Abstract
The transcriptional regulator PlcR and its cognate cell–cell signalling peptide PapR form a quorum-sensing system that controls the expression of extra-cellular virulence factors in various species of the Bacillus cereus group. PlcR and PapR alleles are clustered into four groups defining four pherotypes. However, the molecular basis for group specificity remains elusive, largely because the biologically relevant PapR form is not known. Here, we show that the in vivo active form of PapR is the C-terminal heptapeptide of the precursor, and not the pentapeptide, as previously suggested. Combining genetic complementation, anisotropy assays and structural analysis we provide a detailed functional and structural explanation for the group specificity of the PlcR–PapR quorum-sensing system. We further show that the C-terminal helix of the PlcR regulatory domain, specifically the 278 residue, in conjunction with the N-terminal residues of the PapR heptapeptide determines this system specificity. Variability in the specificity-encoding regions of plcR and papR genes suggests that selection and evolution of quorum-sensing systems play a major role in adaptation and ecology of Bacilli.
INTRODUCTION
Important bacterial processes, such as the production of antibiotics, induction of competence, sporulation and expression of virulence factors, are controlled in a multi-cellular manner via quorum-sensing systems. These communication systems are based on the secretion and recognition of cell–cell signalling molecules. Gram-negative bacteria generally use acyl-homoserine lactones as autoinducers, whereas in Gram-positive bacteria quorum sensing is commonly mediated by peptides or modified peptides. These peptides can act at different levels. In several Gram-positive bacteria, the sensor protein of a two-component system recognizes the inducer, triggering a phosphorylation cascade that targets a cognate regulatory protein. This is the case in the competence system, Com, of Streptococcus pneumoniae and in the virulence system, Agr, of Staphylococcus aureus (1,2). In others, for example the PrgX conjugative system of Enterococcus faecalis, Rap phosphatases of Bacillus subtilis and the PlcR regulator of Bacillus cereus, the autoinducer directly binds the regulatory protein (3,4,16). To gain further insight into the molecular mechanisms of coordinated virulence development by direct peptide recognition we carried out structure/function studies on PlcR and its cell–cell signalling peptide PapR.
The B. cereus group comprises a number of highly related species, which include B. thuringiensis, an insect pathogen, and B. anthracis, the aetiological agent of anthrax. The widespread presence of B. thuringiensis and B. cereus in soil and food, and their close relationship with B. anthracis make this group an important threat to public health, and a potential source of new pathogens. Indeed, B. cereus is generally regarded as a pathogen causing food-borne gastroenteritis (5,6). However, some rare but serious opportunistic non-gastrointestinal infections, for example endophthalmitis and pneumonia, have also been attributed to B. cereus (7–9). Bacillus cereus strains were also recently found to be responsible for severe infections resembling anthrax (10,11).
Several factors are involved in gastrointestinal and non-gastrointestinal diseases associated with B. cereus, which include exported proteins, such as enterotoxins, haemolysins, cytotoxins and various degradative enzymes (5,6). Production of most of these exported virulence factors is activated by PlcR at the onset of stationary phase (12–14). The papR gene encodes a 48-amino acid polypeptide with an N-terminal signal peptide sequence. This signalling peptide is exported and subsequently re-imported into the recipient cell via the oligopeptide permease system (15). During export, PapR is cleaved to liberate the active fragment comprising at least five C-terminal residues. PapR specifically interacts with PlcR inside the cell, allowing PlcR binding to its DNA target sites (16). This triggers a positive feedback on plcR expression and activates the expression of a regulon comprising about 100 genes (12–14,17–19).
PlcR is composed of a N-terminal helix–turn–helix (HTH) DNA-binding domain, and a C-terminal regulatory domain composed of five degenerated tetratricopeptide repeats (TPR) (14,20). TPRs are structurally conserved helical domains involved in protein–protein or protein–peptide interactions that have been identified in a large number of proteins in prokaryotes and eukaryotes (21). Recent crystallographic analysis shows that PapR binds to the centre of the concave side of the TPR domain. PapR recognition appears to trigger an allosteric mechanism that rearranges the HTH domains and thus allows DNA association (20). Structural and phylogenetic analyses further led to the identification of the RNPP family (named after the key members Rap/NprR/PlcR/PrgX) of quorum-sensing proteins. This family appears to comprise all Gram-positive quorum-sensing systems which bind directly to their signalling peptide in the receiver cell.
