Abstract
Human DNA polymerase theta (pol θ or POLQ) is a proofreading-deficient family A enzyme implicated in translesion synthesis (TLS) and perhaps in somatic hypermutation (SHM) of immunoglobulin genes. These proposed functions and kinetic studies imply that pol θ may synthesize DNA with low fidelity. Here, we show that when copying undamaged DNA, pol θ generates single base errors at rates 10- to more than 100-fold higher than for other family A members. Pol θ adds single nucleotides to homopolymeric runs at particularly high rates, exceeding 1% in certain sequence contexts, and generates single base substitutions at an average rate of 2.4 × 10−3, comparable to inaccurate family Y human pol κ (5.8 × 10−3) also implicated in TLS. Like pol κ, pol θ is processive, implying that it may be tightly regulated to avoid deleterious mutagenesis. Pol θ also generates certain base substitutions at high rates within sequence contexts similar to those inferred to be copied by pol θ during SHM of immunoglobulin genes in mice. Thus, pol θ is an exception among family A polymerases, and its low fidelity is consistent with its proposed roles in TLS and SHM.
INTRODUCTION
The human genome encodes numerous DNA polymerases that have key roles in DNA replication and the repair and recombination reactions needed to maintain the integrity of genetic information (1–4). These polymerases are classified by sequence homology into families A, B, X and Y. Family A members include human DNA polymerase γ (pol γ), pol ν and pol θ, the subject of this study. Pol θ is encoded by the POLQ gene that was initially described (5) as an open reading frame in humans with homology to the founding member of family A polymerases, Escherichia coli DNA polymerase I. Subsequent studies (6,7) demonstrated that POLQ encodes a 290-kDa protein with template-dependent DNA polymerase activity. The amino terminus contains seven motifs characteristic of DNA and RNA helicases, although no helicase activity has been reported. The carboxy-terminal half contains the residues required for polymerase activity (6). Recombinant pol θ lacks detectable 3′ exonuclease activity, rendering it incapable of exonucleolytically proofreading any errors that it may make during DNA synthesis. Also, steady-state kinetic studies demonstrate that recombinant human pol θ misinserts incorrect dNTPs at rates that are higher than those of several other family A polymerases (7). Pol θ can also efficiently insert a nucleotide opposite an abasic site or a thymine glycol lesion (7–9), and it can extend the resulting primer terminus. In addition, pol θ can extend a primer terminus generated after another polymerase has inserted a nucleotide opposite a 6–4 photoproduct, a lesion that pol θ itself cannot bypass (7,8). Mice with a T to C base substitution (the chaos1 mouse) or a deletion in the Polq gene (10) have elevated levels of spontaneous and radiation-induced micronuclei in erythrocytes (10). When the chaos1 mutant mouse was crossed with an Atm-deficient mouse, the number of the double homozygous mice was lower than expected (11), indicating that pol θ is important for survival in the absence of ATM-dependent checkpoint signaling. Additional studies suggest that pol θ may participate in cellular responses to DNA damage, possibly including TLS (10,12).
Pol θ has also been suggested to be involved in somatic hypermutation of immunoglobulin genes, although a consistent model for a possible role has not yet emerged. It is highly expressed in lymphoid tissues and regulated in the germinal centers of B cells where SHM takes place (12). In one study, mice expressing catalytically inactive pol θ had reduced SHM at C:G base pairs (13); in another study, pol θ null mice showed a reduction in SHM at both C:G and A:T base pairs accompanied by an increase of G to C transversions (14) and in a third study, pol θ was proposed to be responsible for the residual A:T mutations observed in the absence of pol η (15).
Previous work has shown that mammalian polymerases implicated in TLS and/or SHM, such as the family Y members pol η, pol κ and pol ι, and the family B member pol ζ, all have relatively high error rates when copying undamaged DNA templates (16). Analysis of the error specificity of DNA polymerases can provide insights into the mechanisms by which they avoid or generate errors, and clues about their biological functions. For these reasons, we analyzed the single base deletion, insertion and substitution error rates and specificity of recombinant human pol θ during copying of undamaged DNA.
MATERIALS AND METHODS
Enzymes, reagents and strains
Recombinant full-length human DNA pol θ was purified to near homogeneity as described (6,8). Materials for the M13mp2 fidelity assay were from sources described previously (17). The exonuclease-deficient, large Klenow fragment of E. coli DNA pol I and T4 polynucleotide kinase were purchased from New England BioLabs, Ipswich, MA, USA. [γ-32P]ATP (4500 Ci/mmol) and unlabeled deoxyribonucleotide-triphosphates were from GE Healthcare Biosciences, Piscataway, NJ, USA. Materials for processivity analysis were described previously (18).
M13mp2 fidelity assay
Gap-filling reactions for pol θ (25 μl) contained 20 mM Tris–HCl (pH 7.5), 8 mM MgCl2, 0.1 mM EDTA, 4% glycerol, 80 μg/ml bovine serum albumin and 1 mM each dATP, dCTP, dGTP and dTTP and 0.2 nM M13mp2 (407-nt gap) DNA substrate (from nucleotide –216 through +191 of lacZ gene). Polymerization reactions were initiated by the addition of pol θ (27 nM), incubated at 37°C for 1 h and terminated by adding EDTA to 20 mM. Ten microliters of the reaction mixture were mixed 1:1 with SDS buffer (20 mM Tris pH 8.0, 5 mM EDTA, 5% SDS, 0.5% bromophenol blue and 25% glycerol) and complete gap filling was monitored by agarose gel electrophoresis. Errors were scored as light blue or colorless mutant plaques, while correct synthesis yielded plaques that are dark blue. DNA from independent mutants was sequenced in order to identify the errors made during gap-filling synthesis reactions. For sequence changes that yield light blue and colorless plaques, error rates were calculated by dividing the number of observed mutations of a particular type by the total number of nucleotides synthesized in the lacZ clones, a calculation used previously for particularly inaccurate polymerases (19). As described in the figure legend for Table 1, error rates were also calculated per detectable nucleotide incorporated as described (17).
