Abstract
We describe snap-to-it probes, a novel probe technology to enhance the hybridization specificity of natural and unnatural nucleic acid oligomers using a simple and readily introduced structural motif. Snap-to-it probes were prepared from peptide nucleic acid (PNA) oligomers by modifying each terminus with a coordinating ligand. The two coordinating ligands constrain the probe into a macrocyclic configuration through formation of an intramolecular chelate with a divalent transition metal ion. On hybridization with a DNA target, the intramolecular chelate in the snap-to-it probe dissociates, resulting in the probe ‘snapping-to’ and binding the target nucleic acid. Thermal transition analysis of snap-to-it probes with complementary and single-mismatch DNA targets revealed that the transition between free and target-bound probe conformations was a reversible equilibrium, and the intramolecular chelate provided a thermodynamic barrier to target binding that resulted in a significant increase in mismatch discrimination. A 4–6°C increase in specificity (ΔTm) was observed from snap-to-it probes bearing either terminal iminodiacetic acid ligands coordinated with Ni2+, or terminal dihistidine and nitrilotriacetic acid ligands coordinated with Cu2+. The difference in specificity of the PNA oligomer relative to DNA was more than doubled in snap-to-it probes. Snap-to-it probes labeled with a fluorophore-quencher pair exhibited target-dependent fluorescence enhancement upon binding with target DNA.
INTRODUCTION
Poor nucleic acid hybridization specificity can compromise many areas of research, including the detection of single nucleotide polymorphisms (1–3), the performance of nucleic acid microarrays (4–9) and off-target effects during siRNA silencing (10–13). This problem is particularly relevant when manipulating or detecting short nucleic acid oligomers such as shRNA, siRNA and miRNA, which show immense promise as a new generation of therapeutics. Likewise, synthetic oligonucleotide-based tools are invaluable in biology, chemistry and nanotechnology only if they bind their intended target. These applications highlight the importance of oligonucleotide hybridization specificity and account for the broad interest in developing ultra-specific nucleic acid probes.
Towards synthesizing nucleic acid probes with improved specificity, recent reports have described locked nucleic acids (LNA) (14), modified peptide nucleic acids (PNA) (15,16), molecular beacons (17–19), combinations of these methods (20,21) and other nonlinear oligonucleotide architectures (22,23). PNA probes exhibit increased DNA-binding specificity relative to DNA probes. The absence of electrostatic repulsion between the two backbones of a PNA–DNA heteroduplex increases its stability relative to a DNA–DNA duplex having an identical base sequence. PNA synthesis is analogous to solid-phase peptide synthesis, and the installation of PNA bases, fluorescent probe labels or additional modified monomers can be performed using standard peptide coupling methods (15,21,24–26).
Molecular beacons are stem-and-loop structured probes that undergo a conformational transition on target binding, resulting in increased hybridization specificity (17–19). Stem variations of the DNA duplex in molecular beacons have included other structural motifs such as a L-DNA (27), G-quadruplex (28), DNA/PNA triplex (29) and homo-DNA (30). PNA alone offers increased binding specificity by undergoing a ‘stemless’ conformational transition from a collapsed random coil to a heteroduplex on binding to a DNA target. In addition to enhancing binding specificity, novel structural motifs have been used to improve mismatch discrimination (31–34) and detection (35–38).
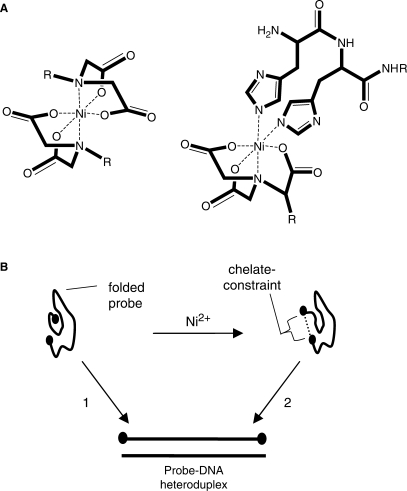
In this report, we describe chelate-constrained probes where the N and C termini of a PNA oligomer are modified with a pair of coordinating ligands that chelate a single metal ion and force the PNA oligomer into a macrocyclic configuration (Figure 1A). The coordinating ligands that were tested included iminodiacetic acid (IDA), nitrilotriacetic acid (NTA) and polyhistidine sequences including dihistidine and hexahistidine. We refer to these probes as ‘snap-to-it’ probes, since on binding with a target, the intramolecular chelate must dissociate and the probe ‘snap-to’ the target (Figure 1B). A similar probe design has been successfully demonstrated by Göritz et al. (39) with terpyridine-modified DNA probes where Zn2+ was used to regulate hybridization with a complementary target. Snap-to-it probes were found to exhibit a marked increase in binding specificity relative to unconstrained oligomers of PNA or DNA having the same base sequence. The intramolecular chelate can be varied independently of Watson–Crick base-pairing of the nucleobase oligomer to control the magnitude of the energy barrier to target hybridization, is facile to introduce through standard synthetic methods, and produces a conformational change on target binding that is readily detected in real time using fluorescence quenching.
Figure 1.
Diagram showing (A) the structural model of a snap-to-it probe at either a bis(iminodiacetate) nickel complex (IDA–IDA, left) or nitrilotriacetate-dihistidine nickel complex (NTA–His2, right), where each forms a hexadentate coordination of ligands about a single divalent transition metal (Ni2+), and ‘R’ represents the PNA oligomer. The dihistidine sequence is shown attached to the N-terminus of the snap-to-it probe. (B) Schematic of snap-to-it probe hybridization with a DNA target in the absence (path 1) or presence (path 2) of divalent ion, where path 2 requires chelate dissociation in order to form the probe-DNA heteroduplex.
