Abstract
DNA methylation and the repair of DNA double-strand breaks (DSBs) are important processes for maintaining genomic integrity. Although DSBs can be produced by numerous agents, they also occur spontaneously as endogenous DSBs (EDSBs). In this study, we evaluated the methylation status of EDSBs to determine if there is a connection between DNA methylation and EDSBs. We utilized interspersed repetitive sequence polymerase chain reaction (PCR), ligation-mediated PCR and combined bisulfite restriction analysis to examine the extent of EDSBs and methylation at long interspersed nuclear element-1 (LINE-1) sequences nearby EDSBs. We tested normal white blood cells and several cell lines derived from epithelial cancers and leukemias. Significant levels of EDSBs were detectable in all cell types. EDSBs were also found in both replicating and non-replicating cells. We found that EDSBs contain higher levels of methylation than the cellular genome. This hypermethylation is replication independent and the methylation was present in the genome at the location prior to the DNA DSB. The differences in methylation levels between EDSBs and the rest of the genome suggests that EDSBs are differentially processed, by production, end-modification, or repair, depending on the DNA methylation status.
INTRODUCTION
Vilenchik and Knudson (1) estimated the existence of endogenous double-strand breaks (EDSBs) and suggested that EDSBs could account for a substantial fraction of oncogenic events in human carcinomas. If EDSBs do not arise uniformly or are not processed at equal rates across the genome, mutation hot spots should be present (1). Our study helps to elucidate if DNA methylation influences EDSBs processing.
Several pieces of evidence suggest that DNA methylation may play an important role in maintaining genomic integrity. Genome-wide decreases in DNA methylation levels commonly occur in cancer (2–5), which leads to higher rates of mutations and genomic instability (6–8). In addition to alterations in the number of chromosomes, hypomethylation can result in chromosomal rearrangements and deletion of DNA, suggesting that DSBs are the intermediate products (6–8). Moreover, because the mutations occur spontaneously, the DSBs should occur endogenously. Studies in ICF syndrome (immunodeficiency, chromosomal instability and facial anomalies) (9), which is characterized by loss-of-function mutations in the cytosine DNA methyltransferase DNMT3B, and Wilm's tumor (10) demonstrated a direct association between loss of DNA methylation and rearrangements in the pericentromeric heterochromatin. Therefore, hypomethylation could lead to spontaneous mutations in cis, which are the epigenetic and genetic events occurring on the same chromosome. Consequently, evaluating methylation status of EDSBs may provide clues to better understanding how DNA methylation helps maintaining genomic integrity.
To determine whether DNA methylation affects EDSBs, we first developed a set of novel techniques to analyze the extent of methylation in genomic EDSBs. These techniques were devised from interspersed repetitive sequence (IRS) polymerase chain reaction (PCR) (11), ligation-mediated (LM) PCR-based assays (12–14) and combined bisulfite restriction analysis (COBRA) for genome-wide methylation level analysis (4,5,15). LMPCR is a commonly used PCR technique designed to analyze locus-specific EDSBs during lymphoid development, such as V(D)J recombination (14,16,17) and hypermutation (18). LMPCR has also been used to detect DNA-associated proteins or chromatin accessibility to such proteins (19,20), and this technique has shown previous utility in genome mapping research (21,22). With these new assays, we evaluated the relationship between DNA methylation and EDSBs.
MATERIALS AND METHODS
Cells, cell lines and culture
Cell lines used were HeLa (cervical cancer), SW480 (colorectal adenocarcinoma), K562 (erythroleukemia), Daudi (B lymphoblast), Jurkat (T-cell leukemia) and Molt4 (T lymphoblast) (ATCC, Manassas, VA, USA). HeLa cells were synchronized at G0 phase by culture in serum deprivation medium, Dulbecco's modified Eagle's medium plus 0.2% fetal bovine serum, for 48 h. HeLa cells in G1/S and S phase cells were synchronized by the thymidine block method, and were cultured with 2 mM thymidine (Sigma-Aldrich, St Louis, MO, USA) to obtain cells at G1/S phase (23). Flow cytometry was used to determine stages of the cell cycle as well as fragmented and apoptotic cells. One millimolar of H2O2 was added for 24 h to induce apoptosis. For radiation treatment, the medium of G0 phase HeLa cells was replaced with 15 ml of ice-cold medium, and cells were exposed to γ- with a 60Co source (Eldorado78).
High-molecular weight DNA preparation
To prepare high-molecular weight (HMW) DNA, 5 × 105 cells were embedded in 1% low-melting point agarose, lysed and digested in 400 μl of 1 mg/ml proteinase K, 50 mM Tris, pH 8.0, 20 mM EDTA, 1% sodium lauryl sarcosine. The plugs were rinsed four times in Tris—EDTA (TE) buffer for 20 min. To polish overhang or cohesive-end EDSBs, T4 DNA polymerase (New England Biolabs, Beverly, MA, USA) and dNTPs were added and later inactivated by adding EDTA to a concentration of 20 mM for 5 min followed by rinsing four times in TE buffer for 20 min. To analyze blunt-end EDSBs, LMPCR was performed without T4 DNA polymerase. The modified LMPCR linkers (24) were prepared from the oligonucleotides 5′-AGGTAACGAGTCAGACCACCGATCGCTCGGAAGCTTACCTCGTGGACGT-3′ and 5′-ACGTCCACGAG-3′. The linkers (50 pmol) were ligated to HMW DNA using T4 DNA ligase (New England Biolabs) at 25°C overnight. DNA was extracted from agarose plugs using a QIAquick gel extraction kit (Qiagen, Basel, Switzerland). For liquid DNA preparation, cells or HMW DNA were incubated in 1% sodium dodecyl sulfate/proteinase K (0.5 mg/ml), at 48°C overnight and subjected to phenol–chloroform extraction and ethanol precipitation. The precipitated DNA was resuspended in 20 µl of TE buffer.