Table 1.
Error Rate for pol θ
| Total changes | Error rate (×10−3)a,b | |
|---|---|---|
| Frameshifts (−1) | 112 | 1.4 |
| Frameshifts (+1) | 270 | 3.3 |
| Base substitutions | 194 | 2.4 |
| Otherc | 29 | – |
| ± >1 basesd | 16 | – |
aError rates are calculated from all observed changes (see Materials and Methods section). As mentioned in the text, mutant frequencies were calculated for two experiments. Experiment 1: mutant frequency was 27.0% (267 mutant plaques from a total of 997). Experiment 2: mutant frequency was 31.0% (873 mutant plaques from a total of 2835). Error rates are calculated from Experiment 1.
bError rates calculated for detectable changes are: frameshifts (−1) 0.9 × 10−3, frameshifts (+1) 1.9 × 10−3, base substitutions 1.4 × 10−3.
cThese errors are further discussed in the text and Table 2.
dAmong the 200 clones sequenced, several clones (13) contained deletions of two or more bases, while three clones contained insertions of two or more bases.
Processivity analysis
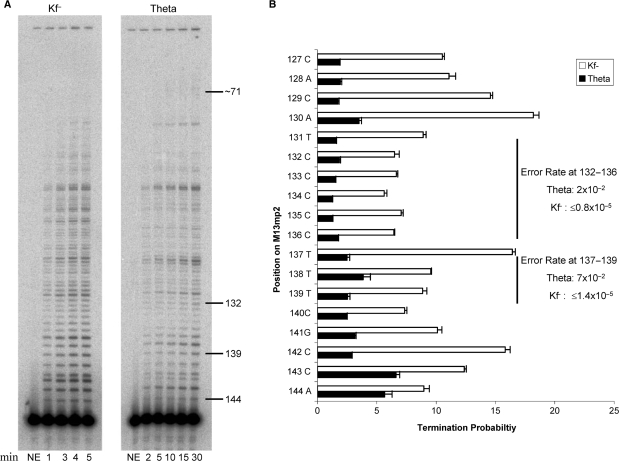
Processivity was monitored using single-stranded M13mp2 DNA primed with a [γ32P]ATP 5′-end labeled DNA oligonucleotide complementary to positions +146 to +164 of the lacZα coding sequence. Reaction mixtures (30 μl) for pol θ contained 20 mM Tris–HCl (pH 7.5), 8 mM MgCl2, 0.1 mM EDTA, 4% glycerol, 80 μg/ml bovine serum albumin and 100 μM each dATP, dCTP, dGTP and dTTP. Reaction mixtures for Klenow fragment exo−pol were as described above for pol θ except 8 mM magnesium acetate was used. Both reaction mixtures contained primed M13mp2 ssDNA substrate in excess (6.6 nM) of pol θ (2.8 nM) or Klenow fragment exo-pol (5.3 × 10−3 nM). Reaction mixtures were incubated at 37°C and 10 µl aliquots were removed at 2, 5, 10, 15 and 30 min for pol θ and at 1, 3, 4 and 5 min for Klenow fragment exo−pol. Samples were mixed in a 1:1 ratio with stop buffer (99% formamide, 5 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue) and DNA products were analyzed by electrophoresis on a 12% denaturing polyacrylamide gel. Products were quantified by phosphorimagery using a Molecular Dynamics Typhoon 9400 and the ImageQuant software. The termination probability for each template position is defined as the ratio of products of a given length to the sum of that product plus all longer DNA products.
RESULTS
Fidelity of human pol θ in the M13mp2 forward mutation assay
The fidelity of human pol θ was determined during synthesis to fill a 407-nt single-stranded gap within a circular duplex M13mp2 DNA substrate. The gap contains the lacZ α-complementation sequence that serves as the target for detecting polymerization errors. From the independent lacZ mutants sequenced, error rates have been calculated for the 12 single base–base mismatches and for addition and deletion errors, all in a variety of sequence contexts (17). This comprehensive description of pol θ error rates and error specificity can then be compared to results obtained with other DNA polymerases using the same approach.
The DNA products of complete gap filling by human pol θ [data not shown, but for example see Figure 3 in ref. (17)] yielded a lacZ mutant frequency of 27% in the first experiment and 31% in a second experiment. These frequencies are much higher than those generated by most other exonuclease-deficient family A polymerases, e.g. 0.57% for exonuclease-deficient Klenow fragment polymerase (20), 0.75% for Thermus aquaticus (Taq) polymerase (21), 1.6% for exonuclease-deficient T7 polymerase (20,21) and 0.62% for exonuclease-deficient pol γ (22). The least accurate family A member studied previously was human pol ν (23), which generated a lacZ mutant frequency of 18%. That value was obtained from reactions conducted at pH 8.8, the pH initially used to study its TLS capacity (24). Here, we decided to perform a pol ν fidelity measurement at pH 7.5, to permit direct comparison to pol θ and the other family A polymerases, all of which were analyzed at neutral pH. Consistent with previous studies showing that Klenow fragment polymerase (25) and Taq pol (21) have higher fidelity at neutral compared to alkaline pH, the products of a pol ν reaction conducted at pH 7.5 yielded a lacZ mutant frequency of 2.3%. Thus, the lacZ mutant frequencies generated by pol θ are more than 10-fold higher than for pol ν or any other exonuclease-deficient family A polymerase examined in this assay, i.e. pol θ is error-prone.
