Abstract
Promyelocytic leukemia nuclear bodies (PML NBs) have been proposed to be involved in tumor suppression, viral defense, DNA repair, and/or transcriptional regulation. To study the dynamics of PML NBs during mitosis, we developed several U2OS cell lines stably coexpressing PML-enhanced cyan fluorescent protein with other individual marker proteins. Using three-dimensional time-lapse live cell imaging and four-dimensional particle tracking, we quantitatively demonstrated that PML NBs exhibit a high percentage of directed movement when cells progressed from prophase to prometaphase. The timing of this increased dynamic movement occurred just before or upon nuclear entry of cyclin B1, but before nuclear envelope breakdown. Our data suggest that entry into prophase leads to a loss of tethering between regions of chromatin and PML NBs, resulting in their increased dynamics. On exit from mitosis, Sp100 and Fas death domain-associated protein (Daxx) entered the daughter nuclei after a functional nuclear membrane was reformed. However, the recruitment of these proteins to PML NBs was delayed and correlated with the timing of de novo PML NB formation. Together, these results provide insight into the dynamic changes associated with PML NBs during mitosis.
INTRODUCTION
The mammalian cell nucleus is a highly compartmentalized and dynamic organelle. Chromatin is organized into chromosome territories (for review, see Cremer et al., 2006), and the majority of nuclear bodies are distributed within the interchromatin space (for review, see Spector, 2001). One class of these bodies is promyelocytic leukemia nuclear bodies (PML NBs), which have also been referred to as nuclear domain 10 (ND 10), Kremer bodies, or PML oncogenic domains (PODs). The number of PML NBs ranges from 1 to 30 nuclear bodies per mammalian nucleus depending on the cell type, the phase of the cell cycle, or the developmental stage (for review, see Maul et al., 2000; Dellaire and Bazett-Jones, 2004). The disruption of PML NBs was first observed in 98% of acute promyelocytic leukemia (APL) patients. A t(15;17) chromosomal translocation between the PML gene and the retinoic acid receptor α (RARα) gene resulted in the production of a PML-RARα fusion protein (de The et al., 1990; Dyck et al., 1994; Weis et al., 1994). This translocation results in the onset of APL and the disruption of PML NBs, and treatment with all-trans-retinoic acid or arsenic trioxide (As2O3) can reverse PML NB disruption and lead to complete tumor remission by degrading the fusion protein (Soignet et al., 1998; Zhong et al., 2000). Other studies have shown that the structure of PML NBs can be regulated by interferon treatment or infection by various viruses (for review, see Regad and Chelbi-Alix, 2001; Everett and Chelbi-Alix, 2007). Moreover, upon exposure of cells to sublethal environmental stresses, e.g., heat shock or CdCl2 treatment, the structure and composition of PML NBs undergoes reorganization (Plehn-Dujowich et al., 2000; Eskiw et al., 2003). Together, these studies suggest that the structure of PML NBs is sensitive to cellular stresses; however, how the structural integrity plays a role in PML NB function is still largely unknown.
Thus far, >50 proteins known to be involved in transcriptional regulation, apoptosis, premature senescence, DNA replication, DNA repair, or antiviral defense have been reported to be localized to PML NBs, suggesting that these nuclear bodies may play important roles in a variety of cellular processes (for review, see Borden, 2002; Bernardi and Pandolfi, 2007). Nevertheless, how these different roles are integrated together in PML NBs is still unclear. Several proteins, such as PML, Sp100, Daxx (Fas death domain–associated protein), and cyclic AMP response element binding-binding protein (CBP), are ubiquitously present in PML NBs. Among them, the PML protein is the major structural component of PML NBs as all the other resident proteins are dispersed in pml−/− mouse nuclei (Wang et al., 1998; Ishov et al., 1999). A recent model suggests two motifs in the PML protein are crucial for the formation of PML NBs as well as the recruitment of component proteins (Shen et al., 2006). SUMO binding motif and SUMOylated RBCC motif, which consists of one RING domain, two B-boxes, and one coiled-coil domain, are responsible for forming a PML protein network, which consecutively recruits other proteins containing either the SUMO binding motif or the SUMO moiety (for review, see Jensen et al., 2001; Shen et al., 2006). The RBCC motif is conserved within the N terminus of all seven major alternatively spliced PML isoforms. It has been shown that stable expression of a single PML isoform in PML null cells yielded a distinct subnuclear structure, suggesting that each isoform may have a specific function (Condemine et al., 2006).
Many of the PML NB-associated proteins, such as p53 and nuclear DNA helicase II (NDH II), have been shown to dynamically associate with or disassociate from these bodies, and the proteins can be sequestered or activated in PML NBs under certain physiological conditions (for review, see Jensen et al., 2001). In addition, several of the PML NB resident proteins, e.g., PML, Sp100, Daxx, and CBP, have also been shown to exhibit high exchange rates by fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching analyses (Wiesmeijer et al., 2002; Everett and Murray, 2005). The dynamic movements of PML NBs have been previously characterized in interphase baby hamster kidney (BHK) cells (Muratani et al., 2002). PML NBs were categorized into three populations according to their dynamics: stationary (25%), limited localized movement (63%), and rapid movement (12%). The stationary and localized movements, similar to the motions of Cajal bodies (Platani et al., 2002), were shown to be independent of the size of PML NBs. However, the rapid moving PML NBs were usually small and typically represented one to two PML NBs per nucleus. Interestingly, the movement of this group of PML NBs was metabolic energy dependent, but transcription independent (Muratani et al., 2002). These data suggested that PML NBs may carry out part of their function(s) by being actively transported within the nucleoplasm.
The most dramatic changes in the composition of PML NBs occur during mitosis. Biochemical and immunocytochemical studies have shown that many resident components of PML NBs, such as Sp100 and Daxx, leave the bodies, whereas PML protein still accumulated in mitotic PML NBs (Everett et al., 1999b). Analysis of a metaphase cell extract showed that the PML protein lost its SUMOylation and became phosphorylated (Everett et al., 1999b). The mitotic PML NBs are fewer and larger than interphase PML NBs. During early G1 phase, some of these mitotic PML NBs remained outside the newly formed daughter nuclei, resulting in cytoplasmic PML NBs (Everett et al., 1999b). A recent study, using two-dimensional time-lapse imaging, showed that PML NBs transformed into mitotic accumulations of PML protein (MAPPs) and became mobile during mitosis (Dellaire et al., 2006). In addition, FRAP analysis showed that there was very little exchange of PML protein in MAPPs. A subset of MAPPs remained associated with chromosomes, and it was suggested to contribute to the reformation of interphase PML NBs in early G1 nuclei because no new PML protein is synthesized during mitosis (Dellaire et al., 2006). Nevertheless, how PML NBs change their dynamic behavior in early prophase and how they reassemble their component proteins into daughter nuclei in early G1 phase is still unclear.
Here, we quantitatively evaluated the dynamics of PML NBs upon entry into and exit from mitosis, by using four-dimensional (4D) live cell imaging and 4D single particle tracking with high spatial and temporal resolution. By developing several U2OS double or triple stable cell lines coexpressing a relatively low level of PML-enhanced cyan fluorescent protein (ECFP) with various marker proteins, we show that PML NBs exhibit a significantly higher percentage of rapid directed movements when cells progressed from prophase to prometaphase. We also identified the window of PML NB dynamic change to occur after nuclear entry of cyclin B1 but before nuclear membrane breakdown. Our data suggest that the increased dynamics of PML NBs upon entry into prophase may result from a loss of tethering to regions of chromatin or the nuclear scaffold. On exit from mitosis, after a functional nuclear membrane was reformed, Sp100 and Daxx entered daughter nuclei first before their recruitment into de novo formed PML NBs. Thus, we suggest that PML NB formation in early G1 phase is initiated by PML protein. In summary, we have demonstrated that PML NBs undergo changes in their dynamic behavior as cells enter into and exit from mitosis.
MATERIALS AND METHODS
cDNA Constructs
The enhanced yellow fluorescent protein (EYFP)-Sp100 construct has been described previously (Muratani et al., 2002). The PML-ECFP construct containing PML isoform VI and a hemagglutinin (HA) tag at the N terminus was provided by T. Tsukamoto (Himeji Institute of Technology, Hyogo, Japan). Importin-β binding (IBB) domain-HcRed was provided by J. Ellenberg and E. Zanin (European Molecular Biology Laboratory, Heidelberg, Germany). EYFP-cyclin B1 and H2A-mCherry were provided by T. Nakamura (Tokyo Medical and Dental University, Tokyo, Japan). Daxx coding sequence was first amplified from a human adult brain cDNA library by polymerase chain reaction (PCR) to generate KpnI and XhoI sites flanking the sequence and was cloned into pCR2.1-TOPO TA cloning vector (Invitrogen, Carlsbad, CA). After sequencing, a correct clone was further subcloned into a modified pEYFP-C1 vector containing an SV2 promoter.
Cell Culture, Transfection, and Stable Cell Lines
Wild-type U2OS and all of the U2OS derived stable cell lines were grown in DMEM (Invitrogen) supplemented with 50 U/ml penicillin, 50 U/ml streptomycin, and 10% fetal bovine serum (HyClone Laboratories, Logan, UT). Cells were grown at 37°C and 5% CO2.
Electroporation (160 V, 950 μF; Gene pulser II, Bio-Rad, Hercules, CA) was performed using MicroPulser Cuvettes (Bio-Rad) with 0.4-cm gap on trypsinized cells resuspended in 200 μl of growth medium and transferred to cuvettes containing 2 μg of fusion protein plasmid plus 20 μg of sheared salmon sperm DNA. The cells were washed once in 2 ml of growth medium and seeded onto either acid-washed coverslips for immunofluorescence analysis or MatTek glass bottom microwells (MatTek, Ashland, MA) for live cell imaging.