MATERIALS AND METHODS
Materials
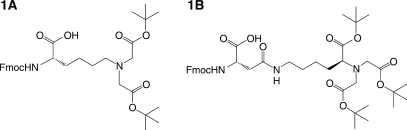
General reagents were purchased from VWR (West Chester, PA, USA) or Sigma–Aldrich (St Louis, MO, USA) and were used without further purification. The coupling agents O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU) and 1-hydroxybenzotriazole (HOBt), and Fmoc-protected amino acid monomers including N-α-Fmoc-N-ε-Dabcyl-l-lysine, and N-α-Fmoc-l-glutamic acid γ-EDANS amide were obtained from Novabiochem (San Diego, CA, USA). The N-α-Fmoc-N-imidazole-trityl-l-histidine monomer was from Bachem (King of Prussia, PA, USA). Fmoc-protected PNA monomers, PAL resin and the spacer 2-(2-(Fmoc-aminoethoxy)ethoxy)acetic acid (Fmoc-AEEA-OH) were obtained from Applied Biosystems (Framingham, MA, USA). The Fmoc-protected monomers of the IDA and NTA chelate moieties (Figure 2) were synthesized at a multigram scale in 4–5 steps, each from commercially available materials. The synthesis of IDA monomer ‘1A’ has been previously described (40,41). The Fmoc-protected NTA derivatized aspartic acid monomer ‘1B’ was synthesized by ring-opening condensation of Fmoc-aspartic anhydride with an NTA derivative of lysine (see below). Probe (‘2’) and target DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA). 1H NMR and 13C NMR were acquired at 300 MHz and 75 MHz, respectively, on a Bruker AV300 instrument in CD3OD or CD3SOCD3.
Figure 2.
Chelating amino acid monomers FmocLys(IDA) (‘1A’) and FmocAsp(NTA) (‘1B’). Carboxyl ligands are protected with tert-butyl protecting groups.
Synthesis of FmocAsp(NTA) (1B)
Fmoc aspartic anhydride (42,43) (2.00 g, 5.93 mmol) was dissolved in 15 ml anhydrous DMF. The solution was added to Nα-Nα-bis[(tert-butyloxycarbonyl)methyl]-L-lysine tert-butyl ester (44) (2.56 g, 5.95 mmol). After stirring for 20 min the solvent was removed in vacuo, and the product was purified on silica gel (99:1 EtOAc:AcOH). The solvent was removed to yield the foamy white solid ‘1B’ (2.6 g, 57%). 1H NMR (500 MHz, CD3OD) δ 7.79 (d, 2 H, J = 7.5 Hz), 7.67 (d, 2 H, J = 7.2 Hz), 7.39 (t, 2 H, J = 7.3 Hz), 7.31 (t, 2 H, J = 7.4 Hz), 4.58 (dd, 1 H, J = 7.4 Hz, 5.3 Hz), 4.27–4.40 (m, 2 H), 4.23 (t, 1 H, J = 7.0 Hz), 3.49 (d, 2 H, J = 17.3 Hz), 3.41 (d, 2 H, J = 17.3 Hz), 3.30 (t, 1 H, J = 7.3 Hz), 3.19 (t, 2 H, J = 6.4 Hz), 2.80 (dd, 1H, J = 15.1 Hz, 5.2 Hz), 2.71 (dd, 1 H, J = 15.1 Hz, 7.6 Hz), 1.56–1.70 (m, 2 H), 1.49–1.56 (m, 2 H), 1.45 (bs, 29 H) p.p.m.; 13C NMR (125 MHz, CD3SOCD3) 171.9, 170.7, 170.4, 169.3, 156.1, 144.3, 127.9, 127.4, 125.7, 120.4, 80.6, 80.2, 65.1, 53.6, 47.1, 30.3, 29.3, 23.4, 23.0 p.p.m.; HRMS: Calcd for (M+Na)+ C41H57N3O11Na 790.3891, found 790.3887. Anal. Calcd for C41H57N3O11: C, 64.13; H, 7.48; N, 5.47; O, 22.92. Found. C, 63.59; H, 7.53; N, 5.38; O, 23.42.
Probe design and synthesis
Unlabeled (‘3–10’) and fluorophore-quencher (‘11–16’) snap-to-it and control probes were synthesized by standard Fmoc solid-phase peptide synthesis methods (Table 1). The Fmoc syntheses of probes ‘3–16’ was accomplished on PAL resin using commercially available Fmoc-protected monomers of PNA (25) and one or more of the monomers N-α-Fmoc-N-imidazole-trityl-l-histidine, N-α-Fmoc-N-ε-Dabcyl-l-lysine, N-α-Fmoc-l-glutamic acid γ-EDANS amide, ‘1A’, or ‘1B’. The PNA segments of the probes were synthesized by Applied Biosystems (Framingham, MA, USA) on receiving a synthesis intermediate linked to PAL resin. The control probe ‘3’ was the unmodified PNA sequence ATC AAC ACT GCA TGT. The snap-to-it probe with the same base sequence, but with terminal IDAs (‘4’) was prepared by coupling monomer ‘1’ at the beginning and end of the PNA oligomer. The tert-butyl ester protecting groups on the IDA moieties were hydrolyzed concurrent with TFA-mediated deprotection and cleavage of the probe from the resin.
Table 1.
Structures and thermal parameters of snap-to-it and control probes 2–16
| Probe | Linear order of elements | CT Tm | MM Tm | ΔTm | CT Tm | MM Tm | ΔTm | ΔΔTm | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −Metal | +Metal | |||||||||||
| 2* | DNA | 51 | 39 | 12 | – | – | – | – | ||||
| Unlabeled series | ||||||||||||
| 3* | P | 65 | 50 | 15 | 65 | 50 | 15 | 0 | ||||
| 4 | I | P | I | 65 | 48 | 17 | 56 | 35 | 21 | 4 | ||
| 5 | I | S | P | S | I | 66 | 50 | 16 | 58 | 38 | 20 | 4 |
| 6 | H2 | P | N | 64 | 48 | 16 | 62 | 44 | 18 | 2 | ||
| 7 | N | P | H2 | 65 | 50 | 15 | 62 | 44 | 18 | 3 | ||
| 8 | H6 | P | N | 65 | 48 | 17 | 60 | 38 | 22 | 5 | ||
| 9* | P | N | 64 | 48 | 16 | 64 | 48 | 16 | 0 | |||
| 10* | P | H2 | 67 | 50 | 17 | 66 | 50 | 16 | −1 | |||
| Fluorophore-quencher series | ||||||||||||
| 11 | E | H2 | P | N | D | 63 | 47 | 16 | 58 | 36 | 22 | 6 |
| 12 | H2 | E | P | N | D | 62 | 46 | 16 | 57 | 36 | 21 | 5 |
| 13 | E | H2 | P | D | N | 62 | 46 | 16 | 57 | 36 | 21 | 5 |
| 14 | H2 | E | P | D | N | 62 | 46 | 16 | 55 | 35 | 20 | 4 |
| 15* | E | P | D | 65 | 48 | 17 | 65 | 49 | 16 | −1 | ||
| 16 | I | E | P | D | I | 59 | 43 | 16 | 59 | 42 | 17 | 1 |
DNA or P = the DNA or PNA oligomer respectively, with the base sequence ATC CCA ACT GCA TGT (underlined forms a C-T mismatch with the single-mismatch 15-mer DNA target). The linear order of the sequence elements in probes 3–16 are drawn left to right from the N-terminus to the C-terminus, corresponding to the 5′ to 3′ direction for DNA. I = Lys(IDA), S = spacer = NH(CH2CH2O)2CH2C(O), H = His, N = Asp(NTA), E = Glu(EDANS), D = Lys(Dabcyl), CT = complementary target, MM = mismatched target, +Metal = Ni2+ in the Unlabeled series and Cu2+ in the Fluorophore-quencher series. *Control probe.