Figure 3.
Processive synthesis by Klenow exo− and pol θ. Reactions were performed as described in Materials and Methods section. (A) Representative phosphor image of the reaction products of processive DNA synthesis resolved on a 12% denaturing polyacrylamide gel. Lanes 2–5 represent primer extension products by Klenow exo− at 1, 3, 4 and 5 min. Lanes 7–11, primer extension products by pol θ at 2, 5, 10, 15 and 30 min. Lanes 1 and 6 represent negative control reactions. The numbers on the right indicate the positions along the lacZ gene. (B) Termination probability per nucleotide at each template position, 144 to 127. Numbers along the Y-axis indicate nucleotide positions along the lacZ gene. The termination probability is defined as the amount of product of a given length divided by the sum of products of that length plus all greater length products. The values plotted are the average of three values for pol θ (for time points 5, 10, and 15 min for pol θ:DNA at one ratio) and an average of three values for Klenow exo− (time points 3, 4 and 5 min at one ratio) and the error bars represent the standard deviation. Error rates for certain sequence contexts (CCCCC at positions 132–136 and TTT at positions 137–139) are shown for both pol θ and Klenow exo– (21).
Error rates and error specificity
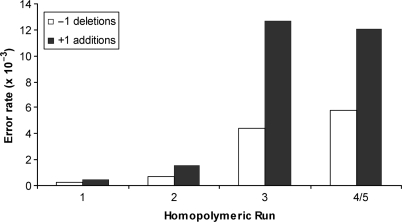
To determine the types and positions of errors made by pol, the sequence of the 407 template bases within the gap was determined for 200 independent lacZ mutants. A total of 621 sequence changes were detected, yielding an average of 3.1 sequence changes per lacZ mutant. Among these were 112 single base deletions, 270 single base additions and 194 single base substitutions (Table 1), 29 other, more complex changes (Table 2) plus 16 deletions or additions of two or more bases (Table 1). The single base changes were distributed throughout the 407 base target sequence and included changes known to result in a colorless or light blue phenotype (Figure 1, mutations in red) and changes that are known to be, or may be, phenotypically silent (Figure 1, mutations in black). When error rates were calculated from these data (as described in Materials and Methods section), overall average rates were 1.4 × 10–3 for single base deletions and 3.3 × 10–3 for single base additions (Table 1). Error rates for single base deletions and additions were sequence context-dependent, increasing as the number of bases within homopolymeric runs increased from one [i.e. a noniterated sequence to two and then three bases (Figure 2)]. There was little or no further increase in error rates in runs of four or five of the iterated bases. The overall average single base substitution error rate was 2.4 × 10–3 and error rates for the 12 individual mismatches varied over at least a 100-fold range, from ≤0.4 × 10–4 for C·dCTP to 42 × 10–4 for the T·dGTP (Table 3). Base substitution errors were also generated at higher than average rates in certain sequence contexts (arrows in Figure 1, and see Discussion section).
Table 2.
Pol θ errors involving more than one base
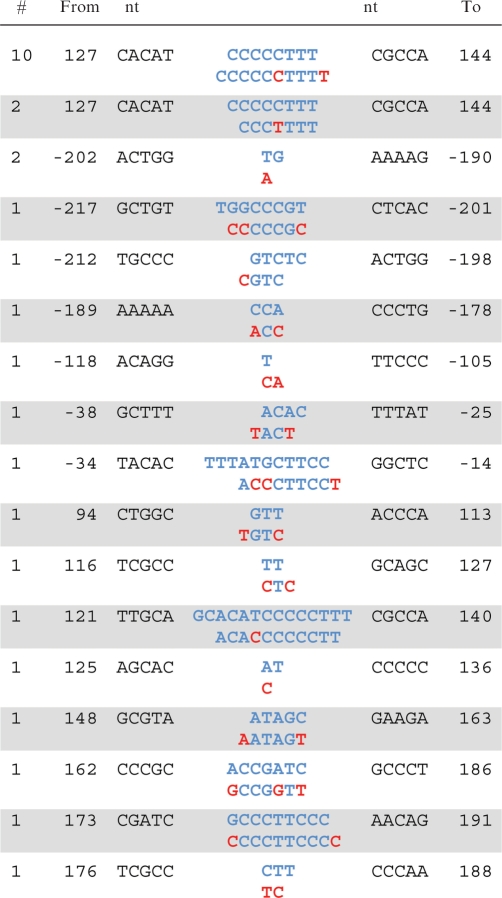 |
For each single event listed, the top sequence shows the original template sequence (Figure 1) including the five nucleotides on the 5′ and 3′ sides of the sequence that was changed. The bottom sequence shows the observed changes highlighted in red. For example, among 200 independent clones, 10 contained an insertion of a C at position 132–136 and a T at position 137–139.
Figure 1.
