Abstract
In animal cells, centrosomes nucleate microtubules that form polarized arrays to organize the cytoplasm. Drosophila presents an interesting paradox however, as centrosome-deficient mutant animals develop into viable adults. To understand this discrepancy, we analyzed behaviors of centrosomes and microtubules in Drosophila cells, in culture and in vivo, using a combination of live-cell imaging, electron microscopy, and RNAi. The canonical model of the cycle of centrosome function in animal cells states that centrosomes act as microtubule-organizing centers throughout the cell cycle. Unexpectedly, we found that many Drosophila cell-types display an altered cycle, in which functional centrosomes are only present during cell division. On mitotic exit, centrosomes disassemble producing interphase cells containing centrioles that lack microtubule-nucleating activity. Furthermore, steady-state interphase microtubule levels are not changed by codepleting both γ-tubulins. However, γ-tubulin RNAi delays microtubule regrowth after depolymerization, suggesting that it may function partially redundantly with another pathway. Therefore, we examined additional microtubule nucleating factors and found that Mini-spindles, CLIP-190, EB1, or dynein RNAi also delayed microtubule regrowth; surprisingly, this was not further prolonged when we codepleted γ-tubulins. Taken together, these results modify our view of the cycle of centrosome function and reveal a multi-component acentrosomal microtubule assembly pathway to establish interphase microtubule arrays in Drosophila.
INTRODUCTION
The microtubule (MT) cytoskeleton is essential for organizing the cytoplasm, polarity establishment, cell motility, morphogenesis, and cell division. Polarized arrays of MTs are nucleated by the centrosome, an organelle consisting of a pair of “mother-daughter” centrioles that recruit and organize a surrounding matrix of pericentriolar material (PCM; Bornens, 2002). Proteomic analyses of centrosomes identified several hundred different proteins, some of which are unique to centrioles (e.g., the structural subunits SAS-4, SAS-6, and SPD-2) or the PCM (e.g., pericentrin; Andersen et al., 2003; Keller et al., 2005; Leidel and Gonczy, 2005a). Within the PCM, pericentrin and other coiled-coil proteins assemble into a scaffold that docks γ-tubulin ring complexes, which nucleate MT growth (Moritz and Agard, 2001). By manipulating centrosome number and position, cells exert precise control over the MT arrays needed for a variety of critical MT-dependent processes.
Centrosomes adhere to a cycle of duplication and function that is intimately coupled to the cell cycle (Tsou and Stearns, 2006). Centrosomes act as dominant MT-organizing and nucleating centers (MTOCs; Schiebel, 2000); however, this activity is modulated in a cell cycle–dependent manner. During interphase, cells possess two paired centrioles that form a single centrosome and organize the interphase MT array. Before mitotic entry the centrosome duplicates once; the two centrosomes then separate from one another and undergo “maturation,” recruiting more PCM to nucleate additional MT growth and facilitate spindle assembly (Glover, 2005). On mitotic exit, each daughter cell receives a single centrosome that reduces its amount of associated PCM (Dictenberg et al., 1998), but remains active as an MTOC. γ-tubulin is thought to be the primary source of microtubule-nucleating activity from the centrosome (Wiese and Zheng, 2006), although it is likely not the only protein capable of this task. For example, developing Drosophila ovaries that harbor homozygous double mutations in both γ-tubulin genes (γTub23C and γTub37C) organize abnormal spindles within mitotic germ cells but are still competent to nucleate MT growth (Tavosanis and Gonzalez, 2003). Likewise, RNA interference (RNAi) of the single γ-tubulin gene in Caenorhabditis elegans leads to defects in spindle bipolarity but does not abolish MT nucleation (Strome et al., 2001; Hannak et al., 2002). These studies suggest that alternative γ-tubulin-independent MT-assembly pathways exist in cells.
The importance and functional versatility of centrosomes is illustrated by their capacity to build several different MT-based machines including cilia, flagella, and mitotic spindles (Rieder et al., 2001). This suggests that centrosomes should be essential for cell function. A recent study tested this hypothesis using Drosophila sas-4 mutants that fail to form centrioles, and therefore centrosomes (Basto et al., 2006). Surprisingly, zygotic mutant embryos developed into viable adults with near normal timing and morphology. Although centrosomes usually organize mitotic spindle poles and were thought to be important for high-fidelity chromosome transmission, dividing sas-4 mutant cells displayed few errors in chromosome segregation, because their chromosomes induced spindle assembly via an acentrosomal pathway (Basto et al., 2006). Similarly, mutant flies that lack Centrosomin, a PCM component essential for centrosome function, undergo normal zygotic development to form viable adults (Megraw et al., 2001). Importantly, these studies raised new questions: how do interphase Drosophila cells nucleate MT growth, organize their cytoplasm, and survive without the polarized MT arrays that centrosomes provide?
Much of our understanding of the cycle of centrosome function in Drosophila is derived from work in early embryos and asymmetrically dividing larval neuroblasts. During the rapid mitotic divisions of the early syncytial embryo, gap phases of the cell cycle (G1 and G2) are absent. Nonetheless, centrosome duplication and function largely follow the canonical cycle observed in most animal cells, as the centrosomes act as MTOCs throughout this cell cycle (Callaini and Riparbelli, 1990). Larval neuroblasts, however, display a novel twist to the classic cycle that is used to more precisely position the mitotic spindle (Rebollo et al., 2007; Rusan and Peifer, 2007). After asymmetric division in these stem cells, the centriole pair separates as expected, but only one centriole (the “dominant centriole”) retains its PCM and MTOC activity during the intervening interphase, whereas the other sheds its PCM and is inactive with respect to MT nucleation. Before entering the next division, the inactive centriole moves to the opposite side of the neuroblast, matures into a functional centrosome and is eventually segregated into the smaller ganglion mother cell. Although these two specialized cell types possess functional centrosomes throughout their respective cell cycles, it is not clear how other Drosophila cells govern their centrosome cycles.
Here, we examine the Drosophila cycle of centrosome function both in cultured cells and within developing animals. Contrary to models of the conventional cycle, we find that Drosophila cells typically utilize centrosomes as MTOCs exclusively during mitosis. As cells exit mitosis, centrosomes disassemble both in cycling cells and within interphase-arrested cells. Furthermore, the generation and arrangement of interphase MTs appears to be independent of centrioles and is not disrupted by γ-tubulin RNAi at steady state. However, interphase MT regrowth assays reveal a “fast” MT-assembly pathway that uses not only γ-tubulin but Mini-spindles, CLIP-190, EB1, and dynein. Our results suggest that Drosophila cells utilize a distinctive “canonical” cycle of centrosome function, in which interphase centrosomes are inactivated, being replaced by an alternative mechanism of organizing the interphase MT array. This provides a mechanistic explanation for the survival of centrosome-deficient flies.
MATERIALS AND METHODS
Cell Culture and Double-stranded RNAi
Drosophila cell culture and RNAi were performed as described (Rogers et al., 2002) with the following modifications. Cells were cultured in Sf900II SFM media (Invitrogen, Carlsbad, CA) without FBS. RNAi was conducted in six-well plates, and cells (50–90% confluency) were treated with 10 μg of double-stranded RNA (dsRNA) in 1 ml of media and replenished with fresh media/dsRNA every day for 7 d. ML-DmD16-c3 cells were obtained from the Drosophila Genome Resource Center (DGRC; Bloomington, IN). Gene-specific primer sequences are shown in Supplementary Table 1. The efficiency of RNAi was determined by Western blotting S2 lysates, where overall protein amounts loaded were normalized and verified using antibodies against α-tubulin. Percent depletion of the target protein was measured using the densitometry functions of ImageJ (NIH; http://rsb.info.nih.gov/ij/).
Immunofluorescence Microscopy
For immunostaining, S2 cells were fixed exactly as described (Rogers et al., 2002) with either −20°C methanol or 10% formaldehyde. Antibodies used in this study were diluted to concentrations ranging from 1 to 20 μg/ml (anti-SAS-6 [our lab], D-PLP [J. Raff, University of Cambridge], DM1a α- tubulin [Sigma, St. Louis, MO], Cnn [R. Jones, UT Southwestern], CP60 and CP190 [M. Moritz, UCSF], E7 α-tubulin [Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA], GTU-88 σ, phosphohistone H3 [Upstate Biotechnology, Lake Placid, N; Cell Signaling Technology, Beverly, MA], green fluorescent protein [GFP; LabVision, Fremont, CA; #MS-1315-PO], anti-Golgi membrane protein [Calbiochem, La Jolla, CA; #345867]). Secondary antibodies (Cy2 and Rhodamine Red; Jackson ImmunoResearch, West Grove, PA) were used at final dilutions of 1:100. Cells were mounted in a 0.1 M propyl galate-glycerol solution. DAPI (Sigma) was used at a concentration of 5 μg/ml. For cold treatments, S2 cells prespread on concanavalin A (conA) dishes were placed on a partially submerged aluminum block in an ethanol-ice bath at 4°C for 1 h and allowed to recover at either 38°C or room temperature, depending on the ambient temperature. For fixed embryo analysis, WT embryos were dechorionated with 50% house hold-bleach and then place in a 1:1 3.7% formaldehyde in phosphate-buffered saline (PBS):heptane for 20 min. Embryos were devitellinized in methanol and then blocked and stained in PBS/1% goat serum/0.1% Triton X-100. Specimens were imaged using either a TE2000-E Nikon microscope (Melville, NY) or a Yokogawa spinning-disk confocal (Perkin Elmer-Cetus, Norwalk, CT) mounted on a TE300 Nikon microscope.
