Abstract
Macroautophagy is a mechanism of degradation of cytoplasmic components in all eukaryotic cells. In macroautophagy, cytoplasmic components are wrapped by double-membrane structures called autophagosomes, whose formation involves unique membrane dynamics, i.e., de novo formation of a double-membrane sac called the isolation membrane and its elongation. However, the precise regulatory mechanism of isolation membrane formation and elongation remains unknown. In this study, we showed that Golgi-resident small GTPase Rab33B (and Rab33A) specifically interacts with Atg16L, an essential factor in isolation membrane formation, in a guanosine triphosphate-dependent manner. Expression of a GTPase-deficient mutant Rab33B (Rab33B-Q92L) induced the lipidation of LC3, which is an essential process in autophagosome formation, even under nutrient-rich conditions, and attenuated macroautophagy, as judged by the degradation of p62/sequestosome 1. In addition, overexpression of the Rab33B binding domain of Atg16L suppressed autophagosome formation. Our findings suggest that Rab33 modulates autophagosome formation through interaction with Atg16L.
INTRODUCTION
Macroautophagy is a well-conserved mechanism in all eukaryotic cells, and it is important for cells to rid the cell of injured or unwanted cytoplasmic material and to degrade normal cellular components in response to energy needs (Levine and Klionsky, 2004). Macroautophagy (simply referred to as autophagy hereafter) is thought to be the major autophagic pathway, and functions not only as a nutrient supply but also as a defense against stresses, including bacterial intrusion, unfolded protein accumulation, and so on (Mizushima, 2007).
In mammalian autophagy, a precursor structure, a crescent-shaped small membrane compartment called isolation membrane core, is present in the initial step in autophagosome formation and it elongates to form characteristic double-membrane structures, called autophagosomes. Finally, autophagosomes fuse with lysosomes to degrade its contents (Yoshimori, 2004).
During the past decade, many genes essential for autophagy (i.e., ATG genes) have been identified by genetic analysis of Saccharomyces cerevisiae (Thumm et al., 1994; Tsukada and Ohsumi, 1993; reviewed in Klionsky et al., 2003), and because most of the yeast ATG genes have mammalian counterparts, the fundamental molecular machinery (or protein–protein interaction cascade) for autophagy is thought to be conserved in all eukaryotic cells (Ohsumi, 2001; Mizushima et al., 2002).
Atg5 interacts with Atg12, a ubiquitin-like protein (Ubl) covalently conjugated to Atg5, and with Atg16L, and these proteins form ∼800-kDa complex through homooligomerization of Atg16L (referred to as Atg5–12/16L complex hereafter). The complex is specifically present on isolation membranes and never present on mature autophagosomes (Mizushima et al., 1998, 2001, 2003). LC3, another Ubl, is conjugated to phosphatidylethanolamine (PE) and localized at elongating isolation membranes and mature autophagosomes (Kabeya et al., 2000). Among mammalian Atg proteins identified thus far, only Atg5–12/16L complex and LC3 are specifically localized on isolation membranes and autophagosomes and thereby thought to function in the regulation of autophagosome formation. Actually, yeast Atg8 (LC3 in mammalian cells) has recently been shown to promote tethering and hemifusion of membranes by in vitro-reconstituted assay (Nakatogawa et al., 2007). However, membranous source of autophagosomes and the molecular mechanism by which the Atg5–12/16L complex (or its individual components) regulates isolation membrane elongation still remain unknown.
In this study, we demonstrated that Rab33B, a member of the Rab small GTPase family that was originally described as a Golgi-resident protein involved in Golgi-to-endoplasmic reticulum (ER) transport (Jiang and Storrie, 2005; Valsdottir et al., 2001; Zheng et al., 1998), directly interacts with Atg16L in a guanosine triphosphate (GTP)-dependent manner and that activation and inactivation of Rab33B modulate autophagy. Based on these results, we discuss the possible role of the interaction between Rab33B and Atg16L in autophagosome formation.
MATERIALS AND METHODS
Isolation and Identification of Glutathione Transferase (GST)-Rab33B–binding Proteins
GST-Rabs were expressed in Escherichia coli JM109 and purified by standard protocols. Glutathione-Sepharose 4B (GE Healthcare, Chalfont St. Giles, United Kingdom) beads coupled with 10 μg of GST-Rab were incubated with a lysate of NIH3T3 cells. Proteins trapped by the beads were analyzed by 10% SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by Coomassie Brilliant Blue (CBB) R-250 staining (Figure 1A). Bands were excised from the gels, and each protein band was in-gel digested with trypsin. The peptides obtained were subjected to matrix-assisted laser desorption ionization (MALDI) mass spectrometry (MS). Proteins were identified by database searching based on their peptide masses.
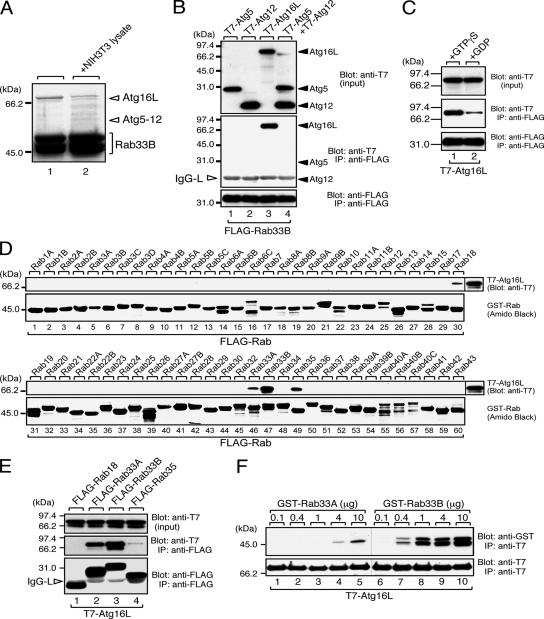
Figure 1.
