Abstract
The Inhibitor of Apoptosis proteins (IAPs) are key repressors of apoptosis. Several IAP proteins contain a RING domain that functions as an E3 ubiquitin ligase involved in the ubiquitin-proteasome pathway. Here we investigated the interplay of ubiquitin-proteasome pathway and RING-mediated IAP turnover. We found that the CARD-RING domain of cIAP1 (cIAP1-CR) is capable of down-regulating protein levels of RING-bearing IAPs such as cIAP1, cIAP2, XIAP, and Livin, while sparing NAIP and Survivin, which do not possess a RING domain. To determine whether polyubiquitination was required, we tested the ability of cIAP1-CR to degrade IAPs under conditions that impair ubiquitination modifications. Remarkably, although the ablation of E1 ubiquitin-activating enzyme prevented cIAP1-CR–mediated down-regulation of cIAP1 and cIAP2, there was no impact on degradation of XIAP and Livin. XIAP mutants that were not ubiquitinated in vivo were readily down-regulated by cIAP1-CR. Moreover, XIAP degradation in response to cisplatin and doxorubicin was largely prevented in cIAP1-silenced cells, despite cIAP2 up-regulation. The knockdown of cIAP1 and cIAP2 partially blunted Fas ligand-mediated down-regulation of XIAP and protected cells from cell death. Together, these results show that the E3 ligase RING domain of cIAP1 targets RING-bearing IAPs for proteasomal degradation by ubiquitin-dependent and -independent pathways.
INTRODUCTION
The Inhibitor of Apoptosis (IAP) gene family encodes proteins that repress the progression of apoptosis (Hunter et al., 2007). Among the IAPs, the X-linked inhibitor of apoptosis (XIAP) suppresses cell death by its potent anticaspase property that is attributed to the baculoviral IAP repeat (BIR) domain. BIR3 domain of XIAP functions as an inhibitor to the initiator caspase-9 and the linker preceding BIR2 acts as an inhibitor to effector caspase-3 and -7 (Hunter et al., 2007). In contrast, the antiapoptotic activity of cIAP1 and cIAP2 has been suggested not to be through direct caspase inhibition, but rather has been postulated to be due to their ability to neutralize Smac, a proapoptotic protein that bears an IAP-binding motif (IBM), which allows it to antagonize IAPs (Wilkinson et al., 2004; Eckelman and Salvesen, 2006). Suppression of the apoptotic cascade by the IAPs represents an important survival factor in cells. The significance of this survival advantage is reflected by the frequent overexpression of IAPs as seen in a diversity of cancer cells as well as in cancer tissues compared with normal (reviewed in Hunter et al., 2007).
Several members of the IAP family harbor a carboxy terminus RING domain that functions as an E3 ubiquitin ligase (Yang et al., 2000). In the ubiquitin pathway, the E3 ubiquitin ligase provides the specificity and catalyzes the transfer of ubiquitin moieties to the substrate (Sun, 2006). In thymocytes, apoptotic stimuli such as glucocorticoid and etoposide can activate E3 ubiquitin ligase of XIAP, causing XIAP autoubiquitination and autodegradation (Yang et al., 2000). As such, stable expression of XIAP that lacks the RING domain is more resistant than wild-type XIAP in apoptosis-mediated degradation (Yang et al., 2000). This resistance to XIAP degradation consequently protects the thymocytes from apoptosis (Yang et al., 2000), suggesting that the RING domain has proapoptotic functions. Similarly, the RING domain of cIAP1 also exhibits proapoptotic activity. During Sindbis virus–induced apoptosis in HEK293 cells, cIAP1 is cleaved to produce a carboxy-terminal fragment that contains the CARD and the RING domain (Clem et al., 2001). The ectopic expression of this cIAP1 carboxy fragment can induce apoptosis in BHK, CHO, and HEK293 cells (Clem et al., 2001). The proapoptotic activity of cIAP1-RING is further evidenced when its forced expression sensitizes melanoma cell lines to cisplatin-induced apoptosis (Silke et al., 2005). Therefore, because overexpression of the RING domain of cIAP1, as well as cIAP2, has been found to down-regulate XIAP in a proteasomal-dependent manner, the sensitization to apoptosis by forced cIAP1-RING expression has been suggested to be due to the reduction in XIAP level (Silke et al., 2005). In spite of these previous overexpression and in vitro studies, it remains unclear whether physiological levels of cIAP1 impacts on XIAP abundance.
To gain further insight into RING-mediated IAP turnover, we investigated the role of cIAP1 in regulating IAP levels by a vector-based expression system and siRNA-mediated knockdown. We show that the RING domain of cIAP1 is capable of down-regulating the RING-bearing cIAP1, cIAP2, XIAP, and Livin, but not NAIP and Survivin that do not contain a RING domain. Although the degradation of cIAP1 and cIAP2 are ubiquitination-dependent, unexpectedly, the down-regulation of XIAP and Livin by cIAP1-RING can proceed independently of polyubiquitination. The importance of cIAP1-RING in IAP turnover under pathophysiological condition was highlighted by XIAP resistance to degradation in the absence of cIAP1 in response to the cytotoxic agents cisplatin and doxorubicin. Interestingly, cIAP1 and cIAP2 are nonredundant with respect to XIAP regulation, because endogenous cIAP2 does not appear to play a role in down-regulating XIAP. Together, these results show that cIAP1, through its E3 ligase domain, can regulate the levels of RING-bearing IAPs by targeting them for proteasomal degradation via ubiquitin-dependent and -independent pathways.
MATERIALS AND METHODS
Cell Culture and Transfections
HeLa cervical carcinoma (ATCC, Manassas, VA), TREx-HeLa (Invitrogen, Carlsbad, CA), C2C12 myoblast (ATCC), and SF295 glioblastoma cells (Neurosurgery Tissue Bank, University of California, San Francisco) were maintained at 37°C and 5% CO2 in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (Invitrogen), penicillin, and streptomycin (Invitrogen). The tet-on TREx-HeLa was maintained with the addition of 5 μg/ml blasticidin. Cells were transfected with 3 μg total plasmid/well, unless otherwise indicated, of a six-well plate using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. When multiple plasmids were used for transfections, the total DNA transfected in all experimental comparisons was always normalized by the inclusion of an empty vector plasmid control (pcDNA3) or one expressing LacZ (pTREx-DEST30-LacZ). For the induction of protein expression from pTREx-DEST30 in TREx-HeLa, 1 μg/ml tetracycline was added to the culture media.