IRS–EDSB–LMPCR
The quantity of IRS–EDSB was measured by real-time PCR using a Lightcycler™ instrument (Roche Applied Science, Indianapolis, IN, USA) with the IRS primers, including long interspersed nuclear elements-1 (LINE-1s or L1s) primers 5′-CTCCCAGCGTGAGCGAC-3′ (outward), 5′-AAGCCGGTCTGAAAAGCGCAA-3′ (inward), Alu, Alu-CL2 5′-ACTGCACTCCAGCCTGGGC-3′ or Tigger1 5′-CTCGCTGAAGGCTCAGATGATC-3′), the linker primer 5′-AGGTAACGAGTCAGACCACCGA-3′ (24), and the Taqman probe homologous to the 3′ linker sequence (6-fam) ACGTCCACGAGGTAAGCT TCCGAGCGA (tamra) (phosphate). Amplification was performed with 0.5 μM of each primer, 0.4 μM Taqman probe, 2 U of HotStarTaq (Qiagen, Valencia, CA, USA), 1× PCR buffer and 10 ng of ligated DNA for up to 40 cycles, with quantification after the extension steps. Two types of control DNA were used. The first was a 100-bp oligonucleotide sequence with the 5′ linker sequence and 3′ homology to L1 oligonucleotide sequences. The second was DNA digested with EcoRV and AluI and ligated to the LMPCR linkers. The amounts of EDSBs were compared with the ligated control digested DNA and reported as L1-EDSB–LMPCR templates per nanogram of DNA.
COBRA–IRS and COBRA–IRS–EDSB
Ligated HMW DNA was modified with bisulfite using a standard protocol (25). Bisulfite-modified DNA was recovered using a Wizard DNA clean-up kit (Promega, Madison, WI, USA) and desulfonated before PCR amplification. For PCR COBRA (15) of L1s (COBRA-L1) (4,5), bisulfite-treated DNA was subjected to 35 cycles of PCR with two primers, B-L1-inward 5′-CGTAAGGGGTTAGGGAGTTTTT-3′ and B-L1-outward 5′-RTAAAACCCTCCRAACCAAATATAAA-3′ (4). Applying a hot-stop technique to prevent heteroduplex amplicons, α32P-labeled-bisulfite-L1-outward oligo was added in the last PCR cycle. The amplicons were doubly digested in a 10 μl reaction volume with 2 U of TaqI and 8 U of TasI in 1× TaqI buffer (MBI Fermentas, Vilnius, Lithuania) at 65°C for 4 h. The PCR is designed to detect unmethylated and methylated L1 sequences of 98 and 80 bp, respectively. The intensity of DNA fragments was measured with a PhosphorImager using Image Quant software (Molecular Dynamics, GE Healthcare, Slough, UK). The LINE-1 methylation level was calculated as the percentage of TaqI intensity divided by the sum of TaqI- and TasI-positive amplicons. For COBRA-L1-EDSB, the B-L1-inward oligo was replaced with B-LMPCR oligo, 5′-GTTTGGAAGTTTATTTTGTGGAT-3′, and 40 PCR cycles were carried out according to the same protocol. Bisulfite-treated Daudi, Jurkat and HeLa DNAs digested with EcoRV and AluI and ligated LMPCR linker were used as positive controls to normalize the inter-assay variation of all COBRA experiments. HeLa DNA without ligation was used as a negative control.
Southern blot hybridization
Southern blot was performed to compare between 5 μg of HpaII- and MspI-digested HeLa DNA. L1-LMPCR amplicons from HMW and AluI–EcoRV-digested HeLa DNA were used as probes. L1-most-outward primer sequence was 5′-TATTCGGCCATCTTGGCTCCT-3′. Competitor DNA, COT-1 DNA, was used to prevent non-specific DNA hybridization, including sequence from L1s. Intensities in the >4 kb regions were measured with a PhosphorImager using Image Quant software (Molecular Dynamics). Semiquantitative methylation percentage was reported as the proportion of probes bound to HpaII-digested DNA to HpaII plus MspI-digested DNA.
Statistical analyses
Statistical significance was determined according to an independent sample t-test, a paired sample t-test or ANOVA using the SPSS program version 11.5 as specified.
RESULTS
Detection of genomic EDSB and methylation
First, we developed a new assay for the detection of EDSBs. This assay is based on a LMPCR, originally designed for the analysis of locus and cell-specific EDSBs (14,16–18). General EDSBs are believed to occur rarely and arbitrarily throughout the genome (1). Using repetitive sequences that randomly scatter throughout the human genome, we can detect genome-wide EDSBs in proximity to these repetitive sequences. We, therefore, combined LMPCR with IRSPCR or inter-Alu PCR (11) using the widely distributed L1s human retrotransposons (26) into a new assay called ‘L1-EDSB-LMPCR’. In this assay, linker oligonucleotides are ligated to EDSBs in HMW DNA preparation and quantitatively analyzed by real-time PCR using an L1 primer and a Taqman probe complementary to the linker (Figure 1A). L1-PCR using a primer in the outward direction will amplify DNA sequences located outside the repetitive sequences. Therefore, the sequences yielded from IRSPCR will represent human genome-wide sequences, including both unique and repetitive sequences (11,21). Increases in the L1-EDSB–LMPCR products corresponded with the amount of control DSBs generated by restriction enzyme digesting DNA. Without ligation, no L1-EDSB–LMPCR could be detected. Finally, significant amounts of EDSBs were detected in all tested cells (Figure 1B).