All U2OS cell-derived stable cell lines were generated by the same protocol. U2OS cells were grown in 100-mm Petri dishes to 70% confluence and transfected with plasmids by electroporation. Cells stably expressing the fusion protein(s) were selected with 1 mg/ml G418, and then single cell cloning was performed to further select a clone, which we named Cp89, with >90% of its population expressing the fusion protein. Double stable cell lines expressing PML-ECFP were generated from Cp89 single stable cell line. Cp89 cells were transfected with the corresponding EYFP-tagged fusion plasmid, which contains the same drug resistance gene. Therefore, 2 d after transfection, EYFP-expressing fluorescent cells were sorted using a FACSVantage SE cell sorter with DiVa option (BD Biosciences, San Jose, CA) by using a helium-neon laser at 488 nm. Sorted cells were further selected by single cell cloning to select a clone with >90% of its population expressing the fusion proteins.
Immunoblotting
For whole cell extracts, cells were resuspended in phosphate-buffered saline (PBS) containing protease inhibitors (Complete EDTA-free; Roche, Basel, Switzerland), and then they were lysed in a equal volume of 2× Laemmli sample buffer to 5 × 106 cells/ml. Cells (5 × 104) were used for immunoblotting. Membranes were blocked in Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE) for 2 h at room temperature (RT). The following primary antibodies were incubated with membranes for 1 h at RT: rabbit anti-PML (1:1000; provided by G. Maul, The Wistar Institute, Philadelphia, PA), rabbit anti-Sp100 (rSpAB, 1:1000; provided by H. Will, Hamburg University, Hamburg, Germany), rabbit anti-lamin B (1:5000; provided by Nilab Chaudhary and Gunter Blobel, Rockefeller University, New York), and monoclonal antibody (mAb) anti-grenn fluorescent protein (GFP) (1:1000; Roche). Membranes were washed with Tris-buffered saline (TBS) containing 0.2% Tween 20 for 10 min ×3, and then incubated with secondary antibodies, IR700 anti-rabbit, and IR800 anti-mouse (Rockland Immunochemicals, Gilbertsville, PA), for 1 h at RT. Membranes were then washed with TBS containing 0.2% Tween 20 for 10 min ×3 and then TBS for 10 min × 1. To visualize immunoblots, membranes were scanned by the Odyssey infrared imager (LI-COR Biosciences), and the images were processed by Odyssey software.
Immunofluorescence
Cells were rinsed in PBS once and fixed for 15 min in 2% formaldehyde in PBS, pH 7.4. After being washed in PBS for 10 min ×3, cells were permeablized 5 min in PBS with 0.2% Triton X-100 and 1% normal goat serum. After being washed in PBS plus 1% goat serum for 10 min ×3, the following primary antibodies were added for 1 h at RT: mAb anti-PML (5E10, 1:10; hybridoma supernatant provided by R. van Driel, University of Amsterdam, Amsterdam, The Netherlands) and rabbit anti-Sp100 (rSpGH, 1:500; provided by H. Will, Hamburg University). Cells were rinsed in PBS with 1% normal goat serum for 10 min ×3, and secondary anti-species–specific antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were added for 1 h at RT. Cells were rinsed in PBS for 10 min ×3 and mounted in mounting medium (90% glycerol in 0.2 M Tris-Cl, pH 8.0, with 1 mg/ml p-phenylenediamine). Fixed cells were observed on a DeltaVision RT system (Applied Precision, Issaqua, WA) equipped with a CoolSNAP hq Monochrome camera (Photometrics, Tucson, AZ), and images were collected using SoftWoRx software (Applied Precision).
Live Cell Imaging and Treatments
Cells were seeded onto MatTek glass-bottomed microwells and grown for 2 d before imaging. Chromatin was visualized either by transient transfection of H2A-mCherry or by staining with Hoechst 33342 for 5 min at the final concentration of 0.2 μg/ml. Before imaging began, cells were changed into Leibovitz's L-15 medium (Invitrogen) supplemented with 30% fetal bovine serum before being mounted onto the stage of a DeltaVision RT microscope (Applied Precision) with a PlanApo 60 × 1.40 numerical aperture objective lens (Olympus America, Center Valley, PA). Temperature was maintained at 37°C using an environmental chamber. The images were collected in different channels with 35- to 50-ms exposure time and deconvolved by using SoftWoRx software. To increase interchromatin space in interphase cells, cells were treated either with 40 nM calyculin A (Durante et al., 1998) or with 570 mOsm hyperosmolar medium (normal live cell medium +10% volume of 10× PBS) (Albiez et al., 2006).
Tracking and Image Analysis
The 4D live cell images were processed by three steps: registration, segmentation and tracking. The first step, registration, was performed by using a custom Macro to control the TurboReg plugin of ImageJ freeware. To remove translational and rotational movement of the nucleus, a rigid registration scheme was applied on the two-dimensional maximally projected chromatin images from the three-dimensional (3D) stacks. The resulting transformations were applied to rest of the 3D channels, both PML and chromatin, from the same 3D stack for each time point (http://bigwww.epfl.ch/thevenaz/turboreg/ and http://rsb.info.nih.gov/ij/).
Before tracking, images were segmented to define trackable particles. Eight steps were used for segmentation: 1) background subtraction, 2) normalization, 3) anisotropic diffusion filtering, 4) structure tensor calculation, 5) logarithm filtering, 6) global maximum entropy thresholding, 7) despeckling, and 8, hole filling. All these steps were performed in ImageJ by using standard ImageJ functions (step 1, 2, 5, 7, and 8) or the freeware plugins, including the edge preserving anisotropic diffusion (step 3), the FeatureJ (step 4), and the Multi-thresholder plugin (step 6). An automated custom ImageJ Macro was used to implement the entire workflow.
The 4D single particle tracking was performed by TIKAL (Bacher et al., 2004). All tracks were manually verified and potential ambiguities were removed in TIKAL's interactive 4D viewer. The length of each track and possible fusion events were counted and recorded. The 3D trajectories were exported in ASCII format into MATLAB (Mathworks, Natick, MA) for data fitting and classification of the movement types. The mean square displacement (MSD) was calculated for each track in each phase, and the MSD plot was fit using the anomalous diffusion equation MSD = 6DΔtα. The anomalous diffusion coefficient (α) was estimated from the curve fitting, and was used to classify three types of movement: α = 0.40 to 0.75, constrained motion; α = 0.75 to 1.25, diffusive motion; and α = 1.25–2.00, directed motion. The percentage of tracks exhibiting a specific movement type in an image set was calculated. The distribution of different movement types from all of the image sets was summarized using a box plot.
To find the correct diffusion coefficients, a model-free maximum-likelihood estimator (MLE) was used as derived in Montiel et al., 2006. Time points (break points) at which the change of diffusion coefficient occurs were found as the maximum in their log-likelihood ratio. The data points of trajectories were first increased by linear interpolation, 20-fold for H2A-mCherry data sets (Figure 6) and 40-fold for NEBD (Figure 5) and cyclin B1 (Figure 8) data sets, before implementing the MLE by a recursive algorithm in MATLAB. The resulting break points were used to partition the particle trajectories into subtracks, which were used to calculate the coefficient of anomalous diffusion (α) by fitting the MSD plot of subtracks with the anomalous diffusion equation.
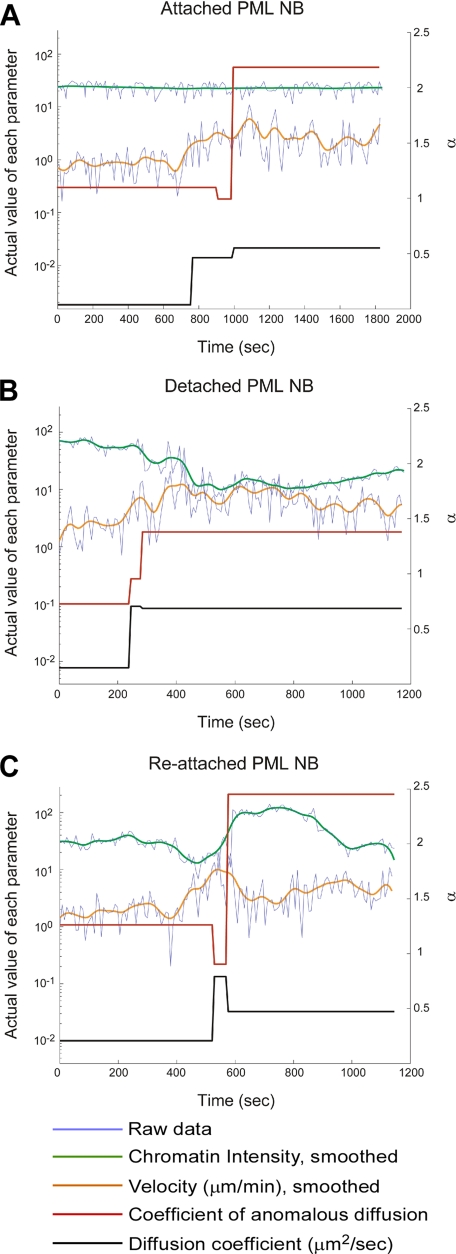
Figure 6.
Correlations between PML NB mobility and chromatin colocalization. Four parameters were measured for each signal particle track and plotted over time. The change of chromatin intensity (I) indicates the change of colocalization between PML NBs and chromatin, and the variation of velocity (v), diffusion coefficient (D), and coefficient of anomalous diffusion (α) indicate the change of PML NB mobility. Three types of correlations can be typically observed in a prophase cell. One example track of each type is shown here. The y-axis on the left is the actual value of I, v, and D in a log scale. The y-axis on the right is α in a linear scale. (A) Attached, a PML NB was attached to chromatin and moved along with chromatin. (B) Detached, a PML NB was released from chromatin and showed increase mobility. (C) Reattached, a PML NB was released from chromatin but then reattached to chromatin at a later time.
