Introduction
Targeting genes in disease has long been a sought-after holy grail, with the concept of gene therapy promising a magic bullet for single gene mutation or deletion disorders. Replacing deficient or non-functional genes with the active form has proven more difficult to achieve than originally hoped. However, a subset of gene therapy termed ‘gene silencing’ is being developed, which is altogether more promising. By utilizing the unique nature of gene-sequence specificity, complementary or antisense molecules can be designed to ‘seek and destroy’ target mRNAs for specific proteins known to be pivotal in the pathogenesis of various disease processes. Two strategies have been employed to silence genes; the first involves the use of single-stranded antisense oligodeoxynucleotides (ASO) and the second uses double-stranded short-interfering RNA molecules (siRNA) otherwise known as RNA interference (RNAi).
Antisense oligodeoxynucleotides (ASO)
With the recent publication by Tillman et al. in the Journal of Pharmaceutical Sciences of the first demonstration of oral delivery of ASO in man [1] comes the possibility of this form of gene therapy becoming part of the normal repertoire of therapeutic options open to the physician in the mid- to long-term future. Zamecnick and Stephenson were the first to recognize the potential of ASO in 1978 when studying inhibition of Rous sarcoma virus replication [2]. Since then, antisense technology has developed into a powerful research tool and has begun to make its mark in the world of clinical therapy.
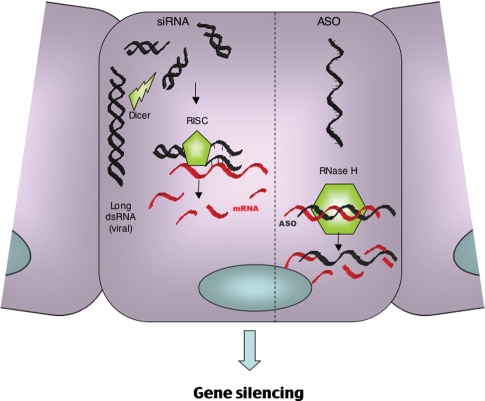
ASO consist of a single strand of 12–22 oligodeoxynucleotides which are complementary to the target mRNA sequence [3]. Binding of the ASO to target mRNA results in steric inhibition of translation by the ribosomal complex but more importantly the induction of RNase H, which cleaves the 3′-O-P-bond of the RNA molecule (Figure 1). This mechanism of action theoretically provides 100% specificity for the target gene, an unachievable goal for most conventional pharmacological agents.
Fig. 1.
The mechanism of action of ASO versus RNA interference.
Since the 1970s, ASO have been used widely as research tools used to investigate mechanisms of disease pathogenesis in vitro, particularly in the field of nephrology. For example, Zhang et al. used a connective tissue growth factor (CTGF) ASO to examine the role of this molecule during epithelial-mesenchymal transition (EMT) in proximal tubular epithelial cells [4], whilst we have employed ASO to discern the individual functions of the isoforms of the Ras monomeric GTPase in human renal fibroblast proliferation [5].
