Abstract
Promyelocytic leukemia protein (PML) nuclear bodies (NBs) are dynamic subnuclear compartments that play roles in several cellular processes, including apoptosis, transcriptional regulation, and DNA repair. Histone deacetylase (HDAC) 7 is a potent corepressor that inhibits transcription by myocyte enhancer factor 2 (MEF2) transcription factors. We show here that endogenous HDAC7 and PML interact and partially colocalize in PML NBs. Tumor necrosis factor (TNF)-α treatment recruits HDAC7 to PML NBs and enhances association of HDAC7 with PML in human umbilical vein endothelial cells. Consequently, TNF-α promotes dissociation of HDAC7 from MEF2 transcription factors and the promoters of MEF2 target genes such as matrix metalloproteinase (MMP)-10, leading to accumulation of MMP-10 mRNA. Conversely, knockdown of PML enhances the association between HDAC7 and MEF2 and decreases MMP-10 mRNA accumulation. Accordingly, ectopic expression of PML recruits HDAC7 to PML NBs and leads to activation of MEF2 reporter activity. Notably, small interfering RNA knockdown of PML decreases basal and TNF-α-induced MMP-10 mRNA accumulation. Our results reveal a novel mechanism by which PML sequesters HDAC7 to relieve repression and up-regulate gene expression.
INTRODUCTION
The promyelocytic leukemia (PML) protein is involved in transcriptional regulation, apoptosis, and oncogenesis (Maul et al., 2000; Borden, 2002; Ching et al., 2005). A fraction of PML is nucleoplasmic and is enriched in discrete nuclear structures, referred to as either Kremer bodies, nuclear domain 10, PML oncogenic domains, or PML nuclear bodies (NBs). PML is capable of regulating transcriptional activity either positively (Tsuzuki et al., 2000; Lin et al., 2003) or negatively (Li and Chen, 2000; Li et al., 2000; Vallian et al., 1998; Wu et al., 2002). The mechanisms by which PML regulates transcription are not completely understood.
Class II histone deacetylases (HDACs), including HDAC4, -5, -7, and -9, are potent transcriptional corepressors (Fischle et al., 1999; Grozinger et al., 1999; Verdel and Khochbin, 1999; Kao et al., 2000, 2002; Zhou et al., 2001; Fischer et al., 2002; Guardiola and Yao, 2002). They associate with myocyte enhancer factor 2 (MEF2) and inhibit its ability to activate target genes such as matrix metalloproteinase (MMP)-10 (Chang et al., 2006), a zinc-dependent matrix metalloprotease known to regulate cell–cell interactions, including those involving smooth muscle and endothelial cells. As such, the ability of MEF2 to activate target genes partly depends on the subcellular distribution of class II HDACs. Nucleocytoplasmic shuttling of class II HDACs is controlled by a highly conserved N-terminal nuclear localization sequence, a C-terminal nuclear export sequence, and phosphorylation (Verdin et al., 2003). Subsequent cytoplasmic retention of HDAC7 is thought to be one mechanism leading to MEF2 activation. In addition to nucleocytoplasmic trafficking, we have found that endogenous HDAC7 exhibits nuclear punctate staining resembling PML NBs, but the functional significance of this localization remains largely unknown. In this study, we have uncovered a novel regulatory pathway by which the proinflammatory cytokine tumor necrosis factor (TNF)-α induces PML expression and enhances the formation of PML NBs. We have also found that induction of PML by TNF-α recruits HDAC7 to PML NBs and derepresses the expression of MEF2 target genes, including MMP-10.
MATERIALS AND METHODS
Plasmids and DNA Constructs
All PML plasmids used in this study are PML4 expression plasmids and are referred to as PML. PML and HDAC7 expression plasmids have been described previously (Xu et al., 2004; Gao et al., 2006). The CMX-HA-PML4 expression plasmid was generated by polymerase chain reaction (PCR) and subcloned into the cytomegalovirus-promoter based CMX-HA (1H) vector (Chakraborty et al., 2006). Truncated PML and HDAC7 cDNAs were PCR amplified using full-length PML4 and HDAC7 as templates. HA-PML (3KR) was generated by site-directed mutagenesis (Strategene, La Jolla, CA). Yellow fluorescent protein-HDAC7, hemagglutinin (HA)-HDAC4, and HA-HDAC5 expression plasmids were generated by PCR. All plasmids were confirmed by sequencing. The MEF2 reporter construct has been described previously (Kao et al., 2001).
Reagents and Antibodies
TNF-α was purchased from Promega (Madison, WI). HDAC7 antibodies have been described previously (Gao et al., 2006). The specificity of anti-HDAC7 antibodies has been determined previously (Gao et al., 2006) and shown by small interfering RNA (siRNA) knockdown experiment (Supplemental Figure S9). The following antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA): α-CBP (sc-369), α-PML (sc-966 & sc-5621), α-actin (sc-1615), α-Daxx (sc-7152), α-MEF2 (sc-313x), α-HDAC1 (sc-7872), α-HDAC2 (sc-7899), α-HDAC3 (sc-11417), α-HDAC4 (sc-11418), and α-HDAC5 (sc-11419). Anti-HA antibodies (2013819) were purchased from Roche Diagnostics (Indianapolis, ID). Anti-tubulin antibodies (T6074), anti-HA antibody-conjugated beads (E6779), anti-FLAG antibody-conjugated beads (F2426), and anti-FLAG antibodies (F3165) were purchased from Sigma-Aldrich (St. Louis, MO). Anti-acetyl-histone H4 antibodies (06-598) were purchased from Millipore (Billerica, MA). Anti-HDAC6 was a generous gift from Dr. X. Zhang (University of South Florida). Anti-N-CoR antibodies were generated and purified in our laboratory. Anti-HA horseradish peroxidase (2013819) was from Roche Diagnostics.