Spectrum of errors generated by human pol θ. The 407 template nucleotides within the single-strand gap of the M13mp2 substrate are shown as five lines of the template sequence. Letters above the target sequence indicate base substitutions. Deletion of a base is depicted by an open triangle, whereas addition of a base is represented by a closed inverted triangle above the target sequence. ‘Red’ characters represent phenotypically detectable changes in the gap region while ‘black’ characters represent phenotypically undetectable changes found in association with detectable changes. ‘Gray’ arrows indicate hot spots of template A or T base substitutions (−36, +19, +113 and +131) and base substitution hot spots (+131 and +136) +1 represents the first transcribed nucleotide of the lacZα-complementation region.
Figure 2.
Pol θ indel error rates as a function of homopolymeric run length. Error rates are the number of all observed single-base deletions and single base-additions (Table 1 and Figure 1) divided by the total number of template nucleotides present in runs of the indicated length indicated in the figure among 200 sequenced lacZ clones generated by pol θ.
Table 3.
Base substitution error rates for pol θ
| Base | Mutation From → To | Mispair Template · dNMP | All observed | Error rate (×10−4)a |
|---|---|---|---|---|
| A | A → G | A · dCMP | 11 | 5.6 |
| A → T | A · dAMP | 9 | 4.5 | |
| A → C | A · dGMP | 7 | 3.5 | |
| T | T → C | T · dGMP | 77 | 42.0 |
| T → A | T · dTMP | 24 | 13.0 | |
| T → G | T · dCMP | 4 | 2.2 | |
| G | G → A | G · dTMP | 16 | 8.4 |
| G → C | G · dGMP | 4 | 2.1 | |
| G → T | G · dAMP | 5 | 2.6 | |
| C | C → T | C · dAMP | 35 | 14.0 |
| C → G | C · dCMP | 0 | ≤ 0.4 | |
| C → A | C · dTMP | 2 | 0.8 |
Processivity
We next determined the processivity of synthesis by pol θ when copying the LacZ template sequence starting at nucleotide number 145 (where +1 is the first transcribed base of LacZ), which is adjacent to hot spots for two different single base additions and two different single base substitutions (arrows in Figure 1). Reactions were performed under conditions of primer-template excess, such that only a small proportion of primers are extended and DNA products therefore primarily reflect one cycle of synthesis (see Materials and methods section). Under these conditions, pol θ generates chains that vary in length from one to more than ∼75 nt (Figure 3A). When individual band intensities were quantified to calculate the probability of termination of processive synthesis at multiple positions, termination probabilities for pol θ ranged from 2% to 7%. At each of these positions, pol θ is less likely to terminate processive synthesis than is exonuclease-deficient Klenow fragment polymerase (Figure 3B), i.e. pol θ is slightly more processive. Termination probabilities at the hot spots for additions (CCCCC at positions 132–136 and TTT at positions 137–139) and substitutions (T to C at position 131 and C to T at position 136) were similar to those observed at other positions. The implications of these results are discussed subsequently.
Pol θ base substitution specificity and a possible role in SHM
During somatic hypermutation of immunoglobulin genes, mammalian pol η is responsible for a large proportion of mutations occurring at A-T base pairs (26–28). Even so, SHM spectra in XPV patients or mice lacking functional pol η still retain a small fraction (5–15%) of mutations at A-T bases (26,28–30) implicating another polymerase in SHM at A-T bases. This is interesting in light of the fact that 132 of 194 single base substitutions (68%) made by pol θ in vitro were generated when copying a template A or T (Table 4), including substitutions at several hot spots (arrows in Figure 1). This specificity prompted the search for a signature mutable motif for pol η-independent substitutions at A-T pairs. Using the largest available spectrum of unselected somatic mutations located in the Jh intronic region from XP-V patients (30), the sequences ADK/MHT and AA/TT (D = A/G/T; K = G/T; M = A/C; H = A/C/T; mutable positions are underlined) were found to be the most likely mutable motifs (Table 4) (31). When these motifs were compared with the distribution of base substitution errors made by pol θ, three hot spots in the pol θ error spectrum significantly correlated with the AA and ADK motifs (Table 4), consistent with a possible contribution of pol θ to somatic mutations in immunoglobulin genes in vivo (13,14). This result is further supported by the absence of correlation between the mutable motifs and spectra of somatic mutations in η–/–θ–/– mice (Table 4).
Table 4.
Mutations in different mutable motifs
| Increase in mutationsa (PW≤Wrandom value) | Pol θ hot spot | ||||||
|---|---|---|---|---|---|---|---|
| Motifs | XP-Vb | Mouse η−/−c | Pol θ in vitro | Mouse η−/−θ−/−d | GCTTTe (5 at –36) | CAATT (6 at 19) | CATCC (11 at 131) |
| ADK / MHT | 4.3 (<0.01) | 2.3 (0.02) | 0.3 (0.80) | 1.0 (0.45) | NO | YES | YES |
| 2.5 (0.02) | |||||||
| AA / TT | 3.7 (<0.01) | 2.8 (0.01) | 0.5 (0.22) | 0.8 (0.71) | YES | YES | NO |
| 3.3 (<0.01) | |||||||
aNumber of mutations in mutable motifs was calculated for the underlined bases (mutable sites). Values listed represent the fold increase in mutations at mutable sites compared with mutations at other sites. Bold italicized numbers represent a significant increase in mutations at mutable motifs (PW Wrandom 0.05), as revealed by using a Monte Carlo procedure (60).
bMutation spectra from Mayorov et al. (31).
cCombined spectra from Martomo et al. and Delbos et al. (61,62).
dMutation spectra from Masuda et al. (16).
eMutable motifs were compared with the distribution of base substitution error rates made by pol theta, three hotspots on the pol theta error spectrum (Figure 1) significantly correlated with the AA and ADK motifs.