Four groups of PlcR–PapR pairs defining four distinct pherotypes were identified: PlcRI, PlcRII, PlcRIII and PlcRIV. These groups have been distinguished by in vivo complementation assays focused on plcR expression in Bacillus strain 407 and by the comparison of 29 PlcR–PapR sequences from various isolated strains representative of the B. cereus group (16,22). Thus for a given strain, PlcR activation depends on PapR originating from the same strain or from a member of the same pherogroup. Some level of specificity appeared to be determined by the first and the fifth residue of the pentapeptide form of PapR. The pentapeptide was identified as the minimal peptide size required for PlcR activation. However, the length of the physiologically relevant PapR remained elusive.
Here, we determined that the mature form of PapR that accumulates in the medium and inside bacterial cells corresponds to PapR C-terminal heptapeptide. We combined structure/function analysis to determine the molecular basis for specificity and cross-reactivity between PlcRs and heptameric PapRs from the four pherotypes.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The acrystalliferous B. thuringiensis 407 Cry− strain (23) was used in this study and designated as Bacillus strain 407. The 407 (plcA′Z) containing a transcriptional fusion between the plcA promoter and the lacZ gene, the PapR null-mutant 407 (plcA′Z ΔpapR) and the sporulating deficient 407 (Δspo0A) strains have been described previously (15,16). The type strain of 7, 12 and 45 B. thuringiensis serotypes originated from the Institut National de la Recherche Agronomique and Institut Pasteur collection. Escherichia coli K-12 strain TG1 was used as a host for the construction of plasmids and cloning experiments. Plasmid DNA for the electroporation of Bacillus was prepared from E. coli strains ET12567 or SCS110 (Stratagene, La Jolla, CA, USA). Plasmids were introduced by electroporation in E. coli and Bacillus strains, as previously described (23,24).
Escherichia coli and the Bacillus were grown in Luria Broth (LB) medium with vigorous shaking (175 r.p.m.) at 37°C (except for PlcR overproduction) or in a sporulation-specific medium (25) for PapR purification. The following antibiotic concentrations were used for bacterial selection: ampicillin 100 µg/ml (for E. coli), kanamycin at 200 µg/ml and erythromycin at 10 µg/ml (for Bacillus).
DNA manipulations
Chromosomal DNA was extracted from Bacillus cells using the Puregene DNA Purification Kit (Gentra Systems, USA). Plasmid DNA was extracted from E. coli using QIAprep spin columns (QIAgen, France). Restriction enzymes (New England Biolabs, USA) and T4 DNA ligase (Invitrogen, USA) were used in accordance with the manufacturer's recommendations. Oligonucleotide primers (Supplementary Table S1) were synthesized by Proligo (Paris, France). PCRs were performed in a thermocycler PTC-100 (MJ Research, Inc., USA). Amplified fragments were purified using the QIAquick PCR purification Kit (QIAgen). Digested DNA fragments were separated on 1% agarose gels after digestion and extracted from gels using the QIAquick gel extraction Kit (QIAgen).
Plasmids used for functional analysis were constructed as follow. Wild-type or modified plcR genes with their own promoters were amplified by PCR using chromosomal DNA and primers described in Supplementary Table S2. PCR fragments were digested by the appropriate enzymes and inserted between the XbaI and HindIII or PstI sites of pHT304 (26). The resulting plasmids and their features are described in Supplementary Table S2. Nucleotide sequences of cloned fragments were sequenced by Genome Express (France).
Construction of the 407 ΔplcR–papR recombinant strain
The thermosensitive allele exchange vector pMADΔplcR–papR was constructed using the following steps. The 5′ and 3′ regions of the plcR–papR locus were amplified by PCR, using primers po1/po2 and po3/orf4 (Supplementary Table S1). The 5′ end was purified as a NcoI/PstI fragment and the 3′ end as an XbaI/BamHI fragment. The kanamycin resistance gene was extracted from pDG783 (27) with PstI and XbaI. The three fragments were ligated with NcoI- and BamHI-digested pMAD (28). Bacillus strain 407 (plcA′Z) was electroporated with the resulting pMADΔplcR–papR plasmid. The plcR–papR locus of this strain was deleted by allele exchange using pMADΔplcR–papR, as previously described (29). The resulting strain was designated 407 (plcA′Z ΔplcR–papR). Integration was verified by PCR and sequencing using primers, vpo1 and vpo2 (Supplementary Table S1), complementary to chromosomal regions flanking the disrupted genes.