Reverse Transcription (RT)-PCR
Total RNA was extracted from human umbilical vein endothelial cells (HUVECs) by using the RNeasy Mini kit (QIAGEN, Valencia, CA) and used as a template for reverse transcriptase with poly(dT) primer (Invitrogen, Carlsbad, CA). PCR reactions were performed on first-strand cDNA with the following primers: PML forward, 5′-GAATCAACGAATGAATGGCT-3′ and PML reverse, 5′-CCAGGGACTCAGAATACAGG-3′; MMP-10 forward, 5′-GCATTTTGGCCCTCTCTTC-3′ and MMP-10 reverse, 5′-CAGGGTATGGATGCCTCTTG-3′; HDAC7 forward, 5′-GCACCCAGCAAACCTTCTAC-3′ and HDAC7 reverse, 5′-AGCCCCTACCTCATCCACAG-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5′-GAAGGTGAAGGTCGGAGT-3′ and GAPDH reverse 5′-GAAGATGGTGATGGGATTTC-3′.
Real-Time PCR
Total RNA was extracted from HUVECs 72 h after transfection of siRNA with the RNeasy Mini kit (QIAGEN) and reverse transcribed into a cDNA pool by using Superscript II reverse transcriptase following the manufacturer's instructions (Invitrogen). The cDNAs were quantified by real-time PCR by using an iCycler (Bio-Rad, Hercules, CA) with the iQ SYBR Green Supermix kit (Bio-Rad) and appropriate primer sets. Forty cycles of PCR were performed with three temperature steps of 95°C for 10 s, 55°C for 20 s, and 72°C for 30 s. Subsequently, the melting curves were examined to ensure the homogeneity of the PCR products. The relative quantities of genes of interest were normalized to GAPDH and compared between samples treated by siPML and control siControl.
Cell Culture, Transfection, and siRNA
HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum, 50 U/ml penicillin G, and 50 μg of streptomycin sulfate at 37°C in 5% CO2. HUVECs were isolated from freshly obtained umbilical cord samples by collagenase digestion of the umbilical vein. HUVECs were a gift from Dr. Matsuyama (Case Western Reserve University, Cleveland, OH) and maintained in endothelial cell basal medium containing endothelial growth medium-2 singlequot growth supplements (Cambrex, East Rutherford, NJ). HUVEC passages 2–5 were used in this study. HUVECs were treated with TNF-α (20 ng/ml) for up to 20 h. Control, PML, and HDAC7 siRNA SMART pools were purchased from Dharmacon RNA Technologies (Lafayette, CO). siRNA duplexes were transfected into HUVECs according to manufacturer's protocol. For mRNA expression levels, total RNA was isolated 48 h after transfection followed by RT-PCR (Invitrogen). Transient transfections and luciferase assays were performed in 48-well culture plates. HeLa cells were transfected using Lipofectamine 2000 (Invitrogen). The amount of DNA was kept constant (0.6 μg for Figure 4A and 0.7 μg for Figure 4, C and E) by addition of pCMX vector. Five hours after transfection, medium was replaced, and the cells were harvested 48 h after transfection followed by luciferase and β-galactosidase (β-gal) activity assays (Promega). Each reaction was performed in triplicate.
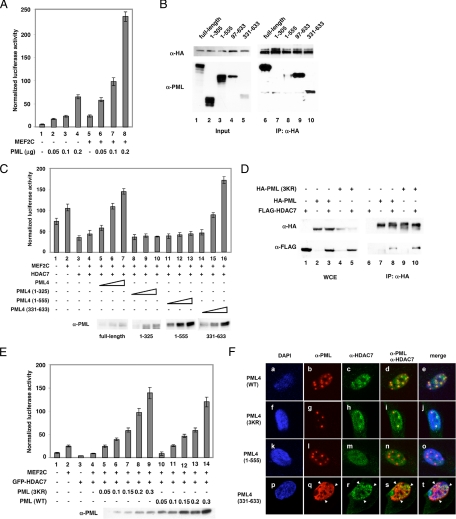
Figure 4.
Coexpression of PML potentiates MEF2 transcription activity. (A) An MEF2 reporter construct (Kao et al., 2001) was cotransfected into HeLa cells with an internal control plasmid, CMX-β-Gal, MEF2C, or PML expression plasmids. Luciferase activity was measured according to our published protocol (Chakraborty et al., 2006). (B) HA-HDAC7 and full-length or truncated PML expression plasmids were cotransfected into HeLa cells. Whole cell extracts were prepared and immunoprecipitated with anti-HA antibodies followed by Western blotting with anti-PML and anti-HA antibodies. (C) Transient transfection reporter assays were performed as described in B with increasing amounts of full-length PML (lanes 5–7) or PML truncations (lanes 8–16) into HeLa cells. Bottom, expression of full-length PML or truncated PML as detected by PML antibodies. A similar trend was observed when HDAC7 was not ectopically expressed (data not shown). (D) Wild-type HA-PML or HA-PML mutant (K65R/K160R/K490R, 3KR) was cotransfected with a FLAG-HDAC7 expression plasmid in HeLa cells. Extracts were prepared and subjected to immunoprecipitation as indicated. Western blotting was performed using the indicated antibodies. (E) Transient transfection assays were carried out as described above. Protein levels of the wild type and PML (3KR) are shown. (F) Wild type HA-PML, HA-PML (3KR), HA-PML (1-555), and green fluorescent protein (GFP)-PML (331-633) were transfected into HUVECs followed by immunostaining with anti-HA [except for GFP (331-633)] and anti-HDAC7 antibodies and confocal microscopy.