Antibodies
Escherichia coli-expressed glutathione S-transferase (GST)- or MPB-SAS-6 (full-length) and D-PLP (amino acids 8–351) proteins were purified on either glutathione-Sepharose or amylose resin (Martinez-Campos et al., 2004). Rabbit polyclonal antisera (ProteinTech, Chicago, IL) were raised against affinity-purified fusion proteins that were also precoupled to Affigel 10/15 (Bio-Rad, Richmond, CA) and used for antibody affinity purification performed by elution with low pH buffer.
Live Cell Microscopy
Endogenous promoters for GFP constructs were made by PCR of genomic regions upstream of γTub23C (761 bp), γTub37C (476 bp), or SAS-6 (208bp) and cloned into pMT vectors (Invitrogen). Stable cell lines were generated using Effectene transfection reagent (Qiagen, Chatsworth, CA)/pCoHygro selection system (Invitrogen) and plated on 0.5 mg/ml conA-coated glass-bottom dishes (MatTek, Ashland, MA) for 1 h before observation. Cells were imaged with a 100× 1.45 NA PLAN APO objective using a TE2000-E Nikon microscope (Melville, NY) equipped with a Cascade 512B cooled CCD camera (Roper Scientific, Tucson, AZ) or a Yokogawa spinning disk confocal (Perkin Elmer-Cetus) mounted on a TE300 Nikon microscope equipped with an ORCA-ER cooled CCD camera (Hamamatsu, Bridgewater, NJ). For in vivo live imaging, embryos of the genotype Gal4(nos-NGT40)/mCherry::SAS6;+/UAS-EB1::GFP were dechorionated with 50% household bleach, covered in halocarbon oil (series 700; Halocarbon Products, River Edge, NJ) and mounted between a no. 1.5 glass coverslip and a gas-permeable membrane (petriPERM; Sigma). Embryos were then imaged using the spinning-disk microscope mentioned earlier. Time-lapse sequences were collected using MetaMorph (Molecular Devices, Sunnyvale, CA). All stable cell lines expressing fluorescent proteins will be made available through the DGRC.
Transmission Electron Microscopy
Cell monolayers grown on polystyrene plates were rinsed with PBS and fixed in 3% glutaraldehyde with 0.1 M sodium cacodylate, pH 7.4, for several hours or overnight. After buffer washes, the monolayers were postfixed for 1 h with 1% osmium tetroxide, 1.25% potassium ferrocyanide, and 0.1 M sodium cacodylate buffer. The cells were dehydrated using increasing concentrations of ethanol, infiltrated, and embedded in Polybed 812 epoxy resin (Polysciences, Warrington, PA). The blocks were sectioned parallel to the substrate at 70 nm using a diamond knife, and the sections were mounted on 200-mesh copper grids followed by staining with 4% aqueous uranyl acetate and Reynolds' lead citrate. Sections were observed with a LEO EM910 transmission electron microscope operating at 80 kV (Leo Electron Microscopy, Cambridge, United Kingdom) and photographed using a Gatan Orius SC1000 CCD Digital Camera and Digital Micrograph 3.11.0 (Gatan, Pleasanton, CA).
High-Throughput Microscopy
S2 cells were seeded in conA–coated 24-well glass-bottom plates (MatTek) for 1 h before fixation, stained (described above), and scanned with either an IC100 Image Cytometer (Beckman Coulter, Fullerton, CA) or an Array Scan VTI (Cellomics) equipped with a 20× 0.5 NA or 40× 0.95 NA objective and an ORCA-ER cooled CCD camera. Images of ∼5000 cells per well were acquired and analyzed using CytoShop v2.1 (Beckman Coulter) or vHCS View (Cellomics, Pittsburgh, PA). Integrated fluorescence intensity measurements were determined from unsaturated images.
RESULTS
Multiple Drosophila Cell Types Lack Functional Centrosomes or MTOCs during Interphase
The canonical model for centrosome function in most animal cells suggests that interphase MTs are nucleated at the centrosome, to provide a polarized array that organizes the cytoplasm (Schiebel, 2000). However, in apparent contradiction to this vital centrosomal function, zygotic centrosome-deficient mutant fly embryos develop into viable adults (Megraw et al., 2001; Basto et al., 2006). We investigated this discrepancy by examining the cycle of centrosome function in Drosophila cells.
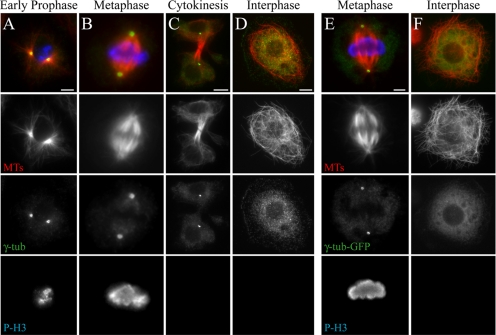
The ability to use RNAi to generate loss-of-function phenotypes in cultured S2 cells makes them a powerful system for studying cytoskeletal cell biology (Rogers et al., 2004a). We initially used antibodies against γ-tubulin to visualize centrosomes, as it is a conserved PCM component and a widely used centrosome marker. As expected, MTOCs appear as hollow spheres of γ-tubulin at the center of MT asters during early prophase, metaphase and throughout cytokinesis (Figure 1, A–C).
Figure 1.
S2 cells lack a γ-tubulin–containing MTOC during interphase. (A–F) S2 cells stained for MTs (red), γ-tubulin (green), and phosphohistone H3 (blue). Representative cells in early prophase (A), metaphase (B and E), telophase (C), and interphase (D and F). (E and F) γ-tubulin-GFP (green)-expressing stable line. Scale, 5 μm.
Surprisingly, anti-γ-tubulin antibodies failed to recognize a discrete MTOC in interphase Drosophila S2 cells. Instead, γ-tubulin immunolocalized in a diffuse pattern throughout the cytoplasm as small punctae (Figure 1 D); these were abolished by γ-tubulin RNAi (Supplemental Figure 1). Consistent with the absence of an MTOC, MTs appeared to be broadly distributed and nonradial, in contrast to the polarized radial arrays in cultured interphase vertebrate cells (Wiese and Zheng, 2006).
As an independent method to examine γ-tubulin distribution, we engineered a stable S2 line expressing γ-tubulin-GFP under control of the gene's endogenous promoter. Stable cell lines expressing γ-tubulin-GFP had mitotic MTOCs equal in number to those observed in wild-type S2 cells stained with anti-γ-tubulin (data not shown). Consistent with the immunostaining, γ-tubulin-GFP concentrated into hollow spheres at spindle poles and was uniformly diffuse throughout the cytoplasm in interphase (Figure 1, E and F).
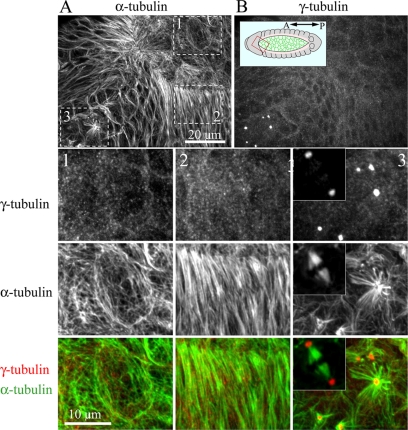
We next examined whether this centrosome behavior was unique to S2 cells. We found that cultured Drosophila D16 cells, an imaginal disk-derived cell line (Ui et al., 1987), also contained γ-tubulin-MTOCs during mitosis but lacked these structures during interphase (Supplemental Figure 2). To examine whether this was also true in vivo, we examined three embryonic cell types during the process of dorsal closure (Figure 2). MT staining revealed that amnioserosal and leading edge cells, two differentiated cell types terminally arrested in interphase, displayed MT arrays lacking a central focus. Similar to cultured cells, MTs in amnioserosal cells were broadly distributed (Figure 2A, panel 1), whereas MT were arranged into elongated bundles that spanned the long-axis of leading edge cells, as previously described (Figure 2A, panel 2; Jankovics and Brunner, 2006). Both cell types lacked a concentrated punctate γ-tubulin staining that would indicate the presence of MTOCs (Figure 2B, panels 1 and 2). In contrast, typical γ-tubulin–labeled centrosomes were observed at mitotic spindle poles within dividing cells of the embryo (Figure 2, panel 3 and inset).
Figure 2.
Drosophila embryos lack a γ-tubulin–containing MTOC during interphase. An embryo in the process of dorsal closure was fixed and stained for α-tubulin (A) and γ-tubulin (B). Inset shows embryo model with red box showing relative orientation of images in A and B. Three different cell populations are boxed in A, corresponding to G2-arrested amnioserosa cells (1), G1-arrested leading-edge cells (2) and epithelial cells undergoing mitosis (3). Boxed regions are displayed at higher magnification. Insets show a bipolar spindle from a mitotic domain of a younger developing embryo with γ-tubulin labeling at the poles.
In the canonical animal cell cycle, centrioles recruit PCM and are active MTOCs throughout the cell cycle. Our observations above suggest either that interphase Drosophila centrioles are absent, which we considered unlikely, or that they fail to recruit PCM and act as MTOCs. To test these hypotheses, we developed tools to examine centrioles in living and fixed cells. We generated a stable S2 line expressing GFP (or mCherry) fused to the fly SAS-6 protein under control of the gene's endogenous promoter. SAS-6 is a conserved structural component of centrioles (Dammermann et al., 2004; Leidel et al., 2005b) and Drosophila sas-6 mutants lack basal bodies at sensory bristles and centrioles within larval brains (Peel et al., 2007). We confirmed that our GFP-SAS-6 construct is a reliable marker for centrioles as it colocalized with D-PLP, a known centriole component (Martinez-Campos et al., 2004; Supplemental Figure 3A). Furthermore, affinity-purified anti-SAS-6 antibodies (generated against full-length recombinant fly SAS-6) labeled spots within γ-tubulin spheres in mitotic cells, as we saw with GFP-SAS-6 (Supplemental Figure 5A). Stable expression of GFP-SAS-6 did not increase interphase centriole number (data not shown).