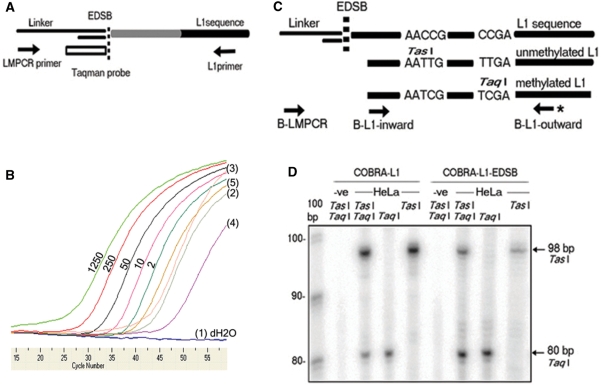
Figure 1.
L1-EDSB–LMPCR and COBRA-L1–EDSB. (A) Schematic representation of L1-EDSB–LMPCR showing L1 sequence and the ligated linker at a nearby EDSB. The white rectangle is a Taqman probe complementary to the LMPCR linker. Arrows are PCR primers. (B) The quantity of EDSBs detected with this method increased directly with the amount of experimentally induced DSBs, and significant amounts of EDSBs were detected in all tested cells but not in cells without ligation. An example of results of real-time L1-EDSB–LMPCR with tests and controls was demonstrated. The values 2, 10, 50, 250 and 1250 are quantities of restriction enzyme-digested (EcoRV and AluI) HeLa genomes ligated with LMPCR linker. dH2O is water. Tested templates are HMW DNA from (1) HeLa without ligation, (2) Daudi blunt-end-EDSBs ligation, (3) Daudi polished-end-EDSBs ligation, (4) Jurkat blunt-end-EDSBs ligation and (5) Jurkat polished-end-EDSBs ligation. (C) Schematic representation of COBRA-L1 and COBRA-L1-EDSB, showing L1 sequence ligated by linker at an EDSB. Arrows are PCR primers, with asterisk indicating α-32P-labeled primer for COBRA. AACCG and CCGA are L1 sequences; when treated with bisulfite and after undergoing PCR, unmethylated AACCG will be converted to AATTG (TasI site) and methylated CCGA to TCGA (TaqI site). (D) A typical example of results from COBRA-L1 and COBRA-L1-EDSB experiments indicating that the intensity between methylated, TaqI and unmethylated, TasI, bands of EDSBs were higher than the matched pair genomes. The arrow at 98 bp indicates TasI-digested unmethylated L1 sequences and the arrow at 80 bp indicates TaqI-digested methylated L1 sequences. −ve is dH2O for COBRA-L1 and non-ligated HMW DNA for COBRA-L1-EDSB. TasI and TaqI are restriction enzymes added in each experiment.
Next, we developed an assay to analyze the methylation level of EDSBs. Previously, we had extensively studied methylation status of L1s in several cancers and normal tissues by COBRA-L1. Treatment with bisulfite converts unmethylated cytosines, but not methylated cytosines, to uracils and then thymines after PCR. Therefore, this bisulfite treatment generates detectable methylation-dependent changes in the restriction pattern of PCR-amplified L1 sequences. Methylation level is then calculated and presented as a percentage of total DNA. We thus combined L1-EDSB–LMPCR with COBRA-L1 by treating linker-ligated DNA with bisulfite before PCR with L1/linker primers and restriction analysis (Figure 1C). With this new ‘COBRA-L1-EDSB’ assay, we can measure the methylation level of L1s near EDSBs, which reflects the methylation level of EDSBs in a genome-wide fashion. The degree of methylation between genomic L1 and L1-EDSB sequences was examined by COBRA-L1 and COBRA-L1–EDSB, respectively (Figure 1D).
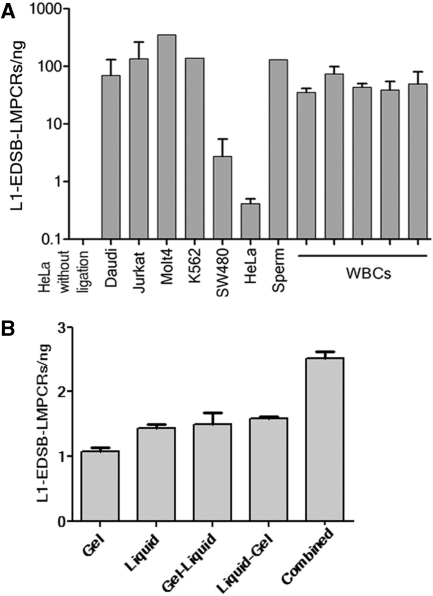
With these new assays, we first evaluated what types of cells possess EDSBs and if the quantity of EDSBs reflects carcinogenic potentials. Using L1-EDSB–LMPCR, significant amounts of EDSBs were detected in all samples from several cancer cell lines, including Daudi, Jurkat, Molt4, K562, SW480 and HeLa cells, as well as in normal cells, including sperm and white blood cells (WBCs) from several individuals (Figure 2A). These data suggest that EDSBs can commonly be found in all cells both normal and cancer.
Figure 2.
Discovery and specificity of EDSB-LMPCR. (A) Using L1-EDSB-LMPCR, significant amounts of EDSBs were detected in all samples, including, cancer cell lines, sperm cells and WBCs from several individuals. (B) L1-EDSB-LMPCR of DNA from HeLa cells using different DNA-extraction methods. Gel, liquid, gel–liquid, liquid–gel and combined are L1-EDSB–LMPCR from DNA prepared in gel, liquid, extracted liquid DNA from gel-embedded HMW DNA, embedded liquid DNA into gel following by HMW DNA preparation protocol and 2 × admixture between 1:1 of HMW DNA and liquid DNA, respectively. Data represent means ± SEM.