Adenovirus, Plasmid DNA Constructs, and Mutagenesis
Adenovirus encoding BCL2 was obtained from Vector Biolabs (Burlingame, CA). pcDNA3-6myc-cIAP1, cIAP, XIAP, Livin, Survivin, and NAIP were previously described (Liston et al., 1996; Arora et al., 2007). The full-length coding sequence of Xenopus E1 (Open Biosystems, Huntsville, AL) was subcloned into pLenti6-directional-TOPO vector (Invitrogen). pCMV-ubiquitin, pCMV-ubiqinitin-K48R, and pCMV-ubiquitin-4K7R were kindly provided by Dr. Z.-X. Jim Xiao (Boston University School of Medicine; Sdek et al., 2005). pCMV-hemagglutinin (HA)-ubiquitin was constructed by inserting sequence that encodes for the HA into pCMV-ubiquitin. Additional ubiquitin mutants were constructed from pCMV-ubiquitin and pCMV-ubiquitin-K48R. XIAP mutants were constructed from pcDNA3-6myc-XIAP. pTREx-cIAP1-CR-H588A was constructed from pTREx-CARD-RING domain of cIAP1 (cIAP1-CR). Other E3 ligase mutants were constructed from their corresponding wild types. Mutagenesis was carried out by using Gene-Tailor Site-Directed Mutagenesis System (Invitrogen). Primer sequences are available upon request. All mutations were confirmed by sequencing.
Transfection of Small Interfering RNA
Annealed double-stranded SMARTpool small interfering RNAs (siRNAs) for E1, ubcH5a, ubcH5b, ubcH5c, ubcH6, siGENOME siRNA for cIAP1 (duplex 10, 5′-AAAGAGAGCCAUUCUGUUCUU), cIAP2 (duplex 2, 5′-UCUAACACAAGAUCAUUGAUU and duplex 9, 5′-AUUCGGUACAGUUCACAUGUU), and nontargeting (NT) luciferase control were purchased from Dharmacon Research (Boulder, CO). Cells were cultured in six-well plates and transfected at 50% confluency with a concentration of 5 nM of each siRNA in using DharmaFECT I Reagent (Dharmacon) according to the manufacturer's protocol. When multiple siRNAs were used for transfections, the total concentration of siRNAs transfected was normalized by the inclusion of the nontargeting control. For E2 experiments in Supplementary Figure S3, plasmids DNA and total 20 nM siRNA were transfected together with LipoFectamine 2000 as described above. In some experiments, cells were exposed to proteasome inhibitor MG132 (Calbiochem, La Jolla, CA), lactacystin (Calbiochem), or ALLN (Sigma, St. Louis, MO).
Induction of Apoptosis
Cisplatin (Sigma), doxorubicin (Sigma), or anti-fas antibody (Upstate Biotechnology, Lake Placid, NY) were used at 20 μM, 10 μM, and 100 ng/ml, respectively. For fas-mediated cell death, cell viability was determined using the WST-1 reagent according to the manufacturer's instructions (Boehringer Mannheim, Laval, QC, Canada).
Protein Preparation and Immunoprecipitation
Cells were collected by centrifugation and lysed in 50 mM Tris-HCl, pH 8.0, containing 1% Triton X-100, 150 mM NaCl, 1 mM NaF, 0.1 mM phenylmethylsulfonyl fluoride, 5 μg/ml pepstatin A, and 10 μg/ml each of leupeptin and aprotinin (lysis buffer), and insoluble cell pellets were collected by centrifugation at 12,000 × g for 30 min at 4°C. The Triton X-100–insoluble pellets were solubilized with sample buffer (62.5 mM, Tris-HCl, pH 6.8, containing 2% SDS, 1% β-mercaptoethanol, and 5% glycerol), and supernatants were collected for protein determination by Bio-Rad Protein Assay (Bio-Rad, Mississauga, ON, Canada) using bovine serum albumin as a standard. For immunoprecipitation, anti-myc antibody–conjugated agarose (Sigma) was used to isolate proteins from Triton X-100 extracts prepared as above. The immunoprecipitates were isolated and separated on SDS-PAGE as previously described (Cheung and Gurd, 2001).
Western Immunoblotting
For immunoblotting, equal amounts of SDS-solubilized samples were separated on polyacrylamide gels and transferred to nitrocellulose as previously described (Cheung and Gurd, 2001). After protein transfer, individual proteins were detected by Western immunoblotting using the following antibodies: E1 (Abcam, Cambridge, MA), FLAG M2 (Sigma), GAPDH (Advanced ImmunoChemical, Long Beach, CA), HA (Sigma), c-myc (Stressgen, San Diego, CA), UbcH5, UbcH6, ubiquitin (Chemicon, Temecula, CA), V5 (Sigma), XIAP (monoclonal, BD Biosciences, San Jose, CA; rabbit polyclonal as described before (Li et al., 2001) and anti-RIAP3 that recognizes XIAP (Aegera Oncology, Montreal, QC, Canada), and vimentin (BD Biosciences). Polyclonal anti-RIAP1 antibodies that recognize both cIAP1 and cIAP2 were generated in the laboratory as described previously (Holcik et al., 2002). Bound primary antibodies were reacted with secondary antibodies conjugated with Alexa Fluor 680 (Molecular Probes) or with IRDye800 (Rockland Biosciences, Gilbertsville, PA), and the infrared fluorescence signals were detected using Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE; Cheung et al., 2006b).