Functional analysis of PapR–PlcR interactions
Bacillus strain 407 (plcA′Z ΔplcR–papR) complemented with plasmids containing wild-type or mutant plcR genes with their own promoter were grown in LB medium without antibiotics at 37°C with vigorous shaking until the onset of the stationary phase T0 (OD600 3 ± 0.3). Each culture was fractionated (12 ml) and different synthetic peptides (Covalab, France) were added to a final concentration of 2 µM to each fraction. These cultures were incubated for 1 h and 2 ml were centrifuged (13 000 r.p.m., 8 min) to perform ß-galactosidase assays as previously described (30). Each assay was repeated at least three times.
PapR purification
Strain 407 Δspo0A in sporulation-specific medium over-expressed the PlcR regulon (31) and it minimizes contamination by medium components; thus, we performed these conditions to obtain sufficient quantity of the extra-cellular form of PapR. Bacteria were grown at 37°C and the supernatant was harvested at the onset of the stationary phase (T0) by centrifugation (15 min, 8000 r.p.m.) and filtered through 0.2 µm-pore size Nalgene filter unit. To test the presence of PapR, samples were dried out, re-suspended in 0.2 ml of LB broth and then added to a 1 ml stationary phase culture of 407 (plcA’Z ▵papR) (3<OD<6). These were incubated for 2 h at 37°C. Then, β-galactosidase assays were performed.
Samples were purified using solid-phase extraction on a SepPak C18 Vac 6cc cartridge (Waters, USA). The PapR sample was eluted with 90% methanol, precipitated for 1 h on ice, centrifuged and the supernatant was dried out. The sample, re-suspended in 100 µl of 0.1% trifluoroacetic acid (TFA) in water, was injected on a Superdex-peptide HR 10/30 column (Amersham, UK) in 30% acetonitrile (ACN), and 0.1% TFA. The active fraction was loaded on a µRPC column (alkyl residues C2/C18, Amersham). The elution was performed in 20-column volume linear gradient ranging from 25 to 35% ACN. The active samples were loaded on a Source 5-RPC column (polystyrene/divinyl benzene matrix, Amersham). The elution was performed in a 10-column volume linear gradient ranging from 25 to 35% ACN. The active fractions were injected on a Satisfaction C8+ column (C8, Cil-Cluzeau, France). The sample was eluted in a 10-column volume linear gradient, ranging from 25 to 35% ACN. The dried PapR fraction was loaded on a Lichrospher C18e (C18, Macherey-Nagel, Germany). The PapR fractions were dried out and re-purified on the µRPC column.
To extract intra-cellular PapR, wild-type 407 cells were grown in LB till T2 (2 h after the onset of the stationary phase). Centrifuged cells were re-suspended in water and were disrupted with glass beads as previously described (30). Cell extract was loaded on the C18 cartridge and PapR was eluted with 90% methanol and dried. The sample, re-suspended in 100 µl of 0.1% TFA, was purified on the Source 5-RPC column. The active fraction was injected on a C8+ column. The identity of the peptide was obtained by MALDI-TOF MS (Voyager DE super STR, Applied Biosystems, Foster City, USA) equipped with a nitrogen laser emitting at 337 nm. The peptide was re-suspended in 10 μl of 10% ACN. One micro litre of peptide was mixed on the stainless steel MALDI plate with 1 μl of CHCA (α-cyano-4-hydroxycinnamic acid, Sigma–Aldrich, Saint Quentin Fallavier, France) at 4 mg/ml in ACN/TFA (50:50; v/v) 0.3% and dried at room temperature. Spectra were recorded in positive reflector mode with 20 kV as accelerating voltage, a delayed extraction time of 130 ns and a 62% grid voltage and they were calibrated using an external calibration composed of human angiotensin II (M + H)+ = 1046.5423 (Sigma, France) and Des Arg1-bradykinin (M + H)+ = 757.3997 (Sigma). Mass spectra were analysed by Data Explorer 4.2 (Applied Biosystems, USA) with the following variables: noise filter/smooth (noise removal of 2), 0.5% base peak intensity, 0.5% maximum peak area and peak resolution of 10 000. Spectral profiles were collected in the mass range 500–2000 Da. All peptide masses were assumed to be monoisotopic and protonated molecular ions (M + H)+. The vMALDI ion source—MS/MS (LTQ Thermo Electron Corporation, San Jose, CA, USA)—was used with an automatic Gain Control (AGC) laser operation in order to maximize the creation of peptide information. Peptide ions were analysed using Xcalibur 1.4 (Thermo Electron). The Data Dependant Acquisition mode that allowed the selection of three precursor ions per survey scan was used. Precursor mass and fragment mass tolerance were 1.4 and 1 Da, respectively.
