Abstract
The spatial and temporal regulation of the interactions among the ∼60 proteins required for endocytosis is under active investigation in many laboratories. We have identified the interaction between monomeric clathrin adaptors and endocytic scaffold proteins as a critical prerequisite for the recruitment and/or spatiotemporal dynamics of endocytic proteins at early and late stages of internalization. Quadruple deletion yeast cells (ΔΔΔΔ) lacking four putative adaptors, Ent1/2 and Yap1801/2 (homologues of epsin and AP180/CALM proteins), with a plasmid encoding Ent1 or Yap1802 mutants, have defects in endocytosis and growth at 37°C. Live-cell imaging revealed that the dynamics of the early- and late-acting scaffold proteins Ede1 and Pan1, respectively, depend upon adaptor interactions mediated by adaptor asparagine-proline-phenylalanine motifs binding to scaffold Eps15 homology domains. These results suggest that adaptor/scaffold interactions regulate transitions from early to late events and that clathrin adaptor/scaffold protein interaction is essential for clathrin-mediated endocytosis.
INTRODUCTION
Clathrin-mediated endocytosis (CME) mediates the internalization of extracellular molecules, plasma membrane lipids, and specific membrane proteins into the cell. Nutrient uptake, membrane remodeling, immune surveillance, and synaptic vesicle recycling all depend on CME to maintain and regulate many of their constituents (Geli and Riezman, 1998; Evans and Owen, 2002; Sorkin, 2004; Szymkiewicz et al., 2004). Thus, CME is an essential process for homeostasis and signaling regulation in all eukaryotic cells.
CME involves >60 cytosolic proteins acting in concert to form a cargo-containing clathrin-coated vesicle (CCV). Current studies focus on elucidating the regulation and mechanisms of spatiotemporal coupling of endocytic proteins, beginning with the selection of cargo proteins at the plasma membrane by clathrin adaptors (Kaksonen et al., 2003, 2005; Newpher et al., 2005). Adaptors recognize sorting signals in the cytoplasmic tail of cargo proteins, such as linear peptide motifs or posttranslational modifications such as ubiquitination (Wendland, 2002; Traub, 2003; Maldonado-Baez and Wendland, 2006). Adaptors also recruit clathrin and other endocytic proteins to sites of endocytosis (Wendland, 2002; Traub, 2003; Owen, 2004). Forming stable cargo–adaptor–clathrin complexes at the plasma membrane is a key, rate-limiting early event in the assembly of CCVs (Ehrlich et al., 2004; Kaksonen et al., 2005; Newpher et al., 2005). Current models predict that these early endocytic complexes trigger the transition from early (cargo-gathering) to late (vesicle scission) events in endocytic internalization, although the mechanism is unknown (Ehrlich et al., 2004; Kaksonen et al., 2005; Merrifield et al., 2005; Naslavsky and Caplan, 2005; Newpher et al., 2005; Newpher and Lemmon, 2006).
Most known adaptors are conserved from yeast to humans (Toret and Drubin, 2007); thus, we used the budding yeast Saccharomyces cerevisiae for this study. Our laboratory has characterized the S. cerevisiae putative adaptor proteins Ent1 and Ent2 (Figure 1A), an essential gene pair that belongs to the epsin protein family (Chen et al., 1998; Wendland et al., 1999; Aguilar et al., 2003). The structural features and the endocytic roles of epsins are conserved (Wendland, 2002); like the other members of this family, Ent1 and Ent2 have a globular epsin amino(N)-terminal homology (ENTH) domain that binds phosphatidylinositol-4,5-bisphosphate at the plasma membrane. Their C termini contain two ubiquitin interaction motifs (UIMs) that bind ubiquitin, two asparagine-proline-phenylalanine (NPF) tripeptide motifs that interact with Eps15 homology (EH) domains, a C-terminal clathrin binding motif (CBM), and regions of unknown function (Figure 1A) (Wendland et al., 1999; Aguilar et al., 2003, 2006).
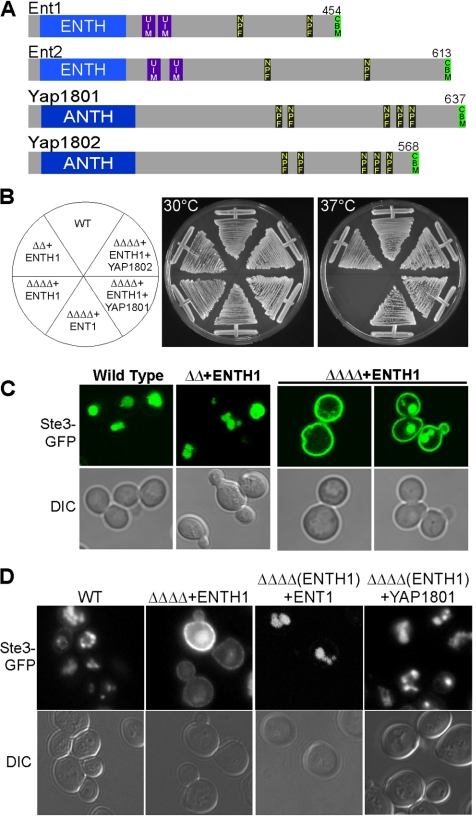
Figure 1.
Yap1801/2 complement growth and endocytosis in the absence of the Epsin C terminus. (A) Ent1, Ent2, Yap1801, and Yap1802 with the conserved lipid binding amino-terminal ENTH and ANTH domains, UIMs, EH domain-binding NPF motifs, and CBMs are shown. (B) ΔΔΔΔ+ENTH1 cells are inviable at 37°C. ent1Δent2Δ (ΔΔ) with the ENT1 ENTH domain (BWY2595) or ent1Δent2Δ yap1801Δyap1802Δ (ΔΔΔΔ) with either full-length ENT1 (BWY2596), ENTH1 (BWY2597), or coexpressing ENTH1 and either of the Yap180 proteins (pBW1344, pBW1345) were grown at 30 or 37°C for 3 d. (C) ΔΔΔΔ+ENTH1 cells exhibit endocytosis defects. WT (SEY6210), ΔΔ+ENTH1 (BWY2595), or ΔΔΔΔ+ENTH1 (BWY2597) cells were transformed with a Ste3-GFP plasmid (pBW0639), grown at 30°C and assessed for Ste3-GFP internalization by live-cell confocal microscopy. (D) Yap1801/2 proteins complement endocytosis in ΔΔΔΔ+ENTH1 cells. Wild-type (WT), ΔΔΔΔ-expressing Ent1 or the ENTH1 domain and ΔΔΔΔ cells coexpressing the ENTH1 domain and either of the Yap180 proteins were tested for localization of chromosomally tagged Ste3-GFP as in Figure 1C.
We showed recently that the minimal region of Ent1/2 required for viability in ent1Δent2Δ (ΔΔ) cells is the conserved ENTH domain (Wendland et al., 1999; Aguilar et al., 2006). The essential function of the ENTH domain is in cell polarity by binding to Cdc42 regulatory factors (Aguilar et al., 2006). Surprisingly, ΔΔ cells expressing the ENTH domain as the only source of epsin (ΔΔ+ENTH) exhibit normal endocytosis. One interpretation of this finding is that the ENTH domain harbors the endocytic functions of epsins. However, the epsin C termini contain all the characterized endocytic protein interaction motifs. This suggests that other adaptor proteins that bind the same partners may compensate for the endocytic role of the yeast epsin C-termini in ΔΔ+ENTH cells.
Two other candidate adaptors are the yeast proteins Yap1801 and Yap1802, the homologues of the mammalian endocytic clathrin assembly proteins CALM/AP180 (Wendland and Emr, 1998; Wendland et al., 1999). Like AP180/CALM, Yap1801/2 have an AP180 amino(N)-terminal homology (ANTH) domain that is structurally and functionally similar to ENTH domains (Ford et al., 2001; Itoh et al., 2001; De Camilli et al., 2002; Stahelin et al., 2003). Yap1801/2 share other similarities with the yeast epsins, including five NPF motifs and a CBM (Figure 1A) (Wendland and Emr, 1998). Ent1/2 and Yap1801/2 also localize to endocytic plasma membrane patches (Wendland and Emr, 1998; Wendland et al., 1999). AP180, a neuronal-specific adaptor protein involved in the endocytic-recycling of synaptic vesicles, binds clathrin and promotes its assembly into cages (Zhang et al., 1998; Ford et al., 2001; Owen et al., 2004; Merrifield et al., 2005; Augustine et al., 2006). CALM is a ubiquitously expressed AP180-related protein that plays a role in endocytosis in non-neuronal cells (Tebar et al., 1999; Wechsler et al., 2003; Owen et al., 2004; Meyerholz et al., 2005).
Yeast two-hybrid experiments and in vitro binding assays showed that NPF motifs in Ent1/2 and Yap1801/2 bind the yeast scaffold proteins Ede1 and Pan1 (Wendland and Emr, 1998; Howard et al., 2002; Aguilar et al., 2003). Ede1 and Pan1 contain EH domains, and they are yeast homologues of the mammalian endocytic scaffolds Eps15 and Intersectin (Miliaras and Wendland, 2004). The NPF motif/EH domain interaction between adaptors and scaffolds is conserved from yeast to humans. Like other multivalent scaffold proteins, Pan1 and Intersectin interact with and organize complexes of multiple endocytic proteins, including factors involved in early and late stages of internalization, and factors that affect actin polymerization, including the Arp2/3 complex and type I myosins (Tang et al., 1997; Yamabhai et al., 1998; Okamoto et al., 1999; Tang et al., 2000; Duncan et al., 2001; Hussain et al., 2001a,b; Miliaras et al., 2004; Barker et al., 2007; Toshima et al., 2007).