DISCUSSION
The results presented here demonstrate that human pol θ synthesizes DNA with moderate processivity and very low fidelity, and that it has a unique error specificity. This combination of properties is unusual, and can be considered in light of mechanisms by which polymerases generate errors and in light of pol θ's; proposed biological functions.
Insertion/deletion errors
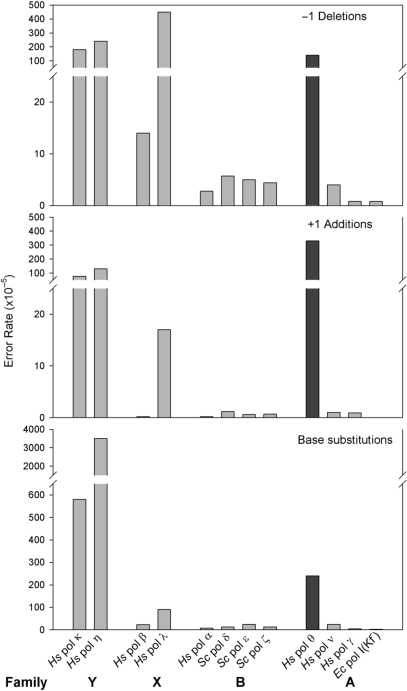
The most unusual and unanticipated aspect of pol θ's; error specificity is the high rate at which single base insertion and deletion errors (indels) are generated. In the M13 forward mutation assay, most DNA polymerases have higher error rates for base substitutions than for single base insertions and deletions, whereas pol θ generates all three types of errors at similar rates (Table 1). In fact, pol θ is among the least accurate polymerases for indels, with an average deletion rate rivaling that of the notoriously inaccurate Y family polymerases pol η and pol κ (Figure 4A), and an average addition rate exceeding that of any DNA polymerase examined in the M13 fidelity assay (Figure 4B).
Figure 4.
Pol θ error rates compared to other DNA polymerases. (A) Single-base deletions (B) Single-base additions (C) Base substitutions. Family Y: hPol κ and hPol η error rates are taken from (20). Family X: hPol β and hPol λ error rates are taken from (57). Family B: hPol α error rates are unpublished results from Kokoska, R.J. and Kunkel, T.A., yPol δ (Exo–) error rates are taken from (58), yPol ɛ (Exo–) error rates are taken from (59) and yPol ζ error rates are taken from (50). Family A: hPol θ and hPol ν data are from this study. For hPol ν, at neutral pH, a mutation frequency of 2.6% was obtained (64 mutant plaques from a total of 6432). Of these, 22 clones were sequenced and as with the previous study (24), these clones show a preference for G to A base substitutions. hPol γ (Exo–) (+p140 and p55) error rates are from (23) and Kf (Exo−) error rates are taken from (21).
What might account for such high indel error rates by pol θ? Family Y polymerases have active sites large enough to simultaneously accommodate two template bases, one of which can be unpaired (32). Because pol θ has indel error rates as high as those of family Y members, it is formally possible that pol θ also has an active site capable of accommodating an unpaired base that is simply skipped (33). The available crystal structures for family A polymerases reveal nascent base pair-binding pockets that snugly accommodate only one template. However, considering the high level of conservation between pol θ and other family A polymerases, the possibility that pol θ can accommodate two bases simultaneously in its active site is not the most likely explanation for its propensity for very high indel error rates.
Our results indicate that the majority of indels generated by pol θ occur within homonucleotide runs (Figure 1). Furthermore, addition and deletion error rates are higher for runs of three to five bases than for two-base runs or noniterated bases (Figure 2). This is a signature for misaligned primer templates (34) containing an extra base in the template strand (for deletions) or in the primer strand (for additions) that is not at the active site but is rather several correct base pairs upstream of the active site (33). It is, therefore, interesting that error rates for additions exceed those for deletions (Figure 2, black versus open bars), and that the error rate for additions is highest for runs of three identical bases and does not increase in longer runs (Figure 2). This pattern suggests that pol θ may preferentially stabilize an extra base in the primer strand located two correct base pairs upstream of the active site.
It is entirely possible that particular residues of pol θ may contribute to stabilizing misaligned intermediates, allowing pol θ to generate single base indels at unusually high rates. Sequence alignments reveal that pol θ contains extra amino acids, designated Inserts 1, 2 and 3 [see Figures 5 and 6 in ref. (7)], that are not found in other family A polymerases. While Insert 3 is unlikely to be near enough to interact with the DNA [see Figure 6 in ref. (7)], Inserts 1 and 2 may be at locations relevant to indel errors. Insert 1 is predicted to be near the tip of the polymerase thumb subdomain that in other family A members interacts with the primer template. This is interesting because deleting the tip of the thumb of Klenow fragment pol increases the error rate for indels, especially single base additions (35). Also, when T7 DNA polymerase lacks thioredoxin, its accessory subunit that interacts with the thumb, it is error-prone for single base indels, especially additions (36). An insert at this position is also found in a plant plastid A-family DNA polymerase (37). Insert 2 is located between conserved amino acids that are known to form secondary structural elements in T7 pol and Taq pol. It is interesting to note that some of these residues interact with the primer strand upstream of the polymerase active site, where the extra base in a misaligned addition intermediate may reside. Interestingly, Inserts 1 and 2 are unique to pol θ in comparison to its closest homolog, whose indel error rates are much lower (Figure 4). Thus, it is conceivable that residues in Insert 1 or 2 of pol θ stabilize misaligned intermediates, allowing pol θ to generate single base indels at unusually high rates. Precedent for this comes from structural studies of family X pol λ (38) showing that specific amino acid side chains interact with an extrahelical template strand base just upstream of the active site, in a manner suggested to preferentially contribute to single base deletions in two-base runs.
