Abstract
The design of molecules that damage a selected DNA sequence provides a formidable opportunity for basic and applied biology. For example, such molecules offer new prospects for controlled manipulation of the genome. The conjugation of DNA-code reading molecules such as polyamides to reagents that induce DNA damages provides an approach to reach this goal. In this work, we showed that a bipyridine conjugate of polyamides was able to induce sequence-specific DNA breaks in cells. We synthesized compounds based on two polyamide parts linked to bipyridine at different positions. Bipyridine conjugates of polyamides were found to have a high affinity for the DNA target and one of them produced a specific and high-yield cleavage in vitro and in cultured cells. The bipyridine conjugate studied here, also presents cell penetrating properties since it is active when directly added to cell culture medium. Harnessing DNA damaging molecules such as bipyridine to predetermined genomic sites, as achieved here, provides an attractive strategy for targeted genome modification and DNA repair studies.
INTRODUCTION
Manipulation of genomic sequences presents a great interest in biotechnology and in therapeutics. One way to reach this goal is to induce targeted DNA damages. It is possible to engineer site-specific damaging agents based on the use of a DNA code-reading domain and of a DNA damaging domain. Such an approach has been successfully applied to target DNA breaks. Protein-based endonucleases, namely zinc-finger nucleases, have been engineered and successfully used in cells (1). On the other hand, synthetic nucleases have demonstrated site-specific cleavage activity in vitro on synthetic or plasmid DNA but to our knowledge not on genomic DNA in cells. Synthetic nucleases that have been described to date are based on triplex-forming oligonucleotides (TFOs) or polyamides as DNA-binding domains and on various cleaving agents (2). Polyamides, composed of N-methylpyrrole and N-methylimidazole have been originally designed by Dervan and collaborators. Such polyamides folded into hairpin structures are compounds that are able to bind in the minor groove and to specifically recognize a short DNA sequence. The recognition rules of DNA by polyamides have been described (3) and strategies aimed at increasing the size of the binding site have been developed. Bis-hairpin polyamides are able to specifically bind to extended target sites (around 12 bp) as compared to standard hairpin polyamides that commonly recognize 6–7 bp. The possibility to functionalize polyamides has been used to impart biological activity of interest. For example, alkylating agents, such as chlorambucil have been linked to hairpin polyamides (4–6) and demonstrated a biological activity (7–11). A few studies have revealed that polyamides can also be used to induce DNA cleavage: nucleases involving phenanthroline derivatives connected to polyamides have shown the capacity to cleave DNA in vitro (12–14). Until now and to our knowledge, no data on damaging activities in cells of the latter artificial nucleases based on oxidation by metals are available in the literature. However, nucleases based on copper coordinated to phenanthroline ligands present many potential advantages for biological applications. Cleavage reactions produce DNA strand breaks without piperidine treatment (15,16) and oxidative cleavage with phenanthroline ligands prevents formation of diffusible reactive oxygen species that are known to be toxic for cells; it has been determined that a presumed oxo-copper intermediate, whose structure remains unknown, is the reactive species (17). In addition, a labile copper pool that can be used by the nuclease in situ is strongly suspected in cells (18) and one study has shown that free phenanthroline cleaves DNA in a cellular context with endogenous copper and reductant (19). Similarly to phenanthroline, bipyridine, a structurally related molecule, is able to induce DNA damage in the presence of copper ions (20,21), although its cleavage properties and mechanism have been less studied.
In this article, we engineered an original conjugate consisting of a bipyridine as a metal ligand linked to polyamides, which is able to specifically cleave its DNA target in cells. For this purpose, we used the bipyridine moiety as a linker between two hairpin polyamides, in order to combine nuclease activity and an extended DNA target site. Different conjugates varying by the attachment point of the polyamides on the bipyridine were tested in vitro to determine, which of them displays the highest specific cleavage activity. The capacity of the selected molecule to reach the cell nucleus and then to induce specific double-strand DNA cleavage intracellularly was then demonstrated.
MATERIALS AND METHODS
Materials and general procedures
Reagents and solvents were purchased from (Sigma-Aldrich–Fluka, Saint Quentin Fallavier, France), VWR and Acros Organics. 2,2′-Bipyridine, 2,2′-bipyridine-3,3′-dicarboxylic acid, 2,2′-bipyridine-4,4′-dicarboxylic acid, 2,2′-bipyridine-5,5′-dicarboxylic acid, 15-aminophenanthroline and phenanthroline-2,9-dicarboxylic acid were from Aldrich. Oligodeoxyribonucleotides were purchased from (Eurogentec, Seraing, Belgium). Polyamides were prepared according to Syniakov et al. (22). 4-Oxo-4-(1,10-phenanthrolin-5-ylamino) butanoic acid was prepared as described previously (23). The polyamide conjugates were analyzed on a Xterra (Milford, USA) C18 column (7 μm—7.8 × 300 mm i.d.) with a gradient of acetonitrile/0.1% TFA (from 0% to 90% up in 40 min) in water/TFA 0.1% at a flow rate of 2 ml/min. Detection at the output of the HPLC column was provided by UV absorbance at 260 and 310 nm. Mass spectrometry analysis and electrospray Q-TOF (ES Q-TOF MS) were performed on a Q-Star instrument (Applied Biosystems, Courtaboeuf, France). In the positive mode analysis, the sample were dissolved in a solution of acetonitrile and water (50/50, v/v) that contained 1% of formic acid. Syntheses of compounds are available in Supplementary Material.
