Abstract
[Fe2L3]4+ (L = C25H20N4) is a synthetic tetracationic supramolecular cylinder (with a triple helical architecture) that targets the major groove of DNA and can bind to DNA Y-shaped junctions. To explore the DNA-binding mode of [Fe2L3]4+, we examine herein the interactions of pure enantiomers of this cylinder with DNA by biochemical and molecular biology methods. The results have revealed that, in addition to the previously reported bending of DNA, the enantiomers extensively unwind DNA, with the M enantiomer being the more efficient at unwinding, and exhibit preferential binding to regular alternating purine–pyrimidine sequences, with the M enantiomer showing a greater preference. Also, interestingly, the DNA binding of bulky cylinders [Fe2(L-CF3)3]4+ and [Fe2(L-Ph)3]4+ results in no DNA unwinding and also no sequence preference of their DNA binding was observed. The observation of sequence-preference in the binding of these supramolecular cylinders suggests that a concept based on the use of metallosupramolecular cylinders might result in molecular designs that recognize the genetic code in a sequence-dependent manner with a potential ability to affect the processing of the genetic code.
INTRODUCTION
Supramolecular chemistry focuses on the design of complex chemical systems from components, which interact through noncovalent intermolecular forces (1). Application of metallosupramolecular chemical design has resulted in a wide range of interesting multicomponent coordination structures generated by self-assembly from suitably designed ligands and specific metal ions (2–4). In the past few years, much effort has been devoted to the construction of new supramolecular assemblies through metal coordination. An example is a synthetic tetracationic supramolecular cylinder (with a triple helical architecture), [Fe2L3]4+ (L = C25H20N4) that targets the major groove of DNA (5–7). A significance of such compounds, particularly those that recognize the genetic code in a sequence-dependent manner, lies in their potential ability to affect the processing of the genetic code. Indeed, synthetic agents that target DNA with recognition through non-covalent surface motifs have the potential to be a powerful new tool for molecular biology and genetics. These facts warrant systematic examinations of the DNA binding of such compounds.
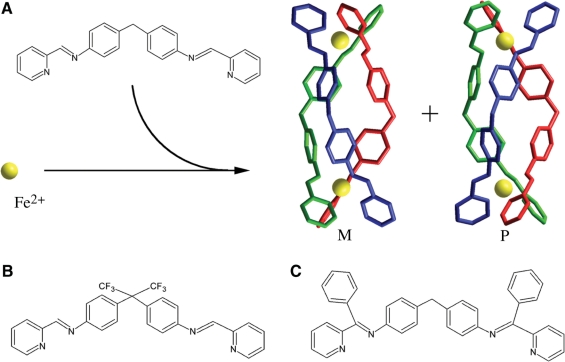
As illustrated in Figure 1A, the complex [Fe2L3]4+ is a chiral Tris-chelate system. This tetracationic helicate (of ∼2 nm length and ∼1 nm diameter) has been shown to bind strongly and noncovalently to major groove of DNA (5,7,8) and can bind at the heart of three-way (Y-shaped) DNA junctions (9–11). The helicate also induces unexpected and quite dramatic intramolecular DNA coiling, giving rise to small coils of DNA (5,7,8). In addition, [Fe2L3]4+ was found to bend DNA by 45° ± 15° per ligand (7).
Figure 1.
The molecular structures of the ligands in the metallosupramolecular cylinders used in this work. (A) Structure of L (L = C25H20N4) and 3D structures of M-[Fe2L3]4+ and P-[Fe2L3]4+; these 3D structures are based on structures obtained by X-ray crystallography (28,29). (B) L-CF3. (C) L-Ph.
By convention the left handed or negatively twisted helicate is labelled (M) and the right handed or positively twisted helicate labelled (P). It has been previously shown (5) that there are differences in DNA binding of the [Fe2L3]4+ enantiomers. The M enantiomer induces more dramatic intramolecular DNA coiling and bending than P enantiomer. To explore DNA-binding mode of [Fe2L3]4+ further, we examined in the present work the interactions of pure enantiomers with DNA by biochemical and molecular biology methods. The results have revealed that, in addition to the previously reported bending of DNA, the enantiomers extensively unwind DNA, with the M enantiomer being the more efficient at unwinding, and exhibit preferential binding to regular alternating purine–pyrimidine sequences, with the M enantiomer showing a greater preference. Also interestingly, the DNA binding of bulky cylinders [Fe2(L-CF3)3]4+ and [Fe2(L-Ph)3]4+ (Figure 1B and C) results in no DNA unwinding and also no sequence preference of their DNA binding was observed.
MATERIALS AND METHODS
Starting materials
The synthesis of the cylinder [Fe2L3]4+ (L = C25H20N4; Figure 1A) has been described previously (5,7,8). The cylinders [Fe2(L-CF3)3]Cl4 (Figure 1B) and [Fe2(L-Ph)3]Cl4 (Figure 1C) also followed these previously outlined procedures. Stock aqueous solutions of the racemic and enantiomeric helicates were prepared and all DNA-binding experiments were carried out in the same way as in previously published article (12). Calf thymus (ct) DNA (42% G + C, mean molecular mass ca. 2 × 107) was also prepared and characterized as described previously (13). Plasmids pUC19 (2686 bp) and pSP73 (2464 bp) were isolated according to standard procedures. Poly(dG-dC) and poly(dA-dT) were obtained from Amersham Pharmacia-Biotech (Piscataway, NJ, USA) and were used without further purification. The polynucleotides were dissolved in 10 mM NaCl and kept frozen until the day of the experiment. The DNA concentrations (moles of bases per liter) of all polynucleotides were determined spectroscopically by using the published molar extinction coefficients at the maximum of the long wavelength absorbance (5). The synthetic oligodeoxyribonucleotides used in this work were purchased from VBC-genomics (Vienna, Austria). The purity of oligonucleotides was verified by high-pressure liquid chromatography (HPLC) or gel electrophoresis. T4 polynucleotide kinase, NdeI, HindIII, Bst1107 I and PvuII restriction endonucleases were purchased from New England Biolabs (Beverly, MA, USA). [γ-32P]-ATP and [α-32P]-dATP were from MP Biomedicals, LLC (Irvine, CA, USA). Acrylamide and bis(acrylamide) were from Merck KgaA (Darmstadt, Germany). The Klenow fragment from DNA polymerase I (exonuclease minus, mutated to remove the 3′–5′ proofreading domain) (KF−) was purchased from Takara (Japan). Agarose was from FMC BioProducts (Rockland, ME, USA). Wizard SV and PCR Clean-Up System used to extract and purify 158-bp DNA fragment (vide infra) was purchased from Promega (Madison, WI, USA). Ethidium bromide (EtBr) was from Merck KGaA and deoxyribonuclease I (DNaseI) was from Roche (Mannheim, Germany).