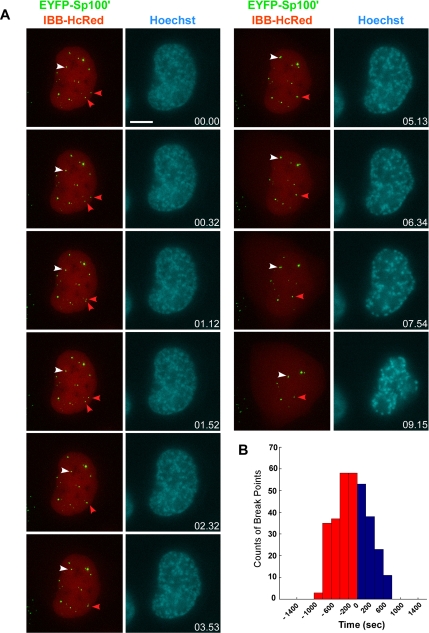
Figure 5.
PML NBs dynamics upon nuclear membrane breakdown. (A) Triple stable cell line, CpYsRi89, stably expressing PML-ECFP, EYFP-Sp100′, and IBB-HcRed. An 8-μm Z-stack of 0.5-μm steps is collected every 8 s for 20 min in the YFP and the HcRed channels. A single Z section at the middle of the cell was taken at every time point in the Hoechst channel as the reference image for DNA. Nuclear membrane breakdown was visualized by the release of the HcRed signal from the nucleus into the cytoplasm. The 3D-projected images show that PML NBs are highly dynamic even before nuclear membrane breakdown. White arrowheads, PML NBs with long-distance displacement. Red arrowheads, the fusion of PML NBs. Bar, 10 μm. (B) The change of diffusion coefficient, calculated by a model-free maximum-likelihood estimator (MLE) algorithm, was used to define break points of a single particle track. Six live cell data sets were collected from the CpYsRi89 triple stable cell line. By visualizing the release of the HcRed signal from the nucleus into the cytoplasm, the time point of NEBD for each data set was defined as time = 0 to synchronize and compare timing between the data sets. The majority of the break points occurred before NEBD.
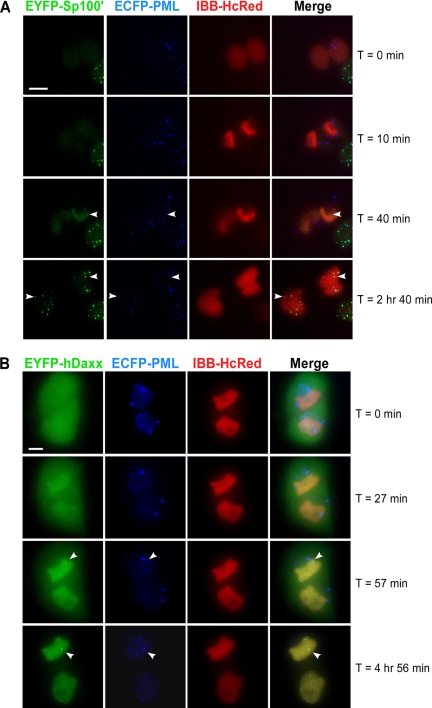
Figure 8.
The formation of PML NBs in G1 cells occurs upon the establishment of a functional nuclear envelope. (A) Double stable cell line, CpYs68, expressing PML-ECFP (blue) and EYFP-Sp100′ (green) was transiently transfected with IBB-HcRed (red). A 20-μm Z-stack of 1.0-μm steps was taken every 1 min for 180 min in three channels. The establishment of a functional nuclear membrane can be visualized by the import of the HcRed signal from the cytoplasm into the nucleus. The 3D-projected images show that the entry of Sp100′ protein into the daughter nuclei occurs after the establishment of a functional nuclear envelope. The recruitment of Sp100′ into the PML NBs occurs much later than the recruitment of Sp100′ protein into the nuclei (white arrowheads). (B) Single stable cell line, Cp89, expressing PML-ECFP (blue) was transiently transfected with EYFP-Daxx (green) and IBB-HcRed (red). A 20-μm Z-stack of 1.0-μm steps was taken every 1.5 min for 300 min in three channels. The establishment of a functional nuclear membrane was visualized by the import of the HcRed signal from the cytoplasm into the nucleus. The 3D-projected images show a long delay of Daxx nuclear entry after the establishment of a functional nuclear envelope. The normal interphase localization of Daxx was reestablished much later, well into G1 phase (white arrowhead). Bar, 10 μm.
The colocalization of PML NBs with chromatin was quantified by the mean fluorescence intensity of the chromatin signal (H2A-mCherry) at the position of a PML NB, which is defined by an area of pixels after segmentation plus one pixel surrounding the area. Velocity was calculated directly from displacements. Displacements were calculated as the Euclidian distances between the time points of particle trajectories.
RESULTS
Development of Stable Cell Lines Expressing PML NB Constituent Proteins
To examine the dynamics of PML NBs in U2OS cells during mitosis, PML NBs were labeled with fluorescently tagged component proteins. A previous study has shown that overexpression of full-length PML isoform IV can induce premature senescence in a p53-dependent manner (Fogal et al., 2000). Therefore, we fused PML isoform VI, which only contains the conserved domains of all nuclear PML isoforms (for review, see Jensen et al., 2001), with ECFP to minimize the isoform-specific effect of ectopically expressed PML protein. A single stable cell line, Cp89, expressing a PML–ECFP fusion protein was derived from wild-type U2OS cells.
It has been reported that transient overexpression of Sp100 generates small Sp100 foci in the nucleoplasm, which do not contain PML protein and are immobile (Wiesmeijer et al., 2002). In addition, during the development of stable cell lines, we noticed that ectopically expressed full-length Sp100 proteins (Supplemental Figure 1A, lanes 3 and 6) generated a large number of small foci at late telophase/early G1 phase before de novo PML NBs formed occurred in daughter nuclei (Supplemental Figure 1B). These small foci were not observed when endogenous Sp100 protein was labeled by anti-Sp100 antibodies in fixed cells. Therefore, to provide the best resemblance of EYFP-Sp100 localization to the endogenous Sp100 protein, we selected a double stable cell line, CpYs68, expressing full-length PML-ECFP and EYFP-Sp100 with a C-terminal deletion of the Sp100 protein (Sp100′). The Sp100′ (1–305 amino acids) encompasses half of the HP1 binding domain (Seeler et al., 1998) but includes the full consensus sequence for SUMOylation (Sternsdorf et al., 1999).
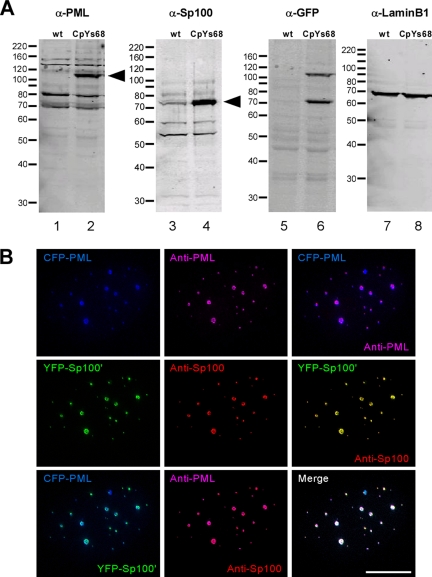
Whole cell extracts from both wild-type (wt) U2OS cells and CpYs68 cells were examined by immunoblotting (Figure 1A). Several PML isoforms (Figure 1A, lanes 1 and 2), and splice variants of Sp100 (Figure 1A, lanes 3 and 4) (Guldner et al., 1999), were expressed in both wt U2OS cells and CpYs68 cells. The arrowheads indicate that the ectopically expressed PML-ECFP (Figure 1A, lane 2) and EYFP-Sp100′ (Figure 1A, lane 4) are of the correct size and correspond to the fluorescent fusion protein (Figure 1A, lanes 5 and 6). Furthermore, CpYs68 cells were fixed and labeled with mouse anti-PML (5E10) and rabbit anti-Sp100 antibodies (rSpGH) and examined by immunofluorescence microscopy. The fusion proteins colocalized with each other (Figure 1B), and all of the Sp100 foci present in the nuclei, either visualized by the EYFP signal or by antibody labeling, colocalized with PML protein. Therefore, the CpYs68 cell line contains PML NBs that are properly labeled by fluorescently tagged PML and Sp100 proteins without overexpressed protein aggregates.
Figure 1.
Characterization of double stable cell line. (A) U2OS cells were stably transfected with PML-ECFP and EYFP-Sp100′. Whole cell extracts from both parental U2OS cells (wt) and double stable cells (CpYs68) were analyzed by immunoblot. Rabbit anti-PML shows several PML isoforms in both wt cells and double stable cells. Rabbit anti-Sp100 antibody also shows several Sp100 isoforms. The arrowheads indicate the bands representing PML-ECFP and EYFP-Sp100′ fusion proteins. (B) Fixed double stable cells were immunolabeled with mouse anti-PML antibody (5E10) and rabbit anti-Sp100 antibody (rSpGH). The projected 3D Z-stacks, collected using a DeltaVision RT microscope, show the colocalization of PML-ECFP and EYFP-Sp100′ fusion proteins with endogenous PML and Sp100 proteins, respectively. Bar, 10 μm.
PML NBs Increase Their Dynamics and Exhibit Rapid Directed Movement Upon Entry into Mitosis
We first tracked and quantitatively characterized the movement of PML NBs during prophase. Cells were stained with Hoechst 33342 to identify prophase cells as visualized by chromosome condensation. All of the PML NBs were visualized by 4D image stacks with high spatial resolution. Image stacks were acquired in the cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) channels to visualize PML NBs that were labeled by both PML-ECFP and EYFP-Sp100′ proteins (Supplemental Movie 1 and Figure 2A). A single Z-section mid-nucleus was acquired in the Hoechst channel as a reference image to register cell movement and to follow the progression of prophase. During the course of imaging, we observed multiple fusion events of PML NBs in ∼85% of the cells that resulted in the formation of MAPPs and a reduction of PML NB number from 16 ± 5 to 7 ± 5 bodies/cell, which is comparable with a previous report (Dellaire et al., 2006). The frequency of MAPP formation per cell is 50% ± 25%. EYFP-Sp100′ remained colocalized with the PML-ECFP protein in MAPPs when cells went into prometaphase and did not leave MAPPs until cells entered metaphase (Figure 2A). In addition, the tracking of PML NBs using the PML-ECFP or EYFP-Sp100′ signals showed no statistically significant difference (Supplemental Figure 2). Therefore, the following 4D live cell data were only acquired in the EYFP-Sp100′ channel to avoid using the shorter wavelength fluorophore, ECFP.