Plasmid
The (+)PPT/luc plasmid (8850 bp) contained the target sequence 5′-T5A4GA4G6A-3′ corresponding to the oligopyrimidine • oligopurine tract named PPT; this sequence is present in the 29 bp duplex used in the present study (Figure 2). The (+)PPT/luc plasmid was previously described in more detail (24).
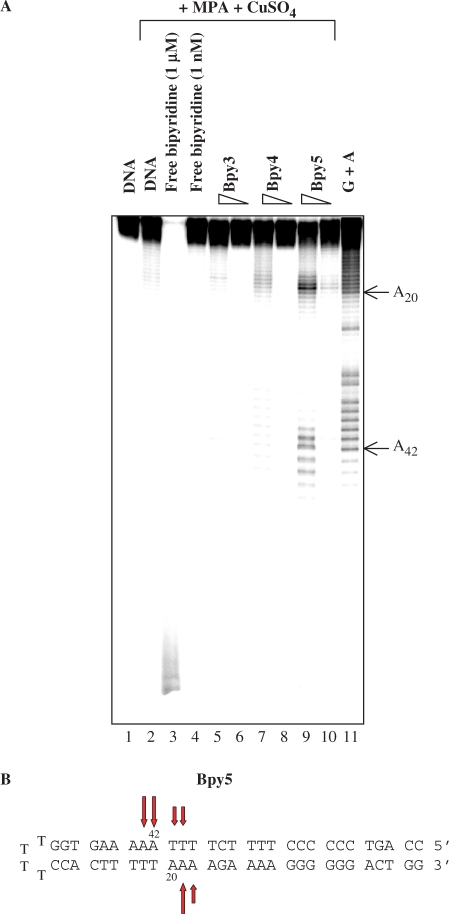
Figure 2.
Targeted cleavage induced by polyamides covalently linked to a bypiridine–copper chelate on a 29-bp intramolecular duplex target. (A) The 5′ radiolabelled duplex was treated, as indicated, by different conjugates at two concentrations (10 nM and 1 µM) in the presence of CuSO4 and MPA. Cleavage products were separated by electrophoresis in denaturing acrylamide gel. (B) The cleavage patterns observed with the conjugates Bpy5 is schematically described. Vertical bars indicate the sites of cleavage observed on each strand; the lengths of the bars represent the relative abundance of the fragments. The location of the adenines A20 and A42 is indicated near the gel.
Cells
The CMV(+)PPT/HeLa cells were derived from HeLa/Tet-on cells (Clontech, Saint-Germain-en-Laye, France) and were previously described (25).
Complex formation and binding affinity between conjugates and target dsDNA
The DNA–polyamides complex formation was followed by gel shift assays with fluorescent or 32P-labeled DNA target fragment and various concentrations of conjugates. The samples containing 2 nM DNA target fragment and various concentrations of conjugates (between 10 µM and 0.01 nM) in 50 mM HEPES pH 7.3, were loaded on 20% nondenaturing polyacrylamide gel and electrophoresis was carried out at 37°C. Radioactivity or fluorescence were detected on PAGE gels by a (Amersham-Pharmacia, Orsay, France) Typhoon instrument and treated with Image Quant software (Molecular Dynamics, Orsay, France). Apparent dissociation constant Kd were obtained by fitting the data with the equation:
where C corresponds to the complex ratio, L corresponds to the initial ligand concentration and D is the initial concentration of the duplex.
In vitro cleavage assays
Synthetic DNA target
A mixture of 32P-labeled and nonlabeled intramolecular duplex (40 nM) was incubated in 50 mM HEPES pH 7.2 and 50 mM NaCl in the presence of 10 µM copper sulfate (CuSO4); the conjugates were added at various concentrations and the reaction was started by addition of 3 mM of mercaptopropionic acid (MPA). The mixtures were incubated for 20 h at 4°C. Reaction was stopped by ethanol precipitation and the reaction products were resolved on a denaturing 15% polyacrylamide 7 M urea gel and analyzed with a Typhoon instrument.