Unwinding of negatively supercoiled DNA
Unwinding of closed circular supercoiled pUC19 plasmid DNA was assayed by an agarose gel mobility shift assay (14). The unwinding angle Φ, induced per cylinder bound to DNA was calculated upon the determination of the cylinder:base ratio at which the complete transformation of the supercoiled to relaxed form of the plasmid was attained. An aliquot of the sample was subjected to electrophoresis on 1% native agarose gel running at 25°C in the dark with TAE (Tris–aceate/EDTA) buffer and the voltage set at 25 V. The gels were then stained with EtBr, followed by photography with transilluminator.
Competition assays
The competition assays were all undertaken with fixed DNA and competitor (EtBr) concentrations and variable helicate. The DNA–EtBr complexes were excited at 546 nm and the fluorescence was measured at 595 nm. To the solution of EtBr and DNA (10 mM Tris pH 7.4, 1 mM EDTA, 1.3 µM EtBr and 3.9 µM DNA) were added aliquots of a 1 mM stock solution of the cylinders and the fluorescence was measured after each addition until the fluorescence was reduced to 50%. In general, the experiments were designed so that the weaker binder was displaced by the stronger one. The apparent binding constants (Kapp) for both enantiomers were calculated from KEB × [EB] = Kapp × [drug], where [EB] is the concentration of EtBr (1.3 µM), [drug] is the concentration of cylinders at a 50% reduction of fluorescence and KEB is known (KEB = 1 × 107 M−1 for ct-DNA; 9.5 × 106 M−1 for poly(dA-dT)2, and 9.9 × 106 M−1 for poly(dG-dC)2) (15).
DNase I footprinting
One subclass of footprinting agents that has been developed for determining the sequence-specific binding of small molecules to DNA comprises enzymes such as deoxyribonuclease I (DNase I) (16). DNase I is an endonuclease that specifically cleaves the O3′–P bond of the phosphodiester backbone of the double-helical DNA substrate. Supercoiled pSP73 plasmid was digested with NdeI or HindIII restriction endonuclease and 3′-end-labeled by treatment with KF− and [α-32P]-dATP. After radioactive labeling, the DNA cleaved first with NdeI was still digested with HindIII, whereas that cleaved first with HindIII was subsequently digested with NdeI. Both ways of the cleavage resulted in 158 and 2306 bp fragments; if the first cleavage was carried out by NdeI the 158 bp fragment was 3′-end-labeled at the bottom strand, whereas if the first cleavage was carried out by HindIII the 158 bp fragment was 3′-end-labeled at the top strand. The 158 bp fragments were purified by 1% agarose gel electrophoresis and isolated from the gel by Promega Wizard SV Gel clean-up system. The solution (9 μl) containing 1.11 × TKMC buffer (10 mM Tris pH 7.9, 10 mM KCl, 10 mM MgCl2 and 5 mM CaCl2), 4.5 × 10−4 M DNA, and cylinder was incubated for 15 min at 25°C. Cleavage was initiated by the addition of 1 μl of 50 μg DNase I/ml and allowed to react for 30 s at room temperature before quenching with 2.5 μl of DNase I stop solution (3 M NH4OAc and 0.25 M EDTA). Optimal enzyme dilutions were established in preliminary calibration experiments. The sample was then precipitated with ethanol, lyophilized, resuspended in a formamide loading buffer. DNA cleavage products were resolved by polyacrylamide (PAA) gel electrophoresis under denaturing conditions (13%/8 M urea PAA gel). The autoradiograms were visualized and quantified by using the bio-imaging analyzer. Assignment of the cleavage to a particular base has been made so that it corresponds to the cleavage of the phosphodiesteric bond on the 5′ side of that base.
Cleavage of 28-bp DNA duplex by restriction endonucleases
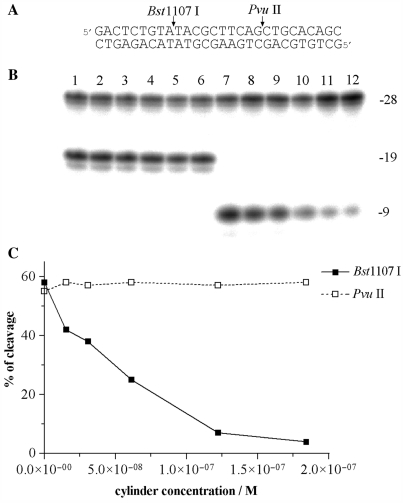
The 28-mer duplex [its nucleotide sequence (Figure 6A) contained restriction cleavage sites for Bst1107 I and PvuII restriction endonucleases] at the concentration of 6.1 × 10−8 M was mixed with the cylinder at various cylinder:duplex ratios and then digested by Bst1107 I or PvuII (1 unit) in the cleavage buffers recommended by the manufacturer. The total volume of the reaction was 10 μl. The products of the cleavage were analyzed by 24% PAA/8M urea denaturing gel electrophoresis.
Figure 6.