The rate and specificity of indel errors are also relevant to how misalignments may form during synthesis by pol θ. As mentioned above, one perhaps unlikely possibility is that the misalignment arises within the polymerase active site, with the unpaired intermediate possibly stabilized by pairing of a correct incoming dNTP with the next template base (39,40). Another possibility is that misalignment initiates with dNTP misinsertion, followed by primer relocation to generate a misaligned intermediate with a correctly paired primer terminus (33). Although these possibilities cannot be excluded, they seem less likely because pol θ indel error rates in certain sequence contexts (Figure 1) are extraordinarily high (e.g. 2–7%, Figure 3B) and often exceed base substitution error rates (Table 3). A third possibility, suggested by earlier studies of other polymerases (33), is that DNA misalignments arise as the polymerase dissociates and/or re-associates with the primer template. In order to test this possibility we examined the processivity of DNA synthesis by pol θ as it copied the lacZ template nucleotides that were hot spots for single base additions at a CCCCC run (position 132–136) and a TTT run (position 137–139). Site-specific termination probabilities at these two hot spots are between 1% and 4% (Figure 3B), similar to the respective 2% and 7% error rates for additions within these runs (Figure 3B). This implies that essentially all cycles of dissociation–reassociation would need to result in misalignments that were ultimately extended, in order to account for the extraordinary pol θ addition rates. This could be so, but would be unprecedented, because previous studies of other polymerases showed that at least 100 dissociation–reassociation cycles were observed for each addition error generated (41). Thus there may be additional opportunities for misalignments to form even during processive synthesis by pol θ. One possibility is suggested by the fact that forming an addition intermediate within a homonucleotide run requires (on average) disruption of one more correct base pair than does formation of a deletion intermediate (42). This energetic difference is consistent with the observation that addition rates of numerous DNA polymerases are consistently lower than are deletion rates [Figure 4 and (33)]. Here, we observe that pol θ is an exception; it is the only polymerase that generates additions at a higher average rate than deletions. One possible explanation is that during synthesis by pol θ, fraying at the primer terminus may involve multiple bases, such that subsequent re-annealing could provide a similar opportunity for forming either an addition or a deletion intermediate. Fraying of multiple base pairs might occur during movement of the primer strand between two separate DNA binding sites. This was previously proposed to account for the difference in indel error rates between the large Klenow fragment of E. coli DNA polymerase I, which can partition the primer strand between the polymerase active site and the exonuclease active site, and a derivative containing the polymerase domain alone, i.e. one completely lacking the 3′ exonuclease domain (20). The idea is nicely articulated in the melting-misalignment model of Maki and colleagues (43,44), who proposed that the tendency of E. coli DNA polymerase III to generate single base additions (albeit at much lower rates than seen here), results from misalignments that arise during partitioning of the primer strand between the polymerase and proofreading exonuclease active sites. Unlike E. coli pol III, recombinant pol θ lacks a detectable proofreading exonuclease activity (6), but the enzyme purified from human HeLa cells has been reported to have an associated 3′ exonuclease (9). It is also possible that the pol θ catalytic subunit itself could have a second DNA-binding site encoded by residues in its large open reading frame, which contains helicase motifs that are unique to pol θ in comparison to other polymerases, including its closest but more accurate homolog, pol ν.
Base substitution errors
The observation that the average base substitution error rate of human pol θ is high (Figure 4C) is consistent with low nucleotide selectivity (7) and promiscuoity in extending mismatched primer termini (8). Pol θ's; closest homolog, pol ν, also generates base substitutions at unusually high rates in polymerization reactions conducted at pH 8.8 (23), a condition where the enzyme has maximum activity (24). Here we compared the fidelity of the two enzymes in reaction mixtures at pH 7.5, and pol θ was about 10-fold less accurate than pol ν (240 × 10−5 versus 24 × 10−5, Figure 4). This difference is seen despite the strong conservation of key residues in and around the putative nascent base pair-binding pockets of pol θ and pol ν (see Figure 7 in Supplementary Data in ref. 45). Pol θ's remarkable infidelity among family A members is further highlighted by comparison to the exonuclease-deficient derivative of Klenow fragment polymerase, which is fully 100-fold more accurate (Figure 4, substitution error rate of 2.5 × 10−5). It is particularly interesting that pol θ is almost as inaccurate as pol κ (Figure 4), a family Y polymerase whose members are characterized by open, solvent accessible active sites. Pol θ generates many of the 12 possible base-base mismatches at high rates (Table 3), including three pyrimidine–pyrimidine mismatches that may be solvated (46). These facts imply that the active site of pol θ may be more solvent accessible than is typical of accurate family A polymerases known to have closed, relatively solvent inaccessible active sites.