The effect of spacing between the PNA oligomer and the chelate moieties was examined by incorporating a -NH(CH2CH2O)2CH2C(O)- spacer into snap-to-it probe ‘5’ using the Fmoc-AEEA monomer. The spacer provided one and a half times the spacing provided between bases by the PNA backbone. Snap-to-it probes ‘6’ to ‘8’ contained the second chelate pair; NTA (made using monomer ‘1B’) paired with polyhistidine. This probe series included both dihistidine (‘6’,‘7’) and hexahistidine motifs (‘8’). Control probes included those with chelating moieties at only a single end (‘9’, ‘10’).
Prompted by the successful application of internal fluorescence detection motifs, such as molecular beacons (19) and light-up probes (45), we synthesized a series of snap-to-it probes with intramolecular fluorophore-quencher pairs designed for real-time fluorescence detection (‘11’–‘16’). Based on the presumed large conformational change between the unbound folded probe and the rigid helical target bound probe, target binding was expected to result in a detectable fluorescence enhancement as a result of the change in proximity between fluorophore and quencher. We prepared a series of snap-to-it probes with the EDANS-Dabcyl label-quencher pair and either the NTA–His2 chelate pair in different linear permutations (‘11’–‘14’), or the IDA–IDA chelate pair (‘16’). The spectral overlap of the EDANS-Dabcyl pair supports fluorescence quenching by fluorescence resonance energy transfer (FRET). A control probe ‘15’ was prepared with the EDANS-Dabcyl label-quencher, but that lacked a chelate pair. The EDANS and Dabcyl moieties were introduced in this series using the commercially available N-α-Fmoc-L-glutamic acid γ-EDANS amide and N-α-Fmoc-N-ε-Dabcyl-L-lysine monomers during solid-phase synthesis.
Cleavage and deprotection of probes ‘3’–‘16’ was accomplished by mixing the resin with 9:1 TFA:m-cresol for 4 h. The probes were precipitated with diethyl ether then centrifuged, then washed twice with ether by vortexing and centrifugation. HPLC purification of the probes was performed with a Waters Delta-Pak C18 column (15 μm, 300 Å, 7.8 × 300 mm) maintained at 50°C and a linear gradient of 5% acetonitrile/0.1% TFA to 50% acetonitrile/0.1% TFA over 45 min. Compound identity and purity were confirmed by LC-MS using a Fisons VG Quattro II mass spectrometer fitted with a Z-spray ESI source (see Supplementary Table S1).
Thermal denaturation
Unlabeled probes were diluted to a final concentration of 1 μM with target DNA (1 μM) in 10 mM potassium phosphate-buffered saline [pH = 7.5, (NaCl) = 100 Mm]. Chelate-constrained samples included 2 μM NiSO4 unless specified otherwise, and unconstrained samples included 5 μM EDTA in a total volume of 3 ml in quartz screw-cap cuvettes (10 mm, Starna Cells, Atascadero, CA, USA). The absorbance was measured at 260 nm on a Cary 3 spectrophotometer with a Peltier temperature controller from 20°C to 80°C with a heating rate of 0.15°C/min followed by a cooling from 80°C to 50°C at –0.06°C/min then 50°C to 20°C at −0.1°C/min. The melting temperature (Tm) was obtained from the first derivative of absorbance at 260 nm measured as a function of temperature, where the Tm is the temperature at which the first derivative is maximum. Probe specificity (ΔTm) was calculated as the difference in the Tm values between the perfectly complementary (5′-ACA TGC AGT GTT GAT-3′) and single-base mismatch (5′-ACA TGC ATT GTT GAT-3′) targets. Unless stated otherwise, the change in specificity (ΔΔTm) was the ΔTm with ion minus the ΔTm in the absence of ion. Heteroduplex destabilization (ΔTm ± metal) was the Tm value of a heteroduplex in the absence of ion minus the Tm value of the same heteroduplex with ion.
3′ Dabcyl labeled probes ‘11’–‘16’ were each dissolved to 300 nM in 10 mM potassium phosphate buffer (pH 9). The solutions were prepared with either EDTA (5 μM) or CuSO4 (300 nM) and either complementary target 5′-FAM-ACA TGC AGT GTT GAT-3′ (45 nM) or mismatch target 5′-FAM-ACA TGC ATT GTT GAT-3′ (45 nM). The fluorescence of 100 μl reaction solutions was measured as a function of temperature using an ABI 7700 Prism spectrofluorometer. The temperature was increased at 1°C per 5 min from 15°C to 80°C, with data collection lasting the final 30 s at each temperature.
Fluorescence spectroscopy
Probes ‘11’–‘15’ were dissolved to 500 nM in 10 mM potassium phosphate buffer (pH = 7.5) or 10 mM Tris (pH = 7.5). The solution was prepared either with EDTA (5 μM), NiSO4 (1 μM), or NiSO4 (1 μM) and target DNA (1 μM). The fluorescence emission spectra of each probe was recorded at λmax ± 30 nm using an SLM 8100 fluorometer under equilibrium conditions.
RESULTS
Thermal denaturation analysis
Snap-to-it and control probes were prepared as described in the ‘Materials and methods’ section in both unlabeled and fluorophore-quencher formats (Table 1). To test for chelate-mediated changes in probe binding specificity, snap-to-it probes and control probes ‘3’–‘16’ were hybridized in the presence and absence of metal ion with a perfectly complementary 15-mer target oligodeoxyribonucleotide, or a single-mismatch 15-mer oligodeoxyribonucleotide containing a G to T change at the central base position. This pair of test targets was selected because it results in the highly destabilizing C-T mismatch, thereby permitting the effect of constraint on specificity to be assessed in the setting of optimal mismatch discrimination of the unconstrained PNA oligomer (46–49).