The use of ASO to target the kidney in vivo has been both challenging and highly rewarding. Unmodified, single-stranded oligonucleotides are rapidly broken down in serum by endogenous nucleases greatly limiting cellular uptake. To overcome this, ASO have a modification of the phosphate backbone whereby non-bridging oxygen molecules are replaced by sulphur molecules, greatly enhancing resistance to nuclease activity. These phosphorothioate ASO have a half-life in serum in the region of 10 h (in comparison to 30–60 min of unmodified forms) and, following parenteral administration, have a systemic bioavailability as high as 90% [6]. Further modifications of the sugar–phosphate backbone of the oligonucleotides can be made to increase their stability and RNA affinity without compromising binding selectivity. Among the available sites for modification, the furanose 2′-position has been demonstrated to offer several advantages [7]. Unfortunately, complete 2′-O-modification of the molecule results in the loss of its ability to activate RNase H. This has led to the development of chimeric oligonucleotides that are formed by combining 2′-O-modified oligonucleotides with regions of 2′-deoxy phosphorothioates. The resulting second-generation ASO both support RNase H activity and demonstrate enhanced nuclease resistance and RNA affinity. Following parenteral administration, these ASO distribute to all peripheral tissues with the highest accumulation being in the liver and kidneys, which have a concentration ratio to plasma of 20:1 and 80:1 respectively after 2 h [8]. Within the kidney, ASO are filtered freely by the glomerulus and reabsorbed by proximal tubule epithelial cells [9] making antisense technology a very attractive tool for the investigation and possibly treatment of renal disease. Cheng et al. were amongst the first to use these tools in an animal model of renal disease. They administered ASO to intracellular adhesion molecule 1 (ICAM-1) intravenously via the tail vein in a mouse model of unilateral ureteric obstruction (UUO) and found a decrease in inflammatory infiltration and extracellular matrix [10]. Similarly, Chen et al. demonstrated ICAM-1 ASO to be very effective in inhibiting the ICAM-1-dependent mechanism of graft infiltration and tissue damage involved in allograft rejection, ischaemic-reperfusion injury and cyclosporin-induced nephrotoxicity [11]. Other renal disease-associated proteins have subsequently been targeted using parenteral ASO, such as connective tissue growth factor (CTGF) [12,13], casein kinase II [14] and TGF-β. Isaka et al. modified the delivery of ASO to target interstitial fibroblasts. They developed an artificial viral envelope (AVE) containing anionic liposomes and proteins from the haemagglutinating virus of Japan (HVJ). The net negative charge on the ASO/AVE complex allows for selective transfection of renal interstitial fibroblasts to the exclusion of proximal tubular cells. Using retrograde ureteric administration, Isaka successfully targeted interstitial fibroblasts with ASO to TGF-β1, in a model of UUO. The ASO-treated obstructed kidneys expressed less collagen I and α-smooth muscle actin and had less interstitial fibrosis [15,16].
Although some animal studies have shown that ASO infusions may lead to complement cascade activation [17,18], these effects appear to be both dose dependent and related to the rate of administration. Subsequent clinical studies using lower doses of ASO have reported minimal toxic effects. Furthermore, subsequent generations of ASO have lower toxicity profiles [19]. Therefore, there has been much interest in pushing forward ASO for clinical therapy. Although currently there is only one ASO licensed for clinical use, Formivirsen (Vitravene®) for AIDS-related CMV retinitis, numerous others are in Phase 2 of clinical development such as ASO to ApoB-100 for the treatment of hypercholesterolaemia, ASO to ICAM-1 in ulcerative colitis and many others (see http://www.isispharm.com).
Short-interfering RNA
In 1990, Richard Jorgensen's plant biology group was the first to note the effects of administration of specific RNA molecules on gene expression [20]. Their attempt to enhance the purple pigmentation in petunias by overexpression of the appropriate transcript paradoxically resulted in the flowers losing their colouring. This phenomenon was termed ‘cosuppression’, though the mechanism of action was not determined until 1998. In their landmark paper, Fire and Mello showed that sequence-specific gene knockdown was possible by microinjection of synthetic double-stranded RNA (dsRNA) in the nematode Caenorhabditis elegans [21]. Furthermore, they showed that the use of dsRNA was over 10 times more potent than either sense or antisense RNA alone, that gene silencing was possible on administration of only a few molecules of dsRNA and that this effect may be passed on to first-generation progeny. The term ‘RNA interference’ was applied to their findings and they were awarded the Nobel Prize for Medicine in October 2006.
