M. P., Hiley S. L., Hughes T., Woolford J. L., Jr. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.