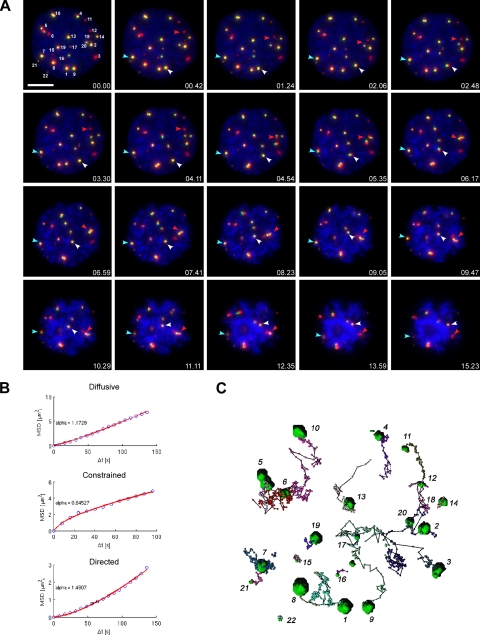
Figure 2.
4D tracking of PML NBs from prophase to prometaphase in living cells. (A) Double stable cells, CpYs68, stably expressing PML-ECFP and EYFP-Sp100′. Chromatin was visualized by Hoechst 33342 staining. A 7-μm Z-stack of 0.5-μm steps were collected in the CFP and YFP channels every 8 s for 26 min. One Z-section at the middle of the cell was collected in the Hoechst channel, at each time point, as the reference image for DNA. The 3D-projected still images of selected time points are shown here. Each PML NB was manually assigned a number and tracked by TIKAL 4D viewer to determine their displacement. The examples of PML NBs with different motions are indicated by colored arrowheads. The red arrowhead, directed motion. White arrowhead, diffusive motion. Blue arrowhead, constrained motion. Red, pseudocolored PML-ECFP. Green, EYFP-Sp100′. Blue, DNA stained by Hoechst 33342. Bar, 10 μm. (B) The trajectories of bodies were exported into MATLAB to calculate the MSD, which is plotted in the y-axis against Δt in the x-axis. The MSD is fitted to the power law MSD = 6DΔtα, with α as the coefficient of anomalous diffusion. A nonlinear relationship between the MSD and Δt indicates nonstandard diffusion. Classification into movement types is performed according to α ∼ 1.5 for directed movement, α ∼ 1 for simple diffusion, and α ∼ 0.5 for constrained diffusion. The representative MSD plot of each movement type is displayed here with the actual data (blue circles) and the fitted curve (red line). (C) Overview of the trajectory of each PML NB in this 4D image data set is summarized by ball-and-stick representations to show the displacement of each body. The initial position of each PML NB is indicated by a green volume.
According to the coefficient of anomalous diffusion (α), the movement of nuclear bodies can be quantitatively categorized into three types, i.e., simple diffusion, constrained diffusion, and directed movement (Eils et al., 2000; Platani et al., 2002). Tracked PML NBs were manually assigned a number (Figure 2A) and checked by eye to correct the automatic tracking. The mean square displacement for each track was then calculated and fitted to the power law MSD = 6DΔt to obtain the value of α. Fitting graphic representations of each movement type is shown in Figure 2B. An overview of the trajectory of each PML NB in this 4D image data set is shown as ball-and-stick representations, and the initial position of each PML NB is indicated by a green volume (Figure 2C). Overall, 40% of PML NBs showed directed movement in prophase compared with 12% in interphase (data not shown; see Muratani et al., 2002); notably, often the fast moving PML NBs adhered to each other to form MAPPs and moved along as one unit (Figure 2A, red arrowhead). As shown in Supplemental Movie 1, the formation of MAPPs did not occur by complete fusion of PML NBs into one large spherical body; but rather by association of bodies resulting in the irregular shape of MAPPs. These data demonstrate that PML NBs increase their dynamics in prophase, and this increase of PML NB dynamics results in the formation of MAPPs.
PML NBs Change Their Dynamics in Early Mitosis
During the tracking of PML NBs in prophase, we noticed that many of these bodies changed their movement type over time as indicated by MSD plots in a multiphasic pattern, which is not usually observed in an interphase cell. In Figure 3A, one example of a PML NB showed constrained movement during the first one-third of the plot but changed to directed movement by the end of the time points measured (Figure 3A, green line). However, by fitting the trajectory to a single curve (Figure 3A, blue line), the r-value was usually poor (r = 0.96). To obtain better fits for MSD plots and to characterize dynamic changes during early mitosis, we subdivided the trajectories into two phases at the transition of prophase to prometaphase when nuclear envelope breakdown (NEBD) occurred. Because a diffused pool of soluble EYFP-Sp100′ can be observed in interphase nuclei and this EYFP-Sp100′ signal dissipated from the nucleus into the cytoplasm during NEBD, we determined the time point of NEBD based on the dissipation of the EYFP signal from the nucleus. Therefore, we used this biphasic model to further dissect the global change of PML NB dynamics. Phase 1 begins from time 0, the beginning of data collection in early prophase, to the time we observed Sp100′ dissipation at prometaphase. Phase 2 starts from the time of Sp100′ dissipation to the end of data collection.
Figure 3.
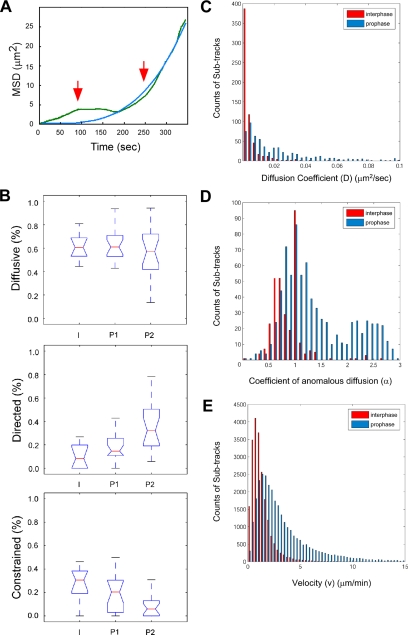
Summary of PML NB movement types in interphase and prophase. (A) An example of a MSD plot, showing a biphasic curve. The green line shows the real data and the blue line shows the fitted curve. The red arrows indicate the two phases. (B) The box plots summarize the movement types of PML NBs from interphase cells (n = 10) and prophase cells (n = 20). The y-axis indicates the percentage of PML NB population that exhibits certain movement types and x-axis indicates the data from different phases, interphase (I), phase 1 of prophase (P1), phase 2 of prophase (P2). The red line indicates the median of the data set. The box indicates 50% of the data points. The notch of the box indicates the significance of the mean values with 95% confidence. If the notch area overlaps between the two sets, the mean values of the two sets are not significantly different. If there is no overlap, the difference is significant. Subtrack distributions of diffusion coefficient (C) and coefficient of anomalous diffusion (D) from interphase cells (red bars) and prophase cells (blue bars). Each track is divided by the breakpoints, which is defined by the change of diffusion coefficient, into subtracks. The α exponent was then calculated for each subtrack. A shift of both D and α value from the interphase cell to the prophase cell data set is observed. (E) Velocity is defined by displacement over time interval at each time point. A shift is also observed from the interphase cell to the prophase cell data set.
In subsequent live cell experiments, we transiently expressed an H2A-mCherry fusion protein to visualize chromatin and to follow the progression of prophase during the course of imaging. In addition, because mCherry has a longer maximum excitation wavelength of 587 nm rather than the 346-nm maximum excitation for Hoechst stain, a full Z-stack, instead of a single Z-section image, can be collected at each time point as a DNA reference. The 4D live cell data sets were then acquired in both the YFP and mCherry channels from early prophase to late prometaphase with 8-s time intervals and 0.5-μm Z-steps. As mentioned, the dynamic movements of PML NBs in interphase nuclei can be categorized into three types: diffusive movement (simple diffusion), directed movement, and constrained diffusion (Muratani et al., 2002). Therefore, we tracked PML NB dynamics in 10 interphase cells and 20 prophase cells, and the results are summarized in Figure 3B. The percentages represent the PML NB population that exhibited each movement type in each cell. The population of PML NBs exhibiting diffusive motion remained constant and had medians of ∼60% during both interphase and prophase. In the interphase cells, a median of 10% of the PML NBs exhibited directed movement, which is in agreement with a previous study (Muratani et al., 2002). Interestingly, in phase 1 of prophase, the directed movement of PML NBs increased to a median of 18%. During phase 2, the median percentage of directed PML NBs further increased from 18 to 30%, whereas the median of the PML NB population showing constrained movement decreased from 20 to 10%. These movement type shifts of the PML NB population are significant as demonstrated by analysis of variance, with p < 0.01. However, the increase of directed PML NBs in phase 2 was contributed not only by the constrained PML NBs but also by the diffusive PML NBs in phase 1. By using this biphasic model, we demonstrated that PML NBs exhibited a global increase in directed movement and a global decrease of constrained movement in early mitosis.