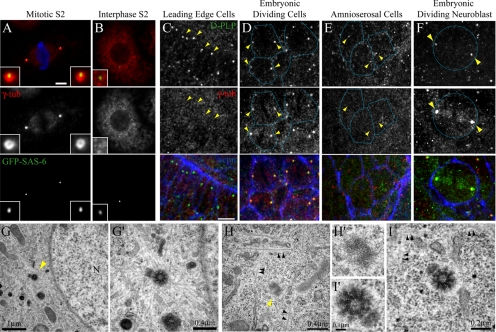
As expected, GFP-SAS-6–marked centrioles recruit PCM in mitotic cells (Figure 3A). During interphase, however, centrioles did not recruit γ-tubulin, as GFP-SAS-6 appeared as discrete spots in the cytoplasm that did not obviously colocalize with γ-tubulin punctae (Figure 3B). To test if this failure to be recruited was unique to γ-tubulin, we examined the distributions of other PCM components including Centrosomin (Cnn), CP190, and CP60 (Oegema et al., 1995; Li and Kaufman, 1996; Butcher et al., 2004). As with γ-tubulin, each protein localized to centrosomes at mitotic spindle poles, but not with centrioles in interphase S2 cells (Supplemental Figure S4). We next assessed whether this failure to recruit PCM during interphase was unique to S2 cells. Instead, failure to recruit PCM during interphase appears to be the standard behavior of Drosophila cells: we observed γ-tubulin recruitment to PLP-labeled centrioles in mitotic but not interphase D16 cells (Supplemental Figure S3B). This behavior was also observed in vivo during normal development: centrioles recruited γ-tubulin in mitotic embryonic cells and in dividing embryonic neuroblasts (Figures 3, D and F; yellow arrowheads mark centriole and γ-tubulin positions, and blue actin-labeling denotes the borders of adjacent cells), whereas centrioles within nondividing leading edge and amnioserosal cells lacked γ-tubulin staining (Figures 3, C and E; yellow arrowheads). In some cycling cells, both in culture and in vivo, small amounts of γ-tubulin staining could be detected on interphase centrioles (data not shown). We hypothesize that this may be due to a gradual recruitment or loss of PCM as centrioles prepare to enter or exit mitosis.
Figure 3.
Centrioles selectively recruit γ-tubulin during mitosis but not interphase. GFP-SAS-6 (green)-expressing S2 cells in metaphase (A) or interphase (B) stained for γ-tubulin (red) and phosphohistone H3 (blue). GFP-SAS-6 localizes to a small spot inside a ring of γ-tubulin during mitosis (A, insets). 100% of mitotic γ-tubulin “spheres” contained GFP-SAS-6 (n = 100). Scale, 5 μm. Embryonic leading edge cells (C), dividing epithelial cells (D), amnioserosa cells (E), and an embryonic dividing neuroblast (E), stained for D-PLP (green), γ-tubulin (red), and actin (blue). Yellow arrowheads mark centriole positions. Cell borders are traced (blue). Scale, 10 μm. Transmission electron micrographs of S2 cells during mitosis (G) and interphase (H and I). A dense MT array radiates from a centriole pair (yellow arrowhead) during prophase (G; N, nucleus), higher magnification shown in G′. MTs (black arrowheads) do not emanate from centrioles during interphase (H and I); higher magnifications are shown in H′ and I′.
The colocalization of γ-tubulin on mitotic but not interphase centrioles suggested that functional centrosomes are not present in interphase cytoplasm, and therefore centrioles would not associate with MTs in a manner characteristic of MTOCs. To determine this relationship in fixed S2 cells, we examined centrioles at higher resolution by electron microscopy. Centrioles were clearly identified as electron-dense structures ∼0.2 μm in length and exhibiting a ring of nine doublet MT bundles, a typical arrangement in Drosophila (Figures 3, G′ and I′; Gonzalez et al., 1998). Centrioles in mitotic cells appeared as doublets where a mature “mother” centriole was in close proximity to smaller “daughter” centriole of variable size (Figure 3G), consistent with their duplication in the previous S-phase. We noticed that in some interphase cells, centrioles appeared as singlets that had not yet duplicated (compare Figure 3, H′ and I′). As expected for functional centrosomes, centriole doublets in mitotic cells were surrounded by a dense network of MTs (prophase cell shown in Figure 3G). In contrast, centrioles in interphase cells did not display this feature and did not appear tightly associated with MTs, although nearby MTs were easily identified (Figures 3, H and I). Thus, based on our analysis of fixed cells, Drosophila displays a unique cycle of centrosome function that directs the inactivation of interphase centrosomes and MTOCs.
Loss of Centrioles Does Not Cause G1 Arrest or Delay
Previous studies demonstrated a critical role for centrosomes in G1 progression through the cell cycle in cultured mammalian cells (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001; Mikule et al., 2007). However, other studies suggested that centrosomes are not required (Uetake et al., 2007). In Drosophila, centriole-deficient larvae develop to adults with near normal timing (Basto et al., 2006), suggesting that, in this system, centrioles do not influence cell cycle progression, but an analysis of the cell cycle had not been performed. Therefore, to test whether fly centrioles regulate cell cycle progression, we depleted S2 cells of SAS-6 using RNAi and examined their cell cycle distributions. Anti-SAS-6 Western blots confirmed that RNAi treatment depleted protein levels by >99% (Supplemental Figure S5B) and effectively eliminated D-PLP–stained centrioles (Supplemental Figure S5C). Although control-treated interphase cells contained a median centriole number of 2.0 centrioles (avg. 3.0 ± 3.2; 654 cells counted), SAS-6–depleted cells contained a median centriole number of 0 (avg. 0.2 ± 0.6; 425 cells counted; Supplemental Figure S5D). Furthermore, SAS-6 depletion slightly increased the mitotic index (3.4 vs. 2.5% in control) and dramatically elevated the frequency of mitotic bipolar spindles with no centrosomes (Supplemental Figure S5E). These phenotypes are very similar to those described for centriole-deficient larval neuroblasts (Martinez-Campos et al., 2004) and are consistent with a role for Drosophila SAS-6 in centriole assembly. If Drosophila SAS-6 and centrioles played an important role in progression through G1, we would expect that loss of SAS-6 would dramatically alter the cell cycle distribution and decrease the number of cells within the G2-phase peak. However, quantitative high-throughput microscopy (HTM) revealed that control and SAS-6 RNAi-treated cells had indistinguishable cell cycle profiles (Supplemental Figure S5F). Therefore, centrioles are likely not essential for interphase cell cycle progression in Drosophila cells. Similar results were observed in S2 cells depleted of Plk4, a kinase required for centriole duplication (Bettencourt-Dias et al., 2005), as well as after SAS-6 small interfering RNA (siRNA) in human cells (Strnad et al., 2007). However, unlike human SAS-6, which is degraded from anaphase until late G1 phase (Strnad et al., 2007), fly SAS-6 is a stable centriole marker throughout the cell cycle (our unpublished results).
Interphase Microtubule Arrays Are Nucleated Independently of Centrioles
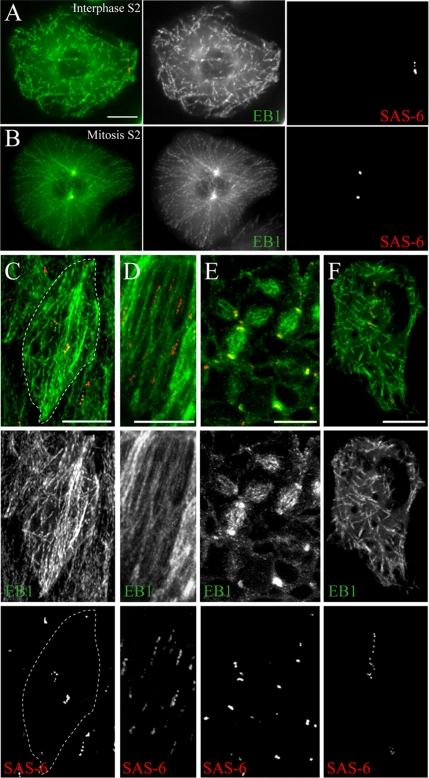
These data suggest that in many Drosophila cell types centrosomes are inactivated in interphase. In the canonical animal cycle of centrosome function, centrosomes serve as interphase MTOCs. To assess relationships between centrioles and MT dynamics, we generated a stable S2 line coexpressing GFP-SAS-6 and EB1-mRFP as well as transgenic flies coexpressing mCherry-SAS-6 and EB1-GFP. EB1 is an MT plus–end tracking protein (+TIP) and can be used to visualize growing MT plus ends (Rogers et al., 2002). We recorded living interphase and mitotic cells by time-lapse microscopy and analyzed the patterns of MT growth relative to centrioles. In S2 cells, EB1-labeled MTs were nucleated at many discrete sites in the cytoplasm of interphase cells and did not emanate from centrioles, which exhibited random movements near the cell periphery (Figure 4A; see Movie 01). In contrast, upon entry into mitosis, MT nucleation originated from centrioles (Figure 4B; see Movie 02). We also examined MT dynamics in several embryonic cell types. In live amnioserosal and leading-edge cells (arrested in G2 and G1 phase, respectively), centriole position oscillated throughout the cell and MT nucleation occurred at sites unrelated to centriole location (Figure 4, C and D; see Movies 03 and 04). Interestingly, in leading edge cells MT organization is not random, but instead is oriented along the long axis of the cell (Figure 4D; Jankovics and Brunner, 2006), but centrosomes do not appear to play a role in this organization. In contrast, EB1 tracks emanated from centrioles at spindle poles within dividing embryonic ectodermal cells (Figure 4E; Movie 05). Thus, in each Drosophila cell type examined, centrioles possess MTOC activity only during mitosis and do not appear to influence interphase MTs. However, as EB1 only labels growing MT plus ends, we cannot rule out the possibility of rapid nucleation and release of some MTs at centrioles.