The mechanism employed by RNAi is thought to be a defensive mechanism against the abnormal presence of double-stranded viral RNA. It is different to that used by ASO and has been conserved over time and is common to all eukaryotes [22]. The process involves initial long dsRNA cleavage by the enzyme Dicer RNase III into short RNA duplexes of 21–23 nucleotides, which are then incorporated into a ribonucleoprotein–endonuclease complex termed ‘RNA Induced Silencing Complex’ (RISC). The siRNA is then unwound and the antisense strand directs the complex to target the specific endogenous RNA sequence. The target RNA transcript is then bound and degraded by the endonuclease activity of RISC (Figure 1). There was an initial reluctance to transfer these findings to mammalian cells since exposure to long strands of dsRNA results in non-specific degradation of all mRNA and inhibition of all protein synthesis. However, Tuschl's group subsequently demonstrated that short-interfering RNA (21-nt) against reporter genes in various mammalian cell lines specifically reduced expression up to 25-fold [23]. Since this time, short-interfering RNA (siRNA) technology has been used widely as a highly specific and powerful tool for the in vitro study of gene function. Its specific mechanism of action makes target site identification and oligo design easier than for ASO as the secondary RNA structure is not an obstacle. In vitro, the duration of knockdown is similar to that of second-generation ASO but the potency maybe significantly greater. Although both siRNA and ASO are highly specific for their target genes, siRNA may induce ‘off target’ effects. This can occur when non-targeted mRNA species share significant homology (>11 bases) with either the sense or antisense strand of the siRNA molecule. Additionally, it has been demonstrated that partial homology (as little as 6–7 bases) between either of the siRNA strands and the 3′ untranslated regions can cause knockdown of many genes within a cell. This mechanism parallels that employed by micro RNA (miRNA) molecules, which are naturally occurring, endogenous gene-regulatory molecules [24,25] and its effects can be difficult to predict.
The major challenge for the use of siRNA, however, is transferring the technology to the in vivo setting. Though these molecules have a biodistribution profile similar to ASO with preferential accumulation in the liver and kidney, they do not readily cross the cell membrane due to their large molecular mass (twice that of single-stranded ASO) and a high negative charge. Unmodified, they have a half-life in serum of a few seconds to a few minutes and are thus rapidly degraded before reaching their target tissues [26] and hence any potency advantage over ASO that they have in vitro is lost. In addition, they can stimulate systemic inflammatory responses by inducing interferon-mediated pathways (though this may be related to the concurrent use of vectors [27]) or by containing newly identified ‘danger motifs’ that bind to certain Toll-like receptors [28]. Local tissue delivery to organs such as the eye and lungs, avoiding a systemic phase, has proven successful in some circumstances and phase 1 trials are taking place into the use of VEGF siRNA in macular degeneration [29,30]. Systemic delivery however remains problematic. In order to increase siRNA delivery to less accessible tissues, researchers have used a variety of different techniques. Hamar et al. used hydrodynamic (large volume and high pressure) injection to deliver siRNA to target the pro-apoptotic protein, Fas in a murine model of acute renal ischaemia-reperfusion injury and were able to demonstrate reduced Fas expression and reduced tubular apoptosis, atrophy and hyaline damage [31]. Alternatively, Hwang et al. injected a short hairpin-RNA-expressing plasmid vector targeting TGF-β1 in a single dose under low pressure, through the renal artery in a mouse model of UUO and demonstrated that collagen I expression in the interstitium was significantly reduced, at least until Day 7 [32]. Similarly, Takabatake et al. injected TGF-β1 siRNA via the renal artery in a rat model of Anti-Thy-1 glomerulonephritis but required the addition of electroporation to achieve a reduction in the expression of TGF-β1 in glomeruli compared to the contralateral kidney by Day 4 [33].
Although these methods have proven successful in animal models, it is difficult to see how they can be translated into a clinical setting. Some groups have followed the lessons learnt from ASO development and have made modifications, such as the addition of a cholesterol moiety or the incorporation of 2′-O-methyl (2′OMe) uridine or guanosine nucleosides into one strand of the siRNA duplex to extend half-life and reduce toxicity [34]. These modifications may yet prove successful in advancing siRNA technology into a therapeutic setting.