Overproduction and purification of PlcR
PlcR proteins were produced and purified as previously described (16)
Anisotropy binding titration
Steady-state fluorescence anisotropy binding titrations were performed at 21°C using a Beacon 2000 Fluorescence Polarization Instrument (Panvera, Corp., Madison, WI, USA). The concentration of the fluorescein-labelled pentapeptide LPFEF (Fl- LPFEF) was always 1.5 nM and the buffer used was 35 mM Tris–HCl, 16 mM sodium phosphate, 257 mM NaCl, 0.9 mM DTT and 0.05 mg/ml BSA, pH 8. The concentration of the non-labelled peptide in the competition assays was 121 µM. The anisotropy values shown correspond to the average of five to eight measurements taken by the instrument after equilibration. Binding data were analysed using Bioeqs software (32). This software uses a numerical solver engine and calculates the free energy of formation of the complexes from the initial elements. Analysis of the binding curves recovered in the absence of competitor peptide was performed using a model in which a single complex (PL*) was postulated. This model assumes that each PlcR monomer (P) bound a single molecule of the labelled peptide (L*). The binding profiles observed in the presence of non-labelled peptide (L) were analysed using a model involving two complexes, PL* and PL. The free energy of formation, and the steady-state anisotropy corresponding to the complex PL* were fixed to the values recovered in the analysis of the curves obtained in the absence of competitor. The error in the determined free energies was estimated using rigorous confidence limit testing at the 67% confidence level, in which the uncertainties arising from parameter correlations were taken into account.
Structural modelling
Structures were built based on the crystal structure of PapR-bound PlcR [group I, PDB entry 2QFC, (20)] using SWISS-MODEL. All modelled complexes were manually adjusted and subjected to structure idealization using REFMAC5 (33).
RESULTS
Purification of the active form of PapR
To determine the length of the processed active peptide, we purified the extra- and the intra-cellular forms of PapR from a group I strain (Bacillus strain 407). Samples were first fractionated by solid-phase extraction (SepPak C18 cartridge). Preliminary characterization indicated that this peptide (thereafter called PapRI) was heat stable (90°C, 20 min) with a molecular mass <1000 Da (it passed through Filtron MicroSep - 1KDa filters). PapRI was purified from the supernatant by one round of gel filtration chromatography, then by five rounds of high-pressure chromatography, leading to a level of purity compatible with direct mass spectrometry analysis. At each step, only one fraction was active.