This assay prepared HMW DNA by in-gel preparation. This technique has been reported to decrease the number of DSBs generated during DNA preparation (27) and did not generate false positives in the LMPCR assay for the analysis of locus-specific EDSBs (14,18,27). To evaluate if in-gel HMW DNA preparation breaks genomic DNA, we compared the yield of the LMPCR from several sources of DNA, including, in-gel HMW DNA, liquid DNA, liquid DNA extracted from in-gel HMW DNA (gel–liquid) and in-gel DNA prepared from liquid DNA (liquid–gel). The quantities of L1-EDSB–LMPCR from liquid DNA were higher than in-gel HMW DNA (Figure 2B). Extracting liquid DNA from a gel increased the amount of LMPCR template over HMW DNA (Figure 2B). This result indicates that more DNA breaks were generated during the liquid DNA preparation process. Embedded liquid DNA into gel following by HMW DNA preparation protocol did not increase the amount of LMPCR template over liquid DNA alone (Figure 2B). Therefore, in-gel preparation did not generate significant additional DNA breaks. Moreover, the quantities of L1-EDSB–LMPCR from gel–liquid and liquid–gel were clearly lower than when L1-EDSB–LMPCR products from HMW DNA and liquid DNA were combined (Combined) (Figure 2B). Therefore, the significant amounts of LMPCR products from HMW DNA were not derived from DNA preparation. This experiment supported the presence of EDSBs.
General characteristics of EDSB–LMPCR
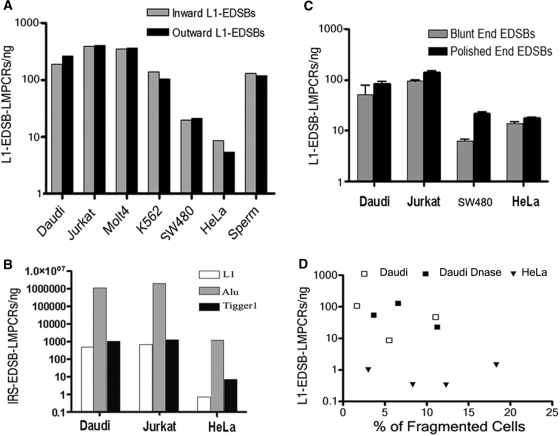
Repetitive sequences have been commonly recognized as locations of general recombination that destabilize human cancer genomes (28,29). It is interesting to investigate if the quantities of EDSBs differ between repetitive and unique sequences. Full length L1 is ∼6 kb (26). L1-EDSB–LMPCR using the L1-inward primer will amplify L1 sequences, while the L1-outward primer should amplify unique sequences near L1 (11). We observed L1-EDSBs at similar levels regardless of the directions of L1 primers used (Figure 3A). The data indicate that the amount of EDSBs does not differ based on the nature of DNA sequences, between L1 repetitive and unique sequences. We found that IRS–LMPCR using primers for other types of repetitive sequences, including Alu and Tigger1, also yielded significant EDSBs in direct proportions to their copy numbers in the human genomes (30) (Figure 3B). L1s are AT rich, whereas Alu sequences are frequently located in CG-rich regions (31). Therefore, EDSBs are widely distributed in the human genome. In addition, because EDSBs are thought to occur rarely and randomly throughout the genome, LMPCR with primers specific to unique sequences yielded no positive locus-specific amplicons (data not shown). EDSB ends are heterogeneous, as significant quantities of L1-EDSB–LMPCR products were obtained, including two EDSB types: blunt- and polished-, blunt plus cohesive, end EDSBs (Figure 3C). Blunt-end DSBs are DNA that both strands terminate in a base pair. A cohesive or overhang is a stretch of unpaired nucleotides in the DNA end (14,16,17).
Figure 3.
General characteristics and distribution of EDSB–LMPCR in the human genome. (A) L1-EDSB–LMPCR, using L1-inward and L1-outward primers, was performed in several cell types, and products were observed at similar levels regardless of the nature of linked EDSB sequences, L1, or unique sequences. Inward L1-EDSB–LMPCR was normalized by the proportion of L1 sequence copies in the human genome (www.ncbi.nlm.nih.gov). (B) Comparison of the amount of EDSB–LMPCRs using L1, Alu and Tigger1 primers indicated EDSBs in direct proportions to their copy numbers in the human genomes. (C) EDSBs were measured from HMW DNA with and without T4 polymerase treatment, respectively. Significant amount of L1-EDSB–LMPCRs of blunt and polished-end EDSBs from cancer cell lines were identified. Therefore, L1-EDSB ends are heterogeneous. (D) The quantity of EDSBs was not related to the proportion of fragmented cells. L1-EDSB–LMPCR quantity related to the percent of fragmented cells, documented by flow cytometry. Daudi DNase represents Daudi cells treated with DNase I before HMW DNA preparation.
The quantity of EDSBs was not related to the proportion of fragmented cells (Figure 3D). Our EDSB–LMPCR protocol, particularly that for epithelial cells, minimizes contamination with apoptotic cells because these dying cells with fragmented DNA (32) usually lose attachment (33) and are thus washed off before cell collection for EDSB analysis. While there was positive L1–EDSB–LMPCR amplification, we were unable to detect any fragmented DNA or apoptotic cells, as determined by LMPCR ladder (34) and flow cytometry (35), respectively (data not shown). Moreover, the apoptotic genome possesses normal levels of methylation and may not be detectable by COBRA-L1–EDSB. Fragmented DNA, as documented by electrophoresis, was collected from floating apoptotic HeLa cells after treatment with 1 mM H2O2 for 24 h (36). COBRA-L1 shows that apoptotic HMW DNA has similar L1 methylation to living cells, but COBRA-L1–EDSB yielded amplicons with multiple sizes that interfere with interpretation (data not shown).