Cleavage of 28-mer duplex in the presence of M-[Fe2L3]4+ by Bst1107 I and PvuII. (A) Sequence of the oligonucleotide duplex and cleavage sites. Labelling was performed with [γ-32P]ATP at 5′-terminus of the upper strand. Cleavage by Bst1107 I and PvuII gives rise to 9-mer and 19-mer products as indicated on the right side of the gel. (B) Autoradiogram of the 24% PAA/8M urea denaturing gel. Lanes 1–6, 28-mer cleaved by PvuII; lanes 7–12, 28-mer cleaved by Bst1107 I; lanes 1 and 7, 28-mer in the absence of the cylinder; lanes 2–6 and 8–12, 28-mer mixed with M-[Fe2L3]4+ at 0.25:1, 0.5:1, 1:1, 2:1 and 3:1 (cylinder:duplex) ratios, respectively. The concentration of the duplex was 6.1 × 10−8 M. (C) Cleavage of the 28-mer duplex as a function of the concentration of the cylinder.
Other physical methods
Absorption spectra were measured with a Varian Cary 4000 UV-VIS spectrophotometer equipped with a thermoelectrically controlled cell holder and quartz cells with the pathlength of 1 cm. Fast protein liquid chromatography purification was carried out on a Pharmacia Biotech FPLC System with a MonoQ HR 5/5 column. The gels were visualized by using the BAS 2500 FUJIFILM bioimaging analyzer, and the radioactivities associated with bands were quantitated with the AIDA image analyzer software (Raytest, Germany). Fluorescence measurements were performed on a Varian Cary Eclipse spectrofluorophotometer using a 1 cm quartz cell at room temperature.
RESULTS
DNA interactions of [Fe2L3]4+ (L = C25H20N4)
Unwinding of negatively supercoiled DNA
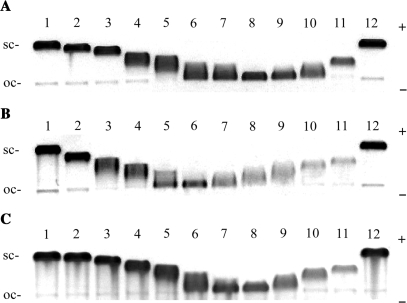
Figure 2 shows electrophoresis gels in which a mixture of relaxed and negatively supercoiled pUC19 plasmid DNA has been treated with increasing amounts of M-[Fe2L3]4+, P-[Fe2L3]4+ and rac-[Fe2L3]4+. The unwinding angle is given by Φ = −18σ/R(c), where σ is the superhelical density and R(c) is the number of cylinders bound per nucleotide at which the supercoiled and relaxed forms co-migrate. The number of cylinders bound per nucleotide was taken to be equal to the mixing ratios based on the assumption that all cylinders present in the sample are completely bound to the DNA. This assumption is substantiated by high values of the apparent binding constants (Kapp) 4.6 × 107 and 7.6 × 107 M−1 determined for binding of the M or P enantiomer (Table 1), respectively to DNA with a random nucleotide sequence (vide infra) (a strong association is characterized by a Kapp > 106 M−1 (17)). The high value of Kapp and the low ratio of cylinder per DNA molecule in the experiment implies that at equilibrium there will be almost no free cylinder. Under the present experimental conditions, σ was calculated to be –0.058 on the basis of the data of cisplatin for which the R(c) was determined in this study and the literature value of Φ = 13° was assumed (14). Using this approach, DNA unwinding angles of 32 ± 3°, 22 ± 3°, and 27 ± 3° were determined for M-[Fe2L3]4+, P-[Fe2L3]4+ and rac-[Fe2L3]4+, respectively.
Figure 2.
Unwinding of negatively supercoiled pUC19 plasmid DNA by M-[Fe2L3]4+ (A), P-[Fe2L3]4+ (B) and rac-[Fe2L3]4+ (C). The plasmid at the concentration of 2 × 10−4 M (this concentration is related to the monomeric nucleotide content) was mixed with increasing concentrations of the cylinders in 10 mM Tris·HCl (pH 7.4) incubated for 30 min at 25°C and analysed on 1% agarose gel. Lanes 1 and 12, control (non-modified DNA); (A) lanes 2–11, cylinder:base ratio = 0.011, 0.017, 0.022, 0.025, 0.028, 0.030, 0.033, 0.039, 0.044, 0.055, respectively; (B) lanes 2–11, cylinder:base ratio = 0.021, 0.032, 0.037, 0.043, 0.048, 0.054, 0.059, 0.064, 0.075, 0.086, respectively; (C) lanes 2–11, rb = 0.008, 0.015, 0.023, 0.027, 0.030, 0.034, 0.038, 0.046, 0.053, 0.061, respectively. The sc and oc (open circle) indicate supercoiled, and relaxed (nicked) forms of plasmid DNA.
Table 1.
Apparent binding constants, Kapp (×106 M−1)
| Compound | ct-DNA | poly(dA-dT)2 | poly(dG-dC)2 |
|---|---|---|---|
| M-[Fe2L3]4+ | 46 | 106 | 77 |
| P-[Fe2L3]4+ | 76 | 142 | 89 |
| [Fe2(L-CF3)3]4+ | 10 | ND | ND |
| [Fe2(L-Ph)3]4+ | 11 | ND | ND |
ND, not determined.
EtBr displacement
The binding strength of the cylinders to DNAs cannot be assessed directly by equilibrium dialysis due to the affinity of the cylinder for the cellulose used in dialysis membranes. The quantification of binding strength of the M and P enantiomers of [Fe2L3]4+ to ct-DNA, poly(dA-dT)2 and poly(dG-dC)2 was therefore investigated by the competition between cylinders and EtBr. Although not ideal (since it probes displacement of an intercalator by a groove binder) the experiment should allow a comparison of the affinity of each enantiomer towards different DNAs. Displacement of EtBr from all studied DNAs was accompanied by a decrease in the fluorescence intensity measured at 595 nm. The apparent binding constants (Kapp) for both enantiomers calculated as described in the experimental part are shown in Table 1.