RESULTS
cIAP1-CARD-RING Down-Regulates RING-bearing IAPs via the Proteasome
The ectopic expression of cIAP1-RING has been found to down-regulate XIAP in a proteasomal-dependent manner (Silke et al., 2005). To extend this observation, we first investigated the ability of cIAP1-RING to regulate other IAP protein levels, by exogenously coexpressing the cIAP1-CR with different full-length human IAP proteins. Overexpression of cIAP1-CR induces the down-regulation of cIAP1, cIAP2, XIAP, and Livin, all of which contain a RING domain, but did not affect the steady-state protein expression level of NAIP and Survivin, both of which are devoid of a RING (Figure 1A). In contrast to the wild-type cIAP1-CR, the E3 mutant cIAP1-CR-H588A did not affect the expression level of any of the IAPs (Figure 1, A–E). To discount the possibility that these IAPs translocated to the insoluble fraction by cIAP1-CR, after extracting cells with the Triton X-100 buffer, we solubilized the pellet with 2% SDS. Although XIAP and Livin were found exclusively in the soluble fraction, cIAP1 and cIAP2 are distributed in both the soluble and insoluble fractions (Supplementary Figure S1). However, all the IAPs examined were down-regulated by cIAP1-CR, regardless of their particular pattern of distribution (Supplementary Figure S1). To verify that cIAP1-RING mediated down-regulation of IAPs is dependent on the proteasome, we examined the effects of proteasome inhibitor MG132. We found that cIAP1-CR–mediated down-regulation of RING-bearing IAPs was at least partially blocked by proteasomal inhibition, with XIAP and Livin showing greater resistance than cIAP1 and cIAP2 (Figure 1, B–E). Together, these results indicate that cIAP1-RING can regulate the abundance of RING-bearing IAPs in a proteasomal-dependent manner.
Figure 1.
cIAP1-CARD-RING mediates proteasomal-dependent down-regulation of RING-bearing IAPs. (A) HeLa cells were transfected with 6myc fusion proteins of different IAPs and with cIAP1-CR or cIAP1-CR-H588A. After 24 h of transfection, proteins were extracted, separated by SDS-PAGE, transferred to nitrocellulose, and subjected to immunoblotting with anti-myc for the detection of 6myc-IAP, anti-FLAG for the detection of cIAP1-CR, and anti-GAPDH. (B–E) HeLa cells were transfected with plasmids encoding 6myc-cIAP1 (B), cIAP2 (C), XIAP (D), or Livin (E) along with cIAP1-CR or cIAP1-CR-H588A. After 19 h of transfection, cells were treated with 2 μM proteasomal inhibitor MG132 or vehicle for an additional 5 h. The inclusion of MG132 protected IAPs from cIAP1-CR–mediated degradation.
cIAP1-mediated Down-Regulation of IAPs Is Independent of the E3 Ligase Function of the Target
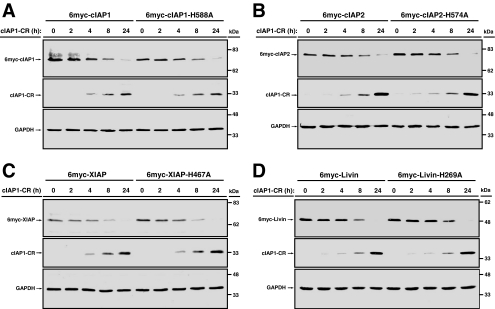
RING-bearing IAPs have been shown to be capable of autoubiquitination (Yang et al., 2000). We next investigated the contribution of transubiquitination by cIAP1-CR compared with autoubiquitination by the individual IAPs in the regulation of their abundance. We mutated the histidine important for E3 ligase activity (Yang et al., 2000) in each IAP and compared the propensity of mutants and wild type toward cIAP1-CR–mediated degradation. Because IAP E3 ligase inactive mutants accumulate in cells, we adjusted the amount of the mutant plasmids to the same expression level as the wild types. We therefore cotransfected pTREx-cIAP1-CR with either 6myc-tagged pcDNA3 constructs of cIAP1 (3 μg), cIAP1-H588A (200 ng), cIAP2 (3 μg), cIAP2-H574A (500 ng), XIAP (4 μg), XIAP-H467A (1 μg), Livin (2 μg), or Livin-H269A (100 ng). All transfections were compensated with a nonexpressing pcDNA3 plasmid by adjusting to the amount used for the wild types. In all four RING-bearing IAPs examined, the status of their E3 ligase was found to be irrelevant to their susceptibility to cIAP1-CR–mediated degradation (Figure 2). These results suggest that transubiquitination alone is sufficient to promote cIAP1-CR–mediated degradation of RING-containing IAPs.
Figure 2.
cIAP1-mediated down-regulation of IAPs is independent of the E3 ligase function of the target. TREx-HeLa cells were transfected with plasmids encoding 6myc-cIAP1 or cIAP1-H588A (A), cIAP2 or cIAP2-H574A (B), XIAP or XIAP-H467A (C), or Livin or Livin-H269A and cIAP1-CR (D). Tetracycline was added to induce the expression of cIAP1-CR for the final 2, 4, 8, and 24 h of a total transfection time of 48 h. Cells were collected at the end of the transfection period and subjected to Western immunoblot analysis.
cIAP1-CR–mediated XIAP and Livin Degradation, But Not cIAP1 and cIAP2, Occurs Independently of E1
The ubiquitin-activating enzyme (E1) catalyzes the activation of ubiquitin carboxy terminus Gly residue in an ATP-dependent step that is a prerequisite for either linkage to the substrate protein's internal Lys residues or to the amine group of the amino terminus residue (Hershko and Ciechanover, 1998; Ciechanover and Ben-Saadon, 2004). We fully anticipated that the E1 enzyme would be an integral part of cIAP1-CR–mediated degradation of IAPs. To test this notion, we used siRNA to knock down E1. After 72 h of exposure to E1 siRNA, both the abundance of E1 protein and the level of endogenous ubiquitination were significantly reduced (Figure 3A). As expected, the ablation of E1 protected cIAP1 and cIAP2 from cIAP1-CR–mediated down-regulation, which was rescued by the expression of exogenous Xenopus E1 (Figure 3, B and C). However, remarkably, under the same E1-negative condition, cIAP1-CR persisted in down-regulating XIAP and Livin (Figure 3, D and E). These results clearly demonstrate that the degradation of XIAP and Livin by cIAP1-CR can occur independently of E1-mediated ubiquitin transfer.