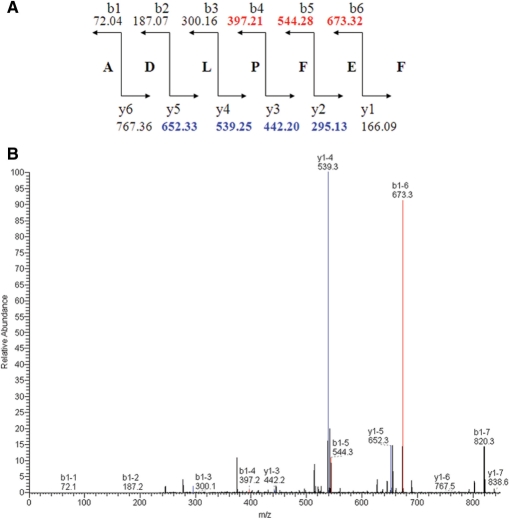
The MALDI-TOF measure of the last active chromatography fraction from the supernatant led to the identification, among several mass peaks, of a (M + H)+ =838.38 Da mass peak (data not shown) corresponding to the theoretical (M + H)+ = 838.39 Da mass of the heptapeptide PapRI (ADLPFEF). In order to disprove the presence of other PapR forms, LC/MS-MS analyses were performed on an active fraction. The results indicated that the mass corresponding to the pentapeptide PapRI (∼652 Da) was not recovered in this fraction. We next characterized the 838.39 Da mass using two approaches: PSD (Post-source Decay) using MALDI-TOF (Voyager DE STR) and the vMALDI ion source – MS/MS (Thermo Electron Corporation). The PSD mass spectrum was analysed using the Data Explorer software (Applied Biosystems). Seven MS/MS masses were determined as fragmentation masses of the sequence, ADLPFEF, according to the theoretical fragmentation of PapRI (Figure 1A). The data from the vMALDI-MS/MS fragmentation were analysed using the Bioworks 3.2 software (Thermo Electron Corporation). The xscore was 1.377 for the monocharge ion measured at (M + H)+ =838.4 Da. Seven masses for the theoretical fragmentation of PapRI (Figure 1A) were recovered (Figure 1B), similar to PSD mass spectrum analysis. These analyses confirmed ADLPFEF as the amino-acid sequence of PapRI.
Figure 1.
Theoretical fragmentation of PapRI and vMALDI-MS/MS mass spectrum. (A) Theoretical fragmentation of the peptide ADLPFEF (PapR7I). (B) vMALDI-MS/MS mass spectrum. Peaks in red correspond to b ion fragmentation expected masses, and peaks in blue to y ion fragmentation.
The intra-cellular active form of the cell–cell signalling peptide was purified after two HPLC steps and the heptapeptide PapRI sequence was found in LC/MS-MS, as a result of the MS/MS profile obtained in PSD or vMALDI-MS/MS. Thus, the mature and active peptide in the extra-cellular and intra-cellular environments corresponds to the last seven amino acids of PapR.
Modelling and affinity of the heptapeptide
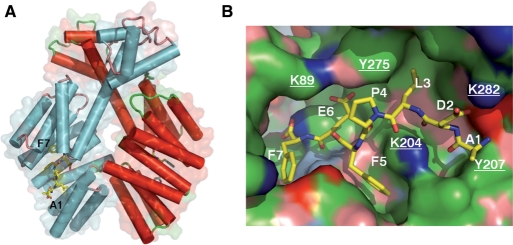
Previously, we have determined the crystal structure of a group I PlcR molecule bound to its cognate pentapeptide LPFEF (PapR5I) (20). We used this structure to establish a molecular model for the complex formed between PlcR and the heptapeptide ADLPFEF (PapR7I) (Figure 2A). In the crystal structure, the groove on PlcR that accommodates PapR5I continues after the N-terminal residue of the pentapeptide. This unoccupied part of the groove is positioned toward the C-terminal helix, and has a length corresponding to the two additional residues of the heptameric PapR (20). Indeed, the two additional residues (AD) of the ADLPFEF heptapeptide (PapR7I) fitted well into the groove extension. In the resulting model, the backbone of the N-terminal PapR7I residues, A, D and L, is anchored to PlcR by hydrogen bonds from K204, Q241 and D244 (Figure 2B). Y207 limits the peptide-binding groove, obstructing access for longer peptides. A salt bridge is possible between PapR7I D and K282 from the PlcR C-terminal helix. This model was used as a molecular basis for our further analyses, and for modelling PlcRII, III and IV and their cognate PapRs.
Figure 2.
Molecular model of PapR7I bound to PlcRI. (A) Atomic model of PlcRI:PapR7I. The structure was modelled based on PDB entry 2QFC. The two protomers of the PlcR dimer are coloured in red and cyan. Helices are represented by tubes, within the transparent molecular surface. PapR7I is shown as a stick model. (B) PlcR is shown as molecular surface and PapR7I as a stick model. Surfaces are coloured: blue, positively charged atoms; red, negatively charged atoms; green, hydrophobic atoms; salmon, polar oxygens; marine, polar nitrogens; yellow, sulphur. Underlined atom labels correspond to PlcR residues.