The melting temperature (Tm), specificity (ΔTm) and change in specificity (ΔΔTm) due to metal for each of probes ‘3–‘16’ are listed in Table 1 (see ‘Thermal denaturation’ subsection under ‘Material and methods’ section for definition of these and other thermal parameters). Relative to a conventional unconstrained DNA probe, we hypothesized that a metal-mediated increase in specificity of a snap-to-it probe would be due to additive effects from the PNA segment in the probe, and the energy barrier provided by the internal chelate.
Specificity of the unlabeled series
To quantitate the contribution to specificity provided by the PNA segment, we compared the thermal denaturation characteristics of control probe ‘2’ (DNA) and probe ‘3’ (PNA), which lack chelates but otherwise have identical base sequences to one another and the other probes in Table 1. The specificity of the PNA control was greater (ΔTm = 15°C) than the DNA control (ΔTm = 12°C) by 3°C. The Tm with a perfectly complementary target was also greater for the PNA control (Tm = 65°C) than the DNA control (Tm = 51°C) by 14°C. The difference in thermal parameters of the controls corresponded closely with previously reported values for DNA and PNA, where an average 15-mer PNA–DNA heteroduplex exhibited a 15°C stabilization in Tm and a 4°C increase in specificity (i.e. ΔΔTm = 4°C) relative to a DNA duplex having the same base sequence (15,24).
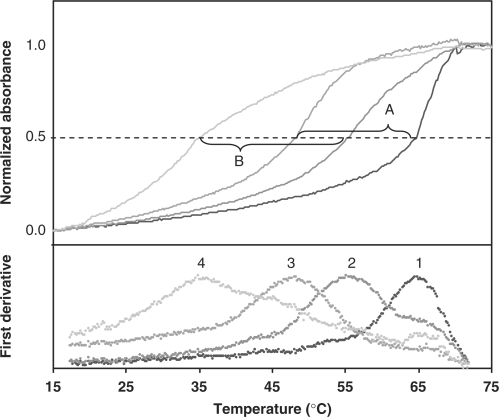
To determine the potential for reversible internal chelation to further increase the specificity of a snap-to-probe beyond that contributed by the PNA segment, we examined the thermal denaturation of snap-to-it probe ‘4’ in the presence and absence of metal ion. Surprisingly, the data revealed a marked additional metal-dependent increase in specificity (ΔΔTm) of 4°C (Figure 3). In the absence of coordinating metal ion, the mismatch heteroduplex had a Tm that was 17°C less than the perfectly matched probe-target heteroduplex. When nickel sulfate was included to impose intramolecular constraint, the mismatched heteroduplex had a Tm that was 21°C lower than the perfectly matched probe-target heteroduplex, resulting in a metal-dependent increase in specificity of 4°C.
Figure 3.
First derivative plots (lower) of thermal denaturation profiles (upper) of IDA–IDA snap-to-it probe ‘4’ hybridized with complementary (curve 1) or single-mismatch (curve 3) DNA in the absence of metal ion (EDTA); or with complementary (curve 2) or single-mismatch (curve 4) DNA in the presence of metal ion (NiSO4). Absorbance was measured at λ = 260 nm. The mismatch selectivity in the absence of metal ion (segment A; ΔTm = 17°C) increased in the presence of metal ion (segment B; ΔTm = 21°C) by 4°C.
It was observed that the specificity imparted to snap-to-probe ‘4’ by nickel was due to preferential destabilization of the mismatched heteroduplex (ΔTm±metal = 13°C) relative to the complementary heteroduplex (ΔTm±metal = 9°C). Snap-to-it probe ‘5’ also exhibited a ΔΔTm of 4°C due to preferential destabilization of the mismatched heteroduplex, indicating that increased spacing between the PNA termini and the chelate had no effect on the specificity of the probe in the presence of divalent metal. These increases in metal-mediated mismatch selectivity of snap-to-it probes ‘4’ and ‘5’ were on the same order of magnitude as that provided by the PNA backbone itself (relative to the DNA backbone), but without a concurrent increase in overall heteroduplex stability.
It is known that transition metal ions may exhibit stabilizing and/or destabilizing effects on a DNA duplex, whereas a PNA heteroduplex is known to be relatively insensitive to salt effects (50–52). To control for the possibility that the observed metal-dependent increase in snap-to-it probe specificity was due to potential ionic effects rather than reversible chelation, snap-to-it and control PNA probes were subjected to thermal denaturation studies both in the presence of a transition metal (to also impose constraint if the probes included partial metal chelators), or in the presence of EDTA to remove any trace metal ions. The thermal analysis of the nonchelating PNA control probe ‘3’ in the presence of EDTA or nickel sulfate indicated that each condition resulted in the same Tm and the same ΔTm (Table 1). Therefore ΔΔTm = 0 for the PNA control probe, confirming that nickel ions alone are ineffective at altering the binding specificity of the PNA–DNA heteroduplex.
Metal ion selection and chelation control
Thermal denaturation analyses were repeated using snap-to-it probe ‘4’ in the presence of EDTA or one of various transition metals (Ni2+, Cu2+, Co2+, Zn2+, Mn2+ or Fe2+) to determine the optimum ion type and ion concentration to obtain maximum snap-to-it probe specificity. The effect of the transition metal cation on target hybridization is likely a combination of factors that include coordination stability with an IDA ligand (53), destabilizing or stabilizing effects on the DNA backbone and/or base-pairing (50,54–56), concentration dependence of chelation, anion effects (among other salt effects, which are generally less substantial when using PNA probes), and coordination geometry.
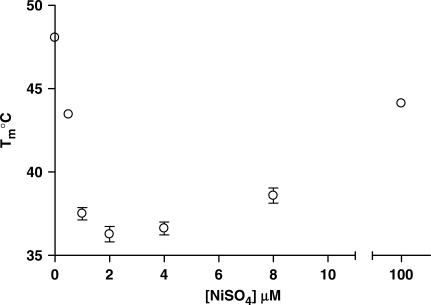
The heteroduplex Tm of snap-to-it probe ‘4’ bound with mismatched DNA target was determined as a function of cation type and concentration. Of all ions tested, nickel ion from nickel sulfate produced the maximum heteroduplex destabilization (ΔTm±metal) equal to 12–13°C at an ion concentration of 2 µM (Figure 4). As the concentration of transition metal ion was increased to a large excess, the Tm approached that measured for unconstrained probes, presumably a consequence of saturating the available chelates and precluding intramolecular probe constraint.
Figure 4.
Melting temperatures for snap-to-it probe ‘4’ hybridization with a single-mismatch DNA 15-mer target as a function of nickel ion concentration. Bars indicate standard error of the mean.