Plasmid target
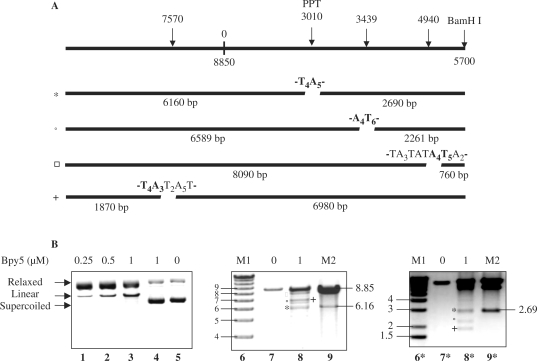
Supercoiled (+)PPT/luc plasmid (SC) was incubated in the presence or in the absence of various concentration of Bpy5 conjugate and 10 µM CuS04 in a buffer containing 50 mM HEPES pH 7.2, 50 mM NaCl, 10 mM MgCl2; the reaction was started by addition of MPA (3 mM), and carried out at 37°C for 17 h. After the cleavage reaction, the cleavage products from the supercoiled plasmid have been digested by BamHI before loading as indicated. Plasmid cleavage by a phenanthroline conjugate of triplex-forming oligonucleotide (OP-TFO) that was targeted to the PPT sequence 5′-T5A4GA4G6A-3′ was used to identify the fragments originating from cleavage occurring at this site: the two fragments have sizes of 6160 bp and 2690 bp when BamHI was used for linearization.
Intracellular cleavage assay
CMV(+)PPT/HeLa cells were treated by Bpy5 bipyridine conjugate or free bipyridine.
Fluorescence immunolabelling
Twenty-four hours after treatment, phosphorylated histone variant H2AX was detected by immunolabeling. Immunolabeling was performed using standard procedure (26) and anti-PhosphoH2AX antiboby (Upstate, monoclonal clone JW301) followed by TRITC-anti-IGg-Fc-specific Mouse antibody (Sigma). In order to stain the nuclei, slides were then incubated with DAPI, then washed and mounted using a Mounting Medium for fluorescence (Vector, Marne La Vallée, France). Observation of immunolabeled cells was performed using a fluorescence microscope (DMR Leica, Rueil-Malmaison, France).
Quantification of DNA damages by real-time PCR
In parallel to immunofluorescence, real-time PCR analyses were performed 24 h after treatment by Bpy5. Analyses were performed with Mx3005P™ Real-Time PCR System (Stratagene, Amsterdam, Nederlands). Primers were designed to amplifiy (A•T)-rich regions located within the same 10 kpb genomic locus. For normalization, primers were designed to amplify a region lacking potential target sites that means (A•T or T•A)n (with n ⩾ 9) sequences (see primer sequences in Supplementary Table 1).
RESULTS
Synthesis of polyamide–bipyridine conjugates and DNA-binding properties
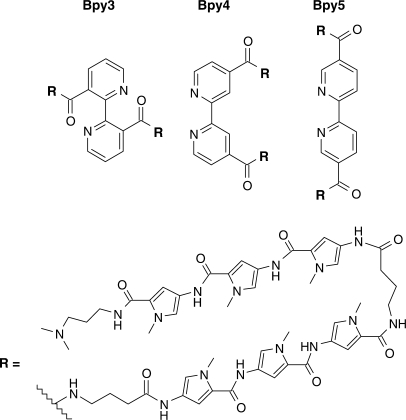
We have recently characterized DNA-binding properties of bis-polyamides, where the two polyamide parts are connected head-to-head by an iminodiacetic acid linker (27); they have revealed affinity for DNA in the nanomolar range and sequence-specific recognition of an extended target of 9 bp when each polyamide was composed of six rings for binding to 4 bp. Here, to exploit these interesting properties, we changed the iminodiacetic acid linker by a bipyridine with the goal to induce DNA break, while maintaining DNA-binding properties (Figure 1). Indeed, such a geometry might allow the coordination of the metal ion close to the DNA minor groove, which is necessary for efficient cleavage. The bipyridine moiety was chosen because of its higher flexibility compared to that of phenanthroline and because of the possibility to easily vary the attachment site of the polyamides: various positions of the heterocycle rings were used in Bpy3, Bpy4 and Bpy5 to determine, which of the molecules are able to maintain a correct hydrogen bonding pattern between the polyamides and the DNA double strand and to induce efficient cleavage as well. All coupling reactions between polyamides and bipyridine were done with standard conditions used for peptide synthesis as described in the Supplementary Material.
Figure 1.
Chemical structures of polyamide–bipyridine conjugates Bpy3, Bpy4 and Bpy5.