L1-EDSB–LMPCR under different conditions
We utilized L1-EDSB–LMPCR on cells under different conditions known to associate with DSBs. Specifically; we used radiation and cell cycle synchronization. Radiation, which directly causes DNA damage, increased L1-EDSB–LMPCR levels directly correlating with the dosages of radiation used to treat the cells (Figure 4A). Nevertheless, there were wide ranges of the amount of EDSBs (Figure 4A). This result may be the consequence of several factors, including DSB end modifications and DSB repair rate. Therefore, even though L1-EDSB–LMPCR can detect radiation-induced DSBs, the technique lacks efficiency in evaluating the precise number of DSBs generated by radiation.
Figure 4.
L1-EDSB–LMPCR under different conditions. L1-EDSB–LMPCR of (A) irradiated HeLa. (B) HeLa cells at G0, G1/S and 0, 3 and 5 h after the release into S phase from thymidine block. Control is without cell synchronization. G0 bore the least amount of EDSBs when compared with control, *P < 0.05 (independent one-tailed t-test). Data represent means ± SEM.
Because EDSBs were hypothesized to be preferentially produced in S phase from the conversion of single-strand lesions (1), we used HeLa cells to assess the frequency of EDSBs and their methylation status during various cell cycle stages: G0, G1/S and S. We found the impact of cell cycle effect is on the borderline of significance and EDSBs can be found in G0 phase (Figure 4B). Interestingly, there was a minor decrease in S phase cells (Figure 4B). Therefore, cell cycle stage is one of the conditions that may alter the amount of EDSBs. Nevertheless, similar to radiation the amount of EDSBs during cell phases may be influenced by several factors in addition to the production rate.
Specificity of COBRA-L1–EDSB
Figure 5A demonstrates a reconstitution experiment of EDSB–LMPCR. Because Daudi cells shows significantly higher level of genomic methylation than HeLa cells, we used DNA prepared from Daudi and HeLa cells as representative DNA with high and low methylation, respectively. We added varying ratios of HeLa and Daudi DNA digested with EcoRV and AluI (ligated to LMPCR linker) into HeLa genomic DNA. As expected, a COBRA-L1–EDSB analysis of the digested DNA, regardless of the presence of the HeLa genome, yielded equivalent levels of methylation to COBRA-L1. Furthermore, the methylation levels in these samples measured by both COBRA-L1 and COBRA-L1–EDSB are increased, which correlates with the proportion of highly methylated Daudi DNA contained in that sample (Figure 5A). The higher levels of methylation, compared to the rest of the genome, are a characteristic of EDSBs but not DSBs randomly generated by DNA shearing during sample preparation. Lower DSB methylation levels of liquid DNA, gel–liquid DNA and liquid–gel DNA were demonstrated when compared to HMW DNA (Figure 5B). In contrast to gDNA prepared from cells cast into low melting point agarose plugs, the preparation protocol for liquid DNA generates DSBs (Figure 2B). Therefore, COBRA-L1–EDSB levels of liquid DNA should be derived from combination between EDSBs and DNA preparation producing DSBs. The lower DSB methylation levels of liquid DNA confirmed that the DNA preparation producing DSBs possess lower methylation levels than EDSBs. Moreover, DSB methylation levels of gel–liquid DNA and liquid–gel DNA are not different from levels of liquid DNA alone (Figure 5B). Therefore, higher methylation levels of COBRA-L1–EDSB, detected from HMW DNA, were unlikely to be derived from HMW DNA preparation.
Figure 5.
Specificity of COBRA-L1–EDSB. (A) A reconstitution experiment of COBRA-L1–EDSB. COBRA-L1 and COBRA-L1–EDSB of the mixture between HeLa and Daudi DNA digested with AluI and EcoRV and ligated to EDSB–LMPCR linker. The 4:0, 3:1, 2:2, 1:3 and 0:4 are proportions of recombinant HeLa:Daudi DNA. COBRA-L1–EDSB 1/10 was the mixture between 2 ng of linker-ligated DNA and 18 ng of HeLa genomic DNA. Because Daudi cells shows significantly higher levels of genomic methylation compared to HeLa cells, we used DNA prepared from Daudi and HeLa cells as representative DNA with high methylation and low methylation, respectively. We added varying ratio of HeLa and Daudi DNA digested with EcoRV and AluI ligated to the LMPCR linker into HeLa genomic DNA. As expected, the methylation levels of the digested DNA, regardless of the presence of the HeLa genome, were measured by COBRA-L1-EDSB and COBRA-L1, and increases in final product correlated with the proportion of highly methylated Daudi DNA. (B) COBRA-L1 and COBRA-L1–EDSB methylation levels in a comparison of DNA preparation methods. Gel, gel–liquid, liquid and liquid–gel refer to DNA analyzed via L1-EDSB-LMPCR from DNA prepared in-gel, extracted liquid DNA from gel embedded HMW DNA, liquid and embedded liquid DNA into gel following by HMW DNA preparation protocol, respectively. COBRA-L1–EDSB methylation levels of HMW DNA were higher than liquid DNA. COBRA-L1–EDSB methylation levels of in-gel DNA prepared from liquid DNA, and liquid DNA prepared from HMW DNA, were lower than HMW but not different from liquid. Therefore, HMW DNA preparation does not cause hypermethylated DSBs. Data represent means ± SEM.