DNase I footprinting
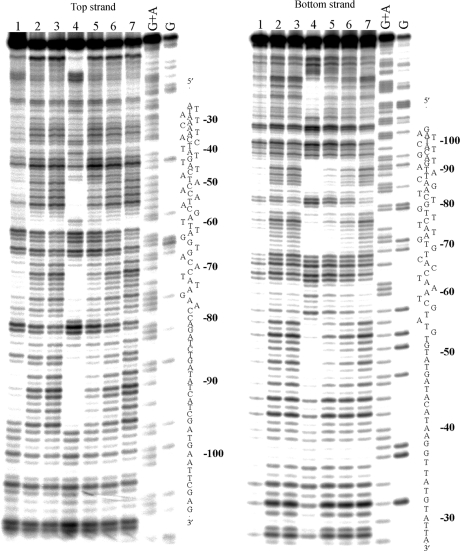
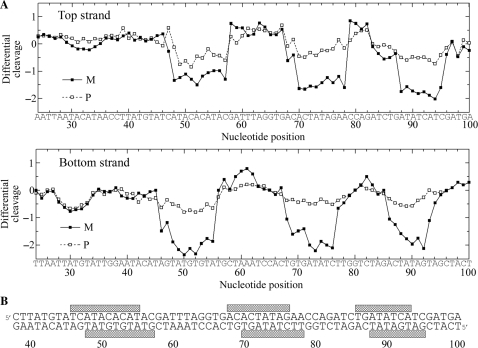
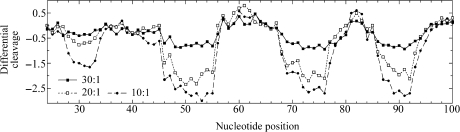
The M and P enantiomers of [Fe2L3]4+ were mixed with the 158 bp restriction fragment of pSP73 at 10:1, 20:1 and 30:1 (base:cylinder) ratios followed by partial cleavage by DNase I. The autoradiograms of the DNA cleavage inhibition patterns are shown in Figure 3. The extent of DNase I cleavage varies along the DNA sequence and the cutting is strongly reduced in several places even in the absence of the cylinders (Figure 3, lanes 7). In the presence of the cylinders, several footprints in the gel can be seen, showing that both enantiomers are capable of recognizing specific DNA sequences. It is also clear that P enantiomer is less specific and that much higher concentration of P-[Fe2L3]4+ must be used to see an extent of inhibition similar to that exhibited by the M enantiomer. In order to obtain a better comparison of the binding specificity of both enantiomers, intensities from gel lanes containing DNA and M or P enantiomer at 20:1 (base:cylinder) ratio (in Figure 3 and from other gels not shown) were measured by densitometry and resulting differential cleavage plots are shown in Figure 4A. Ratio of 20:1 was chosen as an optimum after comparison of differential cleavage plots of M-[Fe2L3]4+ at 10:1, 20:1 and 30:1 ratios (Figure 5). Negative values indicate sites of drug protection from DNase I cleavage and positive values regions of drug-induced enhancement of cleavage. A stretch of DNA of about 70 bp within the 158-mer restriction fragment was sufficiently well resolved to provide quantitative data. Both enantiomers exhibit similar patterns of protection and enhancement, but the extent of protection by P enantiomer is much weaker. Three main sites protected by the M enantiomer are clearly seen. They extend over about 8–10 bp, around positions 52, 73 and 91 and consist mainly of regular alternating purine–pyrimidine sequences.
Figure 3.
Autoradiograms of DNase I footprint of 3′ end labeled top (left) or bottom (right) strand of 158-mer HindIII/NdeI restriction fragment of the plasmid pSP73 in the presence of different concentrations of P-[Fe2L3]4+ (lanes 1–3) and M-[Fe2L3]4+ (lanes 4–6). Lanes 1–3; DNA mixed with P-[Fe2L3]4+ at 10:1, 20:1, 30:1 (base:cylinder) ratios, respectively; lanes 4–6; DNA mixed with M-[Fe2L3]4+ at 10:1, 20:1, 30:1 ratios, respectively; lane 7; DNA in the absence of cylinders; lanes G + A and G correspond to Maxam–Gilbert G + A and G ladders. The nucleotide sequence of the fragment is shown on the right side of the gels and numbers refer to the sequence shown in the corresponding differential cleavage plots in Figure 4A.
Figure 4.
(A) Differential cleavage plots for M-[Fe2L3]4+ (full squares) and P-[Fe2L3]4+ (open squares) induced differences in susceptibility to DNase I digestion on top and bottom strands of the 158-mer HindIII/NdeI fragment of the plasmid pSP73 at 20:1 (base:cylinder) ratio. Vertical scales are in units of ln(fc) – ln(f0), where fc is the fractional cleavage at any bond in the presence of cylinder and f0 is the fractional cleavage of the same bond in the control, given closely similar extents of overall digestion. Positive values indicate enhancement, negative values inhibition. (B) Part of the sequence of 158-mer HindIII/NdeI fragment of the plasmid pSP73 showing preferential binding sites (shown as light bars) of M-[Fe2L3]4+ at 20:1 (base:cylinder) ratio. The binding sites were obtained by shifting the sites of inhibited DNase I cleavage by 2 bp in the 3′ direction.
Figure 5.
Differential cleavage plots for M-[Fe2L3]4+ induced differences in susceptibility to DNase I digestion on bottom strand of the 158-mer HindIII/Nde I fragment of the plasmid pSP73 at 10:1, 20:1 and 30:1 (base:cylinder) ratios.
To identify the sites of drug binding from the sites of inhibited DNase I cleavage, a 3′-shift of about 2–3 bp must be considered because of the bias introduced by the nuclease upon DNA cleavage (18). The resulting binding sites of the M enantiomer are summarized in Figure 4B. Interestingly, closer inspection of the binding sites reveals a symmetric sequence element TATGTGTAT in the first site and palindromic sequences 5′-CTATAG and 5′-GATATC in the second and third binding sites.