We first studied the binding properties of the novel conjugates: we synthesized conjugates where the two polyamide hairpins contained six N-methylpyrrole rings for recognition of 4 (A•T) or (T•A) bp and apparent dissociation constants of conjugate–DNA double strand complexes were determined by gel shift assays using a 29 bp duplex containing a target sequence (see sequence in Figure 2). Apparent dissociation constants of Bpy3, Bpy4 and Bpy5 were obtained in the nanomolar range: 1.7, 2.1 and 5.8 nM, respectively. No modification of the dissociation constants was found in the presence of 10 µM of copper ions, i.e. in conditions of cleavage reactions. These values are comparable to that of compound containing an iminodiacetic acid linker (27) that exhibits a Kd of 4.8 nM under the same experimental conditions (Supplementary Table 2) and encouraged evaluation of DNA cleavage activity.
In vitro DNA cleavage assays and specificity studies
Cleavage activity and specificity of the conjugates were tested on different substrates, a 29 bp duplex (Figure 2) and a plasmid (Figure 3). Experiments were done with mercaptopropionic acid (MPA) as the copper reductant.
Figure 3.
Targeted cleavage of (+)PPT/luc plasmid induced by a bipyridine conjugate of polyamides, Bpy5. (A) Schematic representation of the plasmid target with the location of the cleavage sites and the lengths of the cleavage products. (B) The plasmid was treated as indicated, in the presence of 10 µM CuSO4, 3 mM MPA and cleavage products were analyzed by agarose gel electrophoresis. Lanes 1–3, the plasmid was treated with CuSO4 in the presence of MPA and of increasing concentrations of Bpy5. Lanes 4 and 5, the plasmid was treated with CuSO4 in the absence of MPA and in the presence or not of Bpy5, respectively. Lanes 7* and 8*, samples of lane 5 and 3, respectively were treated by BamHI enzyme. Two migrations of the same gel were presented to allow visualization of all the cleavage products, the longest products (lanes 8 and 9) and the shortest ones (lanes 8* and 9*). DNA markers: (lane 6 and 6*) 1 kb ladder, (Gibco BRL); (lanes 9 and 9*) fragments generated by OP-TFO cleavage on the oligopurine tract 5′-A4GA4G6A-3′, of 6160 and 2690 bp. Lengths are indicated near the gel in kilo base pairs and the cleavage products are indicated with symbols (asterisk, plus, open circle, open square).
First, a 29 bp intramolecular duplex was used as a target and cleavage products were analyzed by denaturing polyacrylamide gel electrophoresis (Figure 2). In spite of their strong interaction with the DNA target, Bpy3 and Bpy4 displayed no significant specific DNA cleavage activities even at a concentration of 1 µM (lanes 5–8). On the contrary, Bpy5 exhibited a targeted DNA cleavage activity (lanes 9 and 10) and the cleavage pattern showed that the cleavage reaction was restricted mainly to adenines at the positions A18–19, A41–44. This result demonstrates that the DNA cleavage was targeted within the 5′-T4A5-3′ sequence corresponding to the expected target site (28). Finally, we showed that the two strands of the DNA duplex are substrates and that in the experimental conditions used, the cleavage yield with Bpy5 on the 29 bp target was around 45%.
To further explore the cleavage specificity and the double-strand break activity of Bpy5, experiments were conducted on a 8850 bp plasmid containing the target sequence present in the 29 bp duplex and the cleavage products were analyzed by agarose gel electrophoresis. Experiments were carried out with the supercoiled plasmid and increasing amounts of Bpy5 (Figure 3, lanes 1–3); Bpy5 treatment induced linearization of the target plasmid, consistent with double-strand break induction. The selectivity of the double-strand cleavage reaction was studied by using the restriction enzyme BamH1 for linearization of the plasmid and analysis of the lengths of the Bpy5 cleavage products. Four major cleavage sites were detected to be sensitive to Bpy5 activity on the plasmid (Figure 3, lane 8 and 8*) and the total double-strand cleavage yield was 40%. Identification of the cleavage sites was made using the molecular mass of the cleaved fragments. As expected, one cleavage site is the 5′-T5A4GA4G6A-3′ sequence that is present on the 29 bp duplex and on the plasmid as well. It was characterized by the use of a phenanthroline–TFO conjugate that specifically recognizes and cleaves the oligopurine tract 5′-A4GA4G6A-3′, leading to 6160 and 2690 bp fragments (Figure 3, lanes 9 and 9*). Based on the recognition code that was established by Dervan and collaborators for hairpin polyamides, the bis-polyamide used in the conjugate should bind to two runs of four contiguous A•T or T•A base pairs. We ran sequence analyses to further characterize the specificity of the polyamides studied here. First, we looked for (A•T or T•A)n with n ⩾ 9 and also for (A•T or T•A)4S(A•T or T•A)4 or (A•T or T•A)4S2(A•T or T•A)4 (with S=C or G) sequences since the bipyridine moiety used as a linker between the two polyamides parts is not known to recognize specific base pairs: 18 (Supplementary Table 2), 41 and 40 sequences were found on the plasmid, respectively. Our data showed that the sequences 5′-T4AWW-3′ or 5′-A4T3WW-3′ (with W=T or A) seem to be the preferred binding sites for the bipyridine conjugate: these sites are consistent with the production of 6589, 2261, 8090, 760, 1870 and 6980 bp long cleavage products. In conclusion, we showed that Bpy5 exhibits a specific cleavage activity only for a category of A•T-rich sequences: this result is consistent with the cleavage pattern observed on the 29 bp target, where the 5′-T4A5-3′ sequence was preferred to the 5′-T4CT4-3′ one and reinforces our previous results obtained with a structurally related bis-polyamide containing an iminodiacetic linker (27), demonstrating a higher sequence specificity for such bis-polyamides compared to that predicted on the basis of recognition rules currently used (3).