EDSBs are ubiquitously hypermethylated
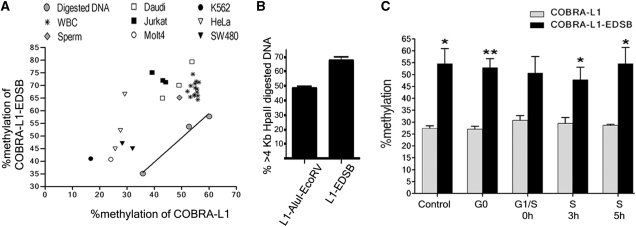
To compare the degree of methylation between L1 and L1-EDSB sequences, we examined by COBRA-L1 and COBRA-L1–EDSB, respectively. We found that, EDSBs are hypermethylated. Specifically, these data show that the EDSBs from all tested cells possess higher methylation levels than the rest of the genome. In our analysis, we included several cancer cell lines, along with normal sperm and WBCs (Figure 6A). In contrast, restriction enzyme-digested DSBs show the same level of methylation compared to genomic DNA (Figure 6A). The COBRA-L1-EDSB levels reflect the rate of EDSB synthesis, EDSB end modification and repair at a given time. Therefore, the EDSB hypermethylation means that methylated and unmethylated DNA may possess distinctive EDSB processing (i.e. repair) pathways.
Figure 6.
Methylation status of EDSBs. (A) EDSBs possess higher methylation levels than the rest of the genome, across all tests. A comparison of COBRA-L1 and COBRA-L1–EDSB among cell types demonstrated universally higher COBRA-L1-EDSB methylation levels than the matched pair COBRA-L1. Digested DNA is from HeLa, Jurkat or Daudi cells, and after being treated with AluI and EcoRV, it was used for a control. Each dot represents an individual test result of COBRA-L1 and COBRA-L1–EDSB. (B) Methylation of EDSBs pre-existed at the break sites. A representation of results from Southern blot hybridization of HpaII and MspI digested DNA demonstrates that EDSBs are located in larger HpaII digested HeLa DNA fragments than experimentally induced methylation-independent DSBs. L1-EDSB are L1-LMPCR probes from HMW DNA, representing EDSBs, while L1–AluI–EcoRV contains AluI plus EcoRV-digested DNA, representing methylation-independent DSBs. The bar graphs reports the percentage of hybridization intensities in the >4 kb regions of HpaII-digested DNA divided by the summation of those of HpaII and MspI-digested DNA. (C) COBRA-L1 and COBRA-L1–EDSB of HeLa cells at G0, G1/S, and 0, 3 and 5 h after the release into S phase from a thymidine block. The control is without cell synchronization. G0 cells contained the most significant hypermethylation level of EDSBs when compared between COBRA-L1 and COBRA-L1-EDSB, *P < 0.05, **P < 0.001 (pair two-tailed t-test). (B) and (C) data represent the mean ± SEM.
The methylation of EDSBs may have pre-existed at the break sites
As shown in Figure 6A, the levels of methylation at EDSBs and the rest of the genomes are positively correlated in all cell lines (P = 0.01; Pearson r = 0.873). We further proved the source of DNA methylation around EDSBs by Southern blot hybridization of HeLa genomic DNA digested with HpaII or MspI. The activity of HpaII is blocked by CpG methylation, while that of the isoschizomer MspI is insensitive to methylation. Therefore, hypermethylated DNA is not digested with HpaII and electrophoretically co-migrates with long DNA fragments. We arbitrarily used the length of >4 kb to represent long DNA fragments concentrated in hypermethylated DNA for subsequent calculations. We determined the percentage of hybridization intensities in the >4 kb regions of HpaII-digested DNA divided by the sum of those digested with HpaII and MspI. The L1-EDSB-LMPCR products hybridized to >4 kb HpaII-digested DNA fragments in higher proportions than the control, restriction enzyme-generated DSB-LMPCR products (Figure 6B). Therefore, EDSBs are hypermethylated, and the methylation was present in the genome at this location before the break itself.
EDSB hypermethylation is replication independent
Because EDSBs can be detected at different levels in all phases of the cell cycle (Figure 4B), we evaluated the level of EDSB methylation during cell cycle progression. EDSBs were hypermethylated in most stages, with G0 being the most significant (Figure 6C). This result implies that the increase levels of methylation at EDSBs are DNA replication independent.
DISCUSSION
Even though molecular characteristics of locus-specific EDSBs during lymphoid development, such as V(D)J recombination (14,16,17) and hypermutation (18), have been described by LMPCR, general EDSBs have not (1). Conventionally, radiation-induced DSBs can be visualized as fragmented DNA that migrate out of cells or chromosomes via electrophoresis, using the comet assay (37) and/or pulsed field gel electrophoresis (38). Nevertheless, because detection using these techniques requires extensive damage of DNA and because EDSBs are rare, the sensitivity is likely not high enough for analyzing EDSBs.
The IRS-EDSB-LMPCR technique is able to detect EDSBs because of the extensive distribution of the IRS sequences and the sensitivity of real-time PCR. This study demonstrates that EDSBs are not only detectable but also widely distributed. Positive results were obtained from all types of selected IRS sequences. Nevertheless, LMPCR may not be able to detect some subset of EDSBs. LMPCR preparation requires ligation of a blunt-end DNA linker to a blunt and phosphorylated DSB end. In this study, we polished cohesive-end EDSBs by T4 polymerase. We found that both blunt and overhang ends are present and that the majority of EDSB ends are blunt. LMPCR fails, however, to detect some complex ends that cannot be polished such as a hairpin loop. Unlike studying V(D)J recombination (17,27), in this study, we did not apply Mung Bean nuclease to screen for these end-types because the enzyme can convert single-strand lesion into DSBs.