Consistent with a shared endocytic role with the epsins, deleting both yeast AP180 genes has no apparent endocytosis or growth defect (Supplemental Figure S1) (Wendland and Emr, 1998; Huang et al., 1999). Here, we show that the yeast epsin and AP180 proteins share a complementary role in endocytosis, and delineate the NPF motif/EH domain interaction between clathrin adaptors and endocytic scaffolds as critical for successful endocytosis.
MATERIALS AND METHODS
Media and Materials
Yeast cells were grown in standard rich (yeast extract-peptone) or synthetic medium (yeast nitrogenous base with amino acids for plasmid selection) and 2% dextrose. We used 750 μg/ml 5-fluoro-orotic acid (5-FOA; United States Biological, Swampscott, MA) in synthetic medium; 250 μg/ml Geneticin (G418; Invitrogen, Carlsbad, CA) was used in rich medium. Bacteria were grown on standard Luria-Bertani media with 50 μg/ml carbenicillin, 30 μg/ml kanamycin, and/or 34 μg/ml chloramphenicol to maintain plasmids. Materials were obtained from Fisher Scientific (Pittsburgh, PA) or Sigma (St. Louis, MO) unless otherwise stated.
Strains and Plasmids
Standard methods were used for growth, sporulation, tetrad dissection, and DNA manipulations and transformations of yeast. The yeast strains used in this study are listed in Supplemental Table 1. The parental quadruple deletion strain (BWY1975) was generated by sporulation and tetrad dissection and confirmed by Western blotting. BWY2503, BWY2506, and BWY2574 (all Pan1-green fluorescent protein[GFP]) strains were generated by C-terminal chromosomal integration of polymerase chain reaction (PCR) products as described in Longtine et al. (1998). The parental quintuple deletion strain (BWY2571) was generated by transformation with a PCR fragment containing homologous untranslated regions of EDE1 surrounding the G418-resistant cassette and selection for growth on G418 plates (Longtine et al., 1998). The plasmids used in this study are listed in Supplemental Table 2. DNA manipulations were performed using standard techniques, employing either T4 DNA polymerase-mediated ligations in Escherichia coli or homologous recombination with overlapping DNA fragments followed by plasmid rescue in S. cerevisiae. Amino acid substitutions were made using QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. All restriction enzymes were purchased from New England Biolabs (Ispwich, MA).
Plasmid Shuffle
ΔΔ, ΔΔΔΔ, 5Δ cells expressing ENT1 or ENT2 from a TRP1 or URA3 plasmid were used for counterselection on 5-fluoroanthranilic acid (5-FAA) or 5-FOA, respectively. Cells cotransformed with ENT1 mutant or truncation URA3 and WT TRP1 plasmids were grown on 5-FAA plates (to evict the TRP1 plasmid) at 30°C for 3 d. Alternatively, cells cotransformed with ENT1 mutant or truncation TRP1 and WT URA3 plasmids were grown on 5-FOA plates (to evict the URA3 plasmid) at 30°C for 3 d.
α-Factor Uptake Experiments
35S-α-factor was purified and α-factor internalization assays were performed using a continuous presence protocol as described previously (Dulic et al., 1991). Cells were grown to early log phase, shifted to 30°C for 15 min, and incubated with α-factor. Samples were taken after incubation at the indicated time and preserved in ice-cold phosphate buffer, pH 6.0 (40 mM KH2PO4 and 6.0 mM K2HPO4) or citrate buffer, pH 1.0 (50 mM Na3C6H5O7·2H2O). Samples were collected on glass filters (VWR, West Chester, PA) and counted on an LS6500 scintillation counter (Beckman Coulter, Fullerton, CA). Data points represent the percentage of α-factor internalized by dividing the counts from the pH 1.0 treated samples (representing only internalized α-factor) by the counts from the corresponding pH 6.0 treated samples (representing total bound α-factor). Each data point is a representative average of three or more assays. Error bars represent the SD.
Ste3-GFP Localization Assay
Cells were transformed with a STE3-GFP plasmid (pBW0639) (Urbanowski and Piper, 2001). For Ste3-GFP visualization, the pH of the culture was adjusted with 10 mM Tris, pH 7.5, 5 min before observation. The cells were then pelleted and resuspended in the same media at 2–5 OD/ml density and imaged. Ste3-GFP images were acquired with an LSM 510 META confocal microscope (Carl Zeiss MicroImaging,, Thornwood, NY) equipped with the appropriate lasers and filter set.
Fluorescence Microscopy
Pan1-GFP images were collected using an Axiovert 135TV inverted microscope (Carl Zeiss MicroImaging) with a Sensicam QE charge-coupled device camera (Cooke, Romulus, MI), Zeiss 100 × 1.4 numerical aperture (NA) Plan-Apochromat objective, motorized filter wheels, fluorescein isothiocyanate filter set (Semrock, Rochester, NY), and IPLab 3.6 (BD Biosciences Bioimaging, Rockville, MD) or SlideBook 4.2 software (Intelligent Imaging Innovations, Denver, CO). Pan1-GFP time-series images were collected with a 750-ms exposure.
Ede1-red fluorescent protein (RFP) images were captured using a 3i Marianas microscope with a Zeiss alpha Plan-Fluar 100 × 1.45 NA objective, Cascade II 512 EM cameras, a 561-nm diode laser, and SlideBook 4.2 software (Intelligent Imaging Innovations). Images were captured with 500-ms exposure, identical camera gain, intensification, and illumination intensity.
For live-cell imaging, cells were grown to early log phase on rich medium plates at 30°C. Cells were placed in 2 μl of minimal media on an uncoated glass coverslip and then inverted onto a glass slide. All imaging was at room temperature.
Image Analysis
Image analysis was performed using the National Institute of Health ImageJ (http://rsb.info.nih.gov/ij/) or SlideBook 4.2 software. Kymographs were generated using ImageJ and the Multiple Kymograph plugin (http://www.embl-heidelberg.de/eamnet/html/kymograph.html). Maximum fluorescence intensity analysis was performed using the SlideBook 4.2 software.
A detailed description of the strains and plasmids used in this work are provided in Supplemental Material.
RESULTS
The Yeast Epsin C Terminus Is Required for Endocytosis in Cells Lacking the Yeast AP180 Proteins
To investigate a redundant function between the yeast epsin and AP180 proteins (Figure 1A), we created a quadruple mutant strain in which ENT1, ENT2, YAP1801, and YAP1802 were deleted. Because ENT1 and ENT2 is an essential gene pair, this mutant strain contained a plasmid encoding wild-type (WT) ENT1 to maintain viability and URA3 as a selectable marker. Plasmid shuffling was used to replace the original ENT1 plasmid with a plasmid encoding the Ent1 ENTH (ENTH1) domain by plating on 5-FOA–containing media at 30°C. On 5-FOA media, cells containing only the ENTH1 plasmid grow, whereas cells with a URA3 plasmid are inviable, allowing selection of cells expressing only the ENTH1 domain.
Due to the vital role endocytosis plays in growth and homeostasis in all eukaryotic cells, many endocytosis mutants grow poorly if at all under stressful or restrictive conditions. To test the requirement for these four putative adaptors for growth and viability, ent1Δent2Δ (ΔΔ) and ent1Δent2Δyap1801Δyap1802Δ (ΔΔΔΔ) mutants with full length Ent1 or the ENTH1 domain were incubated at 30°C (permissive temperature) or 37°C (restrictive temperature). As expected, both ΔΔ+ENTH1 and ΔΔΔΔ+Ent1 were viable at 37°C. In contrast, ΔΔΔΔ+ENTH1 cells were inviable at 37°C (Figure 1B).
We first tested the endocytic role of the epsin C termini by analyzing the trafficking of the transmembrane receptor Ste3 with a C-terminal GFP fusion (Ste3-GFP) by using live-cell fluorescence microscopy. Ste3 is expressed in α mating type cells, and it is the receptor for the a-factor mating pheromone. In the absence of ligand, Ste3 is constitutively internalized to the vacuole and degraded (Urbanowski and Piper, 2001). Ste3-GFP trafficking has been well characterized, and changes in its itinerary indicate a defect in endocytosis; we refer to this as the Ste3-GFP localization assay. Cells expressing Ste3-GFP from a single-copy plasmid were grown at 30°C and observed using confocal microscopy. In both WT and ΔΔ+ENTH1 cells, Ste3-GFP localized primarily to endosomal and vacuolar structures, with low levels found at the plasma membrane (Figure 1C). However, in ΔΔΔΔ+ENTH1 cells, Ste3-GFP accumulated mostly at the plasma membrane, indicative of a delay or blockage in the internalization of the receptor (Figure 1C). We also used another endocytic assay that monitors the ligand-induced internalization of exogenously added 35S-labeled α-factor mating pheromone, as described in Materials and Methods. Approximately 80% of the α-factor was internalized in WT cells versus <30% internalized α-factor in ΔΔΔΔ+ENTH1 cells, providing independent confirmation of the presence of a severe endocytic defect in ΔΔΔΔ+ENTH1 cells (Figure 3B). The inviability at 37°C and the deficient endocytosis of Ste3-GFP and α-factor, even at 30°C in ΔΔΔΔ+ENTH1 cells, confirms that the Yap180 proteins are required for growth under stress conditions and for endocytosis in the absence of the epsin C termini.