Biological implications
The low fidelity of pol θ when copying undamaged DNA is generally consistent with its proposed role in lesion bypass [e.g. of AP sites, (7)], similar to several other DNA polymerases (e.g. pol η) that are implicated in bypass of lesions that perturb DNA helix geometry, which also synthesize undamaged DNA with low fidelity. The high-base substitution error rates of human pol θ and its error specificity in comparison to the SHM specificity at A-T base pairs in humans and mice lacking functional pol η (Table 4) are also consistent with previous reports implicating pol θ in SHM of immunoglobulin genes (13,14,47).
As is typical of family A members, human pol θ is more processive than are certain other polymerases (pol η, pol ι, some bacterial pols) implicated in TLS. In fact, at all template positions examined here (Figure 3B), pol θ is slightly more processive than Klenow fragment polymerase, and can even generate chains exceeding 71 nt in length during a single cycle of processive synthesis. The combination of low fidelity and moderate processivity by human pol θ is shared by both pol κ (48) and by pol ζ (49,50). The latter two enzymes have been implicated in TLS, reputedly as promiscuous mismatch extenders (51). Perhaps relevant to their moderate processivity, pol κ and pol ζ are implicated in DNA transactions that may require filling of gaps of about 30 nt. For example, pol κ has recently been implicated in nucleotide excision repair of UV photoproducts (52) and pol ζ is reported to have a role in gap filling associated with repair of interstrand crosslinks (53,54). In like manner, the processivity of pol θ would make it well suited for specialized transactions requiring more extensive DNA synthesis, e.g. SHM downstream of the initiating deamination event, during a subpathway of base excision repair (55) or nucleotide excision repair, or perhaps during reactions involving the helicase motifs of pol θ. The very low fidelity but moderate processivity of pol θ may also be relevant to the recovery of lacZ mutants containing multiple single base changes within a few nucleotides of each other (Table 2). Such ‘complex’ events are also generated by pol ζ (49) which, as mentioned earlier, shares with pol θ the properties of processive but inaccurate synthesis.
Although the low fidelity and moderate processivity of human pol θ is potentially beneficial for certain specialized DNA transactions, this combination could be adversely mutagenic if pol θ synthesized DNA at the wrong time or place. For example, one study reports that expression of pol θ mRNA is upregulated in human cancers, and that patients expressing high levels of pol θ have poorer clinical prognoses than do patients expressing lower levels of pol θ (12). Given its low fidelity and moderate processivity, pol θ activity may be tightly regulated, which could occur at any of several levels. For example, pol θ is most highly expressed in the germinal centers of splenic B cells where antibody maturation takes place (6,11,12). By analogy to an elegant recent study demonstrating that UmuD and RecA proteins modulate the mutagenic potential and the fidelity of the bacterial family Y DNA polymerase DinB (56), accessory proteins may exist that interact with and regulate pol θ, perhaps decreasing its potential for indel mutagenesis.
ACKNOWLEDGEMENTS
We thank the NIEHS DNA Sequencing Core Facility for expert assistance in the DNA sequence analysis of lacZ mutants, Samuel Berchuck for his contribution in this project (Summers of Discovery) and Drs Katarzyna Bebenek, William C. Copeland, Stephanie Nick McElhinny and Kei-ichi Takata for reading and providing thoughtful comments on the article. This research was supported in part by Research Project Number Z01-ES065070 to TAK the Intramural Research Program of the National Institutes of Health, National Library of Medicine to I.B.R. and National Institute of Environmental Health Sciences to T.A.K., and in part a National Institutes of Health grant (CA101980) to R.D.W., and by the University of Pittsburgh Cancer Institute. Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program, NIH, NIEHS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hanawalt PC. Paradigms for the three rs: DNA replication, recombination, and repair. Mol. Cell. 2007;28:702–707. doi: 10.1016/j.molcel.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 2.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd. ASM Press, Washington, DC; 2006. [Google Scholar]
- 4.Shcherbakova PV, Fijalkowska IJ. Translesion synthesis DNA polymerases and control of genome stability. Front. Biosci. 2006;11:2496–2517. doi: 10.2741/1985. [DOI] [PubMed] [Google Scholar]
- 5.Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. doi: 10.1006/geno.1999.5843. [DOI] [PubMed] [Google Scholar]
- 6.Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair. 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maga G, Shevelev I, Ramadan K, Spadari S, Hubscher U. DNA polymerase theta purified from human cells is a high-fidelity enzyme. J. Mol. Biol. 2002;319:359–369. doi: 10.1016/S0022-2836(02)00325-X. [DOI] [PubMed] [Google Scholar]
- 10.Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol. Cell. Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, et al. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 13.Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, O-Wang J. DNA polymerase theta contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc. Natl Acad. Sci. USA. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, Kurosaki T, O-Wang J. Absence of DNA polymerase theta results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair. 2006;5:1384–1391. doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, O-Wang J. DNA polymerases eta and theta function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J. Biol. Chem. 2007;282:17387–17394. doi: 10.1074/jbc.M611849200. [DOI] [PubMed] [Google Scholar]
- 16.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 18.Abbotts J, Bebenek K, Kunkel TA, Wilson SH. Mechanism of HIV-1 reverse transcriptase. Termination of processive synthesis on a natural DNA template is influenced by the sequence of the template-primer stem. J. Biol. Chem. 1993;268:10312–10323. [PubMed] [Google Scholar]
- 19.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J. Mol. Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 20.Bebenek K, Joyce CM, Fitzgerald MP, Kunkel TA. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 1990;265:13878–13887. [PubMed] [Google Scholar]