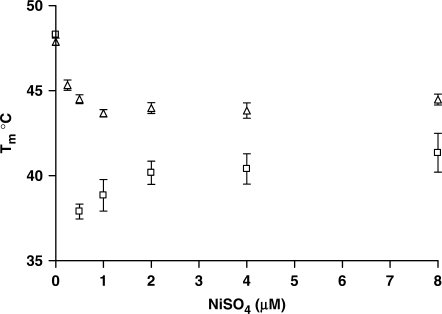
The affinity of polyhistidine segments for divalent cations is known to increase as a function of the number of histidines, and this property was exploited to tailor the strength of the chelate to examine the effect of such changes on the binding properties and ion requirements of the probe (57,58). Thermal denaturation studies of snap-to-it probes ‘6’ and ‘8’ (NTA paired with dihistidine and hexahistidine moieties, respectively) hybridized to mismatched DNA targets were performed as a function of Ni2+ concentration to examine the effect of varying the metal binding affinity of the polyhistidine motif on the optimal metal concentration for mismatch destabilization (Figure 5).
Figure 5.
Melting temperatures for snap-to-it probe ‘6’ (dihistidine motif, clear triangle) and probe ‘8’ (hexahistidine motif, clear square) hybridizations with a single-mismatch DNA 15-mer target as a function of nickel ion concentration. Snap-to-it probe concentration = 500 nM. Bars indicate standard error of the mean.
A Ni2+ concentration equimolar to ‘8’ produced the optimal destabilization of the mismatched hybrid (0.5 µM). Consistent with the greater metal-binding affinity of the hexahistidine motif, snap-to-it probe ‘8’ afforded a significantly greater magnitude of mismatch destabilization compared to the dihistidine motif in snap-to-it probe ‘6’ (i.e. ΔTm±metal = 10°C versus 4°C). Additionally, increases in the metal concentration only slightly above equimolar stoichiometry resulted in the loss of mismatch destabilization of snap-to-it probe ‘8’, but not snap-to-it probe ‘6’. This is consistent with the greater affinity for nickel ion expected at the hexahistidine motif compared to the dihistidine motif. Concentrations of metal greater than those equimolar to snap-to-it probe ‘8’ presumably break the intramolecular chelate by allowing the NTA and hexahistidine chelators to independently bind excess nickel ions. Once the chelate is broken, the intra-molecular constraint that is essential to the enhanced specificity of the probe is lost.
The chelate directionality was found to be inconsequential in discrimination or duplex destabilization, as snap-to-it probes ‘6’ and ‘7’ resulted in indistinguishable ΔΔTm values. PNA probes bearing only a single chelating moiety (probes ‘9’ and ‘10’) exhibited mismatch destabilization equal to the nonchelating control probe ‘3’, suggesting intramolecular, not intermolecular coordination is responsible for the enhanced specificity and mismatch discrimination observed in snap-to-it probes.
Specificity of the fluorophore-quencher series
We synthesized a series of fluorophore-quencher snap-to-it probes designed for possible real-time fluorescence detection of target binding in homogeneous assays or in microarrays (see probes ‘11’ to ‘16’ in Table 1). Based on the presumed large conformational change between the unbound chelated probe and the rigid helical target-bound probe, we hypothesized that fluorophore-quencher snap-to-it probes would offer ‘both’ target-mediated fluorescence enhancement and enhanced metal-mediated binding specificity. In analogous fashion to the specificity studies of the unlabeled series, we tested for chelate-mediated enhancement of binding specificity of the fluorophore-quencher snap-to-it and control probes ‘11’–‘16’ by characterizing their thermal denaturation profiles with perfectly complementary and single-match DNA targets in the presence and absence of metal ion (see Table 1). In contrast to the unlabeled series, copper ion (Cu2+) from copper sulfate proved to be optimal for increasing the specificity (ΔΔTm) of the fluorophore-quencher series conceivably due to differences in ligand-cation coordination.
Snap-to-it probes ‘11’–‘14’ present four possible linear permutations of the same fluorophore (EDANS), quencher (Dabcyl), and chelates (NTA and dishistidine). The thermal denaturation data revealed that these probes had substantially identical metal-dependent increases in specificity (ΔΔTm = 4–6°C), consistent with the linear order of probe elements having little to no effect on probe specificity.
As was observed for the unlabeled probe series, the enhancement in specificity of the fluorophore-quencher probes ‘11’–‘14’ was due to a preferential destabilization of the mismatch heteroduplex (ΔTm±metal = 10–11°C) relative to the complementary heteroduplex (ΔTm±metal = 5–7°C). The earlier observation that copper provided an inferior destabilization of the mismatch heteroduplex relative to nickel in probes containing the IDA chelate provides a possible explanation for the observation that the fluorophore-quencher probe ‘16’ was found to exhibit little metal-dependent increase in specificity.
Consistent with there being no substantial affinity between EDANS and Dabcyl in the fluorophore-quencher snap-to-it probes, control probe ‘15’ bearing only fluorophore and quencher elements exhibited no metal-dependent increase in specificity and thermal parameters that were indistinguishable from the naked PNA control probe ‘3’.
Fluorescence as a function of probe conformation
It is known that PNA molecular beacons do not require a stem-structure in order to exhibit proximity-mediated fluorescence quenching (26). Stemless PNA molecular beacons adopt a folded random coil conformation (i.e. collapsed) in aqueous solution due to hydrophobic surface minimization locating the fluorophore and quencher in close proximity, resulting in considerable contact or FRET-type quenching in the unhybridized state (21).
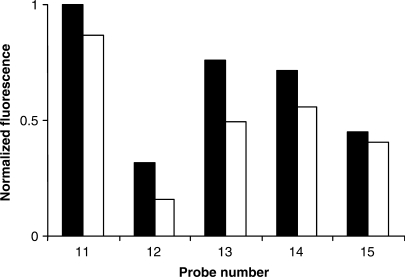
The fluorescence measurements of unhybridized snap-to-it and control probes ‘11’–‘15’ in buffered aqueous solution with either EDTA (unconstrained) or nickel sulfate (chelate-constrained) are depicted in Figure 6. The absolute fluorescence of the probes differed according to the position of the fluorophore and the quencher relative to the chelates. Consistent with the PNA segment driving the probe into a collapsed state, the normalized fluorescence of snap-to-it probes ‘11’–‘14’ was reduced by only 13–51% after the addition of nickel sulfate. A similarly negligible reduction of fluorescence upon the addition of nickel was observed for control probe ‘15’, a probe lacking terminal chelates, perhaps a result of fluorescence quenching by the metal ion itself (59,60).