Even though diffusion coefficients (D) can be calculated from the fitting curve of MSD plots, the change of D values over time cannot be determined this way because we can only obtain one averaged D value from one fitting curve. Therefore, we used an automated algorithm to calculate the change of D values in each particle track throughout the course of imaging. Instead of dividing all of the tracks at the same time point into two phases, we used a model-free MLE (Montiel et al., 2006) to identify at which time point(s) the change of D values occurred in one track. We then used those time point(s) as break point(s) to divide the particle trajectories into subtracks, from which we also obtained the value of α more accurately (see Materials and Methods). Both D and α values of all the subtracks from interphase and from prophase are compared and summarized in Figure 3, C and D, respectively. We also obtained the velocity of PML NBs at each time point by measuring the displacement (Euclidian distance between consecutive time points) over time, which is summarized in Figure 3E. Clearly, in all three histograms, the peak of prophase tracks (blue) expanded to the right from the peak of interphase tracks (red). This indicates that prophase cells have more PML NBs showing an increased diffusion coefficient and high α value compared with interphase cells. In addition, prophase cells have more PML NBs exhibiting large-scale displacements than interphase cells. This indicates that PML NBs dramatically change their dynamics during prophase and especially exhibit more directed motion as we showed previously in the biphasic analysis (Figure 3B).
The change in PML NB movement types may be explained by one or more of the following possibilities. First, because PML NBs may initially be constrained in the crowded nucleus, they may simply increase their movement when the confinement by chromatin is reduced due to chromosome condensation, which creates additional interchromatin space in phase 2. The second possibility is that material exchange between the nucleoplasm and the cytoplasm during NEBD may generate a physical flow that may force PML NBs to move passively. Last, PML NBs may lose their constraints by detaching from chromatin or the nuclear matrix during prophase and therefore increase their overall dynamics. Thus, experiments were designed to distinguish between these three possibilities.
PML NBs Do Not Increase Their Dynamics Upon Increase of Interchromatin Space
First, we wanted to determine whether the increase of interchromatin space can result in increased dynamic movement of PML NBs. Premature chromosome condensation (PCC) was induced by using chemical inhibitors of type 1 and type 2A protein phosphatases (PP1 and PP2A), such as calyculin A (CA) and okadaic acid (OA), to mimic mitotic condensation of chromosomes (Asakawa and Gotoh, 1997). CA and OA treatments have been shown to efficiently induce PCC, usually within an hour of treatment, in >20% of the cells tested (Durante et al., 1998). In addition, hypercondensed chromatin (HCC) by hyperosmolar medium can also reversibly increase interchromatin space (Albiez et al., 2006). Therefore, we used both CA to induce PCC and high salt medium (570 mOsm) to induce HCC independently in the CpYs68 double stable cell line to determine whether an increase of interchromatin space affects the dynamics of PML NBs.
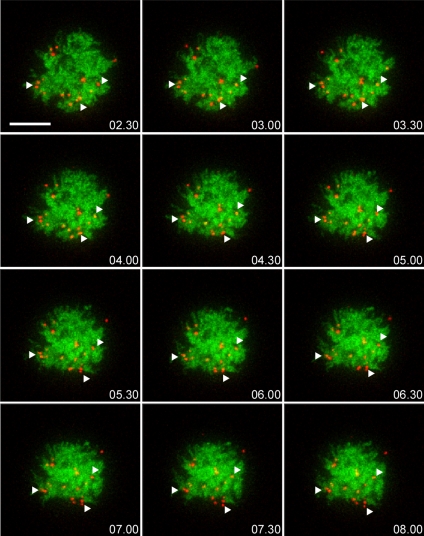
CpYs68 cells were transiently transfected with H2A-mCherry (pseudocolored green in Figure 4) for chromatin visualization. Cells were imaged either 30 min after 40 nM CA treatment or immediately after switching to high salt medium. The 4D live cell image stacks with 1.5-μm steps were collected in the mCherry and CFP channels every 10 s for 8 min (Figure 4 and Supplemental Movie 2). As a result of CA treatment, PML NBs did not change their relative position, and the majority of PML NBs displayed limited movement upon chromosome condensation (Figure 4, arrowheads). It is clear that the dynamic behavior of PML NBs in CA-treated cells is dramatically different from that in normal prophase cells. Few PML NB fusion events were observed, and no long-distance displacement of PML NBs occurred. This further supports the possibility that formation of MAPPs requires the increased dynamics of PML NBs. A similar result was also observed when chromatin was highly condensed after high salt treatment (Supplemental Figure 3, top) and PML NBs exhibited localized movement as shown in the trajectories after tracking (Supplemental Figure 3, bottom). Therefore, PML NBs do not move freely within the increased interchromatin space upon the induction of PCC by CA or HCC by high salt medium.
Figure 4.
PML NB dynamics upon calyculin A induced premature chromosome condensation. A double stable cell line, CpYs68, was transiently transfected with H2A-mCherry construct. Cells were then treated with 40 nM calyculin A for 30 min to induce premature chromatin condensation before 4D live cell imaging by the DeltaVision RT microscope (n = 10). A 24-μm Z-stack of 1.5-μm steps was taken in the CFP and mCherry channels every 10 s for 8 min. The 3D-projected still images from selected time points are shown here. Condensed chromosomes can be visualized by the H2A-mCherry fluorescent signal (pseudocolored in green). PML NBs (pseudocolored in red, the white arrowheads) do not exhibit dynamic movement within the increased interchromatin space. Bar, 10 μm.
The Increase of PML NB Dynamics During Prophase Occurs Before NEBD
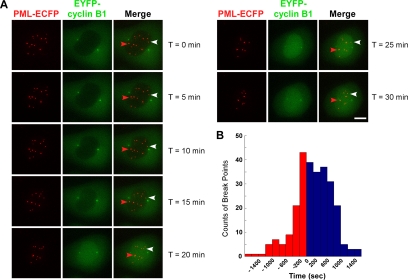
An alternative explanation for the increase of directed PML NB movement during prophase is the physical flow between the nucleoplasm and the cytoplasm. Such changes upon NEBD may be the driving force of PML NB movement in prometaphase. As mentioned, the time of Sp100′ dissipation from the nuclei corresponds to the time of NEBD and was therefore used to roughly divide the prophase data sets in the biphasic analysis. To determine more precisely the timing of NEBD and its temporal relationship with the increase of PML NB dynamics, a construct containing the IBB domain of importin α fused with two copies of HcRed was used (Bubulya et al., 2004; Leung et al., 2004). This IBB fusion protein is actively imported into the nucleus as long as a functional nuclear envelope is maintained. The IBB-HcRed signal is restricted within the nucleus during interphase, but it occurred in the cytoplasm during the transition from prophase to prometaphase, marking the initiation of NEBD (Leung et al., 2004). We therefore generated a triple stable cell line, CpYsRi89, stably expressing PML-ECFP, EYFP-Sp100′, and IBB-HcRed for the 4D live cell analysis.
Cells at the G2/prophase transition were identified by partially condensed chromatin, as visualized by Hoechst DNA staining (Figure 5A). The 4D live cell image stacks with 0.5-μm steps were collected in the HcRed and YFP channels every 8 s for 20 min (Supplemental Movie 3). When chromosome condensation was observed in early prophase, the IBB-HcRed signal was restricted within the nuclei before NEBD occurred, as shown in the still images (Figure 5A). The timing of IBB-HcRed release from the nuclei coincided with the timing of Sp100′ dissipation (data not shown). Even before the IBB-HcRed signal occurred in the cytoplasm, the long-distance displacements (Figure 5A, white arrowheads) and the fusion of PML NBs (Figure 5A, red arrowheads) already occurred. By using the same MLE algorithm, we further quantified the change of PML NB mobility by determining the break points of each track to assess whether the changes occur before or after NEBD. To synchronize the live cell data sets, the time zero for each trajectory was shifted to the time point of the onset of NEBD, which is indicated by the appearance of the IBB-HcRed signal in the cytoplasm. The number of break points that occurred at each time interval relative to NEBD was counted and summarized in a histogram (Figure 5B). Clearly, the majority of PML NBs changed their mobility before NEBD as the peak number of break points occurred before time 0. Therefore, the physical flow between the nucleoplasm and the cytoplasm is unlikely to be the driving force to change PML NB movement types in early mitosis.
PML NBs Are Released from Chromatin in Early Mitosis
Last, we examined whether PML NB mobility was constrained due to the attachment of the PML NBs to chromatin. If this hypothesis is correct, PML NBs that are released from chromatin at early prophase should increase their velocity concomitantly with the loss of colocalization with chromatin. To assess this possibility, we examined the live cell data sets containing H2A-mCherry and EYFP-Sp100′ signals to examine in more detail the interactions between PML NBs and chromatin. The chromatin intensity (mCherry signal) at each PML NB (defined pixel region after segmentation) was calculated in 3D at each time point to quantify the change of colocalization between chromatin and the PML NB. Diffusion coefficient (D), coefficient of anomalous diffusion (α), and velocity (v) were then calculated from the same track and plotted together with chromatin intensity against time (Figure 6).
At least three types of correlations between PML NB mobility and chromatin localization can be observed quantitatively in a typical prophase cell (Figure 6 and Supplemental Figure 4). The first type of PML NBs (attached, 36 ± 10%) increased their mobility while maintaining their colocalization with chromatin (Figure 6A, green line). Interestingly, they often show a moderate increase of velocity (Figure 6A, orange line, <5 μm/min), but they exhibit a much elevated coefficient of anomalous diffusion (Figure 6A, red line, >2). The second type of PML NBs (detached, 44 ± 10%) also showed increased mobility (Figure 6B) but only when they lost their colocalization with chromatin (Figure 6B, green line). In contrast to type 1, type 2 PML NBs exhibit much elevated velocity (Figure 6B, orange line, maximum at 10 μm/min) but a moderate α exponent of ∼1.5 (Figure 6B, red line). This indicates that although mobility of the type 1 PML NBs are constrained by chromatin movement, the type 2 PML NBs move freely at high velocity only after their detachment from chromatin. In addition, we also found a third type of PML NB (fluctuated, 20 ± 8%) that fluctuated between the two states as the value of chromatin intensity and that of velocity changed in opposite direction (Figure 6C, green and orange lines). This type of PML NB was first pulled by the contracting motion of chromosome condensation as they remained attached to chromatin, and the PML NB was then released from chromatin and moved freely. At a later time point, the free-moving PML NB was trapped by chromatin again. These data suggest that mobility of PML NBs is affected by their attachment to chromatin upon entry into mitosis.