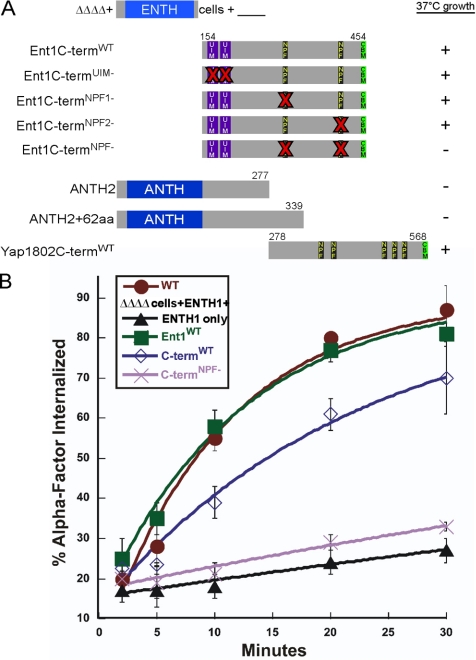
Figure 3.
NPF motifs of clathrin adaptors are required for the endocytic function of the Ent1 C terminus. Schematic of Ent1 and Yap1802 N- and C-terminal truncations, and Ent1 N-terminal truncations lacking only the ENTH domain (Ent1 C-termWT); the ENTH1 domain, ANTH domain, UIMs, NPF motifs, and CBM are indicated. Bold Xs in red indicate sites of point mutations on the Ent1 C-termWT truncation. (A) The NPF motifs are required for ΔΔΔΔ cells growth at 37°C. ΔΔΔΔ+ENTH1 cells (BWY2597) coexpressing Ent1C-termWT (pBW0781), Ent1C-termUIM- (pBW1368), Ent1C-termNPF1- (pBW1362), Ent1C- termNPF2- (pBW1363), Ent1C-termNPF- (pBW1364), the ANTH2 domain (pBW1016), a Yap1802 fragment containing amino acids 1-339 (ANTH2 + 62aa) (pBW1020), and Yap1802CtermWT (pBW1019) in trans were grown at 30 or 37°C for 3 d. Plus (+) and minus (−) signs indicate viability or inviability at 37°C, respectively. Growth plates are shown in Supplemental Figure 3, A and B. (B) NPF motifs are required for the endocytosis function of the Ent1 C-terminus. ΔΔΔΔ cells expressing Ent1 (BWY2497) or coexpressing the ENTH1 and Ent1C-termWT (BWY2738) or Ent1C-termNPF- (BWY2739) were tested for internalization of radiolabeled α-factor at 30°C. Cells were grown on selective media at 30°C. Radiolabeled α-factor was added to mid-log phase cultures at 30°C. Each point represents the average of three or more assays; error bars represent the SD.
The Yap180 Proteins Complement the Endocytic Function of the Yeast Epsin C Terminus
To confirm that the defects in viability and endocytosis observed in ΔΔΔΔ+ENTH1 cells were due to the lack of Yap1801/2, the Yap180 proteins were reintroduced, and growth and endocytosis were tested. Either of the Yap180 proteins restored viability at 37°C and endocytosis of Ste3-GFP of ΔΔΔΔ+ENTH1 cells (Figure 1, B and D; data not shown). These experiments demonstrate that the Yap180 proteins can replace the function of the epsin C termini, suggesting a shared endocytic role for these four putative adaptors.
NPF Motifs Are Required for Endocytosis in Adaptor Mutant Cells
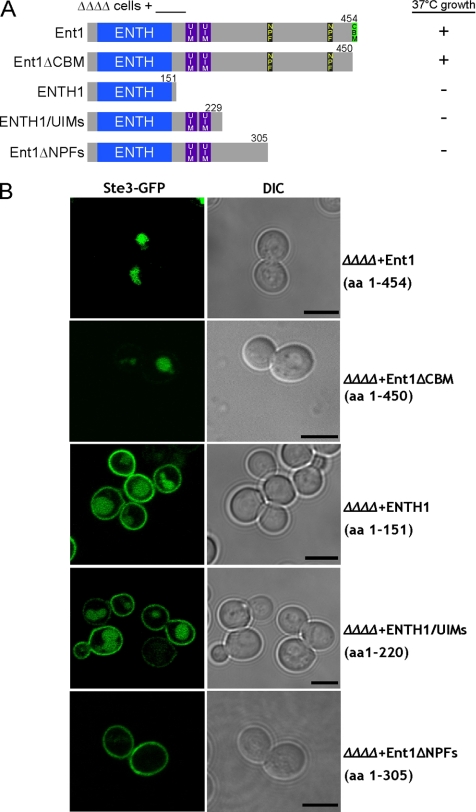
The NPF motifs and the CBM are the two recognizable C-terminal motifs that yeast epsins and Yap180s have in common (Figure 1A). To determine the minimal region of Ent1 that contains overlapping functions with Yap1801/2, ΔΔΔΔ cells with plasmids encoding C-terminal truncations of Ent1 were assayed for endocytosis with the Ste3-GFP localization assay and for growth at 30 and 37°C. After 3 d of growth, only Ent1 C-terminal truncations that included amino acids 306–450 complemented ΔΔΔΔ cells (Figure 2A and Supplemental Figure S2). The same constructs restored endocytosis of Ste3-GFP in ΔΔΔΔ cells (Figure 2B). These experiments indicate that the CBM is dispensable (Baggett et al., 2003) and show that the minimum Ent1 region required for endocytosis and viability in the ΔΔΔΔ cells at 37°C must include the NPF motifs in the absence of the CBM.
Figure 2.
NPF motif-containing region of clathrin adaptors is required for viability and endocytosis in ΔΔΔΔ cells. Schematic of Ent1 C-terminal truncations; the ENTH1 domain, ANTH domain, UIMs, NPF motifs, and CBM are indicated. (A) The NPF motif-containing region required for ΔΔΔΔ cells growth at 37°C. ΔΔΔΔ cells with either full-length Ent1 (BWY2596), an Ent1 C-terminal truncation (BWY2598, BWY2604, and BWY2605), or ENTH1 domain (BWY2597) were grown at 30 or 37°C for 3 d. Plus (+) and minus (−) signs indicate viability or inviability at 37°C, respectively. Growth plates are shown in Supplemental Figure 2. (B) The NPF motif-containing region of Ent1 is required for endocytosis in ΔΔΔΔ cells. Same ΔΔΔΔ cells as in A expressing C-terminal truncations of Ent1 were transformed with a plasmid coding for the plasma membrane protein Ste3 tagged with GFP (pBW0639). Cells were grown at 30°C and assessed for Ste3-GFP localization by using live-cell imaging and confocal microscopy. Bars, 5 μm.
Surprisingly, when a C-terminal Ent1 fragment containing the NPF motifs but lacking the ENTH domain (Figure 3A; Ent1C-termWT) was overexpressed in trans from a second plasmid in the ΔΔΔΔ+ENTH1 cells, both growth and endocytosis were restored (Figure 3B and Supplemental S3A). Endogenous levels of Ent1C-termWT partially restored growth of ΔΔΔΔ+ENTH1 cells at 37°C (Supplemental Figure S3A). Similarly, ΔΔΔΔ+ENTH1 cells overexpressing the Yap1802C-termWT grew at 37°C, unlike the Yap1802 fragment lacking NPF motifs (ANTH2 + 62 amino acids [aa]) (Figure 3A).
The need to overexpress the Ent1C-termWT for full complementation of the growth and endocytosis defects of ΔΔΔΔ+ENTH1 cells provided a sensitized background for testing the importance of the Ent1 NPF motifs in endocytosis. Either or both NPF motifs were mutated to NAA to reduce the interaction with the EH domain of the scaffold proteins Pan1 and Ede1 (de Beer et al., 1998; Aguilar et al., 2003). ΔΔΔΔ+ENTH1 cells were transformed with overexpression plasmids encoding the single Ent1C-termNPF1- or Ent1C-termNPF2- mutants, or the Ent1C-termNPF- double mutant, and growth and endocytosis phenotypes tested. The single NPF-NAA mutants restored 37°C growth of ΔΔΔΔ+ENTH1 cells (Figure 3A and Supplemental Figure S3B). In contrast, the Ent1C-termNPF- double mutant was inviable at 37°C and had impaired α-factor endocytosis, similar to ΔΔΔΔ+ENTH1 cells (Figure 3, A and B). Western blotting confirmed equal levels of expression of the WT and NPF mutant Ent1C-term constructs and localization to cortical patches (Supplemental Figure S3, C and D). Thus, the C-terminal fragment does not require the NPF motifs for association with patches, and the inability to complement ΔΔΔΔ+ENTH1 cells cannot be attributed to its absence from endocytic sites.
The epsin UIMs play an important role in cargo recognition and membrane targeting, but we generally found them to be less important than the NPF motifs for the endocytic function of the epsins in ΔΔΔΔ cells (Polo et al., 2002; Shih et al., 2002; Aguilar et al., 2003; Hicke and Dunn, 2003; Madshus, 2006). First, ΔΔΔΔ+Ent1 C-terminal truncations containing the ENTH domain and the UIMs, but lacking the NPF motifs and the CBM, were inviable at 37°C and defective for endocytosis of Ste3-GFP at 30°C (Figure 2, A and B, and Supplemental Figure S2). Second, the two conserved serines in both UIMs were mutated to aspartic acids in an Ent1C-termWT plasmid, mutations known to significantly reduce ubiquitin binding (Shih et al., 2002; Aguilar et al., 2003); ΔΔΔΔ+ENTH1 cells expressing the Ent1C-termUIM- truncation were viable at 37°C (Figure 3A and Supplemental Figure S3B). These observations suggest that the UIM motifs are not the critical region for viability and endocytosis in adaptor mutant cells.