IRS–EDSB–LMPCR is the first method characterizing EDSBs. Consequently, there is no report of cells with different levels of EDSBs. Therefore, even though it is commonly accepted that in-gel HMW DNA preparation does not significantly affect DNA breaks (27), we performed additional experiments to disregard the possibility of error. The comparison among liquid DNA preparation, HMW DNA and combined experiments suggested that there are no detectable DSBs generated by the HMW DNA preparation process. Finally, cells treated under different conditions, including, serum deprivation and temperature (data not shown), altered EDSB levels. Because the differentially treated cells were processed for HMW DNA simultaneously, the possibility that the same DNA preparation method led to bias shearing of the DNA depending on different prior cellular conditions is remote.
LMPCR detects DSBs directly, while γ-H2AX foci staining aims to detect a cellular response to DSBs. Therefore, L1-EDSB–LMPCR analysis and staining for γ-H2AX foci may not yield the same information. For example, a recent report demonstrated that some genomic regions, particularly heterochromatin, are devoid of radiation-induced γ-H2AX foci (39). Consequently, L1-EDSB–LMPCR and γ-H2AX foci should assess radiation-induced DSBs differently depending on chromatin structures. In this study, we have demonstrated that, even though L1-EDSB-LMPCR products increased after radiation, the correlation to radiation dosage was not as precise as using γ-H2AX foci for detection (40). One explanation for the broad range of results when using L1-EDSB-LMPCR to detect radiation-induced DSBs could be that there is a wide variety of biological processes involved in radiation-induced DSB repairs (41,42).
This study demonstrates that EDSBs are present in all cell types. Moreover, EDSBs normally possess higher levels of methylation compared to the cellular genome, and this methylation pre-exists at the break sites. These findings were not only unprecedented but also not generally expected. Vilenchik and Knudson suggested that the causes of EDSBs are oncogenic events in human carcinomas (1). Moreover, genomic instability can be observed while cancer genomes are hypomethylated (6–8). Therefore, it is tempting to hypothesize that EDSBs should occur more frequently at hypomethylated sequences and, consequently, that COBRA-L1-EDSB should have been hypomethylated. Nonetheless, IRS-EDSB-LMPCR and COBRA-L1-EDSB are methods to measure the extent of methylation at EDSBs during a given time. Therefore, the LMPCR levels reflect not only the rate of EDSB synthesis but also EDSB end modification and repair. Therefore, discovery of DSBs does not exclusively indicate DSB formation, but, the EDSB hypermethylation finding leads us to conclude that methylated and unmethylated DNA possess distinctive EDSB processing (i.e. repair) pathways. If these pathways have different precision, methylated and unmethylated DNA should have unequal rate of spontaneous mutations. In the future, it would be interesting to explore if and how cells process or repair EDSBs depending on DNA methylation. This finding may yield an important clue to prevent global hypomethylation-induced chromosomal rearrangements.
There may be several other mechanisms by which DNA methylation prevents IRS from inducing DNA rearrangements. Repetitive sequences have been commonly recognized as locations of general recombination that destabilizes human cancer genomes, particularly when genetic recombination occurred between different loci (28,29,43). A deletion at Xist alters chromatin conformation, usually associated with DNA methylation, of the inactive X chromosome and this change destabilizes both X chromosomes (44). In the future, it would be intriguing to explore if EDSBs play a role in these mechanisms and whether or not DNA methylation is important in preventing these chromosomal rearrangements.
ACKNOWLEDGEMENTS
We thank Profs Alfred G Knudson, Fox Chase Cancer Center and Stephen Buratowski, Harvard Medical School and Drs Vitrote Sriuranpong, Kanya Suphapeetiporn, Chulalongkorn University and Man Liu, Yale University for critical review and advice. This study was supported by the Thailand Research Fund and Faculty of Graduate School. N.K. and W.P. were supported by the Royal Golden Jubilee Ph.D. grant. Funding to pay the Open Access publication charges for this article was provided by the Thailand Research Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc. Natl Acad. Sci. USA. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 4.Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, Voravud N, Sriuranpong V, Mutirangura A. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 5.Mutirangura A. Quantitative PCR analysis for methylation level of genome: clinical implications in cancer. Asian Biomedicine. 2007;1:121–128. [Google Scholar]
- 6.Lengauer C, Kinzler KW, Vogelstein B. DNA methylation and genetic instability in colorectal cancer cells. Proc. Natl Acad. Sci. USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 8.Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 9.Xu GL, Bestor TH, Bourc'his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 10.Qu GZ, Grundy PE, Narayan A, Ehrlich M. Frequent hypomethylation in Wilms tumors of pericentromeric DNA in chromosomes 1 and 16. Cancer Genet. Cytogenet. 1999;109:34–39. doi: 10.1016/s0165-4608(98)00143-5. [DOI] [PubMed] [Google Scholar]