Figure 3.
cIAP1-CARD-RING mediated degradation of XIAP and Livin, but not cIAP1 and cIAP2, occurs independently of E1. (A) HeLa cells were transfected with nontargeting luciferase siRNA (NT) or ubiquitin-activating enzyme-specific siRNA (E1) for the indicated times. Protein extracts were subjected to Western immunoblot analysis with antibodies against ubiquitin, E1, and GAPDH. (B–E) After 80 h of siRNA transfection, HeLa cells were transfected with 6myc-cIAP1 (B), cIAP2 (C), XIAP (D), or Livin (E) in the presence or absence of LacZ, cIAP1-CR, cIAP1-CR-H588A, and Xenopus E1 for an additional 24 h. Protein extracts were collected and subjected to Western immunoblot analysis. 6myc-proteins, cIAP1-CR, and Xenopus E1 were detected with anti-myc, anti-FLAG, and anti-V5 antibodies, respectively.
Mutation of XIAP Ubiquitination Sites Does Not Affect cIAP1-CR–mediated Degradation
The down-regulation of XIAP by cIAP1-CR in the absence of E1 demonstrates that ubiquitin transfer is unnecessary for RING-mediated XIAP turnover. Corollary to this finding, we predict that XIAP mutations that reduce ubiquitination would have no impact on cIAP1-CR–mediated degradation. The ubiquitination sites of XIAP have been identified previously as Lys322 and Lys328 (Shin et al., 2003). To verify that the mutation of these ubiquitination sites did indeed reduce XIAP ubiquitination, we immunoprecipitated the 6myc-XIAP species and probed eluted material with an anti-HA antibody that recognizes HA-tagged ubiquitin. As expected, the reduction in XIAP ubiquitination was more marked in Lys322Arg and Lys322/328Arg mutants than in the Lys328Arg mutant (Figure 4A; Shin et al., 2003). When we mutated these sites to Arg, either individually or dually, cIAP1-CR–mediated XIAP degradation was unaffected, consistent with the notion that the turnover of XIAP is independent of E1 (Figure 4C). The reduction in XIAP ubiquitination also did not affect steady-state expression of the protein (Figure 4, B and C). Furthermore, a Ser87Asp mutant that mimics the Akt-phosphorylated form of XIAP that displays reduced ubiquitination (Dan et al., 2004) also failed to prevent degradation by cIAP1-CR (Supplementary Figure S2). Together, these results indicate that the reduction in XIAP ubiquitination has no effect on XIAP degradation facilitated by cIAP1-CR.
Figure 4.
Mutation of XIAP ubiquitination sites does not affect cIAP1-CR–mediated degradation. (A) HeLa cells were transfected with the various 6myc fusion proteins of XIAP as well as with pCMV-Ub and pCMV-Ub-HA. Extracts were immunoprecipitated with an anti-myc antibody, followed by immunodetection with anti-HA for HA-ubiquitin or anti-XIAP for determining the relative pulldown of XIAP. (B) Relative expression of 6myc-XIAP in inputs was immunodetected with an anti-myc antibody. (C) HeLa cells were transfected with 6myc fusion proteins of XIAP wild type or mutants and with cIAP1-CR or cIAP1-CR-H588A. After 24 h of transfection, proteins were extracted and subjected to immunoblotting with anti-myc for the detection of 6myc-XIAP, anti-FLAG for the detection of cIAP1-CR and anti-GAPDH.
cIAP1-RING–mediated Down-Regulation of XIAP in the Presence of Dominant Negative Ubiquitin
Thus far, we have demonstrated that the ablation of E1, as well as mutation of XIAP ubiquitination sites, does not impede cIAP1-CR–mediated XIAP turnover. The canonical polyubiquitination-dependent degradation of proteins requires the formation of Lys48 linkage on the substrate (Hershko and Ciechanover, 1998). Our findings presume that blockage of the Lys48 linkage would not impact on XIAP degradation by cIAP1-CR. To examine this notion, we used a plasmid that encodes for an ubiquitin that is mutated at Lys48 (Ub-K48R), as well as one that is mutated at all seven available Lys (Ub-4K7R). This latter construct functions in a dominant negative manner by blocking polyubiquitination but still allows for mono-ubiquitination of proteins (Sheaff et al., 2000; Sdek et al., 2005). We coexpressed 6myc-XIAP and cIAP1-CR along with the wild-type or mutated ubiquitins in HeLa cells. Neither Ub-K48R nor Ub-4K7R prevented XIAP degradation by cIAP1-CR (Figure 5A). We next assessed if the lack of polyubiquitination might affect the rate of XIAP turnover by cIAP1-CR. In the tetracycline-responsive TREx-HeLa cells, we coexpressed 6myc-XIAP with wild-type ubiquitin, Ub-3KR (Lys-to-Arg mutation at positions 29, 48, and 63), or Ub-4K7R, and induced cIAP1-CR expression by the addition of tetracycline for the indicated times (Figure 5B). In spite of the forced expression of these ubiquitin mutants, the rate of cIAP1-CR–mediated down-regulation of XIAP remains unaffected (Figure 5B). In contrast, the down-regulation of cIAP1 by cIAP1-CR was blocked by the ubiquitin mutants (Figure 5C), in accord with the dependency of cIAP1 degradation on polyubiquitination. Taken together, these results provide further support that polyubiquitination is unnecessary for cIAP1-CR–mediated XIAP turnover.
Figure 5.
cIAP1-CR mediates XIAP down-regulation in the presence of dominant negative ubiquitin. (A) HeLa cells were transfected with LacZ, cIAP1-CR, or cIAP1-CR-H588A in the presence of WT-Ub, Ub-K48R, or Ub-4K7R. Cells were collected 24 h after transfection and subjected to Western immunoblot analysis. (B and C) TREx-HeLa cells were transfected with 6myc-XIAP (B) or 6myc-cIAP1, cIAP1-CR (C) and in the presence of WT-Ub, Ub-K48R, Ub-3KR, or Ub-4K7R. Tetracycline was added to induce the expression of cIAP1-CR for the final 4, 8, 18, and 24 h of a total transfection time of 48 h. Cells were collected at the end of the transfection period and subjected to Western immunoblot analysis.