Intracellular DNA damaging activity of Bpy5 polyamide–bipyridine conjugate
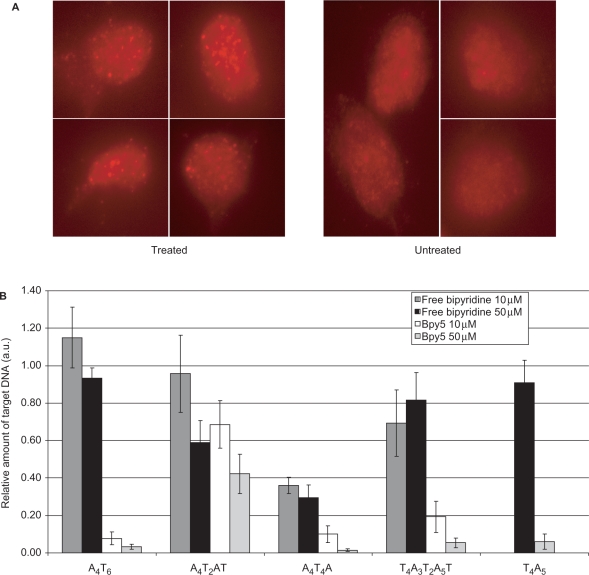
We first evaluated the intracellular DNA damaging activity of the conjugates by immunodetection of the phosphorylated form of the variant histone H2AX, named γH2AX. H2AX protein is phosphorylated at serine 139 following DNA break induction (28) and the detection of γH2AX nuclear foci represents a well-established way to visualize genomic breaks by immunofluorescence. Bpy5 was added to cells and γH2AX foci were examined at increasing times after treatment. Using direct addition in the cell culture medium, a few foci started to be detected 10 h after treatment and the foci number increased to reach its highest level at 24 h (Figure 4A). Almost the same type of results were obtained with the free bipyridine. The data demonstrated that Bpy5 can reach the nucleus and induce genomic breaks in a cellular environment. We next sought to examine whether Bpy5 induced breaks in the genome specifically at its predicted target sequences as observed in vitro on a plasmid substrate. To address this question, we chose to quantify Bpy5-induced breaks at both potential target sites that we identified in vitro (Figure 3), i.e. 5′-T4A3WW-3′ or 5′-A4T3WW-3′ (with W=T or A) and another type of (A•T)-rich sequences with (A•T)n (n ⩾ 9). We quantified damages by real-time PCR using different sets of primers flanking the various (A•T)-rich regions. In this assay, the PCR inhibition reflects the level of DNA modifications induced on both DNA strands in the vicinity of the target site. To evaluate the specificity of Bpy5 conjugate, we compared its effect to that of free bipyridine. Results of PCR inhibition are depicted in Figure 4B. The remaining amplification of DNA from cells treated by Bpy5 varied with the nature of the target sequence. Remarkably, preferential cleavage of 5′-T4A3WW-3′ or 5′-A4T3WW-3′ with W=T or A, which was observed with Bpy5 in vitro did also exist in cells. In contrast, no specific inhibition was observed at the 5′-A4TTAT-3′ sequence since Bpy5 and the free bipyridine exhibited there almost the same activity. Interestingly, for the preferred sequences, a strong inhibition can be obtained with >90% PCR inhibition.
Figure 4.
Intracellular cleavage activity of bipyridine conjugate Bpy5. (A) Detection of DNA breaks by phosphorylated H2AX immunolabeling 24 h after treatment by 10 µM Bpy5. (B) Quantitative PCR on DNA extracted from cells treated by Bpy5 or free bipyridine: DNA analyses were performed 24 h after treatment. PCR quantification of the target DNA was performed, using primers flanking different (A•T)-rich sequences as indicated. The values were normalized by quantification of a control sequence lacking (A•T)-rich sequences (see text for details).