Conclusion
Targeting of specific disease-causing genes using ‘antisense’ mechanisms is highly attractive, particularly for the kidney. Although, over 30 years from the original discovery, there is still only one licensed ASO in clinical use, much of the slow progress has been due to the necessary development of pharmacological modifications to improve ASO efficacy and safety. These next-generation molecules are safe, highly specific and powerful medicines with the potential for both systemic and oral delivery and many are moving forward to the clinical setting at an accelerating pace. RNA interference still has some way to go before drugs utilizing this technology reach the same level of pre-clinical and clinical applications. Though currently they appear most promising for local, non-systemic applications, hopefully with new advances in systemic drug delivery, problems of administration will be overcome and they will eventually add to the growing repertoire of gene silencing therapeutics in the future.
Acknowledgments
J.H.W. is supported by a Clinical Training Fellowship from Kidney Research UK. C.C.S. is supported by a Clinician Scientist Award from the UK Department of Health.
Conflict of interest statement. None declared.
References
- 1.Tillman LG, Geary RS, Hardee GE. Oral delivery of antisense oligonucleotides in man. J Pharm Sci. 2008;97:225–236. doi: 10.1002/jps.21084. [DOI] [PubMed] [Google Scholar]
- 2.Stephenson ML, Zamecnik PC. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci USA. 1978;75:285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loke SL, Stein CA, Zhang XH, et al. Characterization of oligonucleotide transport into living cells. Proc Natl Acad Sci USA. 1989;86:3474–3478. doi: 10.1073/pnas.86.10.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang C, Meng X, Zhu Z, et al. Connective tissue growth factor regulates the key events in tubular epithelial to myofibroblast transition in vitro. Cell Biol Int. 2004;28:863–873. doi: 10.1016/j.cellbi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Sharpe CC, Dockrell ME, Noor MI, et al. Role of Ras isoforms in the stimulated proliferation of human renal fibroblasts in primary culture. J Am Soc Nephrol. 2000;11:1600–1606. doi: 10.1681/ASN.V1191600. [DOI] [PubMed] [Google Scholar]
- 6.Crooke ST, Bennett CF. Progress in antisense oligonucleotide therapeutics. Annu Rev Pharmacol Toxicol. 1996;36:107–129. doi: 10.1146/annurev.pa.36.040196.000543. [DOI] [PubMed] [Google Scholar]
- 7.Tereshko V, Portmann S, Tay EC, et al. Correlating structure and stability of DNA duplexes with incorporated 2′-O-modified RNA analogues. Biochemistry. 1998;37:10626–10634. doi: 10.1021/bi980392a. [DOI] [PubMed] [Google Scholar]
- 8.Sands H, Gorey-Feret LJ, Cocuzza AJ, et al. Biodistribution and metabolism of internally 3H-labeled oligonucleotides: I. Comparison of a phosphodiester and a phosphorothioate. Mol Pharmacol. 1994;45:932–943. [PubMed] [Google Scholar]
- 9.Rappaport J, Hanss B, Kopp JB, et al. Transport of phosphorothioate oligonucleotides in kidney: implications for molecular therapy. Kidney Int. 1995;47:1462–1469. doi: 10.1038/ki.1995.205. [DOI] [PubMed] [Google Scholar]
- 10.Cheng QL, Chen XM, Li F, et al. Effects of ICAM-1 antisense oligonucleotide on the tubulointerstitium in mice with unilateral ureteral obstruction. Kidney Int. 2000;57:183–190. doi: 10.1046/j.1523-1755.2000.00825.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Langer RM, Janczewska S, et al. Methoxyethyl-modified intercellular adhesion molecule-1 antisense phosphorothiateoligonucleotides inhibit allograft rejection, ischemic-reperfusion injury, and cyclosporine-induced nephrotoxicity. Transplantation. 2005;79:401–408. doi: 10.1097/01.tp.0000149505.53886.27. [DOI] [PubMed] [Google Scholar]