Atg16L is a specific Rab33-binding protein. (A) Identification of GST-Rab33B-binding proteins as components of Atg5–12/16L complex by MALDI/MS analysis. Beads coupled with GST-Rab33B were incubated with (lane 2) or without (lane 1) NIH3T3 cell lysates, and proteins bound to the beads were analyzed by SDS-PAGE followed by CBB staining. Except for lane 2 in C, all the binding experiments shown were performed in the presence of GTPγS. The positions of Atg16L and Atg12-conjugated Atg5 are indicated on the right (open arrowheads). (B) Specific interaction between Rab33B and Atg16L. Beads coupled with FLAG-Rab33B were incubated with COS-7 cell lysates containing either T7-Atg5 (lane 1), T7-Atg12 (lane 2), T7-Atg16L (lane 3), or T7-Atg12 and T7-Atg5 (lane 4), and the proteins bound to the beads were analyzed by SDS-PAGE followed by immunoblotting with HRP-conjugated anti-T7 tag antibody (Blot: anti-T7, middle) and anti-FLAG tag antibody (Blot: anti-FLAG, bottom). Input means 1/80 volume of the reaction mixture used for immunoprecipitation (input, top). (C) GTP-dependent interaction between Rab33B and Atg16L. Beads coupled with either GTPγS-bound FLAG-Rab33B (lane 1) or GDP-bound FLAG-Rab33B (lane 2) were incubated with COS-7 cell lysates containing T7-Atg16L, and proteins bound to the beads were analyzed by SDS-PAGE followed by immunoblotting with anti-T7 tag antibody (Blot: anti-T7, middle) and HRP-conjugated anti-FLAG tag antibody (Blot: anti-FLAG, bottom). Input means 1/80 volume of the reaction mixture used for immunoprecipitation (input, top). (D) Rab-binding specificity of Atg16L as determined by GST pull-down assay. Beads coupled with each GST-Rab (Rab1∼43) were incubated with COS-7 cell lysates containing T7-Atg5–12/16L complex, and T7-Atg16L bound to the beads was detected by immunoblotting with HRP-conjugated anti-T7 tag antibody (Blot: anti-T7, top). GST-Rabs were then stained with amido black to ensure that equivalent amounts of GST fusion proteins had been loaded (Amido Black, bottom). (E) Rab-binding specificity of Atg16L as determined by coimmunoprecipitation assay in COS-7 cells. Beads coupled with either FLAG-Rab18, -Rab33A, -Rab33B, or -Rab35 were incubated with COS-7 cell lysates containing T7-Atg16L, and bound Atg16L was detected as described in B. The open arrowheads point to the light chain of IgG used for immunoprecipitation. (F) Direct interaction between Rab33A/B and Atg16L. Beads coupled with purified T7-Atg16L were incubated with the indicated amounts of purified GST-Rab33A (lanes 1–5) or GST-Rab33B (lanes 6–10). Proteins bound to the beads were visualized with HRP-conjugated anti-GST antibody (Blot: anti-GST, top) and anti-T7 tag antibody (Blot: anti-T7, bottom). Note that Rab33B directly interacted with Atg16L with much higher affinity than did Rab33A.
Cell Culture, Transfection, and Drug Treatment
NIH3T3 and HeLaS3 cells were maintained in DMEM (Sigma-Aldrich, St. Louis, MO) containing 10% fetal bovine serum and antibiotics under 5% CO2 at 37°C. Starvation was achieved by washing the cells once with HBSS (Sigma-Aldrich) and transferring them to HBSS for 1 or 2 h. Transfection into NIH3T3 cells was performed using FuGENE6 (Roche Diagnostics, Mannheim, Germany) and Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for DNA and Lipofectamine RNAiMAX (Invitrogen) for siRNA according to the manufacturer's instructions. NIH3T3 cells constitutively expressing FLAG-Rab were generated by retroviral infection (Morita et al., 2000). To increase lysosomal pH, cells were treated with 20 μg/ml nigericin (Sigma-Aldrich) for 20 min. To inhibit lysosomal protease, cells were treated with 10 μg/ml E64d and 10 μg/ml pepstatin A (Peptide Institute, Osaka, Japan) for 17 h. Total lysates from cells treated or not treated with the above-mentioned drugs were analyzed by SDS-PAGE followed by immunoblotting with anti-p62/sequestosome 1 (SQSTM1) rabbit polyclonal antibody (BIOMOL Research laboratories, Plymouth Meeting, PA), anti-LC3 rabbit polyclonal antibody (see below), and anti-actin goat polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Establishment of Stable Cell Lines
To obtain stable cell lines, NIH3T3 cells transfected with an empty vector or a vector possessing a short hairpin RNA (shRNA) containing a rab33b-specific sequence (see below) were selected with 800 μg/ml geneticin (Invitrogen). Surviving cell colonies were picked up and cultured in medium containing 800 μg/ml geneticin.
Immunofluorescence Analysis
Anti-GM130, anti-early endosomal antigen (EEA)1, anti-γ-adaptin mouse monoclonal antibodies were purchased from BD Transduction Laboratories (Lexington, KY). Anti-Atg12 (or Apg12) rabbit polyclonal antibody (Zymed Laboratories, South San Francisco, CA), anti-dsRed/monomeric red fluorescent protein (mRFP) rabbit polyclonal antibody (Medical & Biological Laboratories, Nagoya, Japan), anti-tubulin mouse monoclonal antibody (mAb) (clone B5-1-2; Sigma-Aldrich), anti-giantin rabbit polyclonal antibody (CRP, Cambridge BioScience, Cambridge, United Kingdom), anti-Rab33B mouse mAb (D5; Frontier Science, Ishikari, Japan), anti-green fluorescent protein (GFP) rabbit polyclonal antibody (Clontech, Mountain View, CA), and anti-lysosomal-associated membrane protein (Lamp)-1 mouse mAb (1D4b; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) were also purchased commercially. We used anti-Atg12 antibody to detect isolation membranes, because it stained the punctate structures (i.e., isolation membranes) under starvation conditions alone (data not shown). The fluorescent dye-conjugated secondary antibodies (Alexa Fluor 568-labeled anti-mouse or anti-rabbit immunoglobulin Gs [IgGs]) were from Invitrogen. Anti-T7 tag antibody-conjugated agarose and horseradish peroxidase (HRP)-conjugated anti-T7 tag antibody were from Merck Biosciences Novagen (Darmstadt, Germany). Anti-FLAG M2 Affinity Gel and HRP-conjugated anti-FLAG M2 mouse mAb were from Sigma. Anti-Atg16L rabbit polyclonal antibody was generously provided by Dr. Noboru Mizushima (Tokyo Medical and Dental University, Tokyo, Japan). Anti-LC3 and anti-Rab33B rabbit polyclonal antibodies were produced by using GST-LC3 and GST-Rab33B, respectively, as an antigen, and they were affinity-purified as described previously (Fukuda and Mikoshiba, 1999). The specificity of anti-Rab33B antibody was confirmed by immunoblot analysis (Supplemental Figure S1A), but this antibody was not applicable to immunofluorescence staining (data not shown). Immunostaining was performed as described previously (Fukuda and Itoh, 2004), and the cells stained were examined for fluorescence with a confocal fluorescence microscope (Fluoview 500; Olympus, Tokyo, Japan). To quantitatively measure isolation membrane/autophagosome formation, before fixation cells were incubated in HBSS for 1 h. Isolation membranes and autophagosomes were visualized with anti-Atg12 and anti-LC3 antibody, respectively. The images of the cells were captured at random with the confocal microscope, and the number of the fluorescent dots was counted with MetaMorph software (Molecular Devices, Sunnyvale, CA).