When the concentrations of M and P-[Fe2L3]4+ exceeded 10:1 (base:cylinder) ratios, the DNA cleavage was almost completely inhibited (gels not shown). Previously published atomic force microscopy and linear dichroism results showed that binding of [Fe2L3]4+ (particularly the M enantiomer) to the DNA causes significant intramolecular coiling that can lead to a ‘wrapping up’ of individual DNA molecules. Since no DNA precipitation was observed, the results suggest that such DNA packaging caused by both enantiomers of [Fe2L3]4+ protects DNA fragments from cleavage by DNase I.
Cleavage of 28-bp duplex by restriction endonucleases
In order to further corroborate the binding preference of the M enantiomer for regular alternating purine–pyrimidine sequences, the effect of the cylinder on the cleavage of a 28-bp duplex containing restriction cleavage sites for Bst1107 I and PvuII restriction endonucleases (Figure 6A) was examined. The recognition sequence of Bst1107 I endonuclease (5′-GTATAC) contains regularly alternating purines and pyrimidines, while the recognition sequence of PvuII (5′-CAGCTG) does not. The 28-mer duplex was mixed with increasing concentrations of the M enantiomer and then digested by Bst1107 I or PvuII and the products of the cleavage were analyzed by gel electrophoresis (Figure 6B). The densitometric analysis of the gel (Figure 6C) shows that while the presence of the cylinder had no effect on the cleavage activity of PvuII, activity of the other endonuclease, Bst1107 I, was strongly inhibited in the presence of the M enantiomer.
DNA interactions of bulky cylinders
EtBr displacement
The quantification of binding strength of bulky cylinders [Fe2(L-CF3)3]4+ and [Fe2(L-Ph)3]4+ to ct-DNA was also investigated by the competition between cylinders and EtBr in the same way as in the case of unsubstituted cylinders (vide supra). The apparent binding constants (Kapp) for both bulky cylinders are shown in Table 1.
DNA unwinding
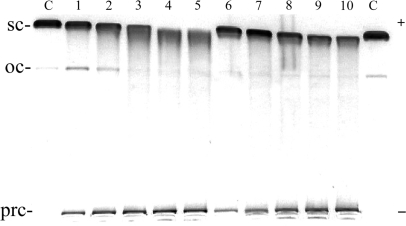
The DNA unwinding by [Fe2(L-CF3)3]4+ and [Fe2(L-Ph)3]4+, was also examined but no unwinding in the region of the cylinder:base ratios used for the parent cylinder was observed (data not shown). If the cylinder:base ratios were further increased up to 0.3 (Figure 7), slight reduction of the mobility of the supercoiled DNA was observed and significant fractions of the DNA left immobilized in the wells as a precipitated DNA (Figure 7, bands marked prc).
Figure 7.
Unwinding of negatively supercoiled pUC19 plasmid DNA by [Fe2(L-CF3)3]4+ (lanes 1–5) and [Fe2(L-Ph)3]4+ (lanes 6–10). The plasmid at the concentration of 2 × 10−4 M (this concentration is related to the monomeric nucleotide content) was mixed with increasing concentrations of the cylinders in 10 mM Tris·HCl (pH 7.4) incubated for 30 min at 25°C and analysed on 1% agarose gel. Lanes C, control (nonmodified DNA); lanes 1–5 and 6–10, cylinder:base ratio was 0.1, 0.15, 0.20, 0.25, 0.30, respectively; sc and oc (open circle) indicate supercoiled and relaxed (nicked) forms of plasmid DNA respectively, and prc indicates precipitated DNA.
DNase I footprinting
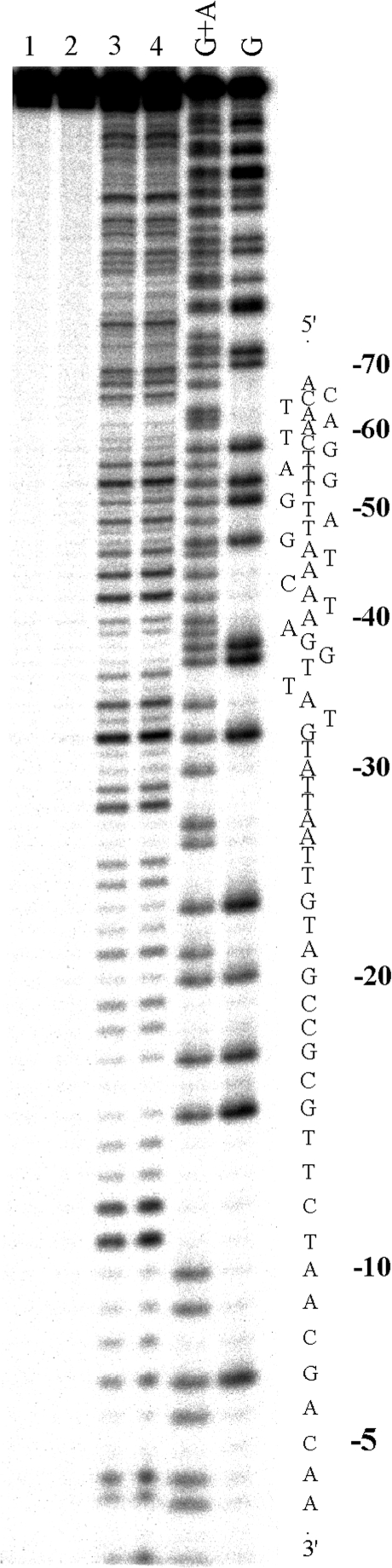
DNase I footprinting of DNA fragment in the presence of bulky cylinders revealed no clear footprints that would indicate sequence specific binding (shown for [Fe2(L-Ph)3]4+ in Figure 8, lane 3). Unlike the parent cylinder the addition of [Fe2(L-CF3)3]4+ or [Fe2(L-Ph)3]4+ to the DNA solution led at higher concentrations to the precipitation of the DNA, which protected the DNA quite efficiently from the cleavage by DNase I (shown for [Fe2(L-Ph)3]4+ in Figure 8, lanes 1 and 2).