Figure 4.
Interphase microtubule nucleation is independent of centrioles and γ-tubulin. (A–F) Projections of GFP-SAS-6 (red) and EB1-mRFP (green) MT growth patterns visualized by merging short segments of a 1-min time-lapse recording into a single flattened image. Tracks of SAS-6 spots in the panels correspond to their movement over time. Acentriolar MT nucleation occurs in interphase S2 cells (A) but is associated with centrioles during mitosis (B). In embryos, MT nucleation (EB1-GFP; green) is not associated with centrioles (mCherry-SAS-6; red) in interphase amnioserosal (C) or leading edge (D) cells but is associated with spindle poles in mitotic domains (E). White dotted-line (C) traces the cell border. (F) A similar pattern of MT growth is observed after γTub23C RNAi. Scale, 5 μm (A, B, and F); 10 μm (C–E).
Steady-State Interphase Microtubule Levels Are Insensitive to γ-Tubulin Depletion
We next investigated the molecular requirements for establishing acentrosomal interphase MT arrays in S2 cells by using RNAi against proteins with known MT-nucleating activity. To begin, we focused on γ-tubulin, the central MT-nucleation factor in eukaryotic cells (Wiese and Zheng, 2006). The Drosophila genome encodes two γ-tubulin genes: γTub23C, the major isotype in S2 cells, and γTub37C, the minor isotype (Raynaud-Messina et al., 2004). In a previous study, RNAi of γTub23C alone did not seem to affect the interphase MT cytoskeleton (Raynaud-Messina et al., 2004). However, these authors were not able to deplete the minor γTub37C and it remained possible that γTub23C and γTub37C function redundantly to nucleate the interphase MTs. Indeed, functional redundancy has been reported for the two γ-tubulins during female germ-cell development and oogenesis (Tavosanis and Gonzalez, 2003). Recently, we discovered that RNAi is much more effective if S2 cells are cultured in the absence of fetal bovine serum and have modified our RNAi procedure (data not shown; see Materials and Methods). Using dsRNA that targets the 3′UTR, γTub23C RNAi was highly effective (below the level of detection) as determined by Western blotting (Figure 5B). Depletion of γTub23C produced the expected mitotic phenotype, elevating the mitotic index by fourfold and produced numerous monopolar spindles (Figure 5C), as previously described (Raynaud-Messina et al., 2004), suggesting that we reduced its levels below a critical threshold. However, interphase cells in the same population still contained MTs, and MT nucleation and growth, as assessed by live cell imaging of EB1-mRFP and GFP-SAS-6, was indistinguishable from wild-type cells (Figure 5F; see Movie 06). Rates of fluorescent EB1 movement revealed that MT growth was similar in control and γTub23C-depleted cells (control 9.9 ± 4.0 μm/s [30 MTs in three cells]; γTub23C 7.9 ± 2.0 μm/s [30 MTs in four cells]).
Figure 5.
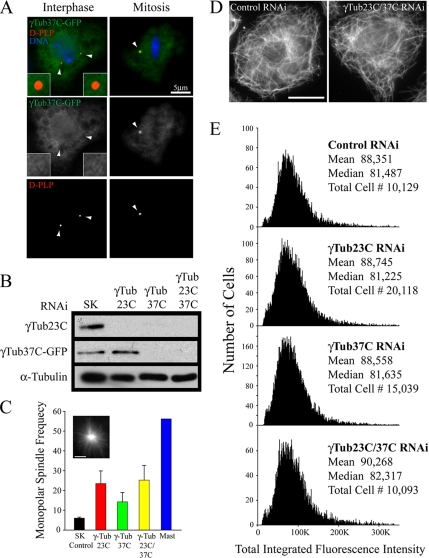
RNAi-mediated codepletion of γTub23C/37C does not affect steady-state MT levels in interphase S2 cells. (A) Immunofluorescence of interphase and mitotic transfected S2 cells expressing γTub37C-GFP (green) and stained for D-PLP (red) and DNA (blue). The right spindle pole in the mitotic cell is not in the focal plane. Arrowheads mark centriole positions. (B) Lysates were prepared from γ-Tub37C-GFP stable-expressing S2 cells after 7 d of control or γ-tubulin RNAi treatments and probed by Western blot using anti-γ-tubulin antibody (anti-α-tubulin was used as a loading control). (C) Bar graph shows the frequency of monopolar spindle formation after 7-d control, γ-tubulin, or MAST RNAi treatments and identified by MT staining (inset; scale, 5 μm). (D) Interphase day 7 control and γTub23C/37C RNAi-treated S2 cells stained for MTs. (E) Distribution histograms of total integrated MT fluorescence intensity in RNAi-treated α-tubulin–immunolabeled S2 cells using quantitative HTM; (y-axis) cell number, (x-axis) arbitrary units. Examples of these cells are shown in D. γ-tubulin RNAi treatments produced no significant difference (p > 0.05) as compared with control RNAi using a nonparametric Kruskal-Wallis one-way analysis of variance followed by a Dunn's post test.
Due to the low abundance of γTub37C, our γ-tubulin antibody did not detect this minor isotype in Western blots of S2 cell lysates. To determine the efficacy of RNAi, we monitored depletion by generating a γTub37C-GFP stable S2 line driven by the gene's endogenous promoter. This approach allowed adequate expression to detect by Western blotting using antibodies against γ-tubulin (Figure 5B) or GFP (data not shown). Like γTub23C, γTub37C-GFP localized to hollow spheres at mitotic spindle poles but was diffuse in the cytoplasm of interphase cells and did not colocalize with centrioles (Figure 5A). After RNAi treatment using a 1.1-kb dsRNA that targets a large segment of the γTub37C ORF, γTub37C-GFP levels were reduced below the level of detection (Figure 5B). Due to the high nucleotide identity (78%) between the two γ-tubulins within this coding region, γTub23C was also efficiently depleted by γTub37C RNAi (Figure 5B). Similar “cross-depletion” of γTub23C was observed in a previous study (Raynaud-Messina et al., 2004). As expected, this treatment elevated the frequency of monopolar spindle formation (Figure 5C), as did the codepletion of γTub23C/37C where dsRNAs against both γTub23C/37C were added to the culture (Figure 5B).
We found that interphase cells depleted of γTub23C or γTub37C individually or together assembled MTs and appeared similar to control-treated cells (Figure 5D). To quantitate changes in MT levels, S2 cells were fixed in glass-bottom 24-well plates, stained for α-tubulin, and scanned using quantitative HTM. Single γ-tubulin “knockdowns” or γTub23C/37C codepleted cells revealed no significant difference in the mass of MT polymer compared with control-treated cells (Figure 5E). Thus, our observations of acentrosomal steady-state MT assembly suggest the presence of a γ-tubulin–independent or redundant mechanism for generating interphase MT arrays.
MT Regrowth Does Not Occur from Interphase Centrioles
To examine the possible sites of MT nucleation with more clarity and to examine the role of γ-tubulin in this process, we cold-treated S2 cells to induce MT depolymerization and analyzed the position of centrioles relative to the sites of regrowth of MTs, by fixing the cells at 0, 2.5, 5, 10, 15, and 30 min time points. When chilled for 1 h, MTs completely depolymerized in both mitotic and interphase S2 cells (Figure 6A, 0 min; data not shown). Within 5 min after a return to room temperature, robust MT regrowth occurred from centrioles in dividing cells (Figure 6A, 5 min cytokinesis). In contrast, MT regrowth in interphase cells did not occur from centrioles; small MTs were first observed by 2.5 min (Supplemental Figure 6A, 2.5 min). By 5 min, MTs appeared at many sites within the cytoplasm, as individual MTs or as small tufts of MTs (Figure 6A). In some cells, some MT foci associated with centrioles but most did not (Supplementary Figure S6A, 5 min). At 10 min, MT foci were more numerous and assembled into an extensive interconnecting MT network throughout the cell (Figure 6A, 10 min). By 15 min, S2 cells displayed a normal interphase array, having dispersed the MT foci that formed in the earlier time points (Figure 6A, 15 min).
Figure 6.
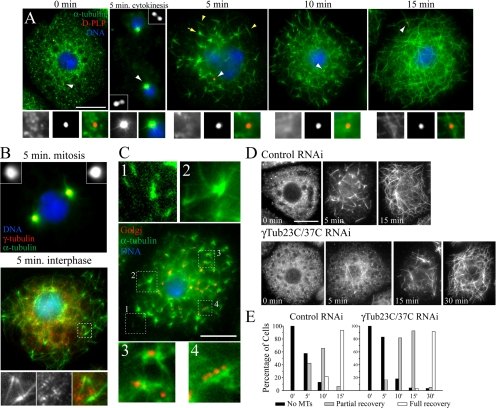
MT regrowth occurs independently of centrioles but is delayed in γ-tubulin–depleted S2 cells. (A) S2 cells were stained for MTs (green), D-PLP centrioles (red), and DNA (blue) at specific time points in a MT regrowth assay. MTs were depolymerized by cold treatment (0 min.) and brought to room temperature to allow polymerization. Cells were fixed at 5, 10, and 15 min. Centrioles (white arrowheads) are shown at higher magnifications in bottom panels (left to right; MTs, D-PLP, merge). A cell in cytokinesis is shown at 5 min with disengaged centrioles (insets). An interphase cell at 5 min contains individual MTs (yellow arrowheads) and tufts of MT foci (yellow arrow). Scale, 10 μm. (B) Mitotic (top) and interphase (bottom) S2 cells after 5 min of MT regrowth stained for MTs (green), γ-tubulin (red, insets), and DNA (blue). The white box (interphase cell) is shown at higher magnification (bottom panel) and denotes a cluster of MT foci (left to right; MTs, γ-tubulin, merge). (C) An S2 cell at 5 min of MT regrowth immunostained for MTs (green), Golgi (red), and DNA (blue). Higher magnifications of (1) individual MTs in the lamella, (2) MT foci, (3) Golgi associated with MT foci, and (4) Golgi punctae along a MT bundle. Scale, 10 μm. (D) Time points show day 7 control and γ-Tub23C/37C RNAi-treated S2 cells stained for MTs during MT regrowth. Scale, 10 μm. (E) Quantitation of MT recovery in the RNAi-treated cells displayed in D.