DISCUSSION
Our results demonstrate for the first time the capacity of molecules containing a metal ligand like bipyridine attached to two hairpin polyamides to induce targeted DNA damage both in vitro and in cultured cells. Polyamides have been successfully used for cell applications, showing interesting biological activity, especially by competing with transcription factors binding [for a recent example, see ref. (29)]. With the goal to increase their efficiency, they have been conjugated to DNA damaging agents. Polyamide–chlorambucil conjugates have been used to induce DNA alkylation at their target site (30) and interfere with gene transcription at the elongation level (7). Phenanthroline derivatives have also been attached to polyamides or other DNA-code reading molecules but only used in vitro. Here, we synthetized an original conjugate of polyamide that exploits the structural feature of the bipyridine moiety to use it as a linker between two polyamide parts. Such engineered nuclease allows both to target extended sequences compared to standard hairpin polyamides and to provoke DNA cleavage at the target site.
To obtain bipyridine conjugates of polyamides with DNA cleaving activity, we have screened for the optimized geometry of such conjugates. All the synthetized conjugates Bpy3, Bpy4 and Bpy5 exhibited dissociation constants in the nanomolar range as observed with an iminodiacetic acid linker. Interestingly, the use of bipyridine as a linker to connect the two polyamides seems appropriate as compared to phenanthroline moieties (see chemical structures in Supplementary Figure 1), since the corresponding phenanthroline conjugate (Phe2) showed a 50- or 100-fold higher dissociation constant compared to Bpy5 or Bpy4, respectively (Supplementary Table 2). This is probably due to the enhanced rigidity of the phenanthroline core, as compared to that of the bipyridine molecule, which can adopt a twisted conformation. Indeed, it has already been shown that the affinity of bis-hairpin polyamides for the target depends on the structure and the size of the linker between the two minor groove binders (31). Concerning the cleavage activities of Bpy3, Bpy4 and Bpy5 conjugates, it strongly depends on the attachment site of the polyamides on the bipyridine moiety. Indeed, in spite of their good affinity, no DNA cleavage was observed with Bpy3 and Bpy4 compounds: the twisted or transoid conformation that could be adopted by the bipyridine for correct interaction of the polyamides in the minor groove might be an explanation, such conformation likely preventing copper coordination and correct interactions with the sugar backbone. For Bpy5, direct cleavage of the target and 3′ shifted pattern were observed, supporting the notion that the mechanism of DNA cleavage by the bipyridine conjugate is quite similar to that described for phenanthroline (17).
Concerning the sequence specificity of Bpy5-induced cleavage, we showed that it is higher than predicted on the basis of DNA recognition by the polyamide parts. Such restriction in the repertoire of target sequences was already described for a bis-polyamide whose structure was close to the one of Bpy5 (27). This increased specificity might be explained by a more constrained structure adopted by this type of bis-hairpin polyamides compared to that of the hairpin polyamide alone. This result supports the fact that the DNA recognition code for bis-polyamides will need to be complemented by systematic investigation of their sequence specificity, as recently done for standard hairpin polyamides (32). For Bpy5, one possible explanation for its preference for short alternated purine/pyrimidine blocks among (A•T)-rich sequences may be the unusual conformation of such sequences (33,34), that favors a DNA bent structure.
The ability of Bpy5 to induce cleavage in cells was first demonstrated by immunofluorescence experiments using H2AX antibodies, although the cleavage mechanism and the nature of lesions remain to be characterized and especially the nature of the metal involved in the cleavage reaction. We also demonstrated that DNA breaks were sequence-specific and corresponded to predicted Bpy5-binding sites. To quantify Bpy5-induced DNA damages, we adapted a PCR-based assay that we have recently developed for quantification of triplex induced cross-links (25). This assay permits to quantify lesions occurring on both strands of DNA and able to block the PCR reaction. Remarkably, the Bpy5 sequence specificity observed on genomic DNA perfectly fits with the one observed on a target plasmid in vitro, with the same sequence preference for short alternated purine/pyrimidine blocks. Finally, DNA lesions were obtained when Bpy5 was directly added to the cell-culture medium, demonstrating their ability to spontaneously reach the nucleus.