Plasmid Construction
The cDNAs encoding the mouse or human Rab proteins and GST-Rab proteins were prepared as described previously (Itoh et al., 2006). The Rab33B cDNA was transferred to the pEF-FLAG tag mammalian expression vector (modified from pEF-BOS) (Fukuda et al., 1994, 1999) or the pEGFP-C1 vector (Clontech). A constitutive active Rab33B-QL (Q92L) mutant and constitutive negative Rab33B-TN (T47N) mutant were produced by two-step polymerase chain reaction (PCR) techniques using the following mutagenic oligonucleotides with an artificial XhoI or ClaI site (underlined) as described previously (Fukuda et al., 1995): 5′-CTCGAGCCCTGCCGTGTCCC-3′ and 5′-CTCGAGCGGTTCAGGAAGAG-3′ for Rab33B-QL and 5′-ATCGATAAGTCAGGCAGTTCTTGCCCAC-3′ and 5′-ATCGATTCTGCGCCGGCCGCTTCC-3′ for Rab33B-TN. The mutant Rab33B fragments were then subcloned into the pEF-FLAG tag vector or pEGFP-C1 vector. The cDNAs encoding Atg5, Atg12, and Atg16Lγ (the longest splicing isoform of Atg16L; simply referred to as Atg16L throughout the text) (Mizushima et al., 2003) were amplified from Marathon-Ready adult mouse brain cDNA (Clontech) by PCR using the following pairs of oligonucleotides with a BamHI or BglII linker (underlined) or with a stop codon (bold) as described previously (Fukuda et al., 1999): 5′-GGATCCATGACAGATGACAAAGAT-3′ and 5′-TCAATCTGTTGGCTGGGGGA-3′ for Atg5; 5′-GGATCCATGTCGGAAGATTCAGAGGT-3′ and 5′-TCATCCCCATGCCTGTGATT-3′ for Atg12; and 5′-GGATCCATGTCGTCGGGCCTGCGCGC-3′ and 5′-TCAAGGCTGTGCCCACAGCA-3′ for Atg16L. These cDNAs were transferred to the pEF-T7 tag mammalian expression vector (Fukuda et al., 1999). Deletion mutants of Atg16L (Figure 3A) were constructed by conventional PCR techniques using the following pairs of oligonucleotides with a BamHI or BglII linker (underlined) or a stop codon (bold): 5′-GGATCCATGTCGTCGGGCCTGCGCGC-3′ and 5′-TCAATCATTCCACGCACCATCATG-3′ for Atg16L-N; 5′-GGATCCAGTCAACTACAAGAAATGGC-3′ and 5′-CTAAGTAGCTGCTCTGCTGAC-3′ for Atg16L-M; and 5′-AGATCTAAGCGACTCTCGCAGCCTGC-3′ and 5′-TCAAGGCTGTGCCCACAGCA-3′ for Atg16L-C; 5′-GGATCCAGTCAACTACAAGAAATGGC-3′ and 5′-TTAGGCCTTCTCAGCCAT-3′ for Atg16L-Mn; 5′-GGATCCCTGGAGACAAACTGCCTG-3′ and 5′-CTAAGTAGCTGCTCTGCTGAC-3′ for Atg16L-Mc. The resultant cDNAs were transferred to the pEF-T7 tag vector, pEF-T7-GST tag vector (Fukuda et al., 2002), or pEGFP-C1 vector. To construct the Rab33B siRNA expression vector, the following oligonucleotides containing a 19-base target site (bold) and a nine-base loop (italics) were annealed and inserted into the BamHI/HindIII site of the pSilencer 2.1-U6 neo (Ambion, Austin, TX) according to the manufacturer's instructions: 5′-GATCCGCGAATTTTGGTGGGAAGTATTCAAGAGATATTTCCCACCAGAATTCGTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAACGAATTCTGGTGGGAAATATCTCTTGAATACTTCCCACCAAAATTCGCG-3′. The knockdown efficiency of the plasmid obtained (referred to as pSilencer-Rab33B) was evaluated by coexpression of pEGFP-C1-Rab33A/B and pSilencer-Rab33B (or a control pSilencer vector) in COS-7 cells (Supplemental Figure S1B). Small interfering RNA (siRNA) against the same site of mouse Rab33B (5′-CGAAUUUUGGUGGGAAGUAdTdT-3′; sense) was also chemically synthesized by B-Bridge International (Mountain View, CA).
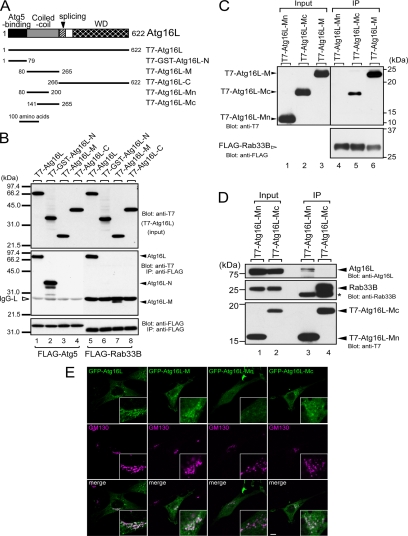
Figure 3.
Rab33B-binding domain of Atg16L. (A) Schematic representation of Atg16L and the truncated mutants used in this study (Mizushima et al., 2003; see text for details). (B) Atg16L interacted with Atg5 via the N-terminal domain (lane 2) and with Rab33B via the coiled-coil domain in the middle of the molecule (lane 7). The binding assays and description of the data are the same as in the legend to Figure 1B. (C) The latter part of Atg16L-M (Atg16L-Mc), not the former part (Atg16L-Mn), interacts with Rab33B. Beads coupled with FLAG-Rab33B were incubated with COS-7 cell lysates containing T7-Atg16L-Mn, T7-Atg16L-Mc, or T7-Atg16L-M. Cell lysates (Input, lanes 1–3) and proteins bound to the beads (IP, lanes 4–6) were analyzed by SDS-PAGE followed by immunoblotting with anti-T7 tag antibody (Blot: anti-T7, top) and HRP-conjugated anti-FLAG tag antibody (Blot: anti-FLAG, bottom). (D) Rab33B-binding site of Atg16L is distinct from the site required for homo-oligomerization. Beads coupled with T7-Atg16L-Mn or T7-Atg16L-Mc were incubated with NIH3T3 cell lysates. Cell lysates (Input, lanes 1 and 2) and proteins bound to the beads (IP; lanes 3 and 4) were analyzed by SDS-PAGE followed by immunoblotting with anti-Atg16L antibody (Blot: anti-Atg16L, top), anti-Rab33B antibody (Blot: anti-Rab33B, middle), and HRP-conjugated anti-T7 antibody (Blot: anti-T7, bottom). The asterisk indicates the light chain of IgG used for immunoprecipitation. (E) Rab33B binding domain of Atg16L is recruited to the Golgi. The NIH3T3 cells transiently expressing GFP-Atg16L, GFP-Atg16L-M, GFP-Atg16L-Mn, or GFP-Atg16L-Mc (green, top) cultured in DMEM were fixed and stained with anti-GM130 antibody (magenta, middle). Merged images are shown in the bottom panels. Bar, 10 μm.