The cognate E2 is essential for the ligase activity of E3-containing proteins and both ubcH5 and ubcH6 have been identified as cognate E2s for the IAPs (Yang and Du, 2004; Wu et al., 2005). We next assessed a potential role for these cognate proteins in cIAP1-CR–mediated XIAP degradation. In TREx-HeLa cells in which ubcH5 and ubcH6 were targeted by siRNA, the coexpression of 6myc-XIAP and wild-type ubiquitin showed reduced level of XIAP ubiquitination as compared with cells that were treated with nontargeting siRNA (Supplementary Figure S3). Consistent with our previous results (Figures 3, 4, and 5), this reduction in XIAP ubiquitination did not interfere with cIAP1-CR–mediated degradation of XIAP (Supplementary Figure S3). These results indicate that E2 cognate proteins are dispensable for proteasomal degradation of XIAP.
Degradation of XIAP during Apoptosis is cIAP1-dependent
To examine RING-mediated metabolism of XIAP in a pathophysiological context, we assessed the dependency of apoptosis-mediated XIAP turnover on the proteasome. In response to cisplatin and doxorubicin, both cytotoxic agents that induce apoptosis, XIAP was down-regulated (Figure 6, A and B). This down-regulation of XIAP, however, were prevented by the proteasome inhibitors ALLN and lactacystin (Figure 6, A and B). To test whether cIAP1 could play a role in XIAP turnover during apoptosis, we knocked down cIAP1 by siRNA before exposure to apoptotic stimuli. XIAP level was decreased in response to cisplatin and doxorubicin; however, in the absence of endogenous cIAP1, but not cIAP2, this decrease was largely prevented (Figure 6, C–E). As expected, the down-regulation of cIAP1 resulted in the up-regulation of cIAP2 protein (Figure 6, C and E; Conze et al., 2005). To determine whether this cIAP2 up-regulation could have affected XIAP level, both cIAP1 and cIAP2 were down-regulated with siRNA before cisplatin or doxorubicin treatment. The silencing of the up-regulated cIAP2 has no additional effect on the level of XIAP (Figure 6, C and E), suggesting that cIAP1 alone mediated the regulation of XIAP expression level. These results indicate that XIAP degradation by cytotoxic drugs is dependent on cIAP1.
Figure 6.
Degradation of XIAP during apoptosis is cIAP1-dependent. (A) HeLa cells were treated simultaneously with cisplatin and ALLN, each at 20 μM, for 24 h. (B) After 16 h of 10 μM doxorubicin exposure, HeLa cells were treated with 10 μM lactacystin for an additional 8 h. (C–F) HeLa cells were transfected with the nontargeting luciferase (NT) and cIAP1-specific and cIAP2-specific (duplexes “2” and “9”) siRNA for 48 h, and then transfected cells were exposed to 20 μM cisplatin (C and D) or 10 μM doxorubicin (E and F) for 24 h. (G) HeLa cells were transfected with the nontargeting luciferase (NT), E1-specific or dually with E1- and cIAP1-specific siRNA for 80 h, and then transfected cells were exposed to 20 μM cisplatin for 24 h. Proteins were extracted and subjected to Western immunoblot analysis. cIAP1 and cIAP2 were recognized by an anti-RIAP1 antibody that cross-reacts with both proteins.
We have demonstrated that endogenous cIAP1 is capable of regulating XIAP protein level during apoptosis (Figure 6, C and E), and we have also shown that cIAP1-CR expression can down-regulate XIAP in the absence of E1 (Figure 3). We next investigated if cIAP1-mediated down-regulation of XIAP during apoptosis could occur in the absence of ubiquitin transfer. In HeLa cells, we down-regulated E1 and cIAP1 and exposed these cells to cisplatin as before. We found that in the absence of E1 alone, the degradation of XIAP persisted as compared with the nontargeting siRNA control, whereas in the absence of both E1 and cIAP1, the protein level of XIAP is again protected (Figure 6G). Similar results were seen with mouse myoblast C2C12 cells (Supplementary Figure S4). Of note, the silencing of cIAP1 in C2C12 cells did not up-regulate cIAP2, unlike in HeLa cells (Figure 6). Together, these results demonstrate that XIAP degradation during apoptosis is dependent on cIAP1 without engaging the ubiquitin transfer system.
Loss of cIAP1 Protects against Fas-mediated Cell Death When the Mitochondrial Pathway Is Blocked by BCL2
Next, we assess whether the physiological level of XIAP, as mediated by cIAP1, affects cell viability. Although the knockdown of cIAP1 protects against XIAP loss in response to cisplatin and doxorubicin, they are poor choices for measuring cell viability affected by the physiological level of XIAP in the absence of cIAP1. Both cisplatin and doxorubicin activate the intrinsic pathway that engages the mitochondria for the release of Smac, an antagonist to the IAPs (Hunter et al., 2007). In the absence of cIAP1, a potential Smac sink is lost (Wilkinson et al., 2004), and hence the release of Smac can negate the effects of a sustained level of XIAP. To circumvent this caveat, we opted to activate the extrinsic apoptotic pathway with CD95/Fas. SF295 glioblastoma is a Fas “type I” tumor cell line for which upon fas ligand engagement, a death-inducing signaling complex (DISC) is formed to activate caspase-8 for the initiation of apoptosis (Scaffidi et al., 1998; Algeciras-Schimnich et al., 2003). In addition to DISC formation in response to Fas ligand, “type I” cells also exhibit a loss of mitochondrial transmembrane potential that promotes the release of Smac (Scaffidi et al., 1998; Du et al., 2000). To restrict apoptosis to the DISC arm of the pathway, we blocked the loss of mitochondrial transmembrane potential by inducing BCL2 expression with an adenoviral vector. To ensure that any effect seen in our system is due to changes in XIAP level but not cIAP2 level, we silenced protein expression of cIAP2 in addition to cIAP1. The knockdown of cIAP1 and cIAP2 partially blunted Fas ligand–mediated down-regulation of XIAP (Figure 7A), and protected SF295 cells from cell death (Figure 7B). These results suggest that cIAP1-mediated degradation of XIAP plays a role in the propagation of apoptosis.
Figure 7.