Timing of Increased PML NB Dynamics During Prophase
Next, we were interested in determining when PML NBs detach from chromatin and in identifying the trigger for the global increase of PML NB dynamics during prophase. It has been shown that the cyclin B1/cyclin-dependent kinase 1 (Cdk1) complex commits the cell to mitosis and promotes metaphase progression by phosphorylating a variety of substrates, including nuclear lamins and condensins (for review, see Marcello et al., 2003). Cyclin B1 forms an active complex with Cdk1 in the cytoplasm and is later translocated from the cytoplasm into the nucleus during late prophase (Jackman et al., 2003), the time during which we observed the global increase of directed PML NB movement. Therefore, we investigated whether the translocation of cyclin B1 correlated with the increase of PML NB dynamics. A new double stable cell line, CpYcb165, was generated to stably express PML-ECFP and EYFP-cyclin B1 based on the starting line, Cp89. Again, transient transfection of H2A-mCherry was used to visualize chromatin and to identify early prophase cells. A 0.5-μm step Z-stack of total 8 μm in the z-axis was collected every 8 s over 30 min in both CFP and YFP channels, with a reference image of one H2A-mCherry Z section in the middle of the cell.
When cells reached the G2/prophase transition, EYFP-cyclin B1 was mainly diffusely distributed throughout the cytoplasm and was excluded from the nucleus. It was also concentrated at the duplicated centrosomes as reported previously (Jackman et al., 2003) (Figure 7A, T = 0 min, and Supplemental Movie 4). In addition, when the two EYFP-cyclin B1 labeled centrosomes separated to the opposite poles of the nucleus, marking the end of G2 phase, the cell usually enters mitosis within 30 min. Therefore, we started image acquisition in this series when the separated centrosomes were observed despite no apparent chromatin condensation at this time point. Nuclear envelope breakdown was visualized by release of cyclin B1 from the nucleus and its distribution throughout the mitotic cell within 6 min. These observations indicated that the EYFP-cyclin B1 fusion protein in our stable cell line behaved as described previously (Hagting et al., 1999).
Figure 7.
PML NB dynamics upon nuclear entry of cyclin B1. (A) Double stable cell line, CpYcb165, stably expresses PML-ECFP and EYFP-cyclin B1. Cells were transiently transfected with H2A-mCherry construct. An 8-μm Z-stack of 0.5-μm steps was collected every 8 s for 30 min in the CFP and YFP channels. The 3D-projected images show that PML NBs exhibited limited movement before cyclin B1 entered the nucleus. After the nuclear entry of cyclin B1, PML NBs became dynamic, and long distance displacement and fusion of PML NBs were observed. White arrowheads, PML NBs with long-distance displacement. Red arrowheads, the fusion of PML NBs. Bar, 10 μm. (B) The change of diffusion coefficient, calculated by a model-free MLE algorithm, was used to define break points of a single particle track. Eight live cell data sets from the CpYcb165 double stable cell line were synchronized to the time point of cyclin B1 nuclear entry (time = 0). The majority of the break points occurred after nuclear entry of cyclin B1.
However, at the beginning of prophase, PML NBs did not significantly change their relative positions (Figure 7A, arrowheads, T = 0 min to T = 10 min). Only when cyclin B1 was translocated into the nucleus did we observe long-range displacements and fusion of PML NBs resulting in the formation of MAPPs (Figure 7A, white and red arrowhead, respectively, T = 15 min to T = 30 min). NEBD occurred near the 30-min time point as indicated by dissipation of cyclin B1 from the nucleus to the cytoplasm. To quantify the change of PML NB mobility in relation to cyclin B1 nuclear entry, we also determined the break points of each track by using the same MLE algorithm described previously. As summarized in Figure 7B, the time 0 was shifted to the time point when the appearance of the EYFP-cyclin B1 signal was observed in the nucleus. Most break points occurring after time 0 indicates that the mobility change of PML NBs occurred along with the nuclear entry of cyclin B1. Sixteen percent of the break points occurred 3 min before time 0, indicating that the trigger of cyclin B1 nuclear entry may also trigger the increase of PML NB dynamics during prophase. Together with the previous results, these data indicate that the increase of PML NB dynamics and the formation of MAPPs occur, for the most part, between cyclin B1 nuclear entry and NEBD.
Dynamics of Fluorescently Tagged Daxx and Sp100 Proteins
From prometaphase to metaphase, PML NB components, such as Sp100 and Daxx, leave newly formed MAPPs and diffuse into the mitotic cytoplasm. The reformation of interphase PML NBs occurs in early G1. A previous study suggested that MAPPs may contribute to PML NB reformation in G1 phase because no new PML protein is synthesized during mitosis, and the PML protein accumulation in MAPPs may be recycled back to the nucleus to form PML NBs in daughter nuclei (Dellaire et al., 2006). Fixed cell studies also suggested that the recruitment of Sp100 and Daxx began in early G1 and that the percentage of PML NBs associated with Sp100 at this stage was higher than that with Daxx, suggesting that Sp100 was recruited before Daxx (Dellaire et al., 2006). However, the recruitment of protein constituents to PML NBs in live G1 cells has not been examined.
The double stable cell line CpYs68 expressing both PML-ECFP and EYFP-Sp100′ was used to examine the reassembly of PML NBs upon exit from mitosis. The 4D live cell images were collected from anaphase through early G1 phase with 1-μm step Z-stack of total 20 μm every 1 min for 3 h. The cells were also transiently transfected with the IBB-HcRed construct to monitor the functional nuclear envelope in daughter nuclei. A functional nuclear envelope was reformed at ∼10 min after the beginning of imaging (Figure 8A and Supplemental Movie 5). Few MAPPs were trapped within the newly formed nuclei, whereas Sp100′ protein was still diffusely distributed throughout the daughter cells. The first indication of Sp100′ protein recruitment into the nucleus was observed at the 25-min time point. However, the recruitment of Sp100′ into the PML NBs (Figure 8A, white arrowheads) occurred much later than the recruitment of Sp100′ protein into the nucleus.
Daxx, another major PML NB component protein, also showed similar recruitment timing. To monitor the recruitment of Daxx during early G1 phase, a double stable cell line, CpYdx56, was generated stably expressing PML-ECFP and EYFP-hDaxx. The 4D live cell images were acquired at telophase and proceeded through early G1 phase with a 1-μm step Z-stack of total 20 μm every 1.5 min for 5 h. At the beginning of the data set, the IBB-HcRed signal was already detectable in daughter nuclei, indicating the reformation of a functional nuclear envelope (Figure 8B and Supplemental Movie 6). However, the recruitment of Daxx into the daughter nuclei did not occur until much later. The first time point at which Daxx protein could be detected in newly formed PML NBs was ∼57 min after the start of imaging. However, it was not until the cells were well into G1 phase that the normal interphase localization of Daxx was reestablished.
The time point of Daxx nuclear recruitment, relative to the reformation of a functional nuclear envelope, is much later than that of Sp100′. However, even after Sp100′ and Daxx proteins were both present in the daughter nuclei, additional time was required to complete their recruitment into newly formed PML NBs as shown by the still images with higher temporal resolution in Supplemental Figure 5. The delay between nuclear entry and recruitment of PML NB component proteins into PML NBs suggests that additional modifications may be needed before the recruitment of PML NB component proteins can occur. By comparing the timing of Sp100′ and Daxx recruitment to the reformation of nuclear envelope, the delayed nuclear entry of Daxx protein suggests a sequential recruitment of PML NB component proteins into early G1 PML NBs.
DISCUSSION
The most dramatic nuclear reorganization occurs during mitosis, the final stage in the cell cycle. Analyzing the disassembly and reassembly of nuclear domains upon entry into and exit from mitosis provides important insight into the formation of nuclear domains under endogenous regulation of mitotic progression (Prasanth et al., 2003; Bubulya et al., 2004; Leung et al., 2004). A previous live cell imaging study has demonstrated, with single particle tracking, that PML NBs exhibit dynamic displacement during mitosis (Dellaire et al., 2006). However, in that study, the authors did not track the dynamic movement of PML NBs with high temporal and spatial resolution, and they were therefore unable to resolve when and how the mobility of PML NBs changes upon mitotic entry. By using 4D live cell imaging with high temporal and spatial resolution, we demonstrate, for the first time, that the change of PML NB dynamics correlates to specific mitotic events.
Traditionally, the classification of mitotic stages was based on morphological changes of the cell during mitosis. As a more detailed molecular map of mitotic progression was developed, Pines and Rieder (2001) proposed a new staging of mitosis based on the activity of defined cell cycle regulators (Pines and Rieder, 2001). They particularly stressed a “point of no return,” when mitotic entry becomes irreversible during late prophase (Rieder and Cole, 1998). This point was named “transition 2,” and it coincides with the activation of a cyclin B/Cdk1 complex, which is the main component of M-phase-promoting factor (for review, see Ohi and Gould, 1999). The activation of cyclin B/Cdk1 is governed by the removal of the two inhibitory phosphates at the N terminus of Cdk1 and the accumulation of cyclin B in the nucleus (for review, see Bassermann et al., 2005). On the sudden accumulation of active cyclin B/Cdk1 in the nucleus, cells pass this “point of no return” and commit to mitosis until conditions change for them to escape from mitosis (Hagting et al., 1998). Our observations indicate that the changes in PML NB dynamics occurs during this point of mitosis.