Protein–Protein Interaction Assays
For immunoprecipitation, cells were grown on 10-cm plates and transfected with appropriate plasmids (10 μg of total DNA) using Lipofectamine 2000 (Invitrogen). After 48 h, cells were washed in phosphate-buffered saline (PBS) and resuspended in radioimmunoprecipitation assay buffer containing protease inhibitors. After incubation on ice for 2 h, the lysed cells were centrifuged at 4°C at 10,000 rpm for 10 min; supernatant was collected and kept at −80°C. These whole cell extracts were incubated with appropriate antibodies for 4 h at 4°C, and then protein A/G beads added to precleared whole cell extracts for 2 h at 4°C. The immunopellets were washed three to four times with 1× PBS, followed by Western blotting with appropriate antibodies. For endogenous immunoprecipitations, HeLa cell extracts were immunoprecipitated with anti-PML and anti-HDAC7 antibodies and probed with anti-HDAC7 antibodies. For HUVECs, cells were treated with TNF-α as described above. Whole cell lysates were prepared and immunoprecipitated with anti-PML antibodies followed by Western blotting with anti-PML or anti-HDAC7 antibodies.
Confocal Microscopy
Transfected cells were fixed in 3.7% paraformaldehyde in PBS for 30 min at room temperature and permeabilized in PBS with the addition of 0.1% Triton X-100 and 10% goat serum for 10 min. The cells were washed three times with PBS and blocked in PBS-goat serum (10%) and 0.1% Tween 20 solution (ABB) for 60 min. Incubation with primary antibodies was carried out for 120 min in ABB. The cells were washed three times in PBS, and the secondary antibodies were added for 30–60 min in the dark in ABB. Coverslips were mounted on slides using Vectashield mounting medium with 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). All confocal images were acquired using an LSM 510 inverted laser-scanning confocal microscope (Carl Zeiss, Thornwood, NY). A 63× numerical aperture of 1.4 oil immersion plan apochromat objective was used for all experiments. For endogenous and transiently transfected PML, images of Alexa Fluor 594 were collected using a 633-nm excitation light from a He/Ne2 laser, a 633-nm dichroic mirror, and 650-nm long pass filter. For endogenous HDAC7, images of Alexa Fluor 488 were collected using a 488-nm excitation light from an argon laser, a 488-nm dichroic mirror and 500- to 550-nm band pass barrier filter. All DAPI-stained nuclear images were collected using a Coherent Mira-F-V5-XW-220 (Verdi 5W) Ti:sapphire laser tuned at 750 nm, a 700-nm dichroic mirror, and a 390- to 465-nm band pass barrier filter. The primary antibodies are described above. The secondary antibodies used were from Invitrogen (anti-mouse and anti-rabbit Alexa Fluor 488, anti-mouse and anti-rabbit Alexa Fluor 594). The applied laser power (in percentage) of the LSM 510 system has a linear range. For quantification of the intensity, if two samples were acquired with similar parameters, e.g., detector gain, scan speed, and so on, varying the laser percentage power can be used to semiquantify the two images in terms of their average pixel intensity. For example, in Figure 2A, the laser power used to acquire the image in panel g was 5 times higher than panel b or l. Thus, the average pixel intensities or the level of proteins seen in panel l is 5 times greater than that seen in panel g (see arrows) in this case.
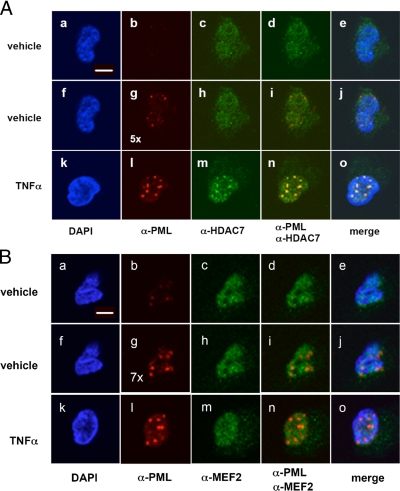
Figure 2.
TNF-α treatment results in the sequestration of HDAC7 to PML NBs. (A) HUVECs were treated with or without TNF-α for 20 h followed by immunostaining with anti-HDAC7 and anti-PML antibodies and confocal microscopy. Note that the intensity of PML NBs in the TNF-α treated samples (l) was 5 times greater than that in the untreated cells (b and g). Unless specified, the laser intensity used in the confocal microscopy was the same as that used in b. The laser intensity used in g was fivefold higher than that in b. HUVECs do not express detectable levels of HDAC4 or HDAC5 (data not shown). (B) Confocal microscopic images of TNF-α treated cells immunostained with anti-PML and anti-MEF2 antibodies. Experiments were carried out according to A.