Loss of cIAP1 protects against Fas-mediated cell death when mitochondrial pathway is blocked by BCL2. (A) SF295 cells were transfected with the nontargeting luciferase (NT) or both cIAP1-specific and cIAP2-specific (duplexes 9) siRNA. After 24 h of siRNA transfection, SF295 cells were transduced with an adenovirus that encodes for BCL2. After 24 h of induction, SF295 cells were treated with 0.1 μg/ml fas for an additional 18 h. Proteins were extracted and subjected to Western immunoblot analysis. (B) SF295 cells treated as in A were subjected to WST-1 viability assay. Bars represent the average ± SEM of four experiments. *p < 0.05; significantly different from vehicle-NT and fas-cIAP1/cIAP2, Student's t test.
DISCUSSION
The proteasomal degradation pathway plays a critical role in the regulation of apoptosis, and members of the IAP protein family occupy a key position in coupling these two essential cellular activities (Ni et al., 2005). During the course of apoptosis, cIAP1 is cleaved to produce two carboxy-terminal fragments that both contain the cIAP1-CR (Clem et al., 2001). Ectopic expression of the RING domain of cIAP1 is proapoptotic (Clem et al., 2001); this property has been suggested to be due to the ability of cIAP1-RING to down-regulate XIAP via the proteasome (Silke et al., 2005). Here, we extend these observations by showing that cIAP1-CR is a negative regulator of RING-bearing IAPs (i.e., cIAP1, cIAP2, XIAP, and Livin), targeting them for degradation, but has no impact on NAIP and Survivin levels, IAPs that lack a RING domain. Furthermore, we find that the RING-mediated degradation of cIAP1 and cIAP2 by cIAP1-CR engages the canonical ubiquitin-proteasome system, but that the down-regulation of XIAP and Livin by cIAP1-CR can follow an alternative ubiquitin-independent mechanism. Although proteasomal inhibition protected all RING-bearing IAPs from cIAP1-CR–mediated degradation, this protection appears to be only partial for cIAP1 and cIAP2, suggesting that a proteasome-independent mechanism could also be involved. Additionally, in spite of the similarity between cIAP1 and cIAP2, they are nonredundant with respect to XIAP turnover. Although endogenous cIAP1 targets XIAP for degradation during cisplatin- and doxorubicin-mediated apoptosis, endogenous cIAP2 is not implicated in XIAP turnover. Importantly, the cIAP1-mediated down-regulation of XIAP protein levels is also involved in the progression of apoptosis of the extrinsic pathway.
We have established that cIAP1 is important for the physiological degradation of XIAP in response to cytotoxic agents in vivo. On exposure to the cytotoxic compounds, protein expression levels of cIAP1, cIAP2, and XIAP were reduced. A previous study has shown that the overexpression of a carrier protein (mbw) that is fused with the cIAP1-RING domain can down-regulate XIAP (Silke et al., 2005). Using siRNA to target endogenous cIAP1, we show that XIAP degradation mediated by cIAP1 occurs after treatment with the cytotoxic agents cisplatin and doxorubicin. When basal cIAP1 expression is repressed, these drugs are unable to effectively cause the removal of XIAP. The upstream events that lead to the down-regulation of full-length cIAP1 and cIAP2 in response to the cytotoxic agents remain unclear, and it is potentially the result of reductions in mRNA levels, protein translation, or posttranslation processing. Posttranslational modifications are conceivable, as seen, for example, in the Sindbis virus–induced cleavage of cIAP1 by caspase-3, which releases the RING-containing carboxyl terminal of cIAP1 (Clem et al., 2001) that can facilitate the down-regulation of XIAP. Collectively, these observations support the concept that IAP-IAP interactions (Dohi et al., 2004; Silke et al., 2005; Arora et al., 2007) play roles in regulating their relative abundance to impact upon the progression of apoptosis.
The RING domain of cIAP1 has E3 ubiquitin ligase activity that promotes both auto-ubiquitination and transubiquitination of other interacting molecules such as Smac and TRAF2 (Yang et al., 2000; Hu and Yang, 2003; Samuel et al., 2006). Remarkably, we find that ubiquitination is not a required factor in the cIAP1-CR–mediated degradation of XIAP and Livin. This finding is in marked contrast to cIAP1 and cIAP2, which do require ubiquitination for their turnover, as expected. Notably, we show that cIAP1-CR–mediated down-regulation of XIAP still occurs under various conditions that are clearly adverse for XIAP ubiquitination, including, in the absence of the ubiquitin-activating E1 and the cognate ubiquitin-conjugating E2, in the presence of the dominant negative Ub-4K7R and in the presence of XIAP ubiquitination sites rendered unavailable by mutagenesis. A recent report shows that the lack of XIAP polyubiquitination fails to affect caspase inhibition, suggesting that polyubiquitination is not the sole determinant of XIAP function (Shin et al., 2003). Our findings support this concept by showing that cisplatin or overexpression of cIAP1-CR is able to down-regulate XIAP in E1-null cells, indicating that an alternative mechanism is available for XIAP down-regulation. Notably, E3 ubiquitin protein ligases have been shown to associate with proteasomal subunits (Xie and Varshavsky, 2002). Thus, it is possible that XIAP and Livin, through RING-RING binding to cIAP1-RING (Silke et al., 2005), are able to bypass polyubiquitination and still be recruited to the proteasome for proteolysis.
The targets of IAP-RING domain E3 ligase include non-IAPs that are components of the NF-κB pathways. For instance, in vitro, the E3 ligase of cIAP2 facilitates the K63-polyubiquitination of RIP1 (Wu et al., 2006), a key event in tumor necrosis factor (TNF)-α–induced NF-κB activation (Chen, 2005). In addition, the E3 RING domain of cIAP1 also promotes the ubiquitination of the RING-bearing TRAF2, an adaptor and a signal transducer of the TNF-α–signaling pathway (Samuel et al., 2006). More recently, the E3 ligase of cIAP1 and cIAP2 was shown to repress the noncanonical NF-κB pathway by promoting proteasomal degradation of NIK (Varfolomeev et al., 2007). To dissect the involvement of cIAP1 in IAP versus non-IAP regulation, such as in the NF-κB pathways, further characterization of RING specificity is required. The carboxy-terminal six residues of cIAP1 were found to be essential for the binding and degradation of XIAP (Silke et al., 2005). Given the conservation of the RING domains within the IAPs, and the divergence between IAP-RING and non-IAP RING finger proteins (i.e., c-cbl, TRAF2, and TRAF4; Silke et al., 2005), it is likely that these six residues of cIAP1 might affect the interactions between cIAP1 and all other RING-bearing IAPs, but not non-IAP RING domain proteins. Moreover, the TRAF2-interacting motif of cIAP1 has been mapped to the BIR1 domain (Samuel et al., 2006). The construction of mutants that restrict cIAP1 regulation, specifically toward IAPs or non-IAPs, will facilitate the delineation of various cIAP1 functions, with respect to NF-κB signaling pathway and apoptosis.