In conclusion, our results establish the biological properties of Bpy5, a polyamide–bipyridine conjugate and encourage further developments of bipyridine conjugates as synthesized here. The possibility to target DNA breaks to specific genomic sites can be exploited for applications aimed at targeted genome modification. Among them, induction of targeted mutations following DNA repair (35) or stimulation of homologous recombination in gene targeting approaches are attractive (1) but the target size of polyamides, even if extended to around 10 bp as achieved here, might be too short and could generate toxic effects. However, for repair studies, it is of high interest to target breaks at determined genomic sites with high efficiency and one advantage to the use of polyamide–bipyridine conjugates is their ability to induce breaks with a different chemistry in contrast to the protein-based nucleases that are available to date (36).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are thankful to Lionel Dubost and Arul Marie for mass spectrometry analysis. The authors thank Loïc Ponger for his help on bioinformatics analysis. This work was supported by grants from La Ligue Nationale Contre le Cancer. F.C. was supported by the French Ministry for Research. P.S. was supported by CNRS. Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pingoud A, Silva GH. Precision genome surgery. Nat. Biotechnol. 2007;25:743–744. doi: 10.1038/nbt0707-743. [DOI] [PubMed] [Google Scholar]
- 2.Zenkova MA. Artificial nucleases. In: Hans Joachim Gross., editor. Nucleic Acids and Molecular Biology, Vol. 13. Springer-Verlag Berlin Heidelberg; 2004. [Google Scholar]
- 3.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 4.Wurtz NR, Dervan PB. Sequence specific alkylation of DNA by hairpin pyrrole-imidazole polyamide conjugates. Chem. Biol. 2000;7:153–161. doi: 10.1016/s1074-5521(00)00085-5. [DOI] [PubMed] [Google Scholar]
- 5.Bando T, Narita A, Saito I, Sugiyama H. Highly efficient sequence-specific DNA interstrand cross-linking by pyrrole/imidazole CPI conjugates. J. Am. Chem. Soc. 2003;125:3471–3485. doi: 10.1021/ja028459b. [DOI] [PubMed] [Google Scholar]
- 6.Bando T, Sasaki S, Minoshima M, Dohno C, Shinohara K, Narita A, Sugiyama H. Efficient DNA alkylation by a pyrrole-imidazole CBI conjugate with an indole linker: sequence-specific alkylation with nine-base-pair recognition. Bioconjug. Chem. 2006;17:715–720. doi: 10.1021/bc060022w. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez D, Chou CJ, Latella L, Zeitlin SG, Ku S, Puri PL, Dervan PB, Gottesfeld JM. A two-hit mechanism for pre-mitotic arrest of cancer cell proliferation by a polyamide-alkylator conjugate. Cell Cycle. 2006;5:1537–1548. doi: 10.4161/cc.5.14.2913. [DOI] [PubMed] [Google Scholar]
- 8.Shinohara K, Narita A, Oyoshi T, Bando T, Teraoka H, Sugiyama H. Sequence-specific gene silencing in mammalian cells by alkylating pyrrole-imidazole polyamides. J. Am. Chem. Soc. 2004;126:5113–5118. doi: 10.1021/ja031673v. [DOI] [PubMed] [Google Scholar]
- 9.Shinohara K, Bando T, Sasaki S, Sakakibara Y, Minoshima M, Sugiyama H. Antitumor activity of sequence-specific alkylating agents: pyrolle-imidazole CBI conjugates with indole linker. Cancer Sci. 2006;97:219–225. doi: 10.1111/j.1349-7006.2006.00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang YD, Dziegielewski J, Chang AY, Dervan PB, Beerman TA. Cell-free and cellular activities of a DNA sequence selective hairpin polyamide-CBI conjugate. J. Biol. Chem. 2002;277:42431–42437. doi: 10.1074/jbc.M207179200. [DOI] [PubMed] [Google Scholar]
- 11.Philips BJ, Chang AY, Dervan PB, Beerman TA. DNA damage effects of a polyamide-CBI conjugate in SV40 virions. Mol. Pharmacol. 2005;67:877–882. doi: 10.1124/mol.104.006254. [DOI] [PubMed] [Google Scholar]
- 12.Pitie M, Burrows CJ, Meunier B. Mechanisms of DNA cleavage by copper complexes of 3-clip-phen and of its conjugate with a distamycin analogue. Nucleic Acids Res. 2000;28:4856–4864. doi: 10.1093/nar/28.24.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitie M, Van Horn JD, Brion D, Burrows CJ, Meunier B. Targeting the DNA cleavage activity of copper phenanthroline and clip-phen to A.T tracts via linkage to a poly-N-methylpyrrole. Bioconjug. Chem. 2000;11:892–900. doi: 10.1021/bc000050t. [DOI] [PubMed] [Google Scholar]
- 14.Liu D, Zhou J, Li H, Zheng B, Yuan G. Site-selective DNA cleavage by a novel complex of copper-conjugate of Phen and polyamide containing N-methylimidazole rings. Bioorg. Med. Chem. Lett. 2006;16:5032–5035. doi: 10.1016/j.bmcl.2006.07.049. [DOI] [PubMed] [Google Scholar]
- 15.Bales BC, Pitie M, Meunier B, Greenberg MM. A minor groove binding copper-phenanthroline conjugate produces direct strand breaks via beta-elimination of 2-deoxyribonolactone. J. Am. Chem. Soc. 2002;124:9062–9063. doi: 10.1021/ja026970z. [DOI] [PubMed] [Google Scholar]