Figure 6.
Normalized fluorescence of the fluorophore-quencher series of snap-to-it probes ‘11’–‘14’ and control probe ‘15’. Solid = EDTA; Clear = nickel sulfate. Measurements were performed in 10 mM potassium phosphate buffer in the absence of target.
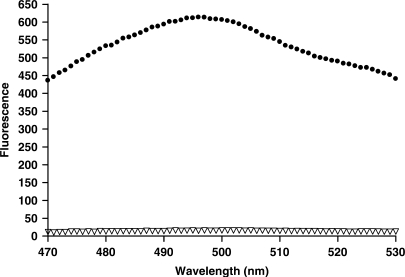
In contrast, the change in snap-to-it probe fluorescence in the presence of target was dramatic, where heteroduplex formation of snap-to-it probe ‘12’ led to a 40-fold increase in fluorescence (Figure 7).
Figure 7.
Fluorescence of snap-to-it probe ‘12’ in 10 mM Tris buffer in the presence of nickel alone (clear triangle), or nickel and DNA target (solid circles).
DISCUSSION
We report here that snap-to-it probes comprising a PNA oligomer constrained by coordinating ligands at each terminus exhibited enhanced target discrimination. PNA oligomers conformationally constrained with either an intramolecular IDA-Ni2+-IDA or NTA-Cu2+-dihistidine chelate hybridized perfectly complementary DNA targets with only a small reduction in Tm relative to unconstrained controls. However, the same snap-to-it probes hybridized to DNA targets containing a mismatch with a significantly reduced Tm compared to unconstrained controls. The net effect was an increase in specificity for binding to the perfectly matched target.
The increase in specificity was additive with that provided by unconstrained PNA relative to unconstrained DNA, and thus from the perspective of unconstrained DNA as a baseline metric represents one of the more robust specificity enhancements afforded by a conformational constraint mechanism (ΔΔTm of 8–10°C relative to unmodified DNA). For example, molecular beacon specificity was first exemplified using closely related stem-and-loop analogues with the fluorophore moiety on the probe sequence, and the quencher moiety on the target sequence. The stem constraint increased the ΔTm of the probe by 13°C relative to unmodified DNA (61). Other studies of molecular beacons containing a probe bound fluorophore and quencher more typically yield ΔΔTm values in the range of 6°C relative to unmodified DNA (18,62).
Another commercially available nucleobase probe with enhanced specificity, ‘minor groove binders’ (MGB), increase the affinity and specificity of PCR probes through the interaction of an isohelical-conjugated tripeptide and an A/T-rich target minor groove. A mismatch in the target minor groove region can increase the probe ΔTm by up to 9°C relative to unmodified DNA (63). A comprehensive study of the mismatch discrimination of LNA–DNA chimeras demonstrated up to a 7°C increase in ΔTm compared with unmodified DNA of the same sequence (14). The enhanced specificity provided by snap-to-it probes thus also compares favorably with other common approaches to enhance probe specificity, and therefore may have utility in homogeneous assays (e.g. RT-PCR), or in microarrays where selectivity is critical.
Coordination of metal ion broadened the probe-target melting transition. Enthalpy changes associated with probe secondary structure may contribute to transition broadening (64,65). Tsourkas et al. (62) reported an analogous result from the analysis of molecular beacons with long stems. The shallow transition may also be an indication of decreased binding cooperativity. The rate of target hybridization was slower for snap-to-it probes than for unconstrained analogues, a result consistent with other pre-organized probes.
Unconstrained PNA has been an important alternative probe chemistry to DNA probes based on its higher binding affinity and increased specificity (24,26). This is an unusual property because these attributes often anticorrelate (22). The enhanced binding specificity of PNA likely arises from intramolecular hydrophobic packing forces that serve to constrain the single-strand molecule without a chelate or stem (18,62). The energy required to unfold the PNA oligomer during target binding must then be overcome in order to form the rigid probe-target duplex. We found that inclusion of an intramolecular chelate as an additional constraint imparted even greater binding specificity to the PNA oligomer. It is interesting to speculate that PNA oligomer unfolding and chelate dissociation are energy barriers that must be traversed sequentially prior to complete target binding. In this respect, snap-to-it probes may mimic the sequential conformational changes (energy barriers) traversed by nucleic acid-binding proteins which are pivotal to their exquisite binding specificity (66–68).
In conclusion, we have demonstrated that PNA-based snap-to-it probes are synthetically accessible using a facile Fmoc-based solid-phase peptide synthesis strategy. We synthesized Fmoc-protected IDA- and NTA-functionalized amino acids for use in solid-phase coupling. Snap-to-it probes were synthesized with terminal IDA functionalities for symmetrical hexadentate chelation with Ni2+, and other divalent transition metal ions. Alternatively, probes were prepared with NTA and (poly)histidine motifs at the termini for unsymmetrical chelation with Cu2+ or Ni2+, and control of chelate affinity through varying the number of consecutive histidine residues. From thermal transition analyses of snap-to-it probes with and without metals, and with either complementary DNA or mismatched DNA, we measured a significant increase in DNA-binding specificity. Finally, we demonstrated that this method of increasing probe specificity is compatible with fluorophore-quencher detection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Alan Mayer for his helpful and critical review of the manuscript. This work was supported by National Institutes of Health (R44 CA94419, R44 CA94612). Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nakatani K. Chemistry challenges in SNP typing. ChemBioChem. 2004;5:1623–1633. doi: 10.1002/cbic.200400161. [DOI] [PubMed] [Google Scholar]
- 2.Engle LJ, Simpson CL, Landers JE. Using high-throughput SNP technologies to study cancer. Oncogene. 2006;25:1594–1601. doi: 10.1038/sj.onc.1209368. [DOI] [PubMed] [Google Scholar]
- 3.Dutt A, Beroukhim R. Single nucleotide polymorphism array analysis of cancer. Curr. Opin. Oncol. 2007;19:43–49. doi: 10.1097/CCO.0b013e328011a8c1. [DOI] [PubMed] [Google Scholar]