Chromatin Immunoprecipitation (ChIP) Assays
This protocol was modified from the Farnham laboratory's protocol (Wells and Farnham, 2002). HUVECs (1 × 106) were used for each immunoprecipitation reaction. HUVECs were cultured in endothelial medium described above for 48 h followed by TNF-α treatment. Cells were harvested at different time points as indicated and rinsed with 1× PBS twice followed by cross-linking with 1% formaldehyde for 10 min at room temperature. Glycine was added to a final concentration of 125 mM for 5 min at room temperature to stop cross-linking. Cells were rinsed with 10 ml of ice-cold 1× PBS and pelleted by centrifugation at 5000 rpm. Cells were resuspended in 10 ml of cell lysis buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 8.0, 85 mM KCl, 0.5% NP-40, 0.5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors mix). After incubation on ice for 10 min, cell nuclei were pelleted by centrifugation at 5000 rpm for 5 min at 4°C. Nuclei were resuspended in nuclei lysis buffer (50 mM Tris-Cl pH 8.1, 10 mM EDTA, 1% SDS, and protease inhibitors mix) and incubated on ice for 10 min. Nuclear lysates were sonicated on ice with a Fisher sonic dismembrator 550 at a setting of 4 for 15 10-s pulses to shear the chromatin to an average of 600-base pair fragments. Lysates were centrifuged at 14,000 rpm at 4°C for 15 min, and the supernatants were precleared with protein A agarose beads (RepliGen, Waltham, MA) for 10 min. Precleared lysates were diluted with IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-Cl, pH 8.1, and 167 mM NaCl) and incubated with individual antibodies on a rotation platform at 4°C overnight. After 12- to 16-h incubation, protein A agarose beads were added and incubated on a rotating platform for 15 min at room temperature to pull down the DNA–protein complexes. The complexes were centrifuged at 14000 rpm at 4°C for 5 min to pellet the beads and washed with 1× dialysis buffer (2 mM EDTA, 50 mM Tris-Cl, pH 8.0, and 0.2% sarkosyl) twice and IP washing buffer (100 mM Tris-Cl, pH 9.0, 500 mM LiCl, 1% NP-40, and 1% deoxycholic acid) at least four times. Ten percent of the no antibody control was saved as total input before washing. DNA–protein complexes were eluted with elution buffer (50 mM NaHCO3 and 1% SDS). This was followed by reverse cross-linking by addition of NaCl to 300 mM and 10 μg of RNase A and incubation at 65°C overnight. The samples were precipitated at −20°C overnight by addition of 2.5-fold of 100% ethanol and then pelleted by centrifugation. The samples were dissolved in 100 μl of 1× TE, 25 μl of 5× proteinase K buffer (50 mM Tris-Cl, pH 7.5, 25 mM EDTA, and 1.25% SDS) and 1.5 μl of proteinase K (10 mg/ml) and incubated in a 45°C water bath for 2 h. Samples were then extracted with phenol-chloroform-isoamyl alcohol (25:24:1) followed by precipitation with 450 mM NaCl, 5 μg of tRNA (Roche Diagnostics), 5 μg of glycogen (Roche Diagnostics), and 2.5-fold of 100% ethanol at −20°C overnight. Pellets were collected by centrifugation and dissolved in 30 μl of 1× TE and analyzed by PCR.
RESULTS
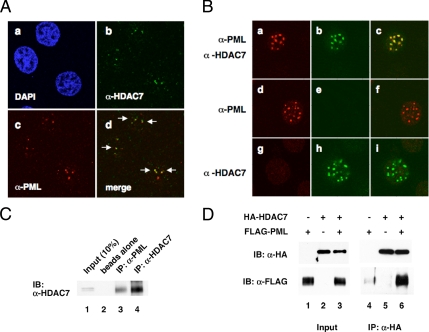
Immunofluorescence and confocal microscopy were used to compare the subcellular distribution of endogenous HDAC7 and PML in HeLa cells. We found that HDAC7 displayed speckled nuclear staining that partially overlapped with PML NBs (Figure 1Ad). A similar observation was made in MDA-MB-231 breast cancer cells (data not shown). We extended these findings by transiently transfecting HeLa cells with a PML expression plasmid. Transfected PML also displayed a punctate nuclear pattern, mimicking the distribution of endogenous PML (Figure 1Ba) and that most endogenous HDAC7 colocalized to PML NBs in the transfected cells (Figure 1Bc). These data indicate that endogenous HDAC7 and PML partially colocalize but that ectopic overexpression of PML results in additional recruitment of HDAC7 to PML NBs (Supplemental Figure S1). We next carried out coimmunoprecipitation experiments and found that endogenous HDAC7 and PML interact in HeLa cells (Figure 1C). This interaction was also detected in Hep2 cells (data not shown) and further confirmed by transient transfection of HDAC7 and PML expression plasmids followed by coimmunoprecipitation (Figure 1D). Moreover, HDAC4 and -5 also interacted with PML and were recruited to PML NBs (Supplemental Figure S2). These data demonstrate that HDAC7 and PML partially colocalize and interact in mammalian cells. We further hypothesize that up-regulation of PML recruits class II HDACs to the PML NBs.
Figure 1.
HDAC7 associates with PML in HeLa cells. (A) Confocal immunofluorescence microscopy was carried out using anti-HDAC7 antibodies and anti-PML antibodies. PML NBs occur as multiple speckles within the nucleus. (B) HeLa cells were transfected with a PML expression plasmid. Immunostaining was carried out to detect transfected PML and endogenous HDAC7. a–c, anti-PML and anti-HDAC7 antibodies were used to stain transfected and endogenous PML and endogenous HDAC7; d–f, only anti-PML antibodies were applied; g–i, only anti-HDAC7 antibodies were applied. Note that transfected PML was expressed at a much higher level than endogenous PML. All images were taken at the same settings. (C) HeLa whole cell extracts were prepared and immunoprecipitated with beads alone, anti-PML, or anti-HDAC7 antibodies followed by Western blotting with anti-HDAC7 antibodies. (D) An HDAC7 expression plasmid was singly or cotransfected with a PML expression plasmid followed by immunoprecipitation and Western blotting with the indicated antibodies.