The high degree of homology between cIAP1 and cIAP2 suggests some level of functional redundancy (Hunter et al., 2007). Because the absence of cIAP1 leads to cIAP2 up-regulation in many cell types, silencing of cIAP1 is a convenient way to study functional redundancy between the two IAPs in vivo. Unexpectedly, we found that this up-regulation of cIAP2 to be insufficient for apoptosis-induced down-regulation of XIAP (Figure 6, C and E). This finding suggests that cIAP2 expression, at a physiological level, is unable to compensate for the lack of cIAP1 with respect to the RING-mediated down-regulation of XIAP during apoptosis. Even though up-regulated cIAP2 is a poor substitute for cIAP1 in RING-mediated degradation, the protein levels of XIAP in cIAP1 knockout mice remain constant in comparison with the wild type (Conze et al., 2005 and our unpublished observations). However, because elevated XIAP expression creates a favorable condition for tumorigenesis (Hunter et al., 2007), this lack of change in XIAP protein levels might reflect an active process during normal development by which an organism maintains homeostasis either by curtailing expression of XIAP in individual cells, or by eliminating cells that express excessive level of XIAP.
As a major factor in tumorigenesis, XIAP has emerged to be an important target in a number of therapeutic strategies that aim to kill tumor cells (Cheung et al., 2006a). Direct genetic evidence demonstrated that chromosome amplification of the 11q21-q23 region, which encompasses both cIAP1 and cIAP2, is seen in a variety of malignancies (Imoto et al., 2002; Dai et al., 2003). In certain context, cIAP1 can be considered an oncogene as it has been found to transform murine cells and produce hepatomas in vivo in cooperation with the oncogene Yap (Zender et al., 2006). As well, Livin is also emerging as a promising therapeutic target for the treatment of malignancy (Chang and Schimmer, 2007). The metabolism of these IAPs is of interest in the field of apoptosis and proliferative and degenerative diseases. There exists a concept in the literature that implicitly assumes that since the RING domain of the IAPs has ubiquitin E3 ligase activity, ubiquitination is a prerequisite for proteasome-mediated IAP degradation, as exemplified by several recent reviews (Ni et al., 2005; Vaux and Silke, 2005; Dean et al., 2007). Our data strongly imply that the RING domain of cIAP1 regulates all RING-bearing IAPs via proteasomal degradation, by ubiquitin-dependent and ubiquitin-independent pathways.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff and students of the Apoptosis Research Centre for valuable discussions. This work was supported by funds from the Canadian Institutes of Health Research (CIHR) Muscular Dystrophy Association (MDA), and the Howard Hughes Medical Institute (HHMI) to R.G.K. H.H.C. is the recipient of a Michael Cuccione Foundation and CIHR Institute of Cancer Research postdoctoral fellowship. S.P. and D.J.M. are recipients of Natural Sciences and Engineering Research Council of Canada postdoctoral fellowships. R.G.K. is a HHMI International Research Scholar and a Fellow of the Royal Society of Canada.
Abbreviations used:
- BIR
baculovirus IAP repeat
- cIAP1
cellular inhibitor of apoptosis 1
- cIAP2
cellular inhibitor of apoptosis 2
- CR
CARD-RING
- DISC
death-inducing signaling complex
- IAP
inhibitor of apoptosis
- siRNA
small interfering RNA
- Ub
ubiquitin
- XIAP
X-linked IAP.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0107) on April 23, 2008.
REFERENCES
- Algeciras-Schimnich A., Pietras E. M., Barnhart B. C., Legembre P., Vijayan S., Holbeck S. L., Peter M. E. Two CD95 tumor classes with different sensitivities to antitumor drugs. Proc. Natl. Acad. Sci. USA. 2003;100:11445–11450. doi: 10.1073/pnas.2034995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora V., Cheung H. H., Plenchette S., Micali O. C., Liston P., Korneluk R. G. Degradation of survivin by the X-linked inhibitor of apoptosis (XIAP)-XAF1 complex. J. Biol. Chem. 2007;282:26202–26209. doi: 10.1074/jbc.M700776200. [DOI] [PubMed] [Google Scholar]
- Chang H., Schimmer A. D. Livin/melanoma inhibitor of apoptosis protein as a potential therapeutic target for the treatment of malignancy. Mol. Cancer Ther. 2007;6:24–30. doi: 10.1158/1535-7163.MCT-06-0443. [DOI] [PubMed] [Google Scholar]
- Chen Z. J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung H. H., Gurd J. W. Tyrosine phosphorylation of the N-methyl-D-aspartate receptor by exogenous and postsynaptic density-associated Src-family kinases. J. Neurochem. 2001;78:524–534. doi: 10.1046/j.1471-4159.2001.00433.x. [DOI] [PubMed] [Google Scholar]
- Cheung H. H., LaCasse E. C., Korneluk R. G. X-linked inhibitor of apoptosis antagonism: strategies in cancer treatment. Clin. Cancer Res. 2006a;12:3238–3242. doi: 10.1158/1078-0432.CCR-06-0817. [DOI] [PubMed] [Google Scholar]
- Cheung H. H., Lynn Kelly N., Liston P., Korneluk R. G. Involvement of caspase-2 and caspase-9 in endoplasmic reticulum stress-induced apoptosis: a role for the IAPs. Exp. Cell Res. 2006b;312:2347–2357. doi: 10.1016/j.yexcr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Clem R. J., Sheu T. T., Richter B. W., He W. W., Thornberry N. A., Duckett C. S., Hardwick J. M. c-IAP1 is cleaved by caspases to produce a proapoptotic C-terminal fragment. J. Biol. Chem. 2001;276:7602–7608. doi: 10.1074/jbc.M010259200. [DOI] [PubMed] [Google Scholar]