- 16.Bales BC, Kodama T, Weledji YN, Pitie M, Meunier B, Greenberg MM. Mechanistic studies on DNA damage by minor groove binding copper-phenanthroline conjugates. Nucleic Acids Res. 2005;33:5371–5379. doi: 10.1093/nar/gki856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson GR, Nazhat NB. Kinetics and mechanism of the reaction of the bis(1,10-phenanthroline)copper(I) ion with hydrogen peroxide in aqueous solution. J. Am. Chem. Soc. 1987;109:1990–1994. [Google Scholar]
- 18.Yang L, McRae R, Henary MM, Patel R, Lai B, Vogt S, Fahrni CJ. Imaging of the intracellular topography of copper with a fluorescent sensor and by synchrotron X-ray fluorescence microscopy. Proc. Natl Acad. Sci. USA. 2005;102:11179–11184. doi: 10.1073/pnas.0406547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burkitt MJ, Milne L, Nicotera P, Orrenius S. 1,10-Phenanthroline stimulates internucleosomal DNA fragmentation in isolated rat-liver nuclei by promoting the redox activity of endogenous copper ions. Biochem. J. 1996;313(Pt 1):163–169. doi: 10.1042/bj3130163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Que BG, Downey KM, So AG. Degradation of deoxyribonucleic acid by a 1,10-phenanthroline-copper complex: the role of hydroxyl radicals. Biochemistry. 1980;19:5987–5991. doi: 10.1021/bi00567a007. [DOI] [PubMed] [Google Scholar]
- 21.Aronovitch J, Godinger D, Samuni A, Czapski G. Ascorbic acid oxidation and DNA scission catalyzed by iron and copper chelates. Free Radic. Res. Commun. 1987;2:241–258. doi: 10.3109/10715768709065289. [DOI] [PubMed] [Google Scholar]
- 22.Sinyakov AN, Feshenko MV, Ryabinin VA. A liquid phase synthesis of DNA-binding polyamides using oligocarboxamide blocks. Russ. J. Bioorg. Chem. 2004;30:98–99. [Google Scholar]
- 23.Ramiro P, Garcia-Fresnadillo D, Guillermo O. Synthesis and characterisation of N-1,10-phenanthrolin-5-ylalkylamides and their photosensitising heteroleptic Ru(II) complexes. Tetrahedron. 2005;61:9478–9483. [Google Scholar]
- 24.Faria M, Wood CD, Perrouault L, Nelson JS, Winter A, White MR, Helene C, Giovannangeli C. Targeted inhibition of transcription elongation in cells mediated by triplex-forming oligonucleotides. Proc. Natl Acad. Sci. USA. 2000;97:3862–3867. doi: 10.1073/pnas.97.8.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunet E, Corgnali M, Cannata F, Perrouault L, Giovannangeli C. Targeting chromosomal sites with locked nucleic acid-modified triplex-forming oligonucleotides: study of efficiency dependence on DNA nuclear environment. Nucleic Acids Res. 2006;34:4546–4553. doi: 10.1093/nar/gkl630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N, et al. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell. Biol. 2002;4:993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- 27.Halby L, Ryabinin VA, Sinyakov AN, Boutorine AS. Functionalized head-to-head hairpin polyamides: synthesis, double-stranded DNA-binding activity and affinity. Bioorg. Med. Chem. Lett. 2005;15:3720–3724. doi: 10.1016/j.bmcl.2005.05.106. [DOI] [PubMed] [Google Scholar]
- 28.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 29.Nickols NG, Dervan PB. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc. Natl Acad. Sci. USA. 2007;104:10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YD, Dziegielewski J, Wurtz NR, Dziegielewska B, Dervan PB, Beerman TA. DNA crosslinking and biological activity of a hairpin polyamide-chlorambucil conjugate. Nucleic Acids Res. 2003;31:1208–1215. doi: 10.1093/nar/gkg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weyermann P, Dervan PB. Recognition of ten base pairs of DNA by head-to-head hairpin dimers. J. Am. Chem. Soc. 2002;124:6872–6878. doi: 10.1021/ja020258k. [DOI] [PubMed] [Google Scholar]
- 32.Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips G.N., Jr., Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl Acad. Sci. USA. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burkhoff AM, Tullius TD. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987;48:935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- 34.Meena SZ, Mulligan C, McLaughlin LW. Removal of a single minor-groove functional group eliminates A-tract curvature. J. Am. Chem. Soc. 2006;128:11756–11757. doi: 10.1021/ja0632310. [DOI] [PubMed] [Google Scholar]
- 35.Kim KH, Nielsen PE, Glazer PM. Site-directed gene mutation at mixed sequence targets by psoralen-conjugated pseudo-complementary peptide nucleic acids. Nucleic Acids Res. 2007;35:7604–7613. doi: 10.1093/nar/gkm666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berkovitch E, Monnat RJ, Kastan MB. Role of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell. Biol. 2007;6:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.