To test for a role of the NPF motifs in a more physiological context, we mutated both NPF motifs in full length Ent1 expressed from the endogenous ENT1 promoter (Ent1NPF-). This mutant was tested for growth at 37°C and for endocytosis. ΔΔΔΔ cells expressing Ent1NPF-, although viable at 37°C and relatively normal for Ste3-GFP localization, exhibited an ∼20–25% reduction in α-factor internalization relative to WT cells, consistent with a role for the NPF motifs in endocytosis (Figure 4, A and B, and Supplemental Figure S4A; data not shown). The milder endocytosis defect observed for the Ent1NPF- full-length protein relative to the Ent1C-termNPF- protein suggests that the cooperation of the different binding determinants in cis facilitates the function of the protein, perhaps through localization and stabilization at nascent endocytic sites (Aguilar et al., 2003, 2006; Aguilar et al., 2006).
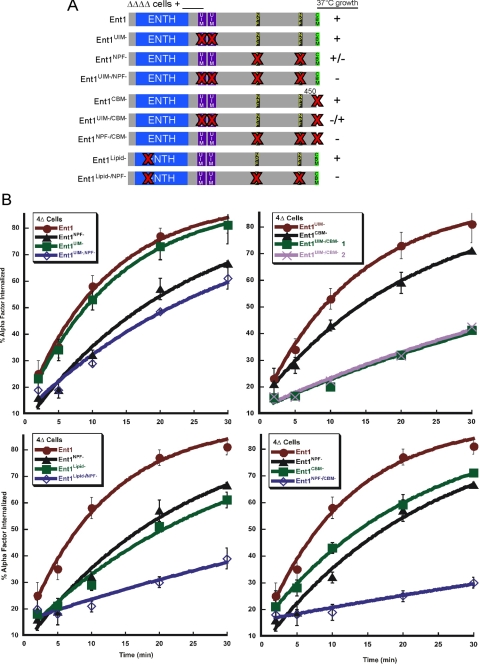
Figure 4.
NPF motifs are critical for the endocytic function of Ent1. (A) Schematic representation of full-length Ent1 point mutant constructs. ΔΔΔΔ cells expressing full-length Ent1 (BWY2596) or the following mutants: Ent1 UIM motif (Ent1UIM-) (BWY2599), Ent1 NPF motif (Ent1NPF-) (BWY2600), Ent1 CBM (Ent1CBM-) (BWY2598), Ent1 UIM and NPF motif (Ent1UIM-/NPF-) (BWY2601), Ent1 UIM and CBM (Ent1UIM-/CBM-) (BWY2602), Ent1 NPF and CBM (Ent1NPF-/CBM-) (BWY2603), Ent1 lipid-binding domain (Ent1Lipid-) (BWY2746), and Ent1 lipid-binding domain and NPF motif (Ent1Lipid-/NPF-) (BWY2747) were tested for viability at 37°C (red bold Xs indicate single amino acid substitutions or deletion of the motif). Cells were grown at 37°C for 3 d. Results are indicated by a +, −, +/− classification (+, viable; −, inviable, and +/−, intermediate growth at 37°C). (B) NPF motifs are critical for the endocytosis function of the Ent1. ΔΔΔΔ cells expressing Ent1 (BWY2497) or Ent1 mutants (BWY2740–BWY2747) were tested for internalization of radiolabeled α-factor at 30°C. Cells were grown on selective media at 30°C. Radiolabeled α-factor was added to mid-log phase cultures at 30°C. Each point represents the average of three independent colonies (except for the Ent1UIM-/CBM- strain (BWY2744); two individual colonies were tested); error bars represent the SD. Ste3-GFP localization assay results are shown in Supplemental Figure 4.
We next tested the proposed cooperativity between the Ent1 C-terminal endocytic motifs in ΔΔΔΔ cells expressing Ent1 containing various combinations of mutations in the lipid-binding pocket of the ENTH domain and the C-terminal UIM, NPF, and CBM motifs. As expected, ΔΔΔΔ+Ent1UIM- cells were viable at 37°C, and they had normal internalization of α-factor and Ste3-GFP at 30°C (Figure 4, A and B, and Supplemental Figure 4A). Mild endocytic defects were found for ΔΔΔΔ cells expressing mutant proteins individually lacking the other motifs (Ent1lipid-, Ent1CBM-, and Ent1NPF- cells; Figure 4, A and B, and Supplemental Figure S4A; data not shown) (Baggett et al., 2003). ΔΔΔΔ cells with the UIMs and the NPFs mutated in combination (Ent1UIM-/NPF-) showed only a minor defect in endocytosis assays, whereas cells with mutated UIMs and CBM (Ent1UIM-/CBM-) or ENTH domain lipid binding pocket and NPFs (Ent1Lipid-/NPF-) were more severely deficient for endocytosis of α-factor and Ste3-GFP, and for growth at 37°C. NPFs and CBM combination mutant cells (Ent1NPF-/CBM-) were the most defective, almost to the same extent as ΔΔΔΔ+ENTH1 cells. GFP-Ent1NPF-/CBM-, the most affected mutant, still localized to cortical patches similarly to GFP-Ent1WT (Supplemental Figure S4B), indicating that the functional deficiency of the mutant proteins cannot be attributed to absence from endocytic sites. Western blotting confirmed that all Ent1 mutant constructs were expressed at equal levels (data not shown). These results support a role for cooperativity between the Ent1 motifs in the function of Ent1, with the NPF motifs as major contributors.
Together, these results are consistent with the NPF motifs being the key functional C-terminal determinants shared by the yeast epsin and AP180 proteins. Our data also imply a critical physiological role for an NPF motif/EH domain interaction between an adaptor and a scaffold protein during endocytosis.
Adaptor Mutant Cells Show Altered Dynamics of the Late-acting Scaffold Protein Pan1
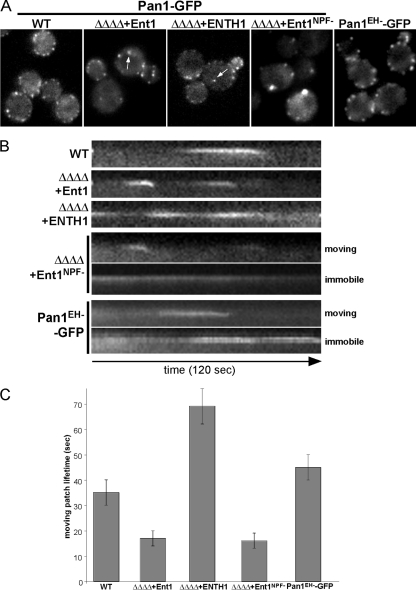
The NPF motifs of the yeast epsin and AP180 proteins interact with the EH domains of the scaffold proteins Ede1 and Pan1 in vitro and in vivo (Wendland and Emr, 1998; Howard et al., 2002; Wendland, 2002; Aguilar et al., 2003). Ede1 and Pan1 are sequentially recruited to sites of endocytosis, and this may depend in part upon binding to NPF motifs in adaptors (Benmerah et al., 2000; Kaksonen et al., 2005). We used our ΔΔΔΔ+ENTH1 and ΔΔΔΔ+Ent1NPF- cells to observe the behaviors of Pan1 and Ede1 in the absence of the epsins and Yap180 proteins; if adaptor binding to scaffolds is important in vivo, then the recruitment and/or dynamic behavior of the scaffolds should be affected.
We used real-time fluorescence microscopy to analyze the spatiotemporal distribution of chromosomally integrated Pan1-GFP in ΔΔΔΔ cells expressing Ent1, Ent1NPF-, or the ENTH1 domain. Based on the assumption that Pan1 association with cortical patches might depend upon its EH domains binding to adaptor NPF motifs, we predicted that Pan1 recruitment to endocytic sites might be inefficient in ΔΔΔΔ+ ENTH1 cells. In all cases, the Pan1-GFP patches localized to the cell cortex as reported previously by others, indicating that the adaptors are not required for the recruitment of Pan1 to the plasma membrane, although we did note a slight reduction in the patch intensity in ΔΔΔΔ+ENTH1 cells (Figure 5A). We also observed occasional cytoplasmic Pan1-GFP patches of unknown identity in ΔΔΔΔ cells expressing either Ent1 or the ENTH1 domain, but never in WT cells (Figure 5A, arrows). These few Pan1-GFP cytoplasmic punctae were freely mobile and never reached the plasma membrane (Supplemental Movies S1, S2, and S3).
Figure 5.
Epsin and AP180 proteins are required for normal dynamics of the late endocytic-scaffold protein Pan1. (A) Pan1-GFP patches show cortical localization in ΔΔΔΔ cells. Single frames of movies of Pan1-GFP in WT (BWY2503), ΔΔΔΔ+Ent1 (BWY2572), ΔΔΔΔ+ENTH1 (BWY2606), ΔΔΔΔ+Ent1NPF- (BWY2608), and pan1Δ expressing Pan1EH-GFP cells (BWY2749) are shown. Images were collected at 1-s intervals for 2 min. Arrows indicate cytoplasmic Pan1-GFP punctae. (B) Most Pan1-GFP patches remain at the cortex in ΔΔΔΔ+ENTH1 cells. Kymographs from single pixel wide lines drawn through Pan1-GFP patches in the indicated cells are shown. The downward deflection of Pan1-GFP seen in WT, ΔΔΔΔ+Ent1 cells and mobile patches in ΔΔΔΔ+Ent1NPF- and pan1Δ+Pan1EH-GFP cells shows the patch is moving toward the cell interior. In ΔΔΔΔ+ENTH1 cells, Pan1-GFP patches do not move inward. ΔΔΔΔ+Ent1NPF- and pan1Δ+Pan1EH-GFP cells exhibit a population of stationary patches similar to the ones observed in ΔΔΔΔ+ENTH1 cells. (C) Deleting the yeast Epsin and AP180 proteins alters the lifetime of Pan1-GFP patches at the cell cortex. Average lifetime ± SD of moving Pan1-GFP patches in WT (35 ± 5 s, n = 30 patches), ΔΔΔΔ+Ent1 (17 ± 3 s, n = 30 patches), ΔΔΔΔ+ENTH1 domain (69 ± 7 s, n = 6 patches), ΔΔΔΔ+Ent1NPF- (16 ± 3 s, n = 15 patches), and pan1Δ+Pan1EH-GFP (45 ± 5 s, n = 15 patches) cells. Patch localization and lifetimes for ΔΔΔΔ+Ent1UIM- and ΔΔΔΔ+Ent1CBM- cells are shown in Supplemental Figure 5.