Previous MT regrowth experiments using cultured fly cells described the appearance of unique MT foci similar to what we observed (Colombié et al., 2006; Cottam et al., 2006). Although individual MTs and foci form independently of interphase centrioles in S2 cells (our findings, Figure 6A), γ-tubulin colocalizes with MT foci that assemble in cold-treated Clone8+ cells (a wing imaginal disk-derived cell line; Cottam et al., 2006). In contrast, we found that in cold-treated S2 cells γ-tubulin did not colocalize with MT foci, although γ-tubulin punctae were apparent in interphase (Figure 6B; bottom panel), and γ-tubulin strongly colocalized with centrioles and active MTOCs in mitotic cells (Figure 6B; top panel).
Next, we examined whether the Golgi apparatus acts as the assembly site of MT foci in cold-treated S2 cells, as noncentrosomal MT nucleation from the Golgi apparatus is described in mammalian cells (Chabin-Brion et al., 2001; Efimov et al., 2007). Golgi in interphase S2 cells appear as numerous discrete punctae widely distributed throughout the cytoplasm (Stanley et al., 1997). Using an mAb that recognizes an integral membrane protein enriched in Golgi membrane fractions, we found that Golgi were restricted to the center of S2 cells that were spread on conA-coated glass. Five min after recovery from cold treatment, MTs were apparent in the thin extended lamella, a region devoid of Golgi (Figure 6C, 1), and some MT foci in the more central region were not associated with Golgi punctae (Figure 6C, 2), suggesting that not all foci nucleate from Golgi. However, Golgi were visible at the center of some MT foci (Figure 6C, 3); this was also observed at an earlier time point (Supplemental Figure S6B; 2.5 min). Golgi also appeared as linear tracks of punctae associated with long MT bundles (Figure 6C, 4). The appearance of most MTs at sites not coincident with Golgi suggests that Golgi is not essential for noncentrosomal MT nucleation in S2 cells. However, we cannot rule out the possibility that Drosophila Golgi has MT-nucleating capability, as some MT association is observed.
MT Regrowth Is Delayed without γ-Tubulin
We next used our assay to directly examine whether γ-tubulin plays a role in the kinetics of MT regrowth and the formation of MT foci. Double γ-Tub23C/37C RNAi did not prevent the assembly of MT foci (Figure 6D). However, γ-tubulin depletion altered the rate of MT recovery in interphase cells (Figure 6D and E). By 15 min, 94% of control RNAi-treated cells fully recovered their MT arrays, whereas only 3% of γ-tubulin depleted cells fully recovered at this time. In the absence of γ-tubulin, cells required an additional 15 min to complete full recovery (Figure 6E). We conclude that γ-tubulin is likely not required for maintaining MT levels at steady state in interphase S2 cells but is required for normal kinetics of MT regrowth after cold treatment. Our findings also suggest that cold-recovering interphase cells utilize a biphasic pathway to assemble acentrosomal MT arrays, where γ-tubulin contributes to an initial fast phase of assembly.
γ-Tubulin, Mini-spindles, CLIP-190, EB1, and Cytoplasmic Dynein Contribute to a Fast MT-Assembly Pathway during MT Regrowth
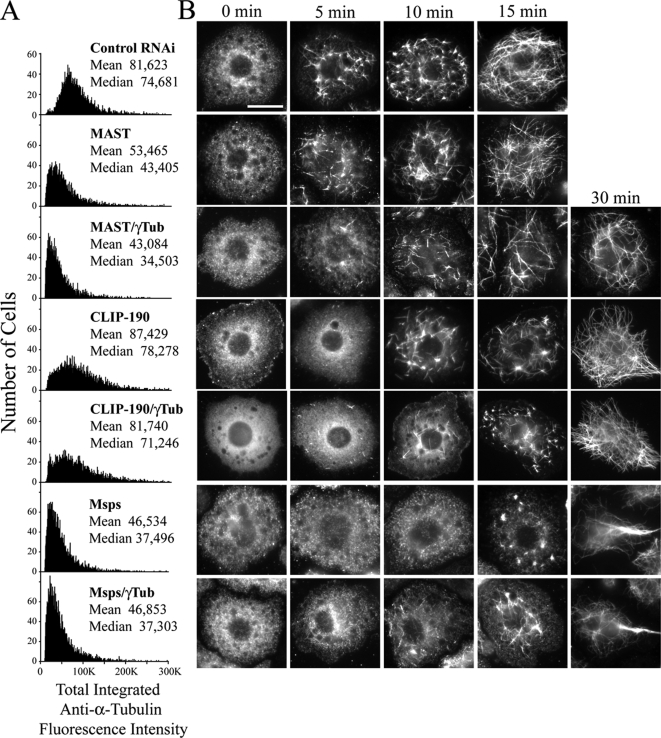
Given our findings that γ-tubulin is dispensable for maintaining steady-state MT levels and that MTs eventually fully recover after cold treatment, we explored other potential candidates that might participate in this putative γ-tubulin–independent or partially redundant mechanism. One potential candidate is CLASP; members of this conserved family of +TIPs regulate MT dynamic instability throughout the cell cycle. Recent work revealed that in human interphase cells, CLASP localizes to the trans-Golgi network (TGN) where it can nucleate a noncentrosomal population of MTs (Efimov et al., 2007). MAST/Orbit is the only fly CLASP. Surprisingly, recent work revealed that while MAST promotes MT stability during interphase, and MAST RNAi reduces MT density as well as the number of MTs that extend to the cell cortex, MAST appears to play no role in MT nucleation in S2 cells, as the kinetics of MT regrowth after cold depolymerization is normal (Sousa et al., 2007). In agreement with this, we found that MAST depletion dramatically elevated monopolar spindle formation (Figure 5C) and eliminated cortical MTs as expected (Supplemental Figure S7). Further, MAST depletion decreased steady-state MT levels (reducing the mean by ∼34%) compared with controls (Figure 7A), validating our quantitative cytometric approach. However, MTs in MAST-depleted S2 cells recovered from cold treatment with normal kinetics, as previously described (Figure 7B; second row; Sousa et al., 2007). These data left open the possibility that MAST and γ-tubulin play redundant roles in nucleating MTs. To test this hypothesis, we used RNAi to simultaneously deplete both proteins from S2 cells. At steady state, these displayed MT levels slightly lower than those found in cells depleted of MAST alone (Figure 7A). Furthermore, MT regrowth in these cells recovered with the same kinetics observed after γ-tubulin RNAi alone: a 15-min delay in full recovery (Figure 7B; third row). Thus, simple redundancy between MAST and γ-tubulin seem unlikely.
Figure 7.
CLIP-190 or Msps RNAi delays MT regrowth in S2 cells but is not observed with either MAST RNAi or codepletion with γ-tubulin. (A) Distribution histograms of total integrated MT fluorescence intensity at steady state in 5000 RNAi-treated α-tubulin–immunolabeled S2 cells using quantitative HTM; (y-axis) cell number, (x-axis) arbitrary units. Approximately 5000 cells were scanned in each treatment, which produced a significant difference (p < 0.05) compared with the control RNAi using a nonparametric Kruskal-Wallis one-way analysis of variance followed by a Dunn's post-test. (B) Time points show representative day 6 RNAi-treated S2 cells stained for MTs during MT regrowth. Each row of micrographs is aligned with the dsRNA(s) that were used in A. Scale, 10 μm.
We next tested another candidate, CLIP-190, the fly homolog of the CLIP-170 family of +TIPs (Dzhindzhev et al., 2005). They possess multiple conserved tubulin-binding CAP-Gly domains that can nucleate MT growth in vitro (Slep and Vale, 2007). Although not as dramatic as MAST RNAi, we found a significant change in steady-state MT levels in CLIP-190–depleted cells compared with controls (Figure 7A). Unlike MAST, however, CLIP-190 RNAi altered the rate of MT regrowth after cold-induced depolymerization and delayed MT recovery by 15 min, similar to γ-tubulin depletion (Figure 7B, fourth row). If γ-tubulin and CLIP-190 function partially redundantly to nucleate MTs, then we expected that codepletion of both proteins would produce a more severe MT assembly phenotype. However, MT steady-state levels were not further reduced by codepletion as compared with CLIP-190 RNAi alone (Figure 7A) and, interestingly, cells recovered from cold treatment with the same kinetics displayed in the single RNAi treatments (Figure 7B; fifth row). Thus, we conclude that CLIP-190 is not redundant with γ-tubulin in maintaining steady-state MT levels and thus may function in the same assembly pathway as γ-tubulin to nucleate a fast phase of MT regrowth.