In Vitro Binding Assays
Coimmunoprecipitation in COS-7 cells, GST pull-down, and immunoblotting analyses were performed as described previously (Fukuda et al., 1999; Kuroda and Fukuda, 2005). Unless otherwise stated, binding assays were performed in the presence of 0.5 mM guanosine 5′-O-(3-thio)triphosphate (GTPγS). The blots and gels shown in this article are representative of at least two independent experiments. In vitro guanine nucleotide exchange was performed as follows: NIH3T3 cells were solubilized in a lysis buffer containing 50 mM HEPES-KOH, pH 7.2, 150 mM NaCl, 10% glycerol, 1% Triton x-100, 2.5 mM EDTA, and 0.5 mM GTPγS or 1 mM guanosine diphosphate (GDP). After 1-h incubation on ice, MgCl2 was added to 10 mM.
RESULTS
Identification of Atg16L as a Specific Effector Molecule for Rab33B
Our exhaustive screening for novel Rab effectors by GST pull-down assay by using GST-Rab1∼43 enable us to identify a 68.9-kDa protein and a 59.0-kDa protein that specifically interact with GST-Rab33B from NIH3T3 cell lysates (Figure 1A). MALDI/MS analysis revealed that the 68.9-kDa protein is Atg16L and the 59.0-kDa protein is Atg12 (calculated molecular mass 20.6 kDa)-conjugated Atg5 (calculated molecular mass 32.4 kDa). Because Atg16L, Atg12, and Atg5 have been shown to form a tight complex (Mizushima et al., 2003), we further investigated whether Rab33B interacts with the Atg5–12/16L complex or its individual components by cotransfection assay in COS-7 cells. As shown in Figure 1B (middle, lane 3), T7-tagged Atg16L (T7-Atg16L) was efficiently coimmunoprecipitated with FLAG-Rab33B, whereas T7-Atg5, T7-Atg12, and T7-Atg5+T7-Atg12 were not at all. In addition, Atg16L preferred GTP-bound Rab33B to GDP-bound Rab33B (Figure 1C). To further determine whether Rab33B interacts with Atg16L in the Atg5–12/16L complex, Rab33B was immunoprecipitated from lysates of NIH3T3 cells constitutively expressing FLAG-Rab33B. Consistent with the results of the GST-pull down assay (Figure 1A), endogenous Atg16L and Atg12-conjugated Atg5 were coimmunoprecipitated with FLAG-Rab33B, and the interaction was unaffected by nutrient conditions (Supplemental Figure S2).
Although Atg16L was not screened as a binding partner of other Rabs under the same experimental conditions (data not shown), we tested the interaction between 60 GST-Rabs and Atg16L to confirm the specificity of the Rab33B–Atg16L interaction. Consistent with the results of the above-mentioned GST pull-down assay from NIH3T3 cell lysates, recombinant T7-Atg16L most strongly interacted with GST-Rab33B and weakly interacted with GST-Rab18, GST-Rab33A, and GST-Rab35 (Figure 1D). Because bacterially produced GST-Rabs sometimes bind various molecules nonspecifically, we also investigated these interactions by cotransfection assay in COS-7 cells. As anticipated, T7-Atg16L was hardly coimmunoprecipitated with FLAG-Rab18 or FLAG-Rab35, whereas T7-Atg16L interacted with both Rab33B and Rab33A (Figure 1E). Furthermore, the interaction between Rab33B and Atg16L must be direct, because interaction between them was readily observed even when purified samples (GST-Rab33B and T7-Atg16L) were used for the binding assay (Figure 1F). The Atg16L was found to bind the purified GST-Rab33B with much higher affinity than it bound the purified GST-Rab33A (more than 10-fold higher affinity; Figure 1F). These results suggest that Atg16L (or the Atg5–12/16L complex) functions as a specific Rab33 effector under physiological conditions.
Localization of Rab33B
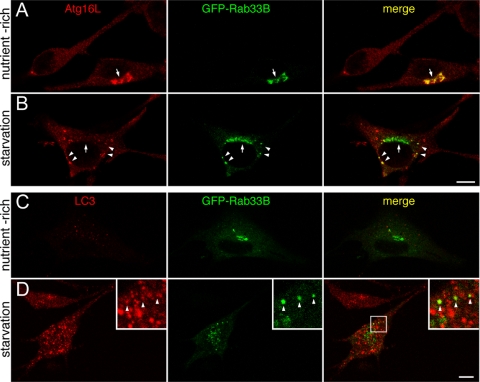
Rab33B was previously shown to be present in the cis-Golgi and to function in Golgi-to-ER retrograde membrane trafficking (Jiang and Storrie, 2005; Valsdottir et al., 2001; Zheng et al., 1998). Consistent with this, Rab33B and giantin (a cis-Golgi marker) were found to be colocalized in human HeLaS3 cells using the mouse monoclonal anti-Rab33B antibody previously published (D5) (Zheng et al., 1998) (Supplemental Figure S3A). Unfortunately, however, this antibody did not work in immunofluorescence analysis of mouse NIH3T3 cells (data not shown), suggesting species-specific recognition by the antibody. On the contrary, an anti-Atg16L serum (Mizushima et al., 2003) provided by Dr. Mizushima stained endogenous Atg16L in NIH3T3 cells (Figure 2, A and B), but it did not in HeLaS3 cells (data not shown). To circumvent this problem, we used GFP (green fluorescent protein)-tagged Rab33B in NIH3T3 cells for subsequent analysis, because GFP-Rab33B was specifically targeted to the Golgi in HeLaS3 cells, NIH3T3 cells, and PC12 cells (Figure 2; data not shown). Under nutrient-rich conditions GFP-Rab33B colocalized with GM130 (a cis-Golgi marker), the same as endogenous Rab33B molecules in HeLaS3 cells, and they did not colocalize with γ-adaptin (a trans-Golgi network marker), EEA1 (an early endosome marker), or Lamp-1 (a lysosome marker) (Supplemental Figure S3, B–E). Although Atg16L is a cytosolic protein under nutrient-rich conditions (Mizushima et al., 2003), endogenous Atg16L was recruited to the Golgi in cells expressing GFP-Rab33B (Figure 2A, arrow). No Golgi-localization of Atg16L was observed in cells not expressing GFP-Rab33B. Rab33A was also able to recruit endogenous Atg16L, but Rab18 or Rab35, which slightly bound Atg16L in GST pull-down assay, was not (Supplemental Figure S4), suggesting that overexpressed GFP-Rab33A or -Rab33B recruits cytosolic Atg16L to the Golgi through a direct interaction with Atg16L. Furthermore, GFP-Rab33B recruited not only Atg16L but also Atg12, which did not bind Rab33B directly, indicating Rab33B is able to recruit Atg5–12/16L complex itself (Supplemental Figure S5; see below). Under starvation conditions punctate Rab33B-positive structures were often observed in the cytoplasm of cells expressing GFP-Rab33B, although the majority of the GFP-Rab33B remained in the Golgi (Figure 2B). Interestingly, these punctate Rab33B-positive structures also contained both endogenous Atg16L and Atg12 (Figure 2B; Supplemental Figure S5) (36.3 ± 3.4% of the GFP-Rab33B–positive dots were also positive for Atg16L, and 12.8 ± 0.02% of the Atg16L-positive dots were also positive for GFP-Rab33B [n > 50 cells]), but, in contrast to the nutrient-rich conditions, no colocalization between GFP-Rab33B and Atg16L was observed in the Golgi. In contrast, very limited colocalization between GFP-Rab33B and LC3, an autophagosome marker (Kabeya et al., 2000), was observed under starvation conditions (Figure 2D) (16.3 ± 1.9% of the GFP-Rab33B–positive dots were also positive for LC3, and 2.9 ± 0.13% of the LC3-positive dots were also positive for GFP-Rab33B [n > 50 cells]), and no colocalization was observed under nutrient-rich conditions (Figure 2C). The preferential colocalization of Rab33B with Atg16L (and Atg12) in NIH3T3 cells is likely to be mediated by the direct interaction between Rab33B and Atg16L, as shown in Figure 1.