Figure 8.
Autoradiogram of 13% PAA/8M urea denaturing gel showing DNase I footprint of 158-mer HindIII/NdeI fragment of the plasmid pSP73 in the presence of different concentrations of [Fe2(L-CF3)3]4+. Lanes 1–3; DNA mixed with [Fe2(L-CF3)3]4+ at 4:1, 10:1 and 40:1 (base:cylinder) ratios, respectively; lane 4; DNA in the absence of the cylinder; lanes G + A and G correspond to Maxam–Gilbert G + A and G ladders. The nucleotide sequence of the fragment is shown on the right side of the gel.
DISCUSSION
The structural features of DNA modified by noncovalently bound supramolecular cylinders have begun to be explored. Their action is exciting because modification of DNA secondary structure by binding of molecules of biological significance is an important aspect of recognition by DNA processing proteins in the cell. The dinuclear iron(II) metallosupramolecular cylinders have been developed as major groove binding agents. It has been previously demonstrated that the cylinders bend and coil DNA (7). The unwinding angle is another valuable biophysical parameter when evaluating the ability of the compound to interact with DNA. It represents the number of degrees by which the DNA is unwound about its helical axis per molecule bound. Thus, one of the interesting questions is whether such a groove binding agent can cause an unwinding of the DNA duplex. The results herein demonstrate that the iron(II) metallosupramolecular cylinders do unwind the DNA duplex. Moreover, compared to other low molecular mass compounds that exhibit nonintercalative DNA binding modes, these cylinders unwind DNA rather extensively. The results herein also show that the M enantiomer is more effective than the P enantiomer at unwinding DNA confirming that the DNA binding of the two enantiomers differs. The higher efficiency of the M enantiomer in unwinding DNA may be a consequence of partial insertion of one of its chelates between DNA bases when adopting a major groove binding mode. No such feature of DNA-binding mode was inferred for the P enantiomer (5). Comparison of the results for the racemate with ct-DNA reveals that its behavior is the ‘average’ of the unwinding angles for the two enantiomers, confirming the previous suggestion (5) that each enantiomer interacts independently from the other one.
It has been shown (19) that local duplex unwinding is an important determinant in the recognition of DNA by various DNA-binding proteins. In addition, this recognition has been shown to correlate with the energetics of the destabilizing effect of DNA-binding agents. This energetics can be estimated crudely using the same approach as used to calculate the free energy required to unwind a 12-bp long DNA fragment containing a single 1,2-GG intrastrand adduct of cisplatin about its helix axis (19). The same calculations were performed, assuming unwinding angles of 32 or 22° for the M and P enantiomer and local twisting within the 8–10 bp long fragment [the 8–10 bp long fragment was taken for these calculations because dinuclear iron(II) metallosupramolecular cylinders bind to short purine–pyrimidine tracts in DNA, 8–10 bp long (vide infra)]. These approximate calculations give a rough estimate of the free energy required to unwind DNA by M or P enantiomer of 8.8–9.6 kJ/mol or 4.2–5.4 kJ/mol, respectively. Interestingly, these values of the free energy are markedly higher than those calculated for DNA unwinding due to the major cross-link of cisplatin (19), which is recognized by a number of damaged DNA-binding proteins (20–23).
A relatively high extent of DNA unwinding due to nonintercalative and noncovalent binding is unprecedented. Thus, unwinding is revealed to be another important conformational alteration induced in DNA by the cylinder in addition to bending. The unwinding angles reported herein add significantly to the body of information on DNA modified by noncovalently bound iron cylinders and offer an opportunity to better understand the structural basis of interactions with DNA modified by supramolecular cylinders and its downstream intracellular effects. We speculate that the high efficiency with which iron cylinders unwind DNA may indicate their potential to affect cellular processes requiring DNA strand separation, such as DNA replication, transcription and/or repair.
Interestingly, bulkier cylinders [Fe2(L-CF3)3]4+ and [Fe2(L-Ph)3]4+ do not unwind DNA (Figure 7). We suggest that negligible efficiency of these cylinders is a consequence of their bulkier shape, which does not fit into the DNA grooves (vide infra).
Competitive binding studies using the fluorescent intercalator EtBr (Table 1) show that both enantiomers exhibit a lower binding affinity for ct-DNA than for synthetic double-stranded DNA copolymers poly(dA-dT)2 and poly(dG-dC)2, suggesting a binding preference for regular purine–pyrimidine tracts rather than for a random nucleotide sequence. Moreover, both enantiomers prefer binding to repetitive AT sequences rather than to GC sequences. It was shown previously (12) that P enantiomer displaces EtBr much more efficiently than M enantiomer from ct-DNA (possessing mostly a random nucleotide sequence). We show in the present work that P enantiomer binds much more efficiently than M enantiomer also to poly(dA-dT)2 and poly(dG-dC)2), i.e. to the regular alternating purine and pyrimidine structure given by these synthetic DNAs. Hence, also these results support the previously reported view (5) that the binding of the P enantiomer of [Fe2L3]4+ and the M enantiomer must be distinct (vide supra).
In order to obtain more details on sequence-selective binding of iron cylinders to DNA, we used DNase I footprinting methodology. DNase I binds to the minor groove of B-DNA and cleaves each strand independently (18,24). The nuclease binds asymmetrically to the minor groove of DNA. Importantly, the local conformation of the DNA at the sites of the contacts of DNase I with DNA phosphates are very important, indirect determinants of sequence-dependent binding to and cutting of DNA by DNase I (25). Steric hindrance from ligand bound to DNA in its minor groove is another obvious factor decreasing affinity and cleavage efficiency of DNase I. Using this footprinting methodology, we demonstrate for the first time that dinuclear iron(II) metallosupramolecular cylinders bind to DNA in a sequence-dependent manner, namely to short purine–pyrimidine tracts in DNA, 8–10 bp long (Figures 3–5). This length roughly corresponds to one molecule of the cylinder bound per turn of the B-DNA double helix, which is in a good agreement with what has been inferred from linear dichroism analyses (5). Interestingly, the M-enantiomer adopts a major groove binding mode so that the observed alterations in the cutting patterns due to its binding to DNA are likely to arise through indirect (conformational) effects. In other words, conformational perturbations induced by the M-enantiomer binding in the major groove are translated into the minor groove, allowing their detection by DNase I probing.