- 11.Nelson DL, Ledbetter SA, Corbo L, Victoria MF, Ramirez-Solis R, Webster TD, Ledbetter DH, Caskey CT. Alu polymerase chain reaction: a method for rapid isolation of human-specific sequences from complex DNA sources. Proc. Natl Acad. Sci. USA. 1989;86:6686–6690. doi: 10.1073/pnas.86.17.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller PR, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 13.Pfeifer GP, Steigerwald SD, Mueller PR, Wold B, Riggs AD. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989;246:810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- 14.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 15.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu C, Roth DB. Characterization of coding ends in thymocytes of scid mice: implications for the mechanism of V(D)J recombination. Immunity. 1995;2:101–112. doi: 10.1016/1074-7613(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 17.Livak F, Schatz DG. Identification of V(D)J recombination coding end intermediates in normal thymocytes. J. Mol. Biol. 1997;267:1–9. doi: 10.1006/jmbi.1996.0834. [DOI] [PubMed] [Google Scholar]
- 18.Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- 19.Pfeifer GP, Tommasi S. In vivo footprinting using UV light and ligation-mediated PCR. Methods Mol. Biol. 2000;130:13–27. doi: 10.1385/1-59259-686-x:13. [DOI] [PubMed] [Google Scholar]
- 20.Espinoza CR, Feeney AJ. Quantifying chromatin accessibility of individual gene family members by combining ligation-mediated PCR with real-time PCR. Biotechniques. 2006;41 doi: 10.2144/000112263. 404, 406, 408. [DOI] [PubMed] [Google Scholar]
- 21.Mutirangura A, Jayakumar A, Sutcliffe JS, Nakao M, McKinney MJ, Buiting K, Horsthemke B, Beaudet AL, Chinault AC, Ledbetter DH. A complete YAC contig of the Prader-Willi/Angelman chromosome region (15q11-q13) and refined localization of the SNRPN gene. Genomics. 1993;18:546–552. doi: 10.1016/s0888-7543(11)80011-x. [DOI] [PubMed] [Google Scholar]
- 22.Mutirangura A, Greenberg F, Butler MG, Malcolm S, Nicholls RD, Chakravarti A, Ledbetter DH. Multiplex PCR of three dinucleotide repeats in the Prader-Willi/Angelman critical region (15q11-q13): molecular diagnosis and mechanism of uniparental disomy. Hum. Mol. Genet. 1993;2:143–151. doi: 10.1093/hmg/2.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bostock CJ, Prescott DM, Kirkpatrick JB. An evaluation of the double thymidine block for synchronizing mammalian cells at the G1-S border. Exp. Cell Res. 1971;68:163–168. doi: 10.1016/0014-4827(71)90599-4. [DOI] [PubMed] [Google Scholar]
- 24.Pornthanakasem W, Mutirangura A. LINE-1 insertion dimorphisms identification by PCR. Biotechniques. 2004;37 doi: 10.2144/04375BM07. 750, 752. [DOI] [PubMed] [Google Scholar]
- 25.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 26.Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat. Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 27.Schlissel MS. Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol. Cell Biol. 1998;18:2029–2037. doi: 10.1128/mcb.18.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kidwell MG, Holyoake AJ. Transposon-induced hotspots for genomic instability. Genome Res. 2001;11:1321–1322. doi: 10.1101/gr.201201. [DOI] [PubMed] [Google Scholar]
- 29.Surtees JA, Argueso JL, Alani E. Mismatch repair proteins: key regulators of genetic recombination. Cytogenet. Genome Res. 2004;107:146–159. doi: 10.1159/000080593. [DOI] [PubMed] [Google Scholar]
- 30.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 31.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 32.Inohara N, Koseki T, Chen S, Benedict MA, Nunez G. Identification of regulatory and catalytic domains in the apoptosis nuclease DFF40/CAD. J. Biol. Chem. 1999;274:270–274. doi: 10.1074/jbc.274.1.270. [DOI] [PubMed] [Google Scholar]
- 33.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 34.Staley K, Blaschke AJ, Chun J. Apoptotic DNA fragmentation is detected by a semi-quantitative ligation-mediated PCR of blunt DNA ends. Cell Death Differ. 1997;4:66–75. doi: 10.1038/sj.cdd.4400207. [DOI] [PubMed] [Google Scholar]
- 35.Sherwood SW, Schimke RT. Cell cycle analysis of apoptosis using flow cytometry. Methods Cell Biol. 1995;46:77–97. doi: 10.1016/s0091-679x(08)61925-1. [DOI] [PubMed] [Google Scholar]
- 36.Ren JG, Xia HL, Just T, Dai YR. Hydroxyl radical-induced apoptosis in human tumor cells is associated with telomere shortening but not telomerase inhibition and caspase activation. FEBS Lett. 2001;488:123–132. doi: 10.1016/s0014-5793(00)02377-2. [DOI] [PubMed] [Google Scholar]
- 37.Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiat. Res. 1990;122:86–94. [PubMed] [Google Scholar]
- 38.Stamato TD, Denko N. Asymmetric field inversion gel electrophoresis: a new method for detecting DNA double-strand breaks in mammalian cells. Radiat. Res. 1990;121:196–205. [PubMed] [Google Scholar]
- 39.Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS ONE. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida K, Yoshida SH, Shimoda C, Morita T. Expression and radiation-induced phosphorylation of histone H2AX in mammalian cells. J. Radiat. Res. 2003;44:47–51. doi: 10.1269/jrr.44.47. [DOI] [PubMed] [Google Scholar]
- 41.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair. 2005;4:639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Wyman C, Ristic D, Kanaar R. Homologous recombination-mediated double-strand break repair. DNA Repair. 2004;3:827–833. doi: 10.1016/j.dnarep.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 43.Tremblay A, Jasin M, Chartrand P. A double-strand break in a chromosomal LINE element can be repaired by gene conversion with various endogenous LINE elements in mouse cells. Mol. Cell Biol. 2000;20:54–60. doi: 10.1128/mcb.20.1.54-60.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diaz-Perez SV, Ferguson DO, Wang C, Csankovszki G, Wang C, Tsai SC, Dutta D, Perez V, Kim S, Eller CD, et al. A deletion at the mouse Xist gene exposes trans-effects that alter the heterochromatin of the inactive X chromosome and the replication time and DNA stability of both X chromosomes. Genetics. 2006;174:1115–1133. doi: 10.1534/genetics.105.051375. [DOI] [PMC free article] [PubMed] [Google Scholar]