To further assess the behavior of Pan1-GFP patches, the lifetime of single patches at the cell cortex was quantitatively analyzed and kymograph representations generated. Consistent with previous reports, Pan1-GFP patches formed at the cell cortex in WT cells with a lifetime of 35 ± 5 s, culminating with inward movement (Figure 5B and 5C) (Kaksonen et al., 2003, 2005). In ΔΔΔΔ+Ent1 cells, 92% of the Pan1-GFP patches formed and dissipated with the typical inward movement, but their lifetime was reduced to 17 ± 3 s (Figure 5B). Surprisingly, kymographs for ΔΔΔΔ+ENTH1 cells showed that at least 75% of Pan1-GFP patches remained stationary at the plasma membrane for the 120 s duration of the experiment; their fluorescence intensity fluctuated, but they never disappeared from the cortex by an inward movement (Supplemental Movie S3). The few Pan1-GFP patches that formed and then disassembled during the time of an experiment (120 s) had an average lifetime of 69 ± 7 s, twice the lifetime of Pan1-GFP in WT cells (Figure 5C). Interestingly, ΔΔΔΔ+Ent1NPF- had both short-lived and stationary populations of Pan1-GFP patches (44% mobile vs. 56% immobile), likely explaining the intermediate endocytosis and growth at 37°C phenotypes (Figure 5B and Supplemental Movie S4). In contrast, ΔΔΔΔ+Ent1UIM- or ΔΔΔΔ+Ent1CBM- cells showed Pan1-GFP dynamics and lifetimes similar to ΔΔΔΔ+Ent1WT cells (Supplemental Figure S5).
If the effect of the adaptor mutants on Pan1 dynamics is direct, it is predicted to be mediated through NPF/EH domain interactions. Thus, we examined the dynamics of GFP-tagged Pan1EH-, in which the critical tryptophan residue in the ligand binding pocket of each EH domain was mutagenized to alanine (de Beer et al., 1998). This mutant Pan1 protein was expressed from its endogenous promoter by using a single copy plasmid in pan1Δ cells as the sole source of Pan1, and these cells grew similarly to WT cells at 37°C (data not shown). This mutant Pan1 localized to cortical patches (Figure 5A), suggesting that the primary targeting of Pan1 to cortical patches can be mediated by additional factors besides the adaptors. Consistent with the behavior of WT Pan1-GFP in ΔΔΔΔ+ENTH1 and ΔΔΔΔ+Ent1NPF- cells, ∼60% of the Pan1EH--GFP patches exhibited greatly extended lifetimes ranging from 75 to >120 s (Figure 5B and Supplemental Movie S6). Pan1EH--GFP cells also had a relatively normal population of patches (∼40%) with slightly extended lifetimes of 45 ± 5 s (Figure 5C). These data suggest that the temporal dynamics of Pan1 depend upon interaction with the adaptors and that the interaction between Pan1 and the clathrin adaptors is critical for the spatial and temporal regulation of clathrin-mediated endocytosis.
Enhanced Cortical Localization of the Early-acting Scaffold Ede1 in Adaptor Mutant Cells
The scaffold protein Ede1 has three N-terminal EH domains that bind NPF motifs, a coil-coiled region, and a ubiquitin-binding ubiquitin associated (UBA) domain at its C terminus (Howard et al., 2002; Shih et al., 2002; Aguilar et al., 2003; Maldonado-Baez and Wendland, 2006). Deleting EDE1 causes a mild defect in endocytosis (Gagny et al., 2000), and Ede1 is an early endocytic coat component (Kaksonen et al., 2005; Toshima et al., 2006). Due to its genetic interactions with Pan1 and its NPF motif/EH domain-mediated interaction with the adaptors (Gagny et al., 2000; Howard et al., 2002; Aguilar et al., 2003), we predicted that the spatiotemporal distribution of Ede1 might also be dependent on the adaptors.
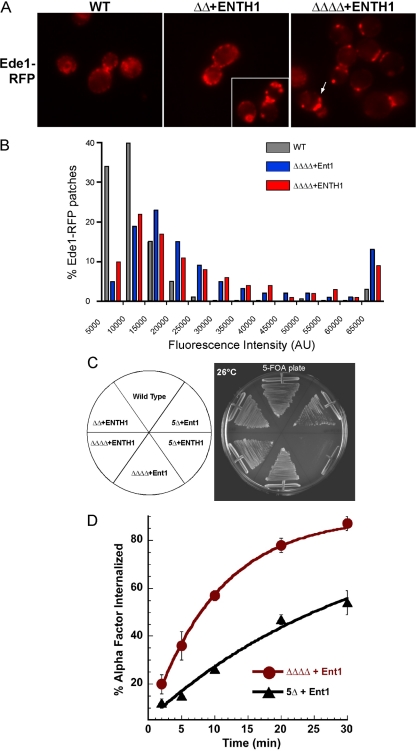
We used live-cell fluorescence microscopy to study the behavior of Ede1 in ΔΔΔΔ cells. ΔΔΔΔ cells with various Ent1 mutants were transformed with a plasmid encoding a C-terminal Ede1-RFP chimera (Ede1-RFP). As previously reported, Ede1 localized to cortical patches at the plasma membrane with 30–180-s lifetimes in WT cells (Figure 6A) (Gagny et al., 2000; Toshima et al., 2006). In ΔΔΔΔ+ENTH1 cells, most patches seemed to have lifetimes >180 s; however, this could be due to less photobleaching due to brighter patch intensity. We noted large and bright patches (patches with fluorescence intensities >20,000 arbitrary units [AU]) in 75% of ΔΔΔΔ+ENTH1 cells (n = 83 cells) versus 19% of WT cells (n = 162 cells). To better quantify this observation, we analyzed the maximum fluorescence intensity of individual Ede1-RFP patches. This analysis showed that ΔΔΔΔ cells had more high intensity (>60,000 AU) Ede1-RFP patches than WT cells (Figure 6B). This number is an underestimate, as only clearly distinct Ede1-RFP patches were quantified, eliminating many of the extremely brightly labeled areas from the analysis.
Figure 6.
The early scaffold Ede1 requires the yeast Epsins and AP180s for normal spatiotemporal dynamics and viability. (A) Ede1-RFP patches localize to the cell cortex in ΔΔΔΔ+ENTH1 cells with altered distribution. Single frames of a movie showing Ede1-RFP (pBW0985) cortical patches in WT (SEY6210), ΔΔ+ENTH1 (BWY2595), and ΔΔΔΔ+ENTH1 cells (BWY2597). Arrow indicates a large, bright patch. (B) ΔΔΔΔ+ENTH1 cells have Ede1-RFP patches with maximum fluorescence intensities higher than WT cells. The maximum fluorescence intensity of individual Ede1-RFP patches was analyzed using Slide-Book 4.2 software (n = 350 patches for each strain). Comparisons of the localization and patch intensity for Ede1-RFP versus Ede1EH--RFP can be found in Supplemental Figure 6, A and B. (C) Ede1 is required for viability at 26°C in ΔΔΔΔ+ENTH1 cells. Quadruple deletion cells (ΔΔΔΔ) (BWY2596 and BWY2597) and quintuple deletion cells (5Δ) with an ENT1 TRP1 or an ENTH1 TRP1 plasmid and a second URA3 plasmid (BWY2571), either empty or encoding ENT2, were grown on 5-FOA containing media for 3 d to evict the URA3 plasmid. (D) Ede1 is required for internalization of ligand-bound Ste2. ΔΔΔΔ and 5Δ (BWY2597 and BWY2574, respectively) cells expressing Ent1 were tested for internalization of radiolabeled α-factor at 30°C. Cells were grown on selective media at 30°C. Radiolabeled α-factor was added to mid-log phase cultures at 30°C. Each point represents the average of three or more assays; error bars represent the SD. Results for the Ste3-GFP localization assay can be found in Supplemental Figure 6C.
To more directly test the hypothesis that NPF motif/EH domain interactions are mediating these effects, we generated a mutant Ede1EH--RFP chimera, in which the critical tryptophan residue in each of the three EH domains was replaced with alanine to abrogate binding to NPF motifs (de Beer et al., 1998; Smythe and Ayscough, 2006). Consistent with our results from adaptor mutant cells, high intensity patches were seen when the localization of the Ede1EH--RFP mutant was examined in WT cells (Supplemental Figure S6, A and B). These results in adaptor mutant cells and in cells in which Ede1 cannot directly bind adaptors could be due to an increase in the number of Ede1-RFP molecules recruited to endocytic sites, a reduced rate of their dissipation, or clustered/aggregated patches as are seen in end3 and ark1/prk1 endocytic mutant cells (Benedetti et al., 1994; Cope et al., 1999). However, we did not observe large clumps of F-actin by rhodamine-phalloidin staining of ΔΔΔΔ+ENTH1 cells grown at 30°C (data not shown), suggesting that the bright Ede1 patches most likely represent an early intermediate in the endocytic process. We conclude that the adaptor proteins are necessary for the normal distribution of the early scaffold Ede1.