Interestingly, the binding preference of the M enantiomer for regular alternating purine–pyrimidine sequences is corroborated by the results demonstrating strong inhibition effect of the M enantiomer on the cleavage activity of the restriction endonuclease whose recognition sequence contains regularly alternating purines and pyrimidines, while the cleavage activity of the restriction endonuclease whose recognition sequence contains a random sequence was unaffected (Figure 6).
The reasons for the sequence selectivity of DNA binding of iron cylinders observed in the present work are unknown. They may result from a number of factors, including specific van der Waals contacts between atoms on the cylinder and the DNA, a greater inherent propensity for distortion of local DNA topology at preferred sequences, which promotes binding by the cylinder. The specificity observed may be also determined more by general DNA shape (major and/or minor groove width and depth, increased helical repeat and bending) than by distinct contacts between the cylinder and DNA. With specific geometry of the DNA grooves in purine–pyrimidine tracts and their slightly increased helical repeat (26,27), these tracts may present dinuclear iron(II) metallosupramolecular cylinder, particularly its M enantiomer, with an environment spatially amenable to binding by this supramolecular agent. To further explore the importance of the precise size and shape of the cylinder and how it affects the recognition of the purine–pyrimidine tracts, bulkier cylinders [Fe2(L-CF3)3]4+ and [Fe2(L-Ph)3]4+ were employed as well. These cylinders are derived from the parent cylinder [Fe2L3]4+, but have additional groups attached to the ligand. This addition results in a dramatic enlargement of the diameter of the cylinders so that it is unlikely that such cylinders would fit into the purine–pyrimidine tracts with gently tuned and specific geometry. Consistent with this, the results shown in Figure 8 (lanes 1 and 2) demonstrate that [Fe2(L-CF3)3]4+ and [Fe2(L-Ph)3]4+ exhibit no sequence-dependent binding to DNA and at higher concentrations just precipitate the DNA as other tetracations. Thus, these results are consistent with the view and support the hypothesis that the size and shape of the cylinder are crucial for recognition of purine–pyrimidine tracts to which they preferentially bind.
This observation of sequence preference in the binding of even these first unsubstituted cylinders indicates some hope and potential that a concept based on the use of metallosupramolecular cylinders might result in molecular designs that recognize the genetic code in a sequence-dependent manner with a potential ability to affect the processing of the genetic code.
ACKNOWLEDGEMENTS
This research was supported by the Academy of Sciences of the Czech Republic (Grants B400040601, IAA400040803, 1QS500040581, KAN200200651, AV0Z50040507, AV0Z50040702), the Ministry of Education of the Czech Republic (MSMT LC06030, ME08017), the Grant Agency of the Ministry of Health of the CR (NR8562-4/2005) and conducted in the context of COST D39 (WGs D39/002/07 and D39/004/06). Funding to pay the Open Access publication charges for this article was provided by the Grant Agency of the Ministry of Health of the CR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lehn J-M. Supramolecular Chemistry – Concepts and Perspective. Wiley-VCH Weinheim; 1995. [Google Scholar]
- 2.Lehn JM. Toward complex matter: supramolecular chemistry and self-organization. Proc. Natl Acad. Sci. USA. 2002;99:4763–4768. doi: 10.1073/pnas.072065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruben M, Ziener U, Lehn JM, Ksenofontov V, Gutlich P, Vaughan GBM. Hierarchical self-assembly of supramolecular spintronic modules into 1D-and 2D-architectures with emergence of magnetic properties. Chem. Eur. J. 2004;11:94–100. doi: 10.1002/chem.200400584. [DOI] [PubMed] [Google Scholar]
- 4.Hannon MJ, Childs LJ. Helices and helicates: beautiful supramolecular motifs with emerging applications. Supramol. Chem. 2004;16:7–22. [Google Scholar]
- 5.Meistermann I, Moreno V, Prieto MJ, Moldrheim E, Sletten E, Khalid S, Rodger PM, Peberdy JC, Isaac CJ, Rodger A, et al. Intramolecular DNA coiling mediated by metallosupramolecular cylinders: differential binding of P and M helical enantiomers. Proc. Natl Acad. Sci. USA. 2002;99:5069–5074. doi: 10.1073/pnas.062634499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moldrheim E, Hannon MJ, Meistermann I, Rodger A, Sletten E. Interaction between a DNA oligonucleotide and a dinuclear iron(II) supramolecular cylinder; an NMR and molecular dynamics study. J. Biol. Inorg. Chem. 2002;7:770–780. doi: 10.1007/s00775-002-0354-2. [DOI] [PubMed] [Google Scholar]
- 7.Hannon MJ, Moreno V, Prieto MJ, Moldrheim E, Sletten E, Meistermann I, Isaac CJ, Sanders KJ, Rodger A. Intramolecular DNA coiling mediated by a metallo-supramolecular cylinder. Angew. Chem., Intl. Ed. 2001;40:879–884. doi: 10.1002/1521-3773(20010302)40:5<879::AID-ANIE879>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 8.Uerpmann C, Malina J, Pascu M, Clarkson GJ, Moreno V, Rodger A, Grandas A, Hannon MJ. Design and DNA binding of an extended triple-stranded metallo-supramolecular cylinder. Chem. Eur. J. 2005;11:1750–1756. doi: 10.1002/chem.200401054. [DOI] [PubMed] [Google Scholar]