A third candidate we examined is Mini-spindles (Msps), the sole fly member of the conserved Dis1/TOG family of MT-associated proteins (MAPs). Like MAST, these are +TIPs that promotes MT growth (van Breugel et al., 2003; Howard and Hymann, 2007). Msps RNAi inhibits MT plus-end growth and increases MT pausing and bundling in interphase S2 cells (Brittle and Ohkura, 2005). Members of this family contain an array of multiple conserved TOG domains that bind αβ tubulin heterodimers capable of inducing MT nucleation in vitro (Slep and Vale, 2007). We observed extensive cortical MT bundling after Msps depletion (Supplemental Figure 7), as well as a decrease in steady-state MT levels (43% reduction in mean polymer mass), but this did not diminish further upon codepletion of γ-tubulin (Figure 7A). These data suggest that Msps and γ-tubulin are not redundant in maintaining MT levels at steady-state. However, as with γ-tubulin depletion, we found that Msps RNAi alone delayed the recovery of MTs after cold treatment by ∼15 min (Figure 7B; sixth row). To examine whether Msps could function in the eventual recovery of MTs that we observed after cold-treating γ-tubulin–depleted cells, we performed Msps/γ-tubulin double RNAi. Surprisingly, these cells displayed a rate of MT regrowth similar to the single RNAi treatments: a delay of 15 min (Figure 7B; seventh row). Because we did not observe a synergistic effect by double Msps/γ-tubulin RNAi, our findings suggest that Msps may also function together with γ-tubulin in a fast MT-nucleation pathway.
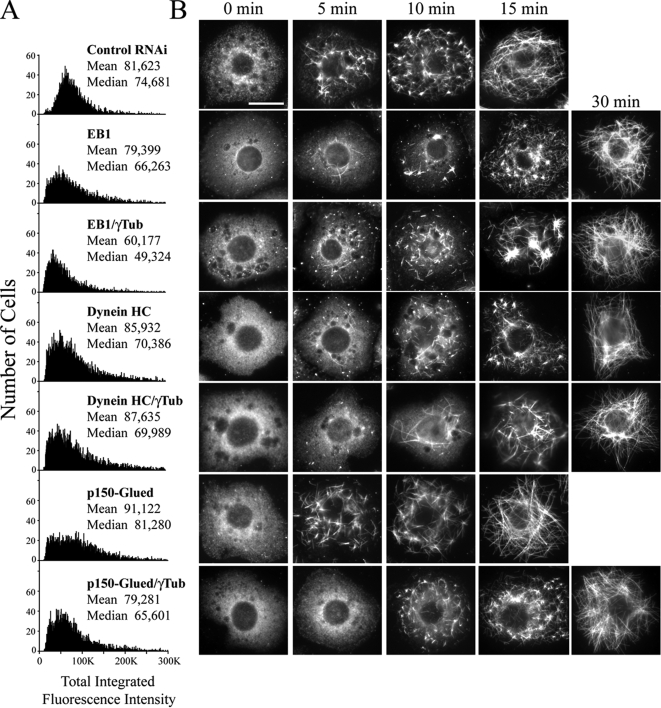
Because our data implicate two +TIPs (CLIP-190 and Msps) in MT assembly, we next examined the contribution of EB1 in our functional assays, as EB1 recruits stabilizing +TIPs to growing MT plus ends (Vaughan, 2005). Moreover, a recent in vitro study demonstrated that Mal3p, the Schizosaccharomyces pombe EB1 homolog, may induce polymer stability by binding along the MT lattice seam (Sandblad et al., 2006). Expectedly, EB1 depletion decreased overall steady-state MT levels, similar to our results with CLIP-190 but not as dramatically as either MAST or Msps RNAi (Figure 8A). We also observed a modest decrease in steady-state MT levels after EB1/γ-tubulin codepletion (Figures 8A). Strikingly, we found that EB1 RNAi alone delayed the kinetics of MT regrowth after cold treatment by 15 min (Figure 8B; second row). However, this delay was not further prolonged after codepletion with γ-tubulin (Figure 8B; third row). Thus, these data suggest that EB1 may also function with γ-tubulin in an initial fast MT-nucleation pathway.
Figure 8.
EB1 or cytoplasmic dynein (DHC) RNAi delays MT regrowth in S2 cells but is not observed with either depletion of the p150Glued component of dynactin or codepletion with γ-tubulin. (A) Distribution histograms of total integrated MT fluorescence intensity at steady state in RNAi-treated α-tubulin–immunolabeled S2 cells using quantitative HTM; (y-axis) cell number, (x-axis) arbitrary units. Approximately 5000 cells were scanned in each treatment that produced a significant difference (p < 0.05) compared with the control RNAi using a nonparametric Kruskal-Wallis one-way analysis of variance followed by a Dunn's post-test. (B) Time points show representative day 6 RNAi-treated S2 cells stained for MTs during MT regrowth. Each row of micrographs is aligned with the dsRNA(s) that were used in A. Scale, 10 μm.
We also examined whether the cytoplasmic dynein MT-based motor complex and its associated adaptor complex, dynactin, participate in interphase MT assembly. Drug inhibition of dynein in fish melanophore cell fragments greatly diminishes the rate of MT nucleation that is needed to build radial acentrosomal arrays in this system, and dynein is effective in MT nucleation in vitro (Malikov et al., 2004). Although dynactin does not nucleate MTs in vitro (Malikov et al., 2004), the p150Glued dynactin component contains a conserved tubulin-binding CAP-Gly motif, also found in CLIP-190, suggesting a possible role in MT nucleation in cells. Consistent with roles in MT stability, we found that either dynein heavy chain (DHC) or p150Glued RNAi reduced steady-state MT levels (Figure 8A), although, as with CLIP-190 RNAi, mean and median levels were not dramatically altered. Codepleting γ-tubulin did not further reduce MT levels (Figure 8A). MT regrowth assays revealed that DHC depletion alone delayed the full recovery of the acentrosomal interphase array by ∼15 min (Figure 8B; fourth row), whereas p150Glued RNAi did not (Figure 8B; sixth row). These results suggest a dynactin-independent function for DHC in Drosophila cells, consistent with the in vitro results of Malikov et al. (2004). As before, we applied the MT regrowth assay to cells codepleted of DHC or p150Glued with γ-tubulin in order to determine their functional relationship in MT nucleation. Both p150Glued/γ-tubulin RNAi and DHC/γ-tubulin displayed a 15-min delay in MT regrowth, similar to DHC or γ-tubulin RNAi alone (Figure 8B; fifth and seventh rows). Thus, these results suggest that DHC, but not dynactin, also contributes to a fast phase of MT nucleation during regrowth.
To determine whether these +TIPs and MT motors play specific roles in MT regrowth, we also examined several additional +TIP or MT-binding proteins for roles in interphase MT regrowth. These included the kinesins Ncd (a minus-end directed motor), conventional kinesin heavy chain (KHC), and the MT-depolymerase Klp10A, as well as the spectroplakin Shortstop (Shot). Ncd, Klp10A, and Shot all display +TIP behavior (Rogers et al., 2004b; Goshima et al., 2005; Mennella et al., 2005; Slep et al., 2005). Depletion of each of these proteins individually did not change rates of MT recovery compared with control treatments (Supplemental Figure 8). Thus, +TIPs do not perform a general role in MT assembly. We note in passing that although the kinetics of MT recovery did not change after KHC RNAi, MT regrowth was restricted to perinuclear regions of the cell (Supplemental Figure S8).
Taken together, our findings suggest that interphase Drosophila cells utilize a biphasic pathway to assemble acentrosomal MT arrays, perhaps by γ-tubulin, CLIP-190, Msps, EB1, and DHC working together to drive an initial fast phase of MT assembly. Removal of any one component inactivates the fast phase and reveals the presence of a partially redundant, slower phase to ensure assembly of the MT array via an uncharacterized mechanism.
DISCUSSION
The Typical Cycle of Centrosome Function in Drosophila Cells
The current model for regulating MT nucleation in animal cells suggests that centrosomes play a key role in generating MT arrays in both mitosis and in interphase. Our analysis revealed that the Drosophila cycle of centrosome function is quite distinct from that of vertebrate cells, where centrosomes act as MTOCs throughout the cell cycle. In Drosophila cells, functional centrosomes are assembled at the onset of mitosis and participate in spindle assembly, but then disassemble upon mitotic exit. Interphase centrioles do not recruit γ-tubulin and, thus, lack the capacity to nucleate and organize MT arrays.
Our results provide mechanistic insight into the recent surprising report that centriole-deficient sas-4 mutant flies develop into viable adults with near normal timing and morphology (Basto et al., 2006). This study raised a fascinating question: how do interphase cells, in this mutant background, survive without the cytoplasmic organization that centrosomes provide? Our results demonstrate that Drosophila cells do not depend on centrosomes to nucleate/organize interphase MTs or to progress through interphase, explaining why an absence of centrosomes has little impact on development.
Our study does not suggest that centrosomes are unimportant, however, only that they are not essential for interphase homeostasis, because they are normally absent during interphase in many cell types. Centrosomes are present during mitosis, however, and are vital for early divisions during cleavage, where they support proper spindle/nuclear spacing within the large syncytium. This was best shown for mutations in the PCM component, Centrosomin; its loss blocks centrosome function, producing spindle assembly defects and maternal-effect lethality (Megraw et al., 1999). Centrosomes are also essential to assemble cilia and flagella; sas-4 mutant adults lack cilia in their mechanosensory neurons, are severely uncoordinated, and die shortly after eclosion (Basto et al., 2006).
Roles for vertebrate centrosomes extend beyond MT organization. Several studies suggest that centrosomes provide an essential scaffold for docking numerous cell cycle regulators (reviewed in Doxsey et al., 2005). This may explain why a cycle of centrosome function that utilizes an interphase centrosome was acquired or maintained in vertebrate evolution, whereas centrioles are not essential for Drosophila cell cycle progression in culture. It would be interesting to manipulate the Drosophila centrosome to mimic the vertebrate cycle, in order to examine the impact of a persistent, functional centrosome on interphase MT- dependent processes at the cellular level as well as overall development of the animal.