- Conze D. B., Albert L., Ferrick D. A., Goeddel D. V., Yeh W. C., Mak T., Ashwell J. D. Posttranscriptional downregulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 in vivo. Mol. Cell Biol. 2005;25:3348–3356. doi: 10.1128/MCB.25.8.3348-3356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., et al. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum. Mol. Genet. 2003;12:791–801. doi: 10.1093/hmg/ddg083. [DOI] [PubMed] [Google Scholar]
- Dan H. C., Sun M., Kaneko S., Feldman R. I., Nicosia S. V., Wang H. G., Tsang B. K., Cheng J. Q. Akt phosphorylation and stabilization of X-linked inhibitor of apoptosis protein (XIAP) J. Biol. Chem. 2004;279:5405–5412. doi: 10.1074/jbc.M312044200. [DOI] [PubMed] [Google Scholar]
- Dean E. J., Ranson M., Blackhall F., Holt S. V., Dive C. Novel therapeutic targets in lung cancer: Inhibitor of apoptosis proteins from laboratory to clinic. Cancer Treat. Rev. 2007;33:203–212. doi: 10.1016/j.ctrv.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Dohi T., et al. An IAP-IAP complex inhibits apoptosis. J. Biol. Chem. 2004;279:34087–34090. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- Du C., Fang M., Li Y., Li L., Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Eckelman B. P., Salvesen G. S. The human anti-apoptotic proteins cIAP1 and cIAP2 bind but do not inhibit caspases. J. Biol. Chem. 2006;281:3254–3260. doi: 10.1074/jbc.M510863200. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Holcik M., Lefebvre C. A., Hicks K., Korneluk R. G. Cloning and characterization of the rat homologues of the inhibitor of apoptosis protein 1, 2, and 3 genes. BMC Genom. 2002;3:5. doi: 10.1186/1471-2164-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Yang X. Cellular inhibitor of apoptosis 1 and 2 are ubiquitin ligases for the apoptosis inducer Smac/DIABLO. J. Biol. Chem. 2003;278:10055–10060. doi: 10.1074/jbc.M207197200. [DOI] [PubMed] [Google Scholar]
- Hunter A. M., LaCasse E. C., Korneluk R. G. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis. 2007;12:1543–1568. doi: 10.1007/s10495-007-0087-3. [DOI] [PubMed] [Google Scholar]
- Imoto I., Tsuda H., Hirasawa A., Miura M., Sakamoto M., Hirohashi S., Inazawa J. Expression of cIAP1, a target for 11q22 amplification, correlates with resistance of cervical cancers to radiotherapy. Cancer Res. 2002;62:4860–4866. [PubMed] [Google Scholar]
- Li J., et al. Human ovarian cancer and cisplatin resistance: possible role of inhibitor of apoptosis proteins. Endocrinology. 2001;142:370–380. doi: 10.1210/endo.142.1.7897. [DOI] [PubMed] [Google Scholar]
- Liston P., et al. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- Ni T., Li W., Zou F. The ubiquitin ligase ability of IAPs regulates apoptosis. IUBMB Life. 2005;57:779–785. doi: 10.1080/15216540500389013. [DOI] [PubMed] [Google Scholar]
- Samuel T., Welsh K., Lober T., Togo S. H., Zapata J. M., Reed J. C. Distinct BIR domains of cIAP1 mediate binding to and ubiquitination of tumor necrosis factor receptor-associated factor 2 and second mitochondrial activator of caspases. J. Biol. Chem. 2006;281:1080–1090. doi: 10.1074/jbc.M509381200. [DOI] [PubMed] [Google Scholar]
- Scaffidi C., Fulda S., Srinivasan A., Friesen C., Li F., Tomaselli K. J., Debatin K. M., Krammer P. H., Peter M. E. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdek P., Ying H., Chang D. L., Qiu W., Zheng H., Touitou R., Allday M. J., Xiao Z. X. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell. 2005;20:699–708. doi: 10.1016/j.molcel.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Sheaff R. J., Singer J. D., Swanger J., Smitherman M., Roberts J. M., Clurman B. E. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- Shin H., Okada K., Wilkinson J. C., Solomon K. M., Duckett C. S., Reed J. C., Salvesen G. S. Identification of ubiquitination sites on the X-linked inhibitor of apoptosis protein. Biochem. J. 2003;373:965–971. doi: 10.1042/BJ20030583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silke J., Kratina T., Chu D., Ekert P. G., Day C. L., Pakusch M., Huang D. C., Vaux D. L. Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc. Natl. Acad. Sci. USA. 2005;102:16182–16187. doi: 10.1073/pnas.0502828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–654. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev E., et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- Vaux D. L., Silke J. IAPs, RINGs and ubiquitylation. Nat. Rev. Mol. Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- Wilkinson J. C., Wilkinson A. S., Scott F. L., Csomos R. A., Salvesen G. S., Duckett C. S. Neutralization of Smac/Diablo by inhibitors of apoptosis (IAPs). A caspase-independent mechanism for apoptotic inhibition. J. Biol. Chem. 2004;279:51082–51090. doi: 10.1074/jbc.M408655200. [DOI] [PubMed] [Google Scholar]
- Wu C. J., Conze D. B., Li T., Srinivasula S. M., Ashwell J. D. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat. Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- Wu C. J., Conze D. B., Li X., Ying S. X., Hanover J. A., Ashwell J. D. TNF-alpha induced c-IAP1/TRAF2 complex translocation to a Ubc6-containing compartment and TRAF2 ubiquitination. EMBO J. 2005;24:1886–1898. doi: 10.1038/sj.emboj.7600649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Varshavsky A. UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat. Cell Biol. 2002;4:1003–1007. doi: 10.1038/ncb889. [DOI] [PubMed] [Google Scholar]
- Yang Q. H., Du C. Smac/DIABLO selectively reduces the levels of c-IAP1 and c-IAP2 but not that of XIAP and livin in HeLa cells. J. Biol. Chem. 2004;279:16963–16970. doi: 10.1074/jbc.M401253200. [DOI] [PubMed] [Google Scholar]
- Yang Y., Fang S., Jensen J. P., Weissman A. M., Ashwell J. D. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- Zender L., et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.