One difficulty in studying endogenous functions of individual class IIa HDACs is that the HDACs interact with common factors and share overlapping functions. Therefore, knocking down one member usually does not generate a phenotype due to functional redundancy. We have found that HUVECs only express high levels of HDAC7; other class IIa HDACs (4, 5, and 9) are not expressed at significant levels (Supplementary Figure S3), providing an ideal system to study the cellular functions and regulation of endogenous HDAC7. Treatment with As2O3 and interferons (IFNs) has been shown to enhance formation of PML NBs in other systems (Koken et al., 1995; Lallemand-Breitenbach et al., 2001). To test whether these treatments stimulated PML NB formation in HUVECs, cells were treated with As2O3, INFγ, or TNF-α for 2 or 20 h followed by Western blotting. Among these, TNF-α was the most potent inducer of PML mRNA and protein levels (data not shown; see below). We found that the intensity of HDAC7 staining in PML NBs increased after TNF-α treatment (Figure 2A, compare c and m). Furthermore, based on intensity, the amount of PML in PML NBs in TNF-α treated cells was five- to sevenfold greater than that in nontreated cells (Figure 2A, b and l). We performed similar experiments and found that in contrast to HDAC7, we did not observe recruitment of MEF2 to PML NBs (Figure 2B). As expected, CBP localized to PML NBs with or without TNF-α treatment (Supplemental Figure S4). Furthermore, HDAC1, -2, -3, -6, or N-CoR did not colocalize to PML NBs after TNF-α treatment.
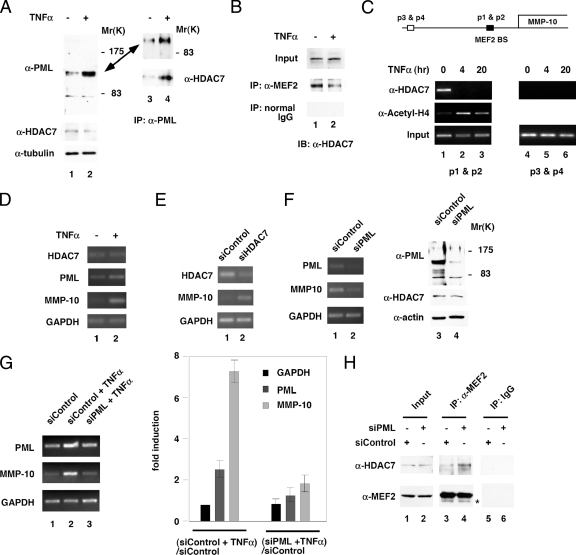
Enhanced formation of PML NBs correlated with significant increases in PML protein levels (Figure 3A, lanes 1 and 2). In contrast, HDAC7 protein levels remained unchanged, which was consistent with the immunostaining data (Figure 3A). This increase in PML protein levels also correlated with an increase in the amount of HDAC7 associated with PML (Figure 3A, lanes 3 and 4). The observation that TNF-α recruits HDAC7 to PML NBs suggests that TNF-α may disrupt the association between HDAC7 and transcription factors such as MEF2. To test whether TNF-α alters the association between HDAC7 and MEF2, we performed coimmunoprecipitations by using anti-MEF2 antibodies and found that TNF-α treatment reduced the amount of HDAC7 bound to MEF2 (Figure 3B). Furthermore, ChIP assays indicate that TNF-α treatment led to loss of association of HDAC7 with the promoter of the MMP-10 gene, a known MEF2 target, and increased acetylation of histone H4 in the vicinity of the MMP-10 promoter (Chang et al., 2006) (Figure 3C, lanes 1–3). As a control, HDAC7 did not associate with a region 2 kb upstream of the MEF2 binding site (lanes 4–6). In contrast, we did not detect an association between PML and the MMP-10 promoter (Supplemental Figure S5). These results suggest that HDAC7 target genes are no longer subject to HDAC7-mediated repression after TNF-α exposure. To test this hypothesis, we used RT-PCR to measure the levels of MMP-10 mRNA. Our data demonstrate that accumulation of the MMP-10 transcript increased in TNF-α-treated HUVECs (Figure 3D). Consistent with previous observations (Chang et al., 2006), siRNA knockdown of HDAC7 also induced expression of MMP-10 mRNA (Figure 3E). To test whether endogenous PML plays a role in the expression of MMP-10 mRNA, we carried out knockdown experiments with siRNA. We found that PML knockdown decreased MMP-10 mRNA accumulation (Figure 3F). These data indicate a positive regulatory function for PML in the expression of the MMP-10 transcript. In contrast, HDAC7 protein levels were not affected. Additionally, PML knockdown significantly attenuated TNF-α-induced up-regulation of MMP-10 (Figure 3G). Concomitantly, knockdown of PML led to enhanced association between MEF2 and HDAC7 (Figure 3H). These data suggest that TNF-α induces expression of MEF2 target genes such as MMP-10, partly by inducing the accumulation of PML, which sequesters HDAC7 from MEF2 association.
Figure 3.