Ede1 Is Essential for Endocytosis and Viability in Adaptor Mutant Cells
Because ΔΔΔΔ+ENTH1 cells were viable and could still internalize some Ste3-GFP at a low rate at the permissive temperature (30°C), we suspected that still other redundant factors may remain that can partially compensate for the absent adaptor proteins. We postulated that Ede1, a putative dual adaptor/scaffold protein related to Eps15 (Miliaras and Wendland, 2004; Sigismund et al., 2005), might partially complement the endocytic and viability function of the adaptors. We deleted EDE1 in ΔΔΔΔ+Ent1 cells to create a quintuple deletion mutant expressing Ent1 (5Δ+Ent1). These cells grew more slowly than WT cells at 30°C, and they had a moderate growth defect at 37°C (data not shown).
To test a role for Ede1 in ΔΔΔΔ+ENTH1 cells, we used plasmid shuffling to swap in an ENTH1 domain plasmid into 5Δ+Ent2 cells. When plated on 5-FOA, 5Δ+ENTH1 cells were inviable under all conditions tested (Figure 6C), indicating that Ede1 is essential in these cells. Because 5Δ+ENTH1 cells were inviable, we tested endocytosis in 5Δ+Ent1 cells. Analysis of the internalization of α-factor and Ste3-GFP localization revealed severe endocytic defects in 5Δ+Ent1 cells at 30°C (Figure 6D and Supplemental Figure S6C). Interestingly, the Ent1CBM- truncation was the only C-terminal Ent1 truncation able to complement 5Δ cells (data not shown), indicating that the CBM is dispensable for viability in these 5Δ cells at 30°C. Together, our results demonstrate that Ede1 also shares an endocytic role with the epsin and Yap180 proteins, consistent with a dual adaptor-scaffold role for Ede1 in yeast.
DISCUSSION
NPF Motifs in the Epsin and Yap180 Proteins Mediate a Shared Endocytic Function
The structural, biochemical, and cellular properties of the yeast and mammalian epsins are conserved. Our study shows that the Yap1801/2 proteins can fulfill the nonessential endocytic role of the epsins (or vice versa), uncovering a previously unknown redundant relationship between these two protein families. We found that the Yap180 proteins mask endocytosis defects in ΔΔ+ENTH1 cells lacking epsin C termini. Like full-length Ent1, GFP-tagged Ent1C-termWT localizes to patches at the plasma membrane in vivo and complements ΔΔΔΔ+ENTH1 cells. Thus, the in trans complementation is most likely via independent action of the fragments (Overstreet et al., 2003), as opposed to the fragments associating with each other. Using C-terminal truncations and point mutants, we mapped the minimal region required for viability and endocytosis in ΔΔΔΔ cells to the NPF motifs, although the UIMs and CBM also played subordinate roles. The need to mutate multiple motifs for the most severe effects is consistent with a cooperative role for multiple functional domains in adaptors; similarly, individual motif mutations in the AP2 endocytic adaptor showed only minor deficits in vivo (Motley et al., 2006). Nonetheless, the data point to the NPF motifs of adaptors providing key interactions in mediating endocytic events.
NPF Motifs Act in Concert with Other Adaptor Motifs
A well-characterized property of the clathrin adaptors is the ability to stimulate clathrin cage assembly in vitro, but this activity is under regulatory control in vivo. In fact, a squid Eps15/AP180 complex stimulates the clathrin cage-assembly activity of squid AP180, and it is mediated by the NPF motif/EH domain interaction (Morgan et al., 2003). Like AP180 and CALM, epsin has also been shown to stimulate clathrin assembly in vitro (Kalthoff et al., 2002). It is tempting to speculate that the severe defects we observed for the Ent1NPF-/CBM- mutant reflect the existence of a stimulatory effect of EH domain interaction with NPF motifs on clathrin assembly during endocytosis in yeast cells, analogous to what was seen for squid Eps15/AP180.
It has been previously found that when mammalian epsin is ubiquitinated, its binding to clathrin is inhibited (Chen et al., 2003). Because UIMs must be able to bind ubiquitin for the UIM-bearing protein to be covalently modified by ubiquitin (Polo et al., 2002), it is possible that the Ent1UIM- point mutants could produce a nonubiquitinated form of the yeast epsins. The minor phenotypic defects we observed with the Ent1UIM- mutant suggest that Ent1 binding to or modification by ubiquitin is not a strict requirement for epsin function, but given the high degree of redundancy we have observed for other aspects of adaptor function, it is likely that future experiments will reveal roles for the UIM domains as well. Although it has not been definitively shown that yeast epsins are ubiquitinated, we have found that they harbor clathrin cage assembly activity in vitro (Baggett, Maldonado-Baez, Prasad, Lafer, and Wendland, unpublished observation). The deleterious effect of mutating both the UIMs and the CBM in yeast epsin could suggest a UIM- or ubiquitin-dependent regulation of adaptor clathrin-assembly activity.
Another interesting genetic interaction we observed was the strong defect when both the lipid-binding pocket and the NPFs were mutated. It is likely that the yeast epsin ENTH domain can bend/destabilize membranes, because it has a predicted amphiphathic helix 0 at its N terminus like mammalian epsins (Ford et al., 2002). It would be interesting if the genetic interaction were due to a greater need for these membrane interactions when adaptor clathrin assembly stimulation is reduced as a result of the NPF mutation. This could be the case if the recent proposal of clathrin assembly being the main driving force for curved membrane regions holds true in yeast (Hinrichsen et al., 2006).
Clearly, although the NPF motifs are the dominant functional motif in the yeast epsin and AP180 proteins, the more severe effects of the C-terminal deletion and combination mutants are indicative of functions beyond just the NPF motifs. Similarly, it is also possible that the EH domains of the scaffolds have other important ligands besides the adaptors; we are currently exploring additional regulatory roles that may be mediated by EH domains.
Clathrin Adaptors Influence the Dynamics of Scaffold Proteins
Our real-time fluorescence microscopy data revealed that the adaptor NPF motif/EH domain scaffold interaction is necessary for the normal spatiotemporal behavior of the endocytic scaffolds Ede1 and Pan1. In the absence of adaptors, both Ede1 and Pan1 patches localized to the plasma membrane, indicating that adaptors are not absolutely required for the initial recruitment of either protein to the cell cortex. EH-independent interactions between Pan1 and its partners Sla1, End3, and the ANTH domain-containing protein Sla2 may provide additional membrane-targeting components for Pan1 (Tang et al., 2000; Toshima et al., 2007). Less is known about other Ede1 partners that could circumvent a role for adaptors in membrane targeting, although a genome-wide approach identified a complex of Ede1 with Sla2 (Gavin et al., 2002; Gavin et al., 2006).
Analysis of Pan1-GFP patches in ΔΔΔΔ+Ent1, ΔΔΔΔ+ENTH1 and ΔΔΔΔ+Ent1NPF- cells revealed two phenotypes for Pan1 patch dynamics. First ΔΔΔΔ+Ent1 and ΔΔΔΔ+Ent1NPF- cells had Pan1 patches with half their normal lifetime, similar to observations of Sla1-GFP and Las17-GFP patches in clathrin mutants. This could be due to a role for clathrin in regulating patch assembly/disassembly (Kaksonen et al., 2005). Lemmon and colleagues have demonstrated that Ent1/2 and Yap1801/2 are required for the cortical localization of clathrin (Newpher et al., 2005), so ΔΔΔΔ cells are also likely to have deficient clathrin recruitment, which may phenocopy some clathrin mutant effects. Second, Pan1-GFP patches in ΔΔΔΔ+ENTH1 and ΔΔΔΔ+Ent1NPF- cells exhibited a population of immobile patches. A subset of Pan1EH--GFP patches exhibited a similar greatly extended lifetime phenotype with no apparent inward movement. These data are consistent with the adaptor NPF/scaffold EH domain interaction leading to a regulatory event necessary for endocytic sites to progress to later stages of internalization that lead to the formation of an endocytic vesicle. Deleting Sla2 (or its coiled-coil region) also impairs the internalization of Pan1 and increases its lifetime (Toshima et al., 2007); however, sla2 mutants increase plasma membrane-associated clathrin pools (Newpher et al., 2005). Thus, there is no simple correlation between clathrin behavior and Pan1 patch lifetimes; clearly, this complex system will benefit from further study.
Ede1 Becomes an Essential Dual Adaptor/Scaffold in Adaptor Mutant Cells
The NPF motif function of the adaptors cannot be fully compensated by other endocytosis proteins, as indicated by the delay in endocytosis at 30°C and its complete block at 37°C in ΔΔΔΔ+ENTH1 cells. However, we showed that Ede1 partially compensates for the epsins and Yap180s, because it becomes essential for viability in ΔΔΔΔ+ENTH1 cells. Ede1 may bypass the clathrin-recruiting function of the adaptors by indirectly recruiting clathrin to endocytic sites through its membership in a Sla2-containing protein complex (Gavin et al., 2002, 2006), because Sla2 interacts directly with clathrin (Baggett et al., 2003; Newpher and Lemmon, 2006; Sun et al., 2006). Ede1 may also bypass the adaptor-dependent ubiquitin functions, because it binds to ubiquitinated plasma membrane proteins through its UBA domain (Shih et al., 2002; Aguilar et al., 2003). The Ede1-related mammalian protein Eps15 is a dual adaptor/scaffold protein that mediates ubiquitinated-cargo internalization, regulates the clathrin-cage assembly activity of AP180, functions as a scaffold protein for Epsin, AP180 and Stonin 2 and interacts with the mammalian Pan1-related protein Intersectin (Polo et al., 2003). Thus, we suggest that that Ede1 is functionally related to the dual adaptor/scaffold protein Eps15 (Morgan et al., 2003; Maldonado-Baez and Wendland, 2006).