- 21.Eckert KA, Kunkel TA. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990;18:3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J. Biol. Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- 23.Arana ME, Takata K, Garcia-Diaz M, Wood RD, Kunkel TA. A unique error signature for human DNA polymerase nu. DNA Repair. 2007;6:213–223. doi: 10.1016/j.dnarep.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 25.Eckert KA, Kunkel TA. Effect of reaction pH on the fidelity and processivity of exonuclease-deficient Klenow polymerase. J. Biol. Chem. 1993;268:13462–13471. [PubMed] [Google Scholar]
- 26.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 27.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat. Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 28.Faili A, Aoufouchi S, Weller S, Vuillier F, Stary A, Sarasin A, Reynaud CA, Weill JC. DNA polymerase eta is involved in hypermutation occurring during immunoglobulin class switch recombination. J. Exp. Med. 2004;199:265–270. doi: 10.1084/jem.20031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng X, Negrete GA, Kasmer C, Yang WW, Gearhart PJ. Absence of DNA polymerase eta reveals targeting of C mutations on the nontranscribed strand in immunoglobulin switch regions. J. Exp. Med. 2004;199:917–924. doi: 10.1084/jem.20032022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayorov VI, Rogozin IB, Adkison LR, Gearhart PJ. DNA polymerase eta contributes to strand bias of mutations of A versus T in immunoglobulin genes. J. Immunol. 2005;174:7781–7786. doi: 10.4049/jimmunol.174.12.7781. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharya P, Grigera F, Rogozin IB, McCarty T, Morse H.C., III, Kenter AL. Identification of murine B cell lines that undergo somatic hypermutation focused to A:T and G:C residues. Eur. J. Immunol. 2008;38:227–239. doi: 10.1002/eji.200737664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Diaz M, Kunkel TA. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb. Symp. Quant. Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Minnick DT, Astatke M, Joyce CM, Kunkel TA. A thumb subdomain mutant of the large fragment of Escherichia coli DNA polymerase I with reduced DNA binding affinity, processivity, and frameshift fidelity. J. Biol. Chem. 1996;271:24954–24961. doi: 10.1074/jbc.271.40.24954. [DOI] [PubMed] [Google Scholar]
- 36.Kunkel TA, Patel SS, Johnson KA. Error-prone replication of repeated DNA sequences by T7 DNA polymerase in the absence of its processivity subunit. Proc. Natl Acad. Sci. USA. 1994;91:6830–6834. doi: 10.1073/pnas.91.15.6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeuchi R, Kimura S, Saotome A, Sakaguchi K. Biochemical properties of a plastidial DNA polymerase of rice. Plant Mol. Biol. 2007;64:601–611. doi: 10.1007/s11103-007-9179-2. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 39.Hashim MF, Schnetz-Boutaud N, Marnett LJ. Replication of template-primers containing propanodeoxyguanosine by DNA polymerase beta. Induction of base pair substitution and frameshift mutations by template slippage and deoxynucleoside triphosphate stabilization. J. Biol. Chem. 1997;272:20205–20212. doi: 10.1074/jbc.272.32.20205. [DOI] [PubMed] [Google Scholar]
- 40.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. Abasic translesion synthesis by DNA polymerase beta violates the ‘A-rule’. Novel types of nucleotide incorporation by human DNA polymerase beta at an abasic lesion in different sequence contexts. J. Biol. Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 41.Bebenek K, Abbotts J, Wilson SH, Kunkel TA. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J. Biol. Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 42.Kunkel TA, Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim. Biophys. Acta. 1988;951:1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 43.Fujii S, Akiyama M, Aoki K, Sugaya Y, Higuchi K, Hiraoka M, Miki Y, Saitoh N, Yoshiyama K, Ihara K, et al. DNA replication errors produced by the replicative apparatus of Escherichia coli. J. Mol. Bio. 1999;289:835–850. doi: 10.1006/jmbi.1999.2802. [DOI] [PubMed] [Google Scholar]
- 44.Seki M, Akiyama M, Sugaya Y, Ohtsubo E, Maki H. Strand asymmetry of +1 frameshift mutagenesis at a homopolymeric run by DNA polymerase III holoenzyme of Escherichia coli. J. Biol. Chem. 1999;274:33313–33319. doi: 10.1074/jbc.274.47.33313. [DOI] [PubMed] [Google Scholar]
- 45.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 46.Kool ET. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 47.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger Iii AJ, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J. Biol. Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 49.Zhong X, Garg P, Stith CM, Nick McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garg P, Stith CM, Majka J, Burgers PM. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase zeta. J. Biol. Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- 51.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 52.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 53.McHugh PJ, Sarkar S. DNA interstrand cross-link repair in the cell cycle: a critical role for polymerase zeta in G1 phase. Cell Cycle. 2006;5:1044–1047. doi: 10.4161/cc.5.10.2763. [DOI] [PubMed] [Google Scholar]
- 54.Shen X, Jun S, O’Neal LE, Sonoda E, Bemark M, Sale JE, Li L. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA) J. Biol. Chem. 2006;281:13869–13872. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- 55.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, et al. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol. Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Godoy VG, Jarosz DF, Simon SM, Abyzov A, Ilyin V, Walker GC. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell. 2007;28:1058–1070. doi: 10.1016/j.molcel.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J. Biol. Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 58.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 59.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J. Biol. Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 60.Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 61.Martomo SA, Yang WW, Wersto RP, Ohkumo T, Kondo Y, Yokoi M, Masutani C, Hanaoka F, Gearhart PJ. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc. Natl Acad. Sci. USA. 2005;102:8656–8661. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]