- 9.Oleksi A, Blanco AG, Boer R, Usón I, Aymamí J, Rodger A, Hannon MJ, Coll M. Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate. Angew. Chem., Intl. Ed. 2006;45:1227–1231. doi: 10.1002/anie.200503822. [DOI] [PubMed] [Google Scholar]
- 10.Cerasino L, Hannon MJ, Sletten E. DNA three-way junction with a dinuclear iron(II) supramolecular helicate at the center: a NMR structural study. Inorg. Chem. 2007;46:6245–6251. doi: 10.1021/ic062415c. [DOI] [PubMed] [Google Scholar]
- 11.Malina J, Hannon MJ, Brabec V. Recognition of DNA three-way junctions by metallosupramolecular cylinders: gel electrophoresis studies. Chem. Eur. J. 2007;13:3871–3877. doi: 10.1002/chem.200700159. [DOI] [PubMed] [Google Scholar]
- 12.Peberdy JC, Malina M, Khalid S, Hannon MJ, Rodger A. Influence of surface shape on DNA binding of bimetallo helicates. J. Inorg. Biochem. 2007;101:1937–1945. doi: 10.1016/j.jinorgbio.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Brabec V, Palecek E. Interaction of nucleic acids with electrically charged surfaces. II. Conformational changes in double-helical polynucleotides. Biophys. Chem. 1976;4:76–92. doi: 10.1016/0301-4622(76)80009-9. [DOI] [PubMed] [Google Scholar]
- 14.Keck MV, Lippard SJ. Unwinding of supercoiled DNA by platinum ethidium and related complexes. J. Am. Chem. Soc. 1992;114:3386–3390. [Google Scholar]
- 15.Wyatt MD, Garbiras BJ, Haskell MK, Lee M, Souhami RL, Hartley JA. Structure-activity relationship of a series of nitrogen mustard- and pyrrole-containing minor groove-binding agents related to distamycin. Anti-Cancer Drug Des. 1994;1:511–529. [PubMed] [Google Scholar]
- 16.Fox KR, Waring MJ. High-resolution footprinting studies of drug-DNA complexes using chemical and enzymatic probes. In: Chairs JB, Waring MJ, editors. Drug Nucleic Acid Interactions.? San Diego/CA: Academic Press Inc; 2001. pp. 412–430. [DOI] [PubMed] [Google Scholar]
- 17.Kemp S, Wheate NJ, Buck DP, Nikac M, Collins JG, Aldrich-Wright JR. The effect of ancillary ligand chirality and phenanthroline functional group substitution on the cytotoxicity of platinum(II)-based metallointercalators. J. Inorg. Biochem. 2007;101:1049–1058. doi: 10.1016/j.jinorgbio.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Fairall L, Rhodes D. A new approach to the analysis of DNase I footprinting data and its application to the TFIIIA/5S DNA complex. Nucleic Acids Res. 1992;20:4727–4731. doi: 10.1093/nar/20.18.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellon SF, Coleman JH, Lippard SJ. DNA unwinding produced by site-specific intrastrand cross-links of the antitumor drug cis-diamminedichloroplatinum(II) Biochemistry. 1991;30:8026–8035. doi: 10.1021/bi00246a021. [DOI] [PubMed] [Google Scholar]
- 20.Kartalou M, Essigmann JM. Recognition of cisplatin adducts by cellular proteins. Mutation Res. 2001;478:1–21. doi: 10.1016/s0027-5107(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 21.Brabec V. DNA modifications by antitumor platinum and ruthenium compounds: their recognition and repair. Prog. Nucleic Acids Res. Mol. Biol. 2002;71:1–68. doi: 10.1016/s0079-6603(02)71040-4. [DOI] [PubMed] [Google Scholar]
- 22.Brabec V, Kasparkova J. Molecular aspects of resistance to antitumor platinum drugs. Drug Resist. Updates. 2002;5:147–161. doi: 10.1016/s1368-7646(02)00047-x. [DOI] [PubMed] [Google Scholar]
- 23.Brabec V, Kasparkova J. Modifications of DNA by platinum complexes: relation to resistance of tumors to platinum antitumor drugs. Drug Resist. Updates. 2005;8:131–146. doi: 10.1016/j.drup.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Fairall L, Harrison SD, Travers AA, Rhodes D. Sequence-specific DNA-binding by a 2 zinc-finger peptide from the Drosophila-melanogaster tramtrack protein. J. Mol. Biol. 1992;226:349–366. doi: 10.1016/0022-2836(92)90952-g. [DOI] [PubMed] [Google Scholar]
- 25.Lahm A, Weston SA, Suck D. Structure of DNase I. In: Eckstein F, Lilley DMJ, editors. Nucleic Acids and Molecular Biology. Berlin, Heidelberg: Springer; 1991. pp. 171–186. [Google Scholar]
- 26.Mah SC, Price MA, Townsend CA, Tullius TD. Features of DNA recognition for oriented binding and cleavage by calicheamicin. Tetrahedron. 1994;50:1361–1378. [Google Scholar]
- 27.Ikemoto N, Kumar R, Ling T, Ellestad G, Danishefsky S, Patel D. calicheamicin-DNA complexes: warhead alignment and saccharide recognition of the minor groove. Proc. Natl Acad. Sci. USA. 1995;92:10506–10510. doi: 10.1073/pnas.92.23.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerckhoffs JMCA, Peberdy JC, Meistermann I, Childs LJ, Isaac CJ, Pearmund CR, Reudegger V, Khalid S, Alcock NW, Hannon MJ, et al. Enantiomeric resolution of supramolecular helicates with different surface topographies. Dalton Trans. 2007:734–742. doi: 10.1039/b614093a. [DOI] [PubMed] [Google Scholar]
- 29.Hannon MJ, Painting CL, Jackson A, Hamblin J, Errington W. An inexpensive approach to supramolecular architecture. Chem. Commun. 1997:1807–1808. [Google Scholar]