Clathrin Adaptor and Scaffold Protein Interaction Regulates the Progression of Endocytosis
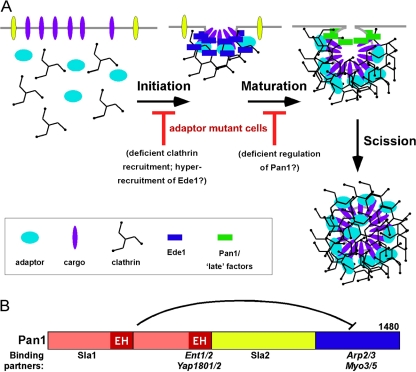
Our data suggest that the adaptors exert a regulatory influence on the endocytic process, perhaps by imposing an order to a sequence of protein complexes, or by acting as or triggering a switch that promotes a transition from early to late events. Based on a plethora of results in the literature and our own findings, we explain the role of the NPF motif/EH domain interaction as follows. The presence of long-lived immotile Pan1-GFP patches in ΔΔΔΔ+ENTH1 or ΔΔΔΔ+Ent1NPF- cells suggest that a stalled intermediate complex accumulates, and that NPF/EH interactions resolve this intermediate to form the next complex(es) in the pathway (Figure 7A). This further suggests that the NPF/EH interaction may either relieve an inhibited state or stimulate the transition of this complex to facilitate vesicle formation.
Figure 7.
The NPF motif/EH domain Adaptor-Scaffold Regulation Model (A) Transitions between the distinct stages of endocytic internalization may be regulated by adaptor/scaffold interactions. In adaptor mutant cells, cargo and clathrin recruitment are compromised and the spatiotemporal dynamics of the early scaffold Ede1 are aberrant, potentially altering the initiation step. The spatiotemporal dynamics of the late scaffold Pan1 are perturbed in adaptor mutant cells; Pan1 is recruited to cortical patches, but it does not exhibit the usual inward movement that marks the invagination/scission step of internalization. That Pan1 is not bound to the adaptors may misregulate Pan1, causing the loss of recruitment of late factors that provide the forces for membrane invagination. (B) Schematic of Pan1 and its major binding partners relevant to this work. The inhibitory interaction between the N terminus (red) and C terminus (blue) is indicated.
Our mechanistic view of the role of NPF–EH interactions in endocytosis focuses on the regulation of Pan1, in part due to its presence at cortical patches after Ede1 has dissociated (Kaksonen et al., 2005). Because Pan1 is the last EH-containing protein present during the endocytic process, it is the best candidate for a factor that could be regulated by NPF interactions. We would like to understand why Pan1-GFP patch inward movement requires the adaptors. Figure 7B shows a schematic of Pan1 with its key binding partners; the N terminus (which binds the adaptors) and the C terminus (which binds and activates Arp2/3 and the type I myosin Myo5) are indicated; Pan1 N terminus inhibition of the C-terminal Arp2/3 activity is also shown (Toshima et al., 2005; Barker et al., 2007). Actin polymerization promotes vesicle invagination and scission (Sun et al., 2006; Toret and Drubin, 2007); thus, we speculate that the adaptors mediate an indirect effect on the stimulation of the actin machinery through interactions with Pan1. More definitive proof of this idea awaits further investigation, including analysis of the dynamics of late-acting components involved in the final stages of endocytosis (actin polymerization, vesicle invagination, and scission) in adaptor mutant cells.
NPF/EH regulation of Pan1 could occur via a direct interaction of adaptors with Pan1 to induce a conformational change that promotes Pan1 C-terminal activities. This could be a direct relief of inhibition; alternatively, an indirect mechanism via another protein(s) may occur. For example, the missing adaptors could reduce clathrin assembly, which could then affect Sla2 activity, a known clathrin-binding protein that is also a Pan1 inhibitor (Newpher et al., 2005; Toshima et al., 2006) Because the adaptors and Sla1 both bind to the N-terminal region of Pan1, the adaptors could also affect the function of Sla1. Interestingly, sla1Δ is lethal in cells with a Pan1 mutant that cannot bind Myo5 (Barker et al., 2007), providing another example of functional connections between the Pan1 N terminus (Sla1 binding) and the Pan1 C terminus (Myo5 binding and actin regulation). Thus, adaptors are good candidates to regulate Pan1, which in turn likely shepherds the transitions of many endocytic proteins between the distinct stages of internalization.
This model also predicts that in the absence of the adaptors or some function they provide (such as cargo recruitment or local clathrin assembly), a prerequisite for the recruitment or regulation of factors that mediate the onset of actin polymerization is also absent. In preliminary studies, we have observed reduced recruitment of the late-acting type I myosin Myo5 in the adaptor mutant cells (our unpublished data). Because neither clathrin nor adaptors are known to bind directly or activate Myo5, it is appealing to speculate that the adaptors mediate a conformational change in Pan1 that triggers a transition so that Pan1 can bind Myo5 and commence the final stages of endocytic internalization. Future work will focus on how the different protein complexes rearrange, how the interactions are regulated, and how the sequence of events is orchestrated during each stage of clathrin-mediated endocytosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Claudio Aguilar (Purdue University), Greg Payne (University of California, Los Angeles), and Rob Piper (University of Iowa) for strains, plasmids, and other reagents. We also thank Claudio Aguilar, Sarah Barker, Michael Edidin, Rejji Kuruvilla, Susan Michaelis, and Linton Traub for helpful discussions and members of the Wendland laboratory for constructive comments on the manuscript. We are grateful to Karen Whitworth, Jennifer Baggett, Sarah Barker, Pavel Bogomiakov, Lan-Yu Yeh, and Sean Kim for help with the making of strains and to Nathan Wright, Keisha Meeks, and Lauren Parlett for technical assistance. We thank Michael McCaffery and Michelle Husain of the Integrated Imaging Center (IIC) for help and advice with light microscopy. This work was supported by National Institutes of Health (NIH) grants R01 GM-60979 (to B.W.), NIH-NCRR 1S10RR022588-01 (IIC shared instrumentation grant; B.W., co-principle investigator), R01 DK0532570 (to L.H.), and a Ford Foundation Diversity Fellowship (to L.M.B.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1019) on April 30, 2008.
REFERENCES
- Aguilar R. C, et al. Epsin N-terminal homology domains perform an essential function regulating Cdc42 through binding Cdc42 GTPase-activating proteins. Proc. Natl. Acad. Sci. USA. 2006;103:4116–4121. doi: 10.1073/pnas.0510513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar R. C., Watson H. A., Wendland B. The yeast epsin Ent1 is recruited to membranes through multiple independent interactions. J. Biol. Chem. 2003;278:10737–10743. doi: 10.1074/jbc.M211622200. [DOI] [PubMed] [Google Scholar]
- Augustine G. J., Morgan J. R., Villalba-Galea C. A., Jin S., Prasad K., Lafer E. M. Clathrin and synaptic vesicle endocytosis: studies at the squid giant synapse. Biochem. Soc. Trans. 2006;34:68–72. doi: 10.1042/BST0340068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggett J. J., D'Aquino K. E., Wendland B. The Sla2p Talin domain plays a role in endocytosis in Saccharomyces cerevisiae. Genetics. 2003;165:1661–1674. doi: 10.1093/genetics/165.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker S. L., Lee L., Pierce B. D., Maldonado-Baez L., Drubin D. G., Wendland B. Interaction of the endocytic scaffold protein Pan1 with the type I myosins contributes to the late stages of endocytosis. Mol. Biol. Cell. 2007;18:2893–2903. doi: 10.1091/mbc.E07-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti H., Raths S., Crausaz F., Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol. Biol. Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A., Poupon V., Cerf-Bensussan N., Dautry-Varsat A. Mapping of Eps15 domains involved in its targeting to clathrin-coated pits. J. Biol. Chem. 2000;275:3288–3295. doi: 10.1074/jbc.275.5.3288. [DOI] [PubMed] [Google Scholar]
- Chen H., Fre S., Slepnev V. I., Capua M. R., Takei K., Butler M. H., Di Fiore P. P., De Camilli P. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- Chen H., Polo S., Di Fiore P. P., De Camilli P. V. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc. Natl. Acad. Sci. USA. 2003;100:14908–14913. doi: 10.1073/pnas.2136625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope M. J., Yang S., Shang C., Drubin D. G. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J. Cell Biol. 1999;144:1203–1218. doi: 10.1083/jcb.144.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T., Carter R. E., Lobel-Rice K. E., Sorkin A., Overduin M. Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain. Science. 1998;281:1357–1360. doi: 10.1126/science.281.5381.1357. [DOI] [PubMed] [Google Scholar]
- De Camilli P., Chen H., Hyman J., Panepucci E., Bateman A., Brunger A. T. The ENTH domain. FEBS Lett. 2002;513:11–18. doi: 10.1016/s0014-5793(01)03306-3. [DOI] [PubMed] [Google Scholar]
- Dulic V., Egerton M., Elguindi I., Raths S., Singer B., Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- Duncan M. C., Cope M., Goode B. L., Wendland B., Drubin D. G. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Boll W., van Oijen A., Hariharan R., Chandran K., Nibert M. L., Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Evans P. R., Owen D. J. Endocytosis and vesicle trafficking. Curr. Opin. Struct. Biol. 2002;12:814–821. doi: 10.1016/s0959-440x(02)00395-0. [DOI] [PubMed] [Google Scholar]