- 4.Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrenreich A. DNA microarray technology for the microbiologist: An overview. Appl. Microbiol. Biotechnol. 2006;73:255–273. doi: 10.1007/s00253-006-0584-2. [DOI] [PubMed] [Google Scholar]
- 6.Hoheisel JD. Microarray technology: beyond transcript profiling and genotype analysis. Nat. Rev. Genet. 2006;7:200–210. doi: 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- 7.Urakawa H, El Fantroussi S, Smidt H, Smoot JC, Tribou EH, Kelly JJ, Noble PA, Stahl DA. Optimization of single-base-pair mismatch discrimination in oligonucleotide microarrays. Appl. Environ. Microbiol. 2003;69:2848–2856. doi: 10.1128/AEM.69.5.2848-2856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mocellin S, Wang E, Panelli M, Pilati P, Marincola FM. DNA array-based gene profiling in tumor immunology. Clin. Cancer Res. 2004;10:4597–4606. doi: 10.1158/1078-0432.CCR-04-0327. [DOI] [PubMed] [Google Scholar]
- 9.Jayapal M, Melendez AJ. DNA microarray technology for target identification and validation. Clin. Exp. Pharmacol. Physiol. 2006;33:496–503. doi: 10.1111/j.1440-1681.2006.04398.x. [DOI] [PubMed] [Google Scholar]
- 10.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 11.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aleman LM, Doench J, Sharp PA. Comparison of siRNA-induced off-target RNA and protein effects. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You Y, Moreira BG, Behlke MA, Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the watson-crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 16.Kumar VA, Ganesh KN. Conformationally constrained PNA analogues: Structural evolution toward DNA/RNA binding selectivity. Acc. Chem. Res. 2005;38:404–412. doi: 10.1021/ar030277e. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 18.Bonnet G, Tyagi S, Libchaber A, Kramer FR. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marras SA, Tyagi S, Kramer FR. Real-time assays with molecular beacons and other fluorescent nucleic acid hybridization probes. Clin. Chim. Acta. 2006;363:48–60. doi: 10.1016/j.cccn.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Yang CJ, Medley CD, Benner SA, Tan W. Locked nucleic acid molecular beacons. J. Am. Chem. Soc. 2005;127:15664–15665. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]
- 21.Seitz O. Solid-phase synthesis of doubly labeled peptide nucleic acids as probes for the real-time detection of hybridization. Angew. Chem. Int. Ed. Engl. 2000;39:3249–3252. doi: 10.1002/1521-3773(20000915)39:18<3249::aid-anie3249>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 22.Demidov VV, Frank-Kamenetskii MD. Two sides of the coin: Affinity and specificity of nucleic acid interactions. Trends Biochem. Sci. 2004;29:62–71. doi: 10.1016/j.tibs.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Kool ET. Recognition of DNA, RNA, and proteins by circular oligonucleotides. Acc. Chem. Res. 1998;31:502–510. doi: 10.1021/ar9602462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shakeel S, Karim S, Ali A. Peptide nucleic acids (PNA) – a review. J. Chem. Technol. Biotechnol. 2006;81:892–899. [Google Scholar]
- 25.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn H, Demidov VV, Coull JM, Fiandaca MJ, Gildea BD, Frank-Kamenetskii MD. Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J. Am. Chem. Soc. 2002;124:1097–1103. doi: 10.1021/ja0041324. [DOI] [PubMed] [Google Scholar]
- 27.Kim Y, Yang CJ, Tan W. Superior structure stability and selectivity of hairpin nucleic acid probes with an l-DNA stem. Nucleic Acids Res. 2007;35:7279–7287. doi: 10.1093/nar/gkm771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourdoncle A, Estevez Torres A, Gosse C, Lacroix L, Vekhoff P, Le Saux T, Jullien L, Mergny JL. Quadruplex-based molecular beacons as tunable DNA probes. J. Am. Chem. Soc. 2006;128:11094–11105. doi: 10.1021/ja0608040. [DOI] [PubMed] [Google Scholar]
- 29.Grossmann TN, Roglin L, Seitz O. Triplex molecular beacons as modular probes for DNA detection. Angew. Chem. Int. Ed. Engl. 2007;46:5223–5225. doi: 10.1002/anie.200700289. [DOI] [PubMed] [Google Scholar]
- 30.Crey-Desbiolles C, Ahn DR, Leumann CJ. Molecular beacons with a homo-DNA stem: improving target selectivity. Nucleic Acids Res. 2005;33:e77. doi: 10.1093/nar/gni076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohler O, Jarikote DV, Seitz O. Forced intercalation probes (fit probes): Thiazole orange as a fluorescent base in peptide nucleic acids for homogeneous single-nucleotide-polymorphism detection. ChemBioChem. 2005;6:69–77. doi: 10.1002/cbic.200400260. [DOI] [PubMed] [Google Scholar]
- 32.Kohler O, Seitz O. Thiazole orange as fluorescent universal base in peptide nucleic acids. Chem. Commun. 2003:2938–2939. doi: 10.1039/b308299g. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto A, Tanaka K, Fukuta T, Saito I. Cytosine detection by a fluorescein-labeled probe containing base-discriminating fluorescent nucleobase. ChemBioChem. 2004;5:958–963. doi: 10.1002/cbic.200400010. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto A, Kanatani K, Saito I. Pyrene-labeled base-discriminating fluorescent DNA probes for homogeneous SNP typing. J. Am. Chem. Soc. 2004;126:4820–4827. doi: 10.1021/ja039625y. [DOI] [PubMed] [Google Scholar]
- 35.Grossmann TN, Seitz O. DNA-catalyzed transfer of a reporter group. J. Am. Chem. Soc. 2006;128:15596–15597. doi: 10.1021/ja0670097. [DOI] [PubMed] [Google Scholar]
- 36.Dose C, Ficht S, Seitz O. Reducing product inhibition in DNA-template-controlled ligation reactions. Angew. Chem. Int. Ed. Engl. 2006;45:5369–5373. doi: 10.1002/anie.200600464. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y, Karalkar NB, Kool ET. Nonenzymatic autoligation in direct three-color detection of RNA and DNA point mutations. Nat. Biotechnol. 2001;19:148–152. doi: 10.1038/84414. [DOI] [PubMed] [Google Scholar]
- 38.Sando S, Kool ET. Quencher as leaving group: efficient detection of DNA-joining reactions. J. Am. Chem. Soc. 2002;124:2096–2097. doi: 10.1021/ja017328s. [DOI] [PubMed] [Google Scholar]
- 39.Goritz M, Kramer R. Allosteric control of oligonucleotide hybridization by metal-induced cyclization. J. Am. Chem. Soc. 2005;127:18016–18017. doi: 10.1021/ja056463s. [DOI] [PubMed] [Google Scholar]