A New Mechanism of Nucleating Interphase Microtubules
Our findings thus support a reassessment of the view that the centrosome is the major MTOC in all interphase cells, producing a polarized radial MT array to organize the cytoplasm of interphase proliferating and migrating animal cells. Other evidence also supports the need for this reassessment. In some cell-types, such as epithelial cells and neurons, MTs are released from centrosomes and incorporated into noncentrosomal MT arrays (Waterman-Storer and Salmon, 1997; Ahmad et al., 1999). Indeed, the MT nucleating capacity of the centrosome, and the fraction of MTs in the cell that originate from the centrosome, can vary dramatically between cell-types and species (Waterman-Storer and Salmon, 1997). Finally, cycling vertebrate cells generally reduce the amount of PCM associated with the centrosome when exiting mitosis (Dictenberg et al., 1998; Rusan and Wadsworth, 2005). Thus, the cycle of centrosome activation/inactivation in Drosophila cells that we describe here may represent an exaggerated version of behavior that is found in at least some vertebrate cells.
If centrosomes do not function in interphase Drosophila cells, then what nucleates MT growth and how is the cytoplasm organized without an MTOC? Using RNAi, we examined the roles of several known and putative MT nucleators in maintaining steady-state MT levels, as well as in the regrowth of acentrosomal MT arrays in interphase S2 cells. The fly genome encodes two different γ-tubulins (Raynaud-Messina et al., 2004): γTub23C, the ubiquitous isotype that plays an important role in mitotic spindle assembly (Sunkel et al., 1995), and γTub37C, which functions in female meiotic spindle assembly (Tavosanis et al., 1997). Double γ-tubulin mutants are severely compromised in various stages of female germ-cell differentiation (Tavosanis and Gonzalez, 2003). Surprisingly, we found that codepletion of both γTub23C and γTub37C did not inhibit production of interphase MT arrays or alter steady-state MT levels. Our findings differ from γ-tubulin loss-of-function in other systems, which generally display severe changes in MT levels. For example, γ-tubulin RNAi in C. elegans embryos inhibits the formation of interphase centrosomal asters and reduces MT levels by 60% during mitosis (Hannak et al., 2002). In the fungi Ustilago maydis and Aspergillus nidulans, loss of γ-tubulin function results in the reduction of cytoplasmic MT number and length (Oakley et al., 1990; Straube et al., 2003). In contrast, conditional γ-tubulin mutants in S. cerevisiae produced fewer nuclear/spindle MTs and abnormally long cytoplasmic MTs (Marschall et al., 1996; Spang et al., 1996).
Acentrosomal MT nucleation is observed in plants and S. pombe (Bartolini and Gundersen, 2006), in human cells from the Golgi apparatus (Efimov et al., 2007), and interphase MT arrays can self-organize in anucleate fission yeast cells lacking centrosomes (Carazo-Salas and Nurse, 2006; Daga et al., 2006). Significantly, however, acentrosomal MT nucleation and organization in these systems differs from Drosophila cells, in that γ-tubulin is required for MT nucleation (Bartolini and Gundersen, 2006).
Although γ-tubulin does not appear to play an essential role in maintaining interphase steady-state MT levels in S2 cells, MT regrowth experiments revealed that γ-tubulin does participate in MT nucleation. In the absence of γ-tubulin, S2 cells subjected to cold-induced MT-depolymerization exhibit a delay in the full recovery of interphase MTs by 15 min, taking twice as long as controls. Similarly, γ-tubulin siRNA in cultured human cells, which normally use centrosomes to nucleate MTs, produced extensive acentrosomal MT arrays after a slight delay in MT regrowth (Lüders et al., 2006), although neither the extent of γ-tubulin depletion nor a measurement of MT levels were determined. Perhaps a γ-tubulin–independent mechanism for MT nucleation is conserved in animal cells (Lüders et al., 2006).
We hypothesize that γ-tubulins function redundantly with an unidentified MT-assembly factor to nucleate acentrosomal MTs. Consistent with this hypothesis, a recent genome-wide RNAi screen in S2 cells identified many genes involved in mitotic spindle assembly, but depletion of only a few individual genes disrupted MT assembly. These were mainly the α/β-tubulins and tubulin-folding proteins (Goshima et al., 2007). To identify redundant molecules, we examined interphase MT arrays in the absence of MAST/Orbit, CLIP-190, Msps, EB1, DHC, or p150Glued, some of which possess MT-nucleating activity (Malikov et al., 2004; Efimov et al., 2007; Slep and Vale, 2007). Only MAST or Msps RNAi substantially reduced steady-state MT levels, potentially via their role in MT stability. Additionally, in most cases, MT levels were not further reduced by simultaneous depletion of γ-tubulin, suggesting that none of these candidates acts redundantly with γ-tubulin to maintain the steady-state level of interphase MT arrays.
However, depletion of γ-tubulin, CLIP-190, Msps, EB1, or DHC all produce a 15-min delay in MT recovery that is not further prolonged if γ-tubulin is codepleted with any one of the remaining assembly factors. Thus, our findings suggest a model in which interphase Drosophila cells use a biphasic pathway to re-establish acentrosomal MT arrays. One speculative possibility is that a multicomponent “functional pathway” of γ-tubulin/CLIP-190/Msps/EB1/DHC collectively drives an initial fast phase of MT nucleation. Theoretically, tubulin heterodimers bound to CLIP-190 and Msps could be recruited, via EB1, to newly formed MT “seeds,” unstable intermediate assembly structures that are partially stabilized by binding DHC. A CLIP-190/Msps/EB1 complex could thus provide additional subunits to elongate a MT seed and also stabilize the formation of a nascent MT plus end. Additional stability could then be provided by γ-tubulin capping a newly formed minus end. Removal of any one of these critical functions would compromise seed stability and MT growth. Obviously, the exact mechanism of MT nucleation for this pathway is unclear as we lack a complete understanding of the MT- stabilizing activity for any one of these proteins. Further, additional components may exist.
Interestingly, inhibition of the fast phase reveals the presence of a slower phase of MT nucleation that provides recovering S2 cells with a redundant mechanism to rebuild the interphase MT array. This back-up mechanism potentially explains why depletion of γ-tubulin has little effect on maintaining steady-state MT levels. The mechanism of slow-phase MT nucleation is unknown but could be accomplished by MT self-assembly, an inherent property of tubulin, or, perhaps, additional MT nucleators may remain to be discovered. Alternatively, the multicomponent pathway that we describe could simply allow for more efficient MT assembly, whereby MTs still assemble after inactivation of the pathway, albeit more slowly, but not necessarily in a biphasic way.
Our data are consistent with MT nucleation at specific sites within the cell, as we observed γ-tubulin–independent formation of small MT foci during the course of a MT regrowth assay, resembling the MTOCs reported in other cold-treated cultured fly cells (Cottam et al., 2006). The Golgi serves as a noncentrosomal surface for MT nucleation in mammalian cells (Chabin-Brion et al., 2001; Efimov et al., 2007). In S2 cells, many of the MT foci are not associated with Golgi, suggesting that they are not the sole source of noncentrosomal MT nucleation. It is possible that MT nucleation occurs off of another membrane-bound organelle, such as the endoplasmic reticulum, that is normally dispersed in the cell through its association with a pre-existing MT network. When MTs are depolymerized, this organelle might retract into a compact structure, with MT foci appearing at this site.
Taken together, our results support a canonical Drosophila cycle of centrosome function that differs from the canonical model in vertebrate cells and illustrates the flexibility of this functional cycle as well as the plasticity of the mechanisms for MT nucleation.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Rogers and Peifer labs for helpful discussions as well as D. Buster, K. Slep, E. Salmon, S. Crews and R. Duronio for critically reading the manuscript. We are grateful to the following individuals for their generous gifts: J. Raff for D-PLP antibody, T. Runfola and J. Brenman for Golgi antibody, R. Jones for Cnn antibody, and M. Moritz for CP60 and CP190 antibody. We also thank Victoria Madden and the UNC MSL-EM division for their help. N.M.R. was supported by a grant from the American Cancer Society (PF-06-108-01-CCG), and the work was supported by grants from the American Heart Association to S.L.R, and National Institutes of Health Grant RO1GM67236 to M.P.
Abbreviations used:
- MT
microtubules
- HTM
high-throughput microscopy
- MTOC
microtubule-organizing center
- PCM
pericentriolar material
- NEB
nuclear envelope breakdown
- γTub
gamma tubulin
- +TIP
plus–end tracking protein
- Msps
mini-spindles
- DHC
dynein heavy chain
- RNAi
RNA interference.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1069) on May 7, 2008.