- Ford M.G.J., Mills I. G., Peter B. J., Vallis Y., Praefcke G.J.K., Evans P. R., McMahon H. T. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Ford M.G.J., Pearse B.M.F., Higgins M. K., Vallis Y., Owen D. J., Gibson A., Hopkins C. R., Evans P. R., McMahon H. T. Simultaneous binding of PtdIns(4,5)P-2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Gagny B., Wiederkehr A., Dumoulin P., Winsor B., Riezman H., Haguenauer-Tsapis R. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J. Cell Sci. 2000;113:3309–3319. doi: 10.1242/jcs.113.18.3309. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Geli M. I., Riezman H. Endocytic internalization in yeast and animal cells: similar and different. J. Cell Sci. 1998;111:1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- Hicke L., Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Hinrichsen L., Meyerhoiz A., Groos S., Ungewickell E. J. Bending a membrane: how clathrin affects budding. Proc. Natl. Acad. Sci. USA. 2006;103:8715–8720. doi: 10.1073/pnas.0600312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. P., Hutton J. L., Olson J. M., Payne G. S. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J. Cell Biol. 2002;157:315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. M., D'Hondt K., Riezman H., Lemmon S. K. Clathrin functions in the absence of heterotetrameric adaptors and AP180-related proteins in yeast. EMBO J. 1999;18:3897–3908. doi: 10.1093/emboj/18.14.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain N. K., et al. Endocytic protein intersectin-I regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol. 2001a;3:927–932. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- Hussain N. K., Jenna S., Glogauer M., Quinn C. C., Wasiak S., Kay B. K., Stossel T. P., Lamarche-Vane N., McPherson P. S. The endocytic protein intersectin-1 regulates actin assemble through a novel mechanism of N-WASP activation. Mol. Biol. Cell. 2001b;12:289A–289A. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- Itoh T., Koshiba S., Kigawa T., Kikuchi A., Yokoyama S., Takenawa T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., Drubin D. G. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Kalthoff C., Alves J., Urbanke C., Knorr R., Ungewickell E. J. Unusual structural organization of the endocytic proteins AP180 and epsin 1. J. Biol. Chem. 2002;277:8209–8216. doi: 10.1074/jbc.M111587200. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Madshus I. H. Ubiquitin binding in endocytosis–how tight should it be and where does it happen? Traffic. 2006;7:258–261. doi: 10.1111/j.1600-0854.2006.00393.x. [DOI] [PubMed] [Google Scholar]
- Maldonado-Baez L., Wendland B. Endocytic adaptors: recruiters, coordinators and regulators. Trends Cell Biol. 2006;16:505–513. doi: 10.1016/j.tcb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Merrifield C. J., Perrais D., Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Meyerholz A., Hinrichsen L., Groos S., Esk P. C., Brandes G., Ungewickell E. J. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic. 2005;6:1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- Miliaras N. B., Park J. H., Wendland B. The function of the endocytic scaffold protein Pan1p depends on multiple domains. Traffic. 2004;5:963–978. doi: 10.1111/j.1600-0854.2004.00238.x. [DOI] [PubMed] [Google Scholar]
- Miliaras N. B., Wendland B. EH proteins–multivalent regulators of endocytosis (and other pathways) Cell Biochem. Biophys. 2004;41:295–318. doi: 10.1385/CBB:41:2:295. [DOI] [PubMed] [Google Scholar]
- Morgan J. R., Prasad K., Jin S. P., Augustine G. J., Lafer E. M. Eps15 homology domain-NPF motif interactions regulate clathrin coat assembly during synaptic vesicle recycling. J. Biol. Chem. 2003;278:33583–33592. doi: 10.1074/jbc.M304346200. [DOI] [PubMed] [Google Scholar]
- Motley A. M., Berg N., Taylor M. J., Sahlender D. A., Hirst J., Owen D. J., Robinson M. S. Functional analysis of AP-2α and μ 2 subunits. Mol. Biol. Cell. 2006;17:5298–5308. doi: 10.1091/mbc.E06-05-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N., Caplan S. C-Terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J. Cell Sci. 2005;118:4093–4101. doi: 10.1242/jcs.02595. [DOI] [PubMed] [Google Scholar]
- Newpher T. M., Lemmon S. K. Clathrin is important for normal actin dynamics and progression of Sla2p-containing patches during endocytosis in yeast. Traffic. 2006;7:574–588. doi: 10.1111/j.1600-0854.2006.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher T. M., Smith R. P., Lemmon V., Lemmon S. K. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Okamoto M., Schoch S., Sudhof T. C. EHSH1/intersectin, a protein that contains EH and SH3 domains and binds to dynamin and SNAP-25–A protein connection between exocytosis and endocytosis? J. Biol. Chem. 1999;274:18446–18454. doi: 10.1074/jbc.274.26.18446. [DOI] [PubMed] [Google Scholar]
- Overstreet E., Chen X., Wendland B., Fischer J. A. Either part of a Drosophila epsin protein, divided after the ENTH domain, functions in endocytosis of delta in the developing eye. Curr. Biol. 2003;13:854–860. doi: 10.1016/s0960-9822(03)00326-9. [DOI] [PubMed] [Google Scholar]
- Owen D. J. Linking endocytic cargo to clathrin: structural and functional insights into coated vesicle formation. Biochem. Soc. Trans. 2004;32:1–14. doi: 10.1042/bst0320001. [DOI] [PubMed] [Google Scholar]
- Owen D. J., Collins B. M., Evans P. R. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Polo S., Confalonieri S., Salcini A. E., Di Fiore P. P. EH and UIM: endocytosis and more. Sci. STKE. 2003;2003:re17. doi: 10.1126/stke.2132003re17. [DOI] [PubMed] [Google Scholar]
- Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- Shih S. C., Katzmann D. J., Schnell J. D., Sutanto M., Emr S. D., Hicke L. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- Sigismund S., Woelk T., Puri C., Maspero E., Tacchetti C., Transidico P., Di Fiore P. P., Polo S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA. 2005;102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe E., Ayscough K. R. Actin regulation in endocytosis. J. Cell Sci. 2006;119:4589–4598. doi: 10.1242/jcs.03247. [DOI] [PubMed] [Google Scholar]
- Sorkin A. Cargo recognition during clathrin-mediated endocytosis: a team effort. Curr. Opin. Cell Biol. 2004;16:392–399. doi: 10.1016/j.ceb.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Stahelin R. V., Long F., Peter B. J., Murray D., De Camilli P., McMahon H. T., Cho W. Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J. Biol. Chem. 2003;278:28993–28999. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]
- Sun Y. D., Martin A. C., Drubin D. G. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev. Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Szymkiewicz I., Shupliakov O., Dikic I. Cargo- and compartment-selective endocytic scaffold proteins. Biochem. J. 2004;383:1–11. doi: 10.1042/BJ20040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Y., Munn A., Cai M. J. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Y., Xu J., Cai M. J. Pan1p, End3p, and Sla1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell. Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebar F., Bohlander S. K., Sorkin A. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol. Biol. Cell. 1999;10:2687–2702. doi: 10.1091/mbc.10.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toret C. P., Drubin D. G. The budding yeast endocytic pathway. J. Cell Sci. 2007;120:1501–1501. doi: 10.1242/jcs.03251. [DOI] [PubMed] [Google Scholar]
- Toshima J., Toshima J. Y., Duncan M. C., Cope M., Sun Y. D., Martin A. C., Anderson S., Yates J. R., Mizuno K., Drubin D. G. Negative regulation of yeast Eps15-like Arp2/3 complex activator, Pan1p, by the Hip1R-related protein, Sla2p, during endocytosis. Mol. Biol. Cell. 2007;18:658–668. doi: 10.1091/mbc.E06-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima J., Toshima J. Y., Martin A. C., Drubin D. G. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat. Cell Biol. 2005;7:246–U244. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- Toshima J. Y., Toshima J., Kaksonen M., Martin A. C., King D. S., Drubin D. G. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc. Natl. Acad. Sci. USA. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L. M. Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. J. Cell Biol. 2003;163:203–208. doi: 10.1083/jcb.200309175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski J. L., Piper R. C. Ubiquitin sorts proteins into the intralumenal degradative compartment of the late-endosome/vacuole. Traffic. 2001;2:622–630. doi: 10.1034/j.1600-0854.2001.20905.x. [DOI] [PubMed] [Google Scholar]
- Wechsler D. S., Engstrom L. D., Alexander B. M., Motto D. G., Roulston D. A novel chromosomal inversion at 11q23 in infant acute myeloid leukemia fuses MLL to CALM, a gene that encodes a clathrin assembly protein. Genes Chromosomes Cancer. 2003;36:26–36. doi: 10.1002/gcc.10136. [DOI] [PubMed] [Google Scholar]
- Wendland B. Epsins: adaptors in endocytosis? Nat. Rev. Mol. Cell. Biol. 2002;3:971–977. doi: 10.1038/nrm970. [DOI] [PubMed] [Google Scholar]
- Wendland B., Emr S. D. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B., Steece K. E., Emr S. D. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamabhai M., Hoffman N. G., Hardison N. L., McPherson P. S., Castagnoli L., Cesareni G., Kay B. K. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J. Biol. Chem. 1998;273:31401–31407. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- Zhang B., Koh Y. H., Beckstead R. B., Budnik V., Ganetzky B., Bellen H. J. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron. 1998;21:1465–1475. doi: 10.1016/s0896-6273(00)80664-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.