- 40.Ruan F, Chen Y, Itoh K, Sasaki T, Hopkins PB. Synthesis of peptides containing unnatural, metal-ligating residues: aminodiacetic acid as a peptide side chain. J. Org. Chem. 1991;56:4347–4354. [Google Scholar]
- 41.Hamachi I, Watanabe J I, Eboshi R, Hiraoka T, Shinkai S. Incorporation of artificial receptors into a protein/peptide surface: a strategy for on/off type of switching of semisynthetic enzymes. Biopolymers. 2000;55:459–468. doi: 10.1002/1097-0282(2000)55:6<459::AID-BIP1021>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 42.Bergmann M, Zervas L. Über ein allgemeines verfahren der peptid-synthese. Ber. Dtsch. Chem. Ges. 1932;65B:1192–1201. [Google Scholar]
- 43.Chen FM, Benoiton NL. N-alkoxycarbonyl-glutamic and aspartic acids. Studies on the activation and cyclodehydration and side-reaction encountered in analysis of glutamic acid using Fmoc-chloride. Int. J. Pept. Protein Res. 1992;40:13–18. [PubMed] [Google Scholar]
- 44.Dorn IT, Neumaier KR, Tampé R. Molecular recognition of histidine-tagged molecules by metal-chelating lipids monitored by fluorescence energy transfer and correlation spectroscopy. J. Am. Chem. Soc. 1998;120:2753–2763. [Google Scholar]
- 45.Svanvik N, Westman G, Wang D, Kubista M. Light-up probes: Thiazole orange-conjugated peptide nucleic acid for detection of target nucleic acid in homogeneous solution. Anal. Biochem. 2000;281:26–35. doi: 10.1006/abio.2000.4534. [DOI] [PubMed] [Google Scholar]
- 46.SantaLucia J, Jr., Hicks D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 47.Jensen KK, Orum H, Nielsen PE, Norden B. Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry. 1997;36:5072–5077. doi: 10.1021/bi9627525. [DOI] [PubMed] [Google Scholar]
- 48.Igloi GL. Variability in the stability of DNA-peptide nucleic acid (PNA) single-base mismatched duplexes: real-time hybridization during affinity electrophoresis in PNA-containing gels. Proc. Natl Acad. Sci. USA. 1998;95:8562–8567. doi: 10.1073/pnas.95.15.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ratilainen T, Holmen A, Tuite E, Nielsen PE, Norden B. Thermodynamics of sequence-specific binding of PNA to DNA. Biochemistry. 2000;39:7781–7791. doi: 10.1021/bi000039g. [DOI] [PubMed] [Google Scholar]
- 50.Eichhorn GL, Shin YA. Interaction of metal ions with polynucleotides and related compounds. XII. The relative effect of various metal ions on DNA helicity. J. Am. Chem. Soc. 1968;90:7323–7328. doi: 10.1021/ja01028a024. [DOI] [PubMed] [Google Scholar]
- 51.Eichhorn GL, Clark P. Interactions of metal ions with polynucleotides and related compounds. V. The unwinding and rewinding of DNA strands under the influence of copper (II) ions. Proc. Natl Acad. Sci. USA. 1965;53:586–593. doi: 10.1073/pnas.53.3.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eichhorn GL. Metal ions as stabilizers or destabilizers of the deoxyriboucleic acid structure. Nature. 1962;194:474–475. doi: 10.1038/194474a0. [DOI] [PubMed] [Google Scholar]
- 53.Perrin DD. Stability Constants of Metal-Ion Complexes Part B: Organic Ligands. Oxford: Pergamon Press; 1979. [Google Scholar]
- 54.Izatt RM, Christensen JJ, Rytting JH. Sites and thermodynamic quantities associated with proton and metal ion interaction with ribonucleic acid, deoxyribonucleic acid, and their constituent bases, nucleosides, and nucleotides. Chem. Rev. 1971;71:439–481. doi: 10.1021/cr60273a002. [DOI] [PubMed] [Google Scholar]
- 55.Pezzano H, Podo F. Structure of binary complexes of mono- and polynucleotides with metal ions of the first transition group. Chem. Rev. 1980;80:365–401. [Google Scholar]
- 56.Cervantes G, Fiol JJ, Terrün A, Moreno V, Alabart JR, Aguilü M, Gümez M, Solans X. Synthesis and characterization of nickel(II) complexes of purine and pyrimidine bases. Crystal and molecular structure of trans-bis(cytosine-O2)bis(ethylenediamine)nickel(II) bis(tetraphenylborate). An unusual metal binding mode of cytosine. Inorg. Chem. 1990;29:5168–5173. [Google Scholar]
- 57.Hochuli E, Bannwarth W, Döbeli H, Gentz R, Stüber D. Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent. Biotechnology. 1988;6:1321–1325. [Google Scholar]
- 58.Orum H, Nielsen PE, Jorgensen M, Larsson C, Stanley C, Koch T. Sequence-specific purification of nucleic acids by PNA-controlled hybrid selection. BioTechniques. 1995;19:472–480. [PubMed] [Google Scholar]
- 59.Varnes AW, Dodson RB, Wehry EL. Interactions of transition-metal ions with photoexcited states of flavins. Fluorescence quenching studies. J. Am. Chem. Soc. 1972;94:946–950. doi: 10.1021/ja00758a037. [DOI] [PubMed] [Google Scholar]
- 60.Zhang LZ, Cheng P. Study of Ni(II) ion-DNA interactions with methylene blue as fluorescent probe. J. Inorg. Biochem. 2004;98:569–574. doi: 10.1016/j.jinorgbio.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 61.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 62.Tsourkas A, Behlke MA, Rose SD, Bao G. Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res. 2003;31:1319–1330. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, et al. 3'-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen C, Wang W, Wang Z, Wei F, Zhao XS. Influence of secondary structure on kinetics and reaction mechanism of DNA hybridization. Nucleic Acids Res. 2007;35:2875–2884. doi: 10.1093/nar/gkm177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loakes D, Hill F, Brown DM, Salisbury SA. Stability and structure of DNA oligonucleotides containing non-specific base analogues. J. Mol. Biol. 1997;270:426–435. doi: 10.1006/jmbi.1997.1129. [DOI] [PubMed] [Google Scholar]
- 66.Lesser DR, Kurpiewski MR, Jen-Jacobson L. The energetic basis of specificity in the Eco RI endonuclease—DNA interaction. Science. 1990;250:776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- 67.van den Broek B, Noom MC, Wuite GJ. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res. 2005;33:2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dupureur CM. NMR studies of restriction enzyme-DNA interactions: role of conformation in sequence specificity. Biochemistry. 2005;44:5065–5074. doi: 10.1021/bi0473758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.