Figure 2.
Colocalization of Rab33B with Atg16L or LC3. (A and B) NIH3T3 cells transiently expressing GFP-Rab33B (green) under nutrient-rich conditions (A) or starvation conditions (B) were fixed and stained with the anti-Atg16L antibody (red). The arrows point to GFP-Rab33B in the Golgi, and the arrowheads point to GFP-Rab33B colocalized with Atg16L. Bar, 10 μm. (C and D) NIH3T3 cells transiently expressing GFP-Rab33B (green) under nutrient-rich conditions (C) or starvation conditions (D) were stained with the anti-LC3 antibody (red). Limited colocalization between GFP-Rab33B and LC3 is shown by the arrowheads (insets, magnified view of the boxed area in the right panel). Bar, 10 μm.
Rab33B Binding Domain in Atg16L Is Distinct from the Domain Required for Atg16L Homo-oligomerization
We performed a deletion analysis to identify the minimal Rab33A/B binding domain of Atg16L by using Rab33B (Figure 3, A-C). Because Atg16L consists of an N-terminal Atg5 binding domain, a coiled-coil domain in the middle region, and C-terminal seven WD-repeats (Mizushima et al., 2003), we constructed three truncated mutants of Atg16L: Atg16L-N, which contained the Atg5 binding domain (amino acids 1–79), Atg16L-M, which contained the coiled-coil domain (amino acids 80–265), and Atg16L-C, which contained the WD repeats (Figure 3A). Because the level of expression of T7-Atg16L-N was much lower than that of the other mutants (data not shown), Atg16L-N was expressed as a GST fusion protein (i.e., T7-GST-Atg16L-N, referred to simply as Atg16L-N below). Interaction between FLAG-Rab33B and T7-tagged Atg16L mutants was investigated by coimmunoprecipitation assays in COS-7 cells. As shown in Figure 3B (middle), the Rab33B binding domain was mapped to the coiled-coil domain of Atg16L (lane 7) and not to the N-terminal Atg5 binding domain (lane 6) or the C-terminal WD repeats (lane 8). Under the same conditions, FLAG-Atg5 specifically bound Atg16-N, but it did not bind Atg16-M or Atg16-C (lanes 2–4). Further truncation of Atg16L-M revealed that Rab33B interacts with the latter part of Atg16L-M (amino acids 141–265; Atg16L-Mc), not the former part of Atg16L-M (amino acids 80–200; Atg16L-Mn) (Figure 3, A and C). Although the coiled-coil region of Atg16L has been shown to be responsible for the homo-oligomerization (Mizushima et al., 2003), Atg16L-Mn interacted only with Atg16L (Figure 3D, lane 3) and Atg16L-Mc mainly with Rab33B (lane 4), indicating that homo-oligomerization site and Rab33B binding site in Atg16L-M should be separate. We also found that GFP-Atg16L-M and GFP-Atg16L-Mc were largely localized in the cytosol but concentrated in the Golgi to some extent, whereas GFP-Atg16L-Mn did not show any clear Golgi localization (Figure 3E). Thus, it is highly possible that the Golgi localization of Rab33B binding domain of Atg16L is mediated by direct interaction with endogenous Rab33B molecules in the Golgi.
GTP-Hydrolysis–deficient Mutant of Rab33B Enhanced LC3 Lipidation
To investigate the possible function of Rab33B in autophagy, we introduced the GTP-hydrolysis–deficient, constitutively active mutant of Rab33B (possessing a glutamine-to-leucine substitution at amino acid position 92; Rab33B-QL) into NIH3T3 cells. Rab33B-QL interacted with Atg16L, whereas the nucleotide-free, constitutive negative mutant (possessing a threonine-to-asparagine-substitution at amino acid position 47; Rab33B-TN) did not (Figure 4A). Under nutrient-rich conditions GFP-Rab33B-QL was predominantly localized in the Golgi (marked by GM130 in blue), the same as wild-type GFP-Rab33B, but it was also localized at Atg12/16L-positive punctate structures in the cytoplasm (Figure 4, B and C; Supplemental Figure S5B). Such Atg12- or Atg16L-positive dots were rarely seen in GFP-Rab33B-TN–expressing cells under nutrient-rich conditions (Figure 4D; Supplemental Figure S5B).
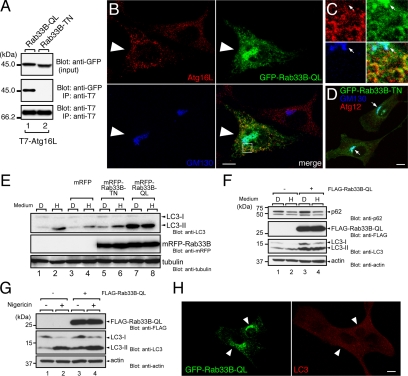
Figure 4.