TNF-α induces dissociation of HDAC7 from MEF2 and association of HDAC7 and PML. (A) HUVECs were treated with or without TNF-α, and whole cell extracts were prepared followed by immunoprecipitation with anti-PML antibodies and Western blotting with the indicated antibodies. Whole cell extracts (lanes 1 and 2) were immunoprecipitated with anti-PML antibodies and probed with anti-PML or anti-HDAC7 antibodies (lanes 3 and 4). One major band migrating at 110–120 kDa and one minor band migrating at 80 kDa were detected. (B) HUVEC whole cell extracts treated with TNF-α were immunoprecipitated with anti-MEF2 antibodies and probed with anti-HDAC7 antibodies. (C) Chromatin immunoprecipitation assays were carried out using anti-HDAC7 and anti-acetyl-histone H4 antibodies as described in Materials and Methods. Lanes 1–3, PCR using primers flanking the MEF2 binding sites (BS). Lanes 4 and 5, PCR using primers 2 kb upstream of the MEF2 BS. (D) HUVECs were treated with or without TNF-α as described in A, and total RNA was isolated followed by RT-PCR using gene-specific primers as indicated. (E) siRNA-mediated knockdown of HDAC7 was performed, and total RNA was isolated followed by RT-PCR using gene-specific primers as indicated. (F) siRNA-mediated knockdown of PML was performed, and total RNA was isolated followed by RT-PCR using gene-specific primers as indicated. (G) siRNA-mediated knockdown of PML was performed followed by TNF-α treatment and total RNA was isolated followed by RT-PCR using gene-specific primers as indicated. Real-time PCR was carried out to measure the mRNA expression of indicated genes. The ratio of GAPDH mRNA for [siControl + TNF-α]/[siControl] was set at 1. -Fold induction is shown. (H) A control or PML siRNA was transfected into HUVECs. Seventy-two hours after transfection, whole cell lysates were prepared followed by coimmunoprecipitation with anti-MEF2 antibodies or control immunoglobulin (IgG). Western blotting were performed using anti-HDAC7 or anti-MEF2 antibodies. IgG heavy chain is shown below MEF2 marked as an asterisk.
To further dissect the mechanism by which PML regulates HDAC7 function, we transiently transfected HeLa cells to examine whether PML is capable of regulating MEF2-mediated transcription. We found that coexpression of PML potentiated activity of a MEF2-responsive reporter in the absence or presence of exogenous MEF2 (Figure 4A). Furthermore, PML overcame HDAC7-mediated transcriptional repression of MEF2 reporter activity (Supplemental Figure S6A). To test whether the ability of PML to regulate MEF2 reporter activity is dependent on class IIa HDACs, we evaluated their effects on an MEF2 reporter. HDAC4, -5, and -7 were all capable of attenuating PML-mediated activation of MEF2 reporter activity, suggesting that PML potentiates MEF2 activity by regulating class II HDACs (Supplemental Figure S6B). We mapped the residues in PML that are essential for interaction with HDAC7 by coimmunoprecipitation experiments and the minimal PML fragment that can activate a MEF2 reporter by transient transfection assays. We found that deletion of amino acids 556–633 significantly reduced the PML:HDAC7 association (Figure 4B, compare lanes 3 and 8), whereas PML (331–633) was sufficient for interaction with HDAC7 (lane 10). Furthermore, amino acids 552–633 did not bind HDAC7 in coimmunoprecipitation experiments (Supplemental Figure S7). Ectopic expression of wild-type PML potentiated transcription activity by MEF2 (Figure 4C, lanes 5–7), whereas PML truncations defective in binding HDAC7 did not (lanes 8–13). Furthermore, the same C-terminal fragment of PML that interacted with HDAC7 was able to activate MEF2 transcription (lanes 14–16). These data suggest that PML interaction with HDAC7 is essential for its ability to potentiate MEF2 transcription activity and that the C-terminal region of PML is required for this interaction.
Sumoylation of PML is thought to contribute to its regulation and formation of PML NBs (Bernardi and Pandolfi, 2007; Heun, 2007). To determine whether sumoylation is essential for PML to interact with HDAC7, we generated a sumoylation-deficient PML mutant, PML (3KR, K65R/K160R/K490R) and tested its association with HDAC7. Although incapable of forming PML NBs, PML (3KR) still forms nuclear aggregates (Zhong et al., 2000). We found that PML (3KR) still associated with HDAC7, indicating that PML sumoylation is dispensable for its association with HDAC7 (Figure 4D). Furthermore, this mutant potently activated MEF2 reporter activity (Figure 4E). We reason that the ability of PML to activate MEF2 reporter activity correlates with the ability of PML to sequester HDAC7. As predicted, transfected wild-type, PML (3KR) and PML (331-633) (Figure 4F, a–j) but not PML (1-555) (Figure 4F, k–o) recruited endogenous HDAC7 to PML-mediated nuclear aggregates in HUVECs. In summary, our data uncover a distinct mechanism by which TNF-α induces MMP-10 mRNA expression in a PML-dependent pathway that involves sequestration of HDAC7.
DISCUSSION
In this study, we investigate the mechanisms by which TNF-α regulates expression of HDAC7 target genes, including MMP-10. Three major findings can be drawn from this study: 1) TNF-α induces PML mRNA and protein accumulation/sumoylation and enhances PML NB formation; 2) TNF-α treatment leads to recruitment of HDAC7 to PML NBs, loss of HDAC7 association with MEF2 transcription factors and the MMP-10 promoter, and induction of MMP-10 mRNA accumulation; and 3) PML knockdown enhances association between HDAC7 and MEF2 and reduces basal expression and TNF-α-induced up-regulation of MMP-10 mRNA.