We first observed that PML NBs become highly dynamic from early prophase to prometaphase. Both the diffusion coefficient (D) and the coefficient of anomalous diffusion (α) of PML NBs are much higher in prophase than in interphase. The overall velocity of PML NB movement during prophase is higher than that observed in interphase as some PML NBs moved with speeds as high as 10 μm/min. Interestingly, when we further categorized PML NB movement by fitting the MSD plot to the equation of anomalous diffusion, several tracks showed a biphasic pattern indicating that PML NBs changed their movement type during the course of imaging. After dividing each image track into two phases at the time point when NEBD occurred, statistics of this biphasic analysis showed a global increase of directed PML NB movement, but a global decrease of constrained PML NB movement in prophase. This indicates that certain constraint(s) on PML NBs were removed during the initiation of mitosis.
By analyzing 4D live cell imaging using a model-free MLE algorithm (Montiel et al., 2006), we identified the timing when PML NBs lost their constraint(s) during mitosis. The increase in PML NB dynamics occurred along with the rapid nuclear entry of cyclin B1 and before NEBD, which represents transition 2 of the mitosis staging paradigm (Pines and Rieder, 2001). Surely, the physical flow created during NEBD is not the driving force of increased PML NB dynamics because long-distance displacement of PML NBs occurred before NEBD. Shortly after the nuclear translocation of cyclin B1, the active cyclin B1/Cdk1 complex phosphorylates different substrates, e.g., lamins and Cdc25, to commit the cells into mitosis (for review, see Takizawa and Morgan, 2000). However, because the initial increase of the break points in Figure 7B is a bit earlier than time 0, it is possible that the trigger(s) for nuclear entry of the cyclin B1/Cdk1 complex may also lead to the removal of the constraints on PML NBs, the change of PML NB dynamics, and MAPP formation during prophase.
One caveat from this set of experiments is that cyclin B1/Cdk1 is not responsible for initial chromosome condensation in early prophase (Hagting et al., 1999). Because PML NBs only increase their dynamics along with the nuclear entry of cyclin B1/Cdk1, this indicates that chromosome condensation, per se, is not sufficient to induce an increase in PML NB dynamics. PML NBs did not show the same type of fast movements in CA-treated cells or in high salt-treated cells, compared with that observed in prophase cells, further supporting the idea that removal of the space constraints on PML NBs is not sufficient to trigger the increase of PML NB dynamics. In addition, PML NBs seemed to be tethered onto prematurely condensed chromatin or HCC and did not change their relative position or fuse during the course of imaging. These observations suggest that the additional interchromatin space created after premature chromosome condensation or high salt treatment does not facilitate the fast movement of PML NBs by virtue of the release of PML NBs from chromatin.
The fast movements of PML NBs are not completely equivalent to the directed motion, because PML NBs exhibiting diffusive movement can also move at high velocity. Interestingly, further analysis of PML NB movement types in CA-treated cells indicated that >60% of the PML NBs exhibited directed movement, and the trajectories of PML NBs showed that most of the tracks moved toward the center of the nucleus, which is probably due to the shrinkage of the cell. This suggests that PML NBs may be potentially dragged by prematurely condensed chromatin. Indeed, when analyzing the localization of PML NBs in CA-treated cells, most of them colocalized with chromatin and remained attached during the course of imaging. An interesting point to consider is that the increase of PML NB dynamics, especially in phase 2, may be partly affected by other dynamic events in mitotic cells. Most likely, the movement of PML NBs after NEBD is affected by condensed chromosomes, which are pushed and dragged toward the metaphase plate by the newly formed spindle. However, this model still does not explain the dynamic increase of PML NB movement before NEBD.
Previous biochemical analysis has indicated that the PML protein is phosphorylated and deSUMOylated during mitosis, and that the phosphorylation can be maintained by calyculin A (Everett et al., 1999b). Moreover, unSUMOylated PML protein cannot recruit PML NB components into PML NBs, but it can still form immature PML protein aggregations (Dellaire et al., 2006; Shen et al., 2006). These studies indicate that posttranslational modification of PML protein may play an important role in changing PML NB dynamics and MAPP formation during mitosis. However, it is not clear which kinase(s) directly targets PML protein during mitosis. Although one report has shown that PML protein can be directly phosphorylated by casein kinase 2 at Ser517, which leads to PML protein degradation through a ubiquitin-mediated pathway (Scaglioni et al., 2006), our own observations of PML NB dynamics in prophase may help to direct studies examining the role of posttranslational modification on PML NB dynamics.
An association between PML NBs and chromatin has been suggested in many studies (Shiels et al., 2001; Wang et al., 2004; Dellaire et al., 2006). Because the resolution of the light microscope is limited, we cannot distinguish whether the chromatin association of PML NBs is due to the attachment of PML NBs to specific gene loci, or, instead, to a chromatin-containing “framework.” Previous studies indicates that chromatin is necessary to maintain the integrity and stability of PML NBs, and upon stress and nuclease treatments, PML NBs increase their mobility immediately (Maul et al., 1995; Eskiw et al., 2004; for review, see Ching et al., 2005). Electron spectroscopic imaging (ESI) suggests that PML NBs may have a direct connection with chromatin (Eskiw et al., 2003; Dellaire et al., 2006). A subpopulation of PML NBs are also known to associate with specific chromosomal regions, such as major histocompatibility complex (MHC) gene clusters (Shiels et al., 2001), telomeres (Yeager et al., 1999), and centromeres (Everett et al., 1999a). One recent study showed that PML NBs, in conjunction with special AT-rich sequence binding protein 1, can alter the chromatin loop structure and regulate transcription of the MHC I locus (Kumar et al., 2007). Because most of these studies were performed on fixed cells, it is important to show how these chromatin associations of PML NBs affect PML NB dynamics in living cells.
Because 4D live cell imaging of H2A-mCherry and EYFP-Sp100′ presents a 3D view of chromatin organization at high temporal and spatial resolution, we analyzed the correlation between chromatin association and the velocity of PML NBs during prophase. The PML NBs that exhibited directed movement in phase 2 can be clearly distinguished into two types. One type of PML NB moves freely without chromatin association, and the other type seems to move with chromatin. Our data further suggested an association of PML NBs with chromatin, and the loss of chromatin association leads to increased mobility of PML NBs. Interestingly, Platani et al. (2002) have demonstrated that ATP depletion can affect the dynamics of Cajal bodies (CBs), and they suggested that tethering of CBs onto chromatin requires ATP (Platani et al., 2002). It will be interesting to further investigate whether PML NBs are also tethered to chromatin through the same mechanism as CBs. Together with previous observations, we suggest that PML NBs may increase their dynamics by disassociation from chromatin upon entry into mitosis.
The formation of PML NBs requires proper posttranslational modifications of PML protein (Sternsdorf et al., 1999), whose regulation can alter PML NB structure and composition during mitosis (Everett et al., 1999b). It has also been proposed that the formation of mature PML NBs requires several steps, including aggregation of PML protein through its RING domain, formation of PML protein networks through its SUMOylated moiety and its SUMO interacting motif, and the recruitment of PML-associated proteins (Shen et al., 2006). Our data have demonstrated that there is a delay between the nuclear entry of PML NB constituents and their recruitment into PML NBs, e.g., Sp100 and Daxx, suggesting that certain rate-limiting steps may prevent the maturation of PML NBs in early G1 cells. In addition, Sp100 and Daxx are recruited into PML NBs at different stages, from telophase to G1 phase. Even though Sp100 and Daxx recruitment follow the recruitment of PML protein, suggesting that PML protein may be the seed for PML NB formation, we cannot exclude the possibility that other unknown components may initialize the formation of PML NBs because other studies have shown PML NB-like structures can be formed by PML NB components under certain circumstances (Topisirovic et al., 2002; Everett et al., 2006). When we followed MAPPs during the telophase/G1 transition, we did observe that some MAPPs were trapped in the newly formed nuclei as described previously by Dellaire et al., 2006. However, many PML NBs are formed de novo at different sites in daughter nuclei, and the de novo formation of PML NBs seems to be synchronized at a specific time. Further analysis is necessary to determine whether PML NBs form at specific nuclear sites and whether their formation is triggered by specific postmitotic events.
In summary, by using quantitative 4D live cell analysis, we elucidated and characterized changes in the dynamic movements of PML NBs during mitosis. Our data showed for the first time that the majority of PML NBs changed their dynamics in early prophase at or around the timing of nuclear entry of cyclin B1 but before NEBD. This change in dynamics was correlated with a loss of chromatin association. At the end of mitosis, the association of PML NB constituent proteins with de novo formed PML NBs was consistent with a sequential recruitment model.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Spector laboratory for critical review of the manuscript and helpful discussion. The following individuals provided constructs or reagents that were critical to the studies: PML-ECFP (T. Tsukamoto), mCherry (R. Tsien, UCSD, San Diego, CA), IBB-HcRed (J. Ellenberg and E. Zanin), and EYFP-cyclin B1 and H2A-mCherry (T. Nakamura). We also thank M. Reichenzeller (German Cancer Research Center, Heidelberg, Germany) for critical review of the manuscript. This work was supported by grant 42694 from National Institutes of Health/National Institute of General Medical Sciences (to D.L.S.) and the Endocyte EU project (to R.E.).
Abbreviations used:
- CA
calyculin A
- HCC
hypercondensed chromatin
- IBB
importin-βbinding
- MAPP
mitotic accumulations of PML protein
- MLE
maximum-likelihood estimator
- MSD
mean square displacement
- NEBD
nuclear envelope breakdown
- PMLNB
promyelocytic leukemia nuclear body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0035) on May 14, 2008.