Effect of the constitutive active mutant of Rab33B on autophagy. (A) The constitutive active mutant Rab33B interacted with Atg16L. Beads coupled with T7-Atg16L were incubated with COS-7 cell lysates containing either GFP-Rab33B-QL + 0.5 mM GTPγS (lane 1) or GFP-Rab33B-TN + 1 mM GDP (lane 2). The lysates (top, input) and proteins bound to the beads (middle, IP) were analyzed by SDS-PAGE followed by immunoblotting with anti-GFP antibody (Blot: anti-GFP, top and middle panels) and HRP-conjugated anti-T7 tag antibody (Blot: anti-T7, bottom). Input means 1/80 volume of the reaction mixture used for immunoprecipitation (input, top). (B) GFP-Rab33B-QL induced Atg16L-positive punctate structures in the cytoplasm. NIH3T3 cells transiently expressing GFP-Rab33B-QL (green, large arrowheads) were stained with the anti-Atg16L antibody (red) and anti-GM130 antibody (blue). Bar, 10 μm. (C) High magnification of the boxed area in B. Colocalization of Rab33B-QL and Atg16L was evident in the punctate structures in the cytoplasm, but not in the Golgi. The arrows point to the position of the Golgi. (D) NIH3T3 cells transiently expressing GFP-Rab33B-TN (green) were stained with the anti-GM130 antibody (blue) and anti-Atg12 antibody (red). In contrast to the GFP-Rab33B-QL in (B), GFP-Rab33B-TN was exclusively localized in the Golgi (arrows). Bar, 10 μm. (E) Induction of LC3-lipidation by mRFP-Rab33B-QL. Cell lysates from HEK293 cells transiently expressing nothing (lanes 1 and 2), mRFP (lanes 3 and 4), mRFP-Rab33B-TN (lanes 5 and 6), or mRFP-Rab33B-QL (lanes 7 and 8) were analyzed by SDS-PAGE followed by immunoblotting with anti-LC3 antibody (Blot: anti-LC3, top panel), anti-mRFP antibody (Blot: anti-mRFP, middle panel), and anti-α-tubulin antibody (Blot: anti-tubulin, bottom panel). (F) Accumulation of p62 in FLAG-Rab33B-QL-expressing cells. Lysates from NIH3T3 cells (lanes 1 and 2) and NIH3T3 cells constitutively expressing FLAG-Rab33B-QL (lanes 3 and 4) under nutrient-rich conditions (lanes 1 and 3, medium “D”) or starvation conditions (lanes 2 and 4, medium “H”) were analyzed by SDS-PAGE followed by immunoblotting with anti-p62 antibody (Blot: anti-p62, top panel), anti-FLAG tag antibody (Blot: anti-FLAG, second panel from the top), anti-LC3 antibody (Blot: anti-LC3, third panel form the top), and anti-actin antibody (Blot: anti-actin, bottom panel). (G) Effect of nigericin. Lysates from NIH3T3 cells (lanes 1 and 2) and NIH3T3 cells constitutively expressing FLAG-Rab33B-QL (lanes 3 and 4) under nutrient-rich conditions in the presence of (lanes 2 and 4) or in the absence of (lanes 1 and 3) 20 μg/ml nigericin were analyzed by SDS-PAGE followed by immunoblotting with anti-FLAG tag antibody (Blot: anti-FLAG, top), anti-LC3 antibody (Blot: anti-LC3, middle), and anti-actin antibody (Blot: anti-actin, bottom). (H) GFP-Rab33B-QL did not induce LC3-positive punctate structures in the cytoplasm under nutrient-rich conditions. NIH3T3 cells transiently expressing GFP-Rab33B-QL (green, arrowheads) were stained with the anti-LC3 antibody (red). Bar, 10 μm.
To evaluate the effect of Rab33B-QL on autophagy, LC3 lipidation (i.e., the amount of LC3-II) was investigated first (Kabeya et al., 2000). Transient expression of mRFP-Rab33B-QL markedly increased the amount of LC3-II regardless of nutrient conditions (Figure 4E, top, lanes 7 and 8), but neither transient expression of mRFP nor mRFP-Rab33B-TN increased it (lanes 3–6). Similarly, constitutive expression of FLAG-Rab33B-QL increased the amount of LC3-II even under nutrient-rich conditions (Figure 4F, compare lanes 1 and 3 in the third panel from the top). To determine whether the observed effect was caused by up-regulation of autophagosome formation or blockage of autophagic degradation (Mizushima and Yoshimori, 2007), accumulation of LC3-II in the presence of H+/K+ ionophore nigericin was investigated. As shown in Figure 4G, the amount of LC3-II additionally accumulated (middle, compare lanes 3 and 4), indicating that the accumulation of LC3-II induced by the expression of Rab33B-QL is unlikely to be attributable to blockage of autophagic degradation. Although immunoblotting showed that the amount of LC3-II was clearly increased in Rab33B-QL–expressing cells even under nutrient-rich conditions (Figure 4E), surprisingly the immunofluorescence analysis did not reveal any typical large LC3 dots in the cytoplasm (Figure 4H), in contrast to the starvation-induced LC3 dots (Figure 2D).
Next, we investigated the effect of Rab33B-QL on the amount of p62/SQSTM1, an LC3- and ubiquitin-binding protein and a target of macroautophagy (Komatsu et al., 2007; Pankiv et al., 2007). Under nutrient-rich conditions more p62/SQSTM1 accumulated in NIH3T3 cells constitutively expressing FLAG-Rab33B-QL than in control NIH3T3 cells (Figure 4F, top, compare lanes 1 and 3), indicating that Rab33B-QL inhibits constitutive autophagy to some extent. Under starvation conditions, however, p62/SQSTM1 was efficiently degraded in both NIH3T3 cells expressing FLAG-Rab33B-QL and control NIH3T3 cells (Figure 4F, top, compare lanes 2 and 4), indicating that Rab33B-QL inhibits basal-level autophagy, but not starvation-induced autophagy. Because expression of other Rab isoforms, including FLAG-Rab18-QL and FLAG-Rab35-QL, did not induce accumulation of p62/SQSTM1 or LC3-II (data not shown), the observed effects are likely to be a specific function of the Rab33 isoform.
Functional Ablation of Rab33B by Dominant-Negative Construct of Atg16L and RNA Interference (RNAi)
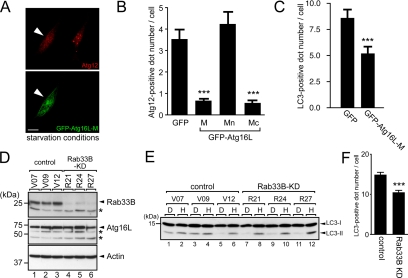
We finally investigated the function of endogenous Rab33B by two independent approaches. First, we used the Rab33B binding domain of Atg16L in an attempt to block the function of Rab33B (and also Rab33A) by interrupting the interaction between endogenous Rab33B and Atg16L. As anticipated, transiently expressed GFP-Atg16L-M and Mc, but not GFP-Atg16-Mn, significantly decreased the number of Atg12-positive dots (Figure 5, A and B) and of LC3-positive dots (Figure 5C). Further truncation study of Atg16L revealed that the Rab33B binding activity and autophagy-inhibiting activity of Atg16L were highly correlated (Supplemental Figure S6). These results suggest that the interaction between Rab33 and Atg16L is important for autophagosome formation.
Figure 5.
Functional ablation of Rab33B by dominant-negative construct of Atg16L and RNAi. (A) Expression of Atg16L-M inhibits starvation-induced isolation membrane formation. NIH3T3 cells transiently expressing GFP-Atg16L-M (green, bottom) under starvation conditions were fixed and stained with anti-Atg12 antibody (red, top). The arrowheads indicate NIH3T3 cells expressing GFP-Atg16L-M. Bar, 10 μm. (B) Decreased number of Atg12-positive isolation membranes in cells expressing Atg16L-M and Atg16L-Mc, but not Atg16L-Mn, under starvation conditions. The cells cultured in HBSS for 1 h were fixed and stained with anti-Atg12 antibody. The number of Atg12-positive dots in the cells was counted. Bars represent the means ± SE of representative data. ***p < 0.001 (Mann–Whitney test). Similar results were obtained in two independent experiments. (C) Decreased number of LC3-positive dots in cells expressing Atg16L-M under starvation conditions. The cells cultured in HBSS for 1 h were fixed and stained with anti-LC3 antibody. The number of LC3-positive dots in the cells was counted. Bars represent the means ± SE of representative data. ***p < 0.001 (Mann–Whitney test). Similar results were obtained in two independent experiments. (D) Establishment of the Rab33B KD cell lines. The lysates from Rab33B KD cells (R21, R24, and R27, lanes 4–6) and control cells (V07, V09, and V12, lanes 1–3) were probed with anti-Rab33B (top), anti-Atg16L (middle), and anti-actin antibody (bottom). The asterisks presumably correspond to the nonspecific bands. (E) LC3-lipidation in Rab33B-KD cells under starvation conditions. The lysates from control (lanes 1–6) or Rab33B-KD cells (lanes 7–12) transferred into DMEM (D; lanes 1, 3, 5, 7, 9, and 11) or HBSS (H; lanes 2, 4, 6, 8, 10, and 12) were analyzed by immunoblotting using anti-LC3 antibody. (F) Number of LC3-positive autophagosomes in control and Rab33B-KD cells under starvation conditions. The cells were cultured in HBSS for 1 h, fixed, and stained with anti-LC3 antibody. The numbers of LC3-positive dots in the cells were counted. Bars represent the means ± SE of data from three independent cell lines (control lines V07, V09, and V12 and Rab33B-KD lines R21, R24, and R27). ***p < 0.001 (Mann-Whitney test).