- 12.Guha M, Xu ZG, Tung D, et al. Specific down-regulation of connective tissue growth factor attenuates progression of nephropathy in mouse models of type 1 and type 2 diabetes. Faseb J. 2007;21:3355–3368. doi: 10.1096/fj.06-6713com. [DOI] [PubMed] [Google Scholar]
- 13.Yokoi H, Mukoyama M, Nagae T, et al. Reduction in connective tissue growth factor by antisense treatment ameliorates renal tubulointerstitial fibrosis. J Am Soc Nephrol. 2004;15:1430–1440. doi: 10.1097/01.asn.0000130565.69170.85. [DOI] [PubMed] [Google Scholar]
- 14.Yamada M, Katsuma S, Adachi T, et al. Inhibition of protein kinase CK2 prevents the progression of glomerulonephritis. Proc Natl Acad Sci USA. 2005;102:7736–7741. doi: 10.1073/pnas.0409818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isaka Y, Akagi Y, Kaneda Y, et al. The HVJ liposome method. Exp Nephrol. 1998;6:144–147. doi: 10.1159/000020515. [DOI] [PubMed] [Google Scholar]
- 16.Isaka Y, Tsujie M, Ando Y, et al. Transforming growth factor-beta 1 antisense oligodeoxynucleotides block interstitial fibrosis in unilateral ureteral obstruction. Kidney Int. 2000;58:1885–1892. doi: 10.1111/j.1523-1755.2000.00360.x. [DOI] [PubMed] [Google Scholar]
- 17.Farman CA, Kornbrust DJ. Oligodeoxynucleotide studies in primates: antisense and immune stimulatory indications. Toxicol Pathol. 2003;31(Suppl):119–122. doi: 10.1080/01926230390174995. [DOI] [PubMed] [Google Scholar]
- 18.Levin AA. A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim Biophys Acta. 1999;1489:69–84. doi: 10.1016/s0167-4781(99)00140-2. [DOI] [PubMed] [Google Scholar]
- 19.Jason TL, Koropatnick J, Berg RW. Toxicology of antisense therapeutics. Toxicol Appl Pharmacol. 2004;201:66–83. doi: 10.1016/j.taap.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 20.Napoli C, Lemieux C, Jorgensen R. Introduction of a Chimeric Chalcone Synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 22.Zamore PD. Ancient pathways programmed by small RNAs. Science. 2002;296:1265–1269. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- 23.Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 24.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birmingham A, Anderson EM, Reynolds A, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 26.Layzer JM, McCaffrey AP, Tanner AK, et al. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Z, Li J, He F, et al. Cationic lipids enhance siRNA-mediated interferon response in mice. Biochem Biophys Res Commun. 2005;330:755–759. doi: 10.1016/j.bbrc.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 28.Hornung V, Guenthner-Biller M, Bourquin C, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 29.Bitko V, Musiyenko A, Shulyayeva O, et al. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 30.Reich SJ, Fosnot J, Kuroki A, et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis. 2003;9:210–216. [PubMed] [Google Scholar]
- 31.Hamar P, Song E, Kokeny G, et al. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc Natl Acad Sci USA. 2004;101:14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang M, Kim HJ, Noh HJ, et al. TGF-beta1 siRNA suppresses the tubulointerstitial fibrosis in the kidney of ureteral obstruction. Exp Mol Pathol. 2006;81:48–54. doi: 10.1016/j.yexmp.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Takabatake Y, Isaka Y, Mizui M, et al. Exploring RNA interference as a therapeutic strategy for renal disease. Gene Ther. 2005;12:965–973. doi: 10.1038/sj.gt.3302480. [DOI] [PubMed] [Google Scholar]
- 34.Judge AD, Bola G, Lee AC, et al. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]