REFERENCES
- Ahmad F. J., Yu W., McNally F. J., Baas P. W. An essential role for katanin in severing microtubules in the neuron. J. Cell Biol. 1999;145:305–315. doi: 10.1083/jcb.145.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;4:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Bartolini F., Gundersen G. G. Generation of noncentrosomal MT arrays. J. Cell Sci. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods G. C., Khodjakov A., Raff J. W. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M. K., Carmo N., Balloux F., Callaini G., Glover D. M. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and MT anchoring mechanisms. Curr. Opin. Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Brittle A. L., Ohkura H. Minispindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 2005;24:1387–1396. doi: 10.1038/sj.emboj.7600629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher R. D., Chodagam S., Basto R., Wakefield J. G., Henderson D. S., Raff J. W., Whitfield W. G. The Drosophila centrosome-associated protein CP190 is essential for viability but not for cell division. J. Cell Sci. 2004;117:1191–1199. doi: 10.1242/jcs.00979. [DOI] [PubMed] [Google Scholar]
- Callaini G., Riparbelli Centriole and centrosome cycle in the early Drosophila embryo. J. Cell Sci. 1990;97:539–543. doi: 10.1242/jcs.97.3.539. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Nurse P. Self-organization of interphase microtubule arrays in fission yeast. Nat. Cell Biol. 2006;8:1102–1107. doi: 10.1038/ncb1479. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion K., Marceiller J., Perez F., Settegrana C., Drechou A., Durand G., Poüs C. The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombié N., Verollet C., Sampaio P., Moisand A., Sunkel C., Bourbon H. M., Wright M., Raynaud-Messina B. The Drosophila gamma-tubulin small complex subunit Dgrip84 is required for structural and functional integrity of the spindle apparatus. Mol. Biol. Cell. 2006;17:272–282. doi: 10.1091/mbc.E05-08-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam D. M., Tucker J. B., Rogers-Bald M. M., Mackie J. B., Macintyre J., Scarborough J. A., Ohkura H., Milner M. J. Non-centrosomal microtubule-organising centres in cold-treated cultured Drosophila cells. Cell Motil. Cytoskelet. 2006;63:88–100. doi: 10.1002/cm.20103. [DOI] [PubMed] [Google Scholar]
- Daga R. R., Lee K. G., Bratman S., Salas-Pino S., Chang F. Self-organization of microtubule bundles in anucleate fission yeast cells. Nat. Cell Biol. 2006;8:1108–1113. doi: 10.1038/ncb1480. [DOI] [PubMed] [Google Scholar]
- Dammermann A., Muller-Reichert T., Pelletier L., Habermann B., Desai A., Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Dictenberg J. B., Zimmerman W., Sparks C. A., Young A., Vidair C., Zheng Y., Carrington W., Fay F. S., Doxsey S. J. Pericentrin and gamma-tubulin form a protein complex and are organized into a lattice at the centrosome. J. Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S., Zimmerman W., Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N. S., Rogers S. L., Vale R. D., Ohkura H. Distinct mechanisms govern the localization of Drosophila CLIP-190 to unattached kinetochores and microtubules plus-ends. J. Cell Sci. 2005;118:3781–3790. doi: 10.1242/jcs.02504. [DOI] [PubMed] [Google Scholar]
- Efimov, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. M. Polo kinase and progression through M phase in Drosophila: a perspective from the spindle poles. Oncogene. 2005;24:230–237. doi: 10.1038/sj.onc.1208279. [DOI] [PubMed] [Google Scholar]
- Gonzalez C., Tavosanis G., Mollinari C. Centrosomes and microtubule organization during Drosophila development. J. Cell Sci. 1998;111:2697–2706. doi: 10.1242/jcs.111.18.2697. [DOI] [PubMed] [Google Scholar]
- Goshima G., Nédélec F., Vale R. D. Mechanisms for focusing mitotic spindle poles by minus-end-directed motor proteins. J. Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., Vale R. D., Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E., Oegema K., Kirkham M., Gonczy P., Habermann B., Hyman A. A. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is γ-tubulin dependent. J. Cell Biol. 2002;157:591–602. doi: 10.1083/jcb.200202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe E. H., Miller F. J., Cham M., Khodjakov A., Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- Howard J., Hymann A. A. Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 2007;19:31–50. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Jankovics F., Brunner D. Transiently reorganized microtubules are essential for zippering during dorsal closure in Drosophila melanogaster. Dev. Cell. 2006;11:375–385. doi: 10.1016/j.devcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Keller L. C., Romijn E. P., Zamora I., Yates JR., 3rd, Marshall W. F. Proteomic analysis of isolated Chlanydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 2005;21:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C. L. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Gonczy P. Centrosome duplication and nematodes: recent insights from an old relationship. Dev. Cell. 2005a;9:317–325. doi: 10.1016/j.devcel.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 2005b;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Li K., Kaufman T. C. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell. 1996;85:585–596. doi: 10.1016/s0092-8674(00)81258-1. [DOI] [PubMed] [Google Scholar]
- Lüders J., Patel U. K., Stearns T. GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated MT nucleation. Nat. Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- Malikov V., Kashina A., Rodionov V. Cytoplasmic dynein nucleates microtubules to organize them into radial arrays in vivo. Mol. Biol. Cell. 2004;15:2742–2749. doi: 10.1091/mbc.E03-10-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall L. G., Jeng R. L., Mulholland J., Stearns T. Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J. Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Campos M., Basto R., Baker J., Kernan M., Raff J. W. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 2004;165:673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T. L., Li K., Kao L. R., Kaufman T. C. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- Megraw T. L., Kao L. R., Kaufman T. C. Zygotic development without functional mitotic centrosomes. Curr. Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Mennella V., Rogers G. C., Rogers S. L., Buster D. W., Vale R. D., Sharp D. J. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 2005;7:235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- Mikule K., Delaval B., Kaldis P., Jurcyzk A., Hergert P., Doxsey S. Loss of centrosome integrity induces p38–p53-p21-dependent G1-S arrest. Nat. Cell Biol. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- Moritz M., Agard D. A. Gamma-tubulin complexes and microtubule nucleation. Curr. Opin. Struct. Biol. 2001;11:174–181. doi: 10.1016/s0959-440x(00)00187-1. [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Oakley C. E., Yoon Y., Jung M. K. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Oegema K., Whitfield W. G., Alberts B. The cell cycle-dependent localization of the CP190 centrosomal protein is determined by the coordinate action of two separable domains. J. Cell Biol. 1995;131:1261–1273. doi: 10.1083/jcb.131.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel N., Stevens N. R., Basto R., Raff J. W. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 2007;15:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud-Messina B., Mazzolini L., Moisand A., Cirinesi A. M., Wright M. Elongation of centriolar microtubule triplets contributes to the formation of the mitotic spindle in gamma-tubulin-depleted cells. J. Cell Sci. 2004;117:5497–5507. doi: 10.1242/jcs.01401. [DOI] [PubMed] [Google Scholar]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H., Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Faruki S., Khodjakov A. The centrosome in vertebrates: more than a MT-organizing center. Trends Cell Biol. 2001;11:413–419. doi: 10.1016/s0962-8924(01)02085-2. [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Rogers G. C., Sharp D. J., Vale R. D. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Schwimmer T. A., Ems-McClung S. C., Walczak C. E., Vale R. D., Scholey J. M., Sharp D. J. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004a;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Wiedemann U., Häcker U., Turck C., Vale R. D. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr. Biol. 2004b;14:1827–1833. doi: 10.1016/j.cub.2004.09.078. [DOI] [PubMed] [Google Scholar]
- Rusan N. M., Wadsworth P. Centrosome fragments and microtubules are transported asymmetrically away from division plane in anaphase. J. Cell Biol. 2005;168:21–28. doi: 10.1083/jcb.200409153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N. M., Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandblad L., Busch K. E., Tittmann P., Gross H., Brunner D., Hoenger A. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 2006;127:1415–1424. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Spang A., Geissler S., Grein K., Schiebel E. Gamma-tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J. Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel E. Gamma-tubulin complexes: binding to the centrosome, regulation and MT nucleation. Curr. Opin. Cell Biol. 2000;12:113–118. doi: 10.1016/s0955-0674(99)00064-2. [DOI] [PubMed] [Google Scholar]
- Slep K. C., Rogers S. L., Elliott S. L., Ohkura H., Kolodziej P. A., Vale R. D. Structural determinants for EB1-mediated recruitment of APC and spectroplakins to the microtubule end. J. Cell Biol. 2005;168:587–598. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep K. C., Vale R. D. Structural basis of microtubule plus end tracking by XMAP215, Clip-170 and EB1. Mol. Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A., Reis R., Sampaio P., Sunkel C. E. The Drosophila CLASP homolog, Mast/Orbit regulates the dynamic behavior of interphase microtubules by promoting the pause state. Cell Motil. Cytoskelet. 2007;64:605–620. doi: 10.1002/cm.20208. [DOI] [PubMed] [Google Scholar]
- Stanley H., Botas J., Malhotra V. The mechanism of Golgi segregation during mitosis is cell type-specific. Proc. Natl. Acad. Sci. USA. 1997;94:14467–14470. doi: 10.1073/pnas.94.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A., Brill M., Oakley B. R., Horio T., Steinberg G. Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Mol. Biol. Cell. 2003;14:642–657. doi: 10.1091/mbc.E02-08-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S., Powers J., Dunn M., Reese K., Malone C. J., White J., Seydoux G., Saxton W. Spindle dynamics and the role of γ-tubulin in early Caenorhabditis elegans embryos. Mol. Biol. Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel C. E., Gomes R., Sampaio P., Perdigao J., Gonzalez C. Gamma-tubulin is required for the structure and function of the microtubule organizing centre in Drosophila neuroblasts. EMBO J. 1995;14:28–36. doi: 10.1002/j.1460-2075.1995.tb06972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavosanis G., Llamazares S., Goulielmos G., Gonzalez C. Essential role for γ-tubulin in the acentriolar female meiotic spindle in Drosophila. EMBO J. 1997;16:1809–1819. doi: 10.1093/emboj/16.8.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavosanis G., Gonzalez C. γ-Tubulin function during female germ-cell development and oogenesis in Drosophila. Proc. Natl. Acad. Sci. USA. 2003;100:10263–10268. doi: 10.1073/pnas.1731925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M. F., Stearns T. Controlling centrosome number: licenses and blocks. Curr. Opin. Cell Biol. 2006;18:74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Uetake Y., Loncarek J., Nordberg J. J., English C. N., La Terra S., Khodjakov A., Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui K., Ueda R., Miyake T. Cell lines from imaginal discs of Drosophila melanogaster. In Vitro Cell Dev. Biol. 1987;23:707–711. doi: 10.1007/BF02620984. [DOI] [PubMed] [Google Scholar]
- van Breugel M., Drechsel D., Hyman A. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J. Cell Biol. 2003;161:359–369. doi: 10.1083/jcb.200211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan K. T. TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J. Cell Biol. 2005;171:197–200. doi: 10.1083/jcb.200509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C. M., Salmon E. D. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J. Cell Biol. 1997;139:417–434. doi: 10.1083/jcb.139.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. MT nucleation: gamma-tubulin and beyond. J. Cell Sci. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.