We have discovered a novel mechanism by which class II HDACs can be kept in an inactive form in the nucleus through induction or overexpression of PML. Class II HDACs are potent transcriptional corepressors; therefore, induction of PML increases expression of HDAC7 target genes such as MMP-10. In HUVECs, this occurs when TNF-α signaling induces PML expression and association between PML and HDAC7. This PML-mediated regulation is distinct from the known mechanism by which phosphorylation controls the subcellular localization of class II HDACs through sequestration in the cytoplasm (Verdin et al., 2003). We also note that knockdown of PML down-regulates MMP-10 mRNA levels even without TNF-α treatment, suggesting that PML can sequester HDAC7 in the absence of TNF-α (Figure 3). Indeed, PML and HDAC7 interact without TNF-α treatment (Figures 1 and 3). Interestingly, we also found that overexpression of HDAC7 also increases the interaction between PML and HDAC7 (Supplemental Figure S8), suggesting that PML or HDAC7 expression levels may regulate their association. Consistent with this model, HDAC7 knockdown significantly up-regulates MMP-10 mRNA expression (Figure 3E). Accordingly, we predict that a subset of genes induced by HDAC7 knockdown can also be down-regulated by PML knockdown. PML and HDAC7 also interact in other cell lines such as Hep2 and MDA-MB-231 (data not shown), so it is likely that such a regulatory pathway is of general importance.
Consistent with our observations that TNF-α induces PML-dependent up-regulation of HDAC7 target genes, overexpression of PML potently activates a MEF2 reporter activity and recruits HDAC7 to PML NBs, suggesting that PML may sequester HDAC7 from associating with the MEF2 binding site, thus relieving repression. A recent publication by Block et al. (2006) has shown that, when fused to a DNA binding domain, a small fraction of PML fusion protein colocalizes with the reporter construct that harbors the binding site. This result suggests a direct effect of PML on the reporter activity. However, several observations argue against this model. First, in Block's paper, the authors tethered the Tet repressor, TetR, with PML to generate HA-TetR-PML. This fusion directly binds to the Tet operator present in the reporter construct. Nonetheless, only a small fraction of the cotransfected reporter construct colocalized with PML NBs. The majority of the reporter construct did not. Therefore, the reporter activity is unlikely to be directly affected by PML. In our assay system, PML does not tether to a DNA binding domain. We do not think that PML will target the MEF2/HDAC7 binding site in the reporter construct. Second, it has been proposed that PML NBs do not contain nucleic acid (Boisvert et al., 2000). Third, a paper published by Wang et al. (2004) concluded that PML NBs form in nuclear compartments of high transcriptional activity, but they do not directly regulate transcription of genes in these compartments. Furthermore, there is a strong correlation between the association of PML mutants with HDAC7 and the ability of PML mutants to activate a MEF2 reporter activity, indicating that HDAC7 association is critical for the ability of PML to activate MEF2 reporter activity (Figure 4). Last, we show that knockdown of PML enhanced the association between MEF2 and HDAC7 (Figure 3H), suggesting that PML and MEF2 compete for HDAC7 binding. Based on these observations, we believe that PML NBs do not directly activate MEF2-mediated transcription.
A PML mutant that cannot be sumoylated, PML4 (3KR), is capable of forming subnuclear aggregates but not proper PML NBs, when it is expressed in several cell lines, including HUVEC, HeLa, MDA-MB-231, and PML−/− mouse embryonic fibroblasts (Figure 4F; data not shown). We also found that HDAC7 is recruited to these nuclear aggregates. Thus, the ability of PML to interact with HDAC7 and activate MEF2 reporter is independent of its sumoylation, which is consistent with our findings (Figure 4). This result indicates that these residues are not critical for activation of MEF2 reporter activity. Further investigations are required to address this issue.
We have identified PML as a key effector of the proinflammatory cytokine TNF-α. Our data also show that other proinflammatory stimuli such as IFNs and bacterial lipopolysaccharide are capable of inducing formation of PML NBs (data not shown). Similar to TNF-α, these stimuli showed a sustained effect on the formation of PML NBs. This regulation is distinct from the mechanism by which As2O3-induced transient PML NB formation (data not shown; Zhu et al., 1997; Wang et al., 1998). The sustained effect of TNF-α on PML NBs could have important physiological consequences in HUVECs. Consistent with our observations, PML is highly expressed in inflammatory tissues (Terris et al., 1995). It remains to be investigated whether PML controls expression of other genes through a similar mechanism and whether PML−/− animals develop defects in inflammatory responses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. D. Samols, E. Stavnezer, and M. Snider for comments on the manuscript. We also thank K. Stanya and E. Reineke for input. H.-Y.K. is an American Cancer Society Research Scholar. This project is supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases grant R01 DK-62985 and by American Cancer Society grant (RSG GMC-106736) to H.-Y.K. and by the Confocal Microscopy Core Facility of the Comprehensive Cancer Center of Case Western Reserve University and University Hospitals of Cleveland (NIH P30 CA43703-12). K.-S.C. is supported by National Institutes of Health grant R01 CA-099963.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1203) on May 7, 2008.