Second, we attempted to knockdown endogenous Rab33 molecules by RNA interference technology. Because we previously checked that Rab33A protein was undetectable in NIH3T3 cells by using our anti-Rab33A–specific antibody (Tsuboi and Fukuda, 2006; Supplemental Figure S7), we first knocked down endogenous Rab33B molecules by transient transfection of specific siRNA against Rab33B. However, the siRNA treatment did not yield complete knockdown of Rab33B, and we did not observe any effect on autophagosome formation (Supplemental Figure S8). Next, we established Rab33B-knockdown (KD) NIH3T3 cell lines stably expressing the specific shRNA against Rab33B. All three Rab33B-KD cell lines we established (referred to as R21, R24, and R27) exhibited a significant reduction of Rab33B protein (but still not complete knockdown) in comparison with the control cells (referred to as V07, V09, and V12) (Figure 5D) without affecting the expression level of Atg16L, Atg12-conjugated Atg5, actin, or LC3 (Figure 5, D and E; data not shown). Although the number of autophagosomes monitored by LC3 staining was slightly but significantly decreased in Rab33B-KD cells (Figure 5F), we did not observe any clear difference in LC3 lipidation between the control cells and Rab33B-KD cells (Figure 5E). In addition, no significant difference in accumulation of LC3-II and p62/SQSTM1 between control cells and Rab33B-KD cells was observed even in the presence of nigericin or lysosomal protease inhibitors (data not shown).
DISCUSSION
Although autophagosome formation is a dynamic membrane trafficking event, i.e., forming isolation membrane core, elongating it, and finally enclosing cytosol by a double-membrane sphere called autophagosome (Yoshimori, 2004), the relationship between Atg proteins and membrane trafficking regulators is largely unknown. In the present study, we for the first time discovered that one of the Atg proteins, Atg16L, specifically interacts with Rab-type small GTPases, Rab33A and B, in a GTP-dependent manner (Figure 1) and that expression of the Rab33B binding domain of Atg16L strongly inhibits autophagosome formation (Figure 5). We also found that activation of Rab33B (i.e., expression of GTP-locked Rab33B-QL) induces lipidation of LC3 (i.e., increase of LC3-II) and accumulation of p62/SQSTM1 even in the absence of a starvation signal (Figure 4). These findings suggest that Rab33 acts as a modulator in autophagosome formation through interaction with Atg16L.
The Atg5–12/16L complex is generally believed to be essential for the elongation step of isolation membranes (Mizushima et al., 2001; Mizushima et al., 2003). Rab33 interacts with Atg16L without affecting the integrity of Atg5–12/16L complex, indicating that the Atg5–12/16L complex itself function as an effector of Rab33. Because overexpressed GFP-Rab33A/B recruited the complex to membranes in the Golgi, where the complex is not localized natively, one possible function of Rab33 is to recruit (or anchor) the Atg5–12/16L complex to the surface of membranous structures. In addition, expression of the Rab33B-QL mutant accelerated LC3-lipidation independently of the starvation signal and lysosomal degradation. Our results, together with a recent report that yeast Atg12-conjugated Atg5 promotes conjugation between Atg8 and PE in vitro (Hanada et al., 2007), suggest that Rab33B promotes LC3-PE conjugation to recruit the Atg5–12/16L complex to the Rab33B-localized membrane in mammalian cells.
Rab33B-QL induced accumulation of p62 as well as LC3-II, suggesting that expression of Rab33B-QL inhibits autophagy. Their accumulation cannot be attributable to the blockade of fusion between autophagosomes and lysosomes, because nigericin treatment increased LC3-II in Rab33B-QL–expressing cells. Despite increasing the amount of LC3-II (lipid-conjugated form of LC3; Figure 4, E and F), no typical large LC3-positive dots were observed in Rab33B-QL–expressing cells in the immunofluorescence analysis (Figure 4H), suggesting that the LC3-II induced by Rab33B-QL might be localized diffusely within the cell, in contrast to the exclusive localization of LC3-II at autophagosomes in the control cells. We therefore speculate that Rab33B-QL recruits Atg5–12/16L complex to incorrect membranes and induces ectopic LC3-lipidation, which would cause inhibition of autophagosome formation. Alternatively, GTPase activity of Rab33B may be required to form large mature autophagosomes.
Although the Rab33B binding domain of Atg16L strongly inhibited autophagosome formation (Figure 5, A–C), depletion of Rab33B by stable expression of specific siRNA had little effect on autophagosome formation (Figure 5, E and F). Such discrepancy may be explained by the following notion. Under our experimental conditions, Rab33B knockdown was incomplete (Figure 5D), and residual Rab33B molecules that have not been knocked down (or small amount of Rab33A) may promote autophagosome formation. Interestingly, Hosokawa et al. (2006) has recently shown by Atg5 tetracycline-off cell lines that autophagy normally occurs even in the presence of undetectable level of Atg5 protein by immunoblotting, indicating that complete suppression of Atg5 is needed for inhibition of autophagy. Complete knockout of both Rab33A and Rab33B proteins is necessary to address this issue.
Another possible explanation for the discrepancy is that Rab33 is not essential for autophagosome formation under starvation conditions. Although the Rab33B binding activity and autophagy-inhibiting activity of Atg16L are highly correlated (Supplemental Figure S6), the possibility that Atg16L-Mc inhibits autophagosome formation through interaction with unidentified factor(s) other than Rab33B cannot be ruled out. If that were true, Rab33B may be indirectly involved in autophagosome formation through modulating the interaction between Atg16L and such factor(s). Alternatively, Rab33B may be involved in different types of autophagy other than macroautophagy. It would be interesting to investigate whether Rab33 is involved in other types of autophagosome formation, including exclusion of infectious bacteria, antigen presentation on major histocompatibility complex class II molecules, or clearance of insoluble aggregates in neural cells (reviewed in Mizushima, 2007).