REFERENCES
- Albiez H., et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14:707–733. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]
- Asakawa Y., Gotoh E. A method for detecting sister chromatid exchanges using prematurely condensed chromosomes and immunogold-silver staining. Mutagenesis. 1997;12:175–177. doi: 10.1093/mutage/12.3.175. [DOI] [PubMed] [Google Scholar]
- Bacher C. P., Reichenzeller M., Athale C., Herrmann H., Eils R. 4-D single particle tracking of synthetic and proteinaceous microspheres reveals preferential movement of nuclear particles along chromatin-poor tracks. BMC Cell Biol. 2004;5:45. doi: 10.1186/1471-2121-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassermann F., Peschel C., Duyster J. Mitotic entry: a matter of oscillating destruction. Cell Cycle. 2005;4:1515–1517. doi: 10.4161/cc.4.11.2192. [DOI] [PubMed] [Google Scholar]
- Bernardi R., Pandolfi P. P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Borden K. L. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell Biol. 2002;22:5259–5269. doi: 10.1128/MCB.22.15.5259-5269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubulya P. A., Prasanth K. V., Deerinck T. J., Gerlich D., Beaudouin J., Ellisman M. H., Ellenberg J., Spector D. L. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 2004;167:51–63. doi: 10.1083/jcb.200404120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching R. W., Dellaire G., Eskiw C. H., Bazett-Jones D. P. PML bodies: a meeting place for genomic loci? J. Cell Sci. 2005;118:847–854. doi: 10.1242/jcs.01700. [DOI] [PubMed] [Google Scholar]
- Condemine W., Takahashi Y., Zhu J., Puvion-Dutilleul F., Guegan S., Janin A., de The H. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 2006;66:6192–6198. doi: 10.1158/0008-5472.CAN-05-3792. [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer M., Dietzel S., Muller S., Solovei I., Fakan S. Chromosome territories–a functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- de The H., Chomienne C., Lanotte M., Degos L., Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature. 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- Dellaire G., Bazett-Jones D. P. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays. 2004;26:963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- Dellaire G., Eskiw C. H., Dehghani H., Ching R. W., Bazett-Jones D. P. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J. Cell Sci. 2006;119:1034–1042. doi: 10.1242/jcs.02817. [DOI] [PubMed] [Google Scholar]
- Durante M., Furusawa Y., Gotoh E. A simple method for simultaneous interphase-metaphase chromosome analysis in biodosimetry. Int. J. Radiat. Biol. 1998;74:457–462. doi: 10.1080/095530098141320. [DOI] [PubMed] [Google Scholar]
- Dyck J. A., Maul G. G., Miller W. H., Jr., Chen J. D., Kakizuka A., Evans R. M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Eils R., Gerlich D., Tvarusko W., Spector D. L., Misteli T. Quantitative imaging of pre-mRNA splicing factors in living cells. Mol. Biol. Cell. 2000;11:413–418. doi: 10.1091/mbc.11.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskiw C. H., Dellaire G., Bazett-Jones D. P. Chromatin contributes to structural integrity of promyelocytic leukemia bodies through a SUMO-1-independent mechanism. J. Biol. Chem. 2004;279:9577–9585. doi: 10.1074/jbc.M312580200. [DOI] [PubMed] [Google Scholar]
- Eskiw C. H., Dellaire G., Mymryk J. S., Bazett-Jones D. P. Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J. Cell Sci. 2003;116:4455–4466. doi: 10.1242/jcs.00758. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Chelbi-Alix M. K. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Earnshaw W. C., Pluta A. F., Sternsdorf T., Ainsztein A. M., Carmena M., Ruchaud S., Hsu W. L., Orr A. A dynamic connection between centromeres and ND10 proteins. J. Cell Sci. 1999a;112:3443–3454. doi: 10.1242/jcs.112.20.3443. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Lomonte P., Sternsdorf T., van Driel R., Orr A. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 1999b;112:4581–4588. doi: 10.1242/jcs.112.24.4581. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Murray J. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 2005;79:5078–5089. doi: 10.1128/JVI.79.8.5078-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R. D., Rechter S., Papior P., Tavalai N., Stamminger T., Orr A. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 2006;80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogal V., Gostissa M., Sandy P., Zacchi P., Sternsdorf T., Jensen K., Pandolfi P. P., Will H., Schneider C., Del Sal G. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000;19:6185–6195. doi: 10.1093/emboj/19.22.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldner H. H., Szostecki C., Schroder P., Matschl U., Jensen K., Luders C., Will H., Sternsdorf T. Splice variants of the nuclear dot-associated Sp100 protein contain homologies to HMG-1 and a human nuclear phosphoprotein-box motif. J. Cell Sci. 1999;112:733–747. doi: 10.1242/jcs.112.5.733. [DOI] [PubMed] [Google Scholar]
- Hagting A., Jackman M., Simpson K., Pines J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr. Biol. 1999;9:680–689. doi: 10.1016/s0960-9822(99)80308-x. [DOI] [PubMed] [Google Scholar]
- Hagting A., Karlsson C., Clute P., Jackman M., Pines J. MPF localization is controlled by nuclear export. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., 3rd, Maul G. G. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M., Lindon C., Nigg E. A., Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- Jensen K., Shiels C., Freemont P. S. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- Kumar P. P., Bischof O., Purbey P. K., Notani D., Urlaub H., Dejean A., Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- Leung A. K., Gerlich D., Miller G., Lyon C., Lam Y. W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A. I. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello A., Ferrari A., Pellegrini V., Pegoraro G., Lusic M., Beltram F., Giacca M. Recruitment of human cyclin T1 to nuclear bodies through direct interaction with the PML protein. EMBO J. 2003;22:2156–2166. doi: 10.1093/emboj/cdg205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul G. G., Negorev D., Bell P., Ishov A. M. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- Maul G. G., Yu E., Ishov A. M., Epstein A. L. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J. Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- Montiel D., Cang H., Yang H. Quantitative characterization of changes in dynamical behavior for single-particle tracking studies. J. Phys. Chem. B. 2006;110:19763–19770. doi: 10.1021/jp062024j. [DOI] [PubMed] [Google Scholar]
- Muratani M., Gerlich D., Janicki S. M., Gebhard M., Eils R., Spector D. L. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 2002;4:106–110. doi: 10.1038/ncb740. [DOI] [PubMed] [Google Scholar]
- Ohi R., Gould K. L. Regulating the onset of mitosis. Curr. Opin. Cell Biol. 1999;11:267–273. doi: 10.1016/s0955-0674(99)80036-2. [DOI] [PubMed] [Google Scholar]
- Pines J., Rieder C. L. Re-staging mitosis: a contemporary view of mitotic progression. Nat. Cell Biol. 2001;3:E3–E6. doi: 10.1038/35050676. [DOI] [PubMed] [Google Scholar]
- Platani M., Goldberg I., Lamond A. I., Swedlow J. R. Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 2002;4:502–508. doi: 10.1038/ncb809. [DOI] [PubMed] [Google Scholar]
- Plehn-Dujowich D., Bell P., Ishov A. M., Baumann C., Maul G. G. Non-apoptotic chromosome condensation induced by stress: delineation of interchromosomal spaces. Chromosoma. 2000;109:266–279. doi: 10.1007/s004120000073. [DOI] [PubMed] [Google Scholar]
- Prasanth K. V., Sacco-Bubulya P. A., Prasanth S. G., Spector D. L. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T., Chelbi-Alix M. K. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 2001;20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Cole R. W. Entry into mitosis in vertebrate somatic cells is guarded by a chromosome damage checkpoint that reverses the cell cycle when triggered during early but not late prophase. J. Cell Biol. 1998;142:1013–1022. doi: 10.1083/jcb.142.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglioni P. P., Yung T. M., Cai L. F., Erdjument-Bromage H., Kaufman A. J., Singh B., Teruya-Feldstein J., Tempst P., Pandolfi P. P. A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell. 2006;126:269–283. doi: 10.1016/j.cell.2006.05.041. [DOI] [PubMed] [Google Scholar]
- Seeler J. S., Marchio A., Sitterlin D., Transy C., Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels C., Islam S. A., Vatcheva R., Sasieni P., Sternberg M. J., Freemont P. S., Sheer D. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 2001;114:3705–3716. doi: 10.1242/jcs.114.20.3705. [DOI] [PubMed] [Google Scholar]
- Soignet S. L., et al. Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. N. Engl. J. Med. 1998;339:1341–1348. doi: 10.1056/NEJM199811053391901. [DOI] [PubMed] [Google Scholar]
- Spector D. L. Nuclear domains. J. Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen K., Reich B., Will H. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- Takizawa C. G., Morgan D. O. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr. Opin. Cell Biol. 2000;12:658–665. doi: 10.1016/s0955-0674(00)00149-6. [DOI] [PubMed] [Google Scholar]
- Topisirovic I., Capili A. D., Borden K. L. Gamma interferon and cadmium treatments modulate eukaryotic initiation factor 4E-dependent mRNA transport of cyclin D1 in a PML-dependent manner. Mol. Cell Biol. 2002;22:6183–6198. doi: 10.1128/MCB.22.17.6183-6198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Shiels C., Sasieni P., Wu P. J., Islam S. A., Freemont P. S., Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. G., Rivi R., Delva L., Konig A., Scheinberg D. A., Gambacorti-Passerini C., Gabrilove J. L., Warrell R. P., Jr., Pandolfi P. P. Arsenic trioxide and melarsoprol induce programmed cell death in myeloid leukemia cell lines and function in a PML and PML-RARalpha independent manner. Blood. 1998;92:1497–1504. [PubMed] [Google Scholar]
- Weis K., Rambaud S., Lavau C., Jansen J., Carvalho T., Carmo-Fonseca M., Lamond A., Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Wiesmeijer K., Molenaar C., Bekeer I. M., Tanke H. J., Dirks R. W. Mobile foci of Sp100 do not contain PML: PML bodies are immobile but PML and Sp100 proteins are not. J. Struct. Biol. 2002;140:180–188. doi: 10.1016/s1047-8477(02)00529-4. [DOI] [PubMed] [Google Scholar]
- Yeager T. R., Neumann A. A., Englezou A., Huschtscha L. I., Noble J. R., Reddel R. R. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- Zhong S., Salomoni P., Ronchetti S., Guo A., Ruggero D., Pandolfi P. P. Promyelocytic leukemia protein (PML) and Daxx participate in a novel nuclear pathway for apoptosis. J. Exp. Med. 2000;191:631–640. doi: 10.1084/jem.191.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.