REFERENCES
- Bernardi R., Pandolfi P. P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Block G. J., Eskiw C. H., Dellaire G., Bazett-Jones D. P. Transcriptional regulation is affected by subnuclear targeting of reporter plasmids to PML nuclear bodies. Mol. Cell. Biol. 2006;26:8814–8825. doi: 10.1128/MCB.00636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F. M., Hendzel M. J., Bazett-Jones D. P. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K. L. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 2002;22:5259–5269. doi: 10.1128/MCB.22.15.5259-5269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Reineke E. L., Lam M., Li X., Liu Y., Gao C., Khurana S., Kao H. Y. alpha-Actinin 4 potentiates myocyte enhancer factor-2 transcription activity by antagonizing histone deacetylase 7. J. Biol. Chem. 2006;281:35070–35080. doi: 10.1074/jbc.M602474200. [DOI] [PubMed] [Google Scholar]
- Chang S., Young B. D., Li S., Qi X., Richardson J. A., Olson E. N. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Ching R. W., Dellaire G., Eskiw C. H., Bazett-Jones D. P. PML bodies: a meeting place for genomic loci? J. Cell Sci. 2005;118:847–854. doi: 10.1242/jcs.01700. [DOI] [PubMed] [Google Scholar]
- Fischer D. D., Cai R., Bhatia U., Asselbergs F. A., Song C., Terry R., Trogani N., Widmer R., Atadja P., Cohen D. Isolation and characterization of a novel class II histone deacetylase, HDAC10. J. Biol. Chem. 2002;277:6656–6666. doi: 10.1074/jbc.M108055200. [DOI] [PubMed] [Google Scholar]
- Fischle W., Emiliani S., Hendzel M. J., Nagase T., Nomura N., Voelter W., Verdin E. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- Gao C., Li X., Lam M., Liu Y., Chakraborty S., Kao H. Y. CRM1 mediates nuclear export of HDAC7 independently of HDAC7 phosphorylation and association with 14-3-3s. FEBS Lett. 2006;580:5096–5104. doi: 10.1016/j.febslet.2006.08.038. [DOI] [PubMed] [Google Scholar]
- Grozinger C. M., Hassig C. A., Schreiber S. L. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardiola A. R., Yao T. P. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J. Biol. Chem. 2002;277:3350–3356. doi: 10.1074/jbc.M109861200. [DOI] [PubMed] [Google Scholar]
- Heun P. SUMOrganization of the nucleus. Curr. Opin. Cell Biol. 2007;19:350–355. doi: 10.1016/j.ceb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Kao H. Y., Downes M., Ordentlich P., Evans R. M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Kao H. Y., Lee C. H., Komarov A., Han C. C., Evans R. M. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J. Biol. Chem. 2002;277:187–193. doi: 10.1074/jbc.M108931200. [DOI] [PubMed] [Google Scholar]
- Kao H. Y., Verdel A., Tsai C. C., Simon C., Juguilon H., Khochbin S. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 2001;276:47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- Koken M. H., Linares-Cruz G., Quignon F., Viron A., Chelbi-Alix M. K., Sobczak-Thepot J., Juhlin L., Degos L., Calvo F., de The H. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chen J. D. PML and the oncogenic nuclear domain in regulating transcriptional repression. Curr. Opin. Cell Biol. 2000;12:641–644. doi: 10.1016/s0955-0674(00)00144-7. [DOI] [PubMed] [Google Scholar]
- Li H., Leo C., Zhu J., Wu X., O'Neil J., Park E. J., Chen J. D. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. Y., Lai M. Z., Ann D. K., Shih H. M. Promyelocytic leukemia protein (PML) functions as a glucocorticoid receptor co-activator by sequestering Daxx to the PML oncogenic domains (PODs) to enhance its transactivation potential. J. Biol. Chem. 2003;278:15958–15965. doi: 10.1074/jbc.M300387200. [DOI] [PubMed] [Google Scholar]
- Maul G. G., Negorev D., Bell P., Ishov A. M. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- Terris B., Baldin V., Dubois S., Degott C., Flejou J. F., Henin D., Dejean A. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 1995;55:1590–1597. [PubMed] [Google Scholar]
- Tsuzuki S., Towatari M., Saito H., Enver T. Potentiation of GATA-2 activity through interactions with the promyelocytic leukemia protein (PML) and the t(15;17)-generated PML-retinoic acid receptor alpha oncoprotein. Mol. Cell. Biol. 2000;20:6276–6286. doi: 10.1128/mcb.20.17.6276-6286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallian S., Chin K. V., Chang K. S. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol. Cell. Biol. 1998;18:7147–7156. doi: 10.1128/mcb.18.12.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A., Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentation-dependent chromatin modifiers. J. Biol. Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- Verdin E., Dequiedt F., Kasler H. G. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- Wang J., Shiels C., Sasieni P., Wu P. J., Islam S. A., Freemont P. S., Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. G., Rivi R., Delva L., Konig A., Scheinberg D. A., Gambacorti-Passerini C., Gabrilove J. L., Warrell R. P., Jr., Pandolfi P. P. Arsenic trioxide and melarsoprol induce programmed cell death in myeloid leukemia cell lines and function in a PML and PML-RARalpha independent manner. Blood. 1998;92:1497–1504. [PubMed] [Google Scholar]
- Wells J., Farnham P. J. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods. 2002;26:48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]
- Wu W. S., Xu Z. X., Ran R., Meng F., Chang K. S. Promyelocytic leukemia protein PML inhibits Nur77-mediated transcription through specific functional interactions. Oncogene. 2002;21:3925–3933. doi: 10.1038/sj.onc.1205491. [DOI] [PubMed] [Google Scholar]
- Xu Z. X., Zhao R. X., Ding T., Tran T. T., Zhang W., Pandolfi P. P., Chang K. S. Promyelocytic leukemia protein 4 induces apoptosis by inhibition of survivin expression. J. Biol. Chem. 2004;279:1838–1844. doi: 10.1074/jbc.M310987200. [DOI] [PubMed] [Google Scholar]
- Zhong S., Muller S., Ronchetti S., Freemont P. S., Dejean A., Pandolfi P. P. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- Zhou X., Marks P. A., Rifkind R. A., Richon V. M. Cloning and characterization of a histone deacetylase, HDAC9. Proc. Natl. Acad. Sci. USA. 2001;98:10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Koken M. H., Quignon F., Chelbi-Alix M. K., Degos L., Wang Z. Y., Chen Z., de The H. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA. 1997;94:3978–3983. doi: 10.1073/pnas.94.8.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.