Our findings may provide important clues that will lead to identification of the membrane source of isolation membranes/autophagosomes. The localization of Rab33 and the coiled-coil domain of Atg16L in the Golgi strongly suggests the involvement of the Golgi in autophagosome formation. However, we do not think that the autophagosomes are formed by Golgi-derived vesicles/membranes alone, because some reports actually claim that isolation membranes and autophagosome membranes are composed of membranes derived from rough ER (Dunn, 1990a; Dunn, 1990b; Furuno et al., 1990; Ueno et al., 1991). Consistent with this notion, Rab24, an ER-resident Rab, has been shown to be present in autophagosomes, although its precise function remains unknown (Munafó and Colombo, 2002). Furthermore, it has been reported that one of the Atg proteins, Atg9, a membrane protein with unknown function, shuttled between trans-Golgi network and late endosomes (Young et al., 2006). Therefore, we think that isolation membranes and autophagosome membranes may be made up of membranes from several different sources, including the Golgi. Intensive study is needed to answer this question.
In summary, we have demonstrated that a Golgi-resident small GTPase Rab33 interacts with Atg5–12/16L, an essential complex for autophagosome formation, in a GTP-dependent manner and that expression of the Rab33B binding domain of Atg16L inhibits autophagosome formation. So far as we know, this study is the first study to report a direct link between Atg proteins and Rab proteins (general membrane trafficking proteins conserved in all eukaryotes), and we believe that our finding is the first clue in elucidating the dynamic membrane trafficking that occurs during autophagy, specifically, the cross talk between autophagosome formation and Golgi-derived membrane trafficking.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Noboru Mizushima (Tokyo Medical and Dental University) for kindly donating anti-Atg16L antibody, Dr. Toshio Kitamura (The University of Tokyo) for kindly donating Plat-E cells and retroviral vectors, and Shunsuke Kimura (Osaka University) for valuable discussions. We are grateful to the Research Resources Center of RIKEN Brain Science Institute for DNA sequencing analysis and mass spectrometry. This work was supported in part by Ministry of Education, Culture, Sports, and Technology of Japan (grants 18022048, 18050038, 18057026, 18207015, and 19044003 to M.F. and 19790242 to T.I.), by the Naito Foundation, by the Takeda Science Foundation, by the Gushinkai Foundation, and by the Uehara Memorial Foundation (all to M.F.).
Abbreviations used:
- GFP
green fluorescent protein
- GST
glutathione transferase
- HRP
horseradish peroxidase
- PE
phosphatidylethanolamine
- SQSTM1
sequestosome 1
- Ubl
ubiquitin-like protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1231) on April 30, 2008.
REFERENCES
- Dunn W. A., Jr. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J. Cell Biol. 1990a;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W. A., Jr. Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J. Cell Biol. 1990b;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Aruga J., Niinobe M., Aimoto S., Mikoshiba K. Inositol-1,3,4,5-tetrakisphosphate binding to C2B domain of IP4BP/synaptotagmin II. J. Biol. Chem. 1994;269:29206–29211. [PubMed] [Google Scholar]
- Fukuda M., Itoh T. Slac2-a/melanophilin contains multiple PEST-like sequences that are highly sensitive to proteolysis. J. Biol. Chem. 2004;279:22314–22321. doi: 10.1074/jbc.401791200. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Kanno E., Mikoshiba K. Conserved N-terminal cysteine motif is essential for homo- and heterodimer formation of synaptotagmins III, V, VI, and X. J. Biol. Chem. 1999;274:31421–31427. doi: 10.1074/jbc.274.44.31421. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Kojima T., Aruga J., Niinobe M., Mikoshiba K. Functional diversity of C2 domains of synaptotagmin family: mutational analysis of inositol high polyphosphate binding domain. J. Biol. Chem. 1995;270:26523–26527. doi: 10.1074/jbc.270.44.26523. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Kuroda T. S., Mikoshiba K. Slac2-a/melanophilin, the missing link between Rab27 and myosin Va, Implications of a tripartite protein complex for melanosome transport. J. Biol. Chem. 2002;277:12432–12436. doi: 10.1074/jbc.C200005200. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Mikoshiba K. A novel alternatively spliced variant of synaptotagmin VI lacking a transmembrane domain: implications for distinct functions of the two isoforms. J. Biol. Chem. 1999;274:31428–31434. doi: 10.1074/jbc.274.44.31428. [DOI] [PubMed] [Google Scholar]
- Furuno K., Ishikawa T., Akasaki K., Lee S., Nishimura Y., Tsuji H., Himeno M., Kato K. Immunocytochemical study of the surrounding envelope of autophagic vacuoles in cultured rat hepatocytes. Exp. Cell Res. 1990;189:261–268. doi: 10.1016/0014-4827(90)90245-6. [DOI] [PubMed] [Google Scholar]
- Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohusmi Y. The ATG12-ATG5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Hara Y., Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Itoh T., Satoh M., Kanno E., Fukuda M. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells. 2006;11:1023–1037. doi: 10.1111/j.1365-2443.2006.00997.x. [DOI] [PubMed] [Google Scholar]
- Jiang S., Storrie B. Cisternal rab proteins regulate Golgi apparatus redistribution in response to hypotonic stress. Mol. Biol. Cell. 2005;16:2586–2596. doi: 10.1091/mbc.E04-10-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Komatsu M., et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Kuroda T. S., Fukuda M. Identification and biochemical analysis of Slac2-c/MyRIP as a Rab27-, myosin Va/VIIa-, and actin-binding protein. Methods Enzymol. 2005;403:431–444. doi: 10.1016/S0076-6879(05)03038-7. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion, molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Sugita H., Yoshimori T., Ohsumi Y. A new protein conjugation system in human: the counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Ohsumi Y., Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct. Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Morita S., Kojima T., Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Munafó D. B., Colombo M. I. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 2002;3:472–482. doi: 10.1034/j.1600-0854.2002.30704.x. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D. H. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tsuboi T., Fukuda M. Rab3A and Rab27A cooperatively regulate the docking step of dense-core vesicle exocytosis in PC12 cells. J. Cell Sci. 2006;119:2196–2203. doi: 10.1242/jcs.02962. [DOI] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Ueno T., Muno D., Kominami E. Membrane markers of endoplasmic reticulum preserved in autophagic vacuolar membranes isolated from leupeptin-administered rat liver. J. Biol. Chem. 1991;266:18995–18999. [PubMed] [Google Scholar]
- Valsdottir R., Hashimoto H., Ashman K., Koda T., Storrie B., Nilsson T. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 2001;508:201–209. doi: 10.1016/s0014-5793(01)02993-3. [DOI] [PubMed] [Google Scholar]
- Young A.R.J., Chan E.Y.W., Hu X. W., Köchl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- Zheng J. Y., Koda T., Fujiwara T., Kishi M., Ikehara Y., Kakinuma M. A novel Rab GTPase, Rab33B, is ubiquitously expressed and localized to the medial Golgi cisternae. J. Cell Sci. 1998;111:1061–1069. doi: 10.1242/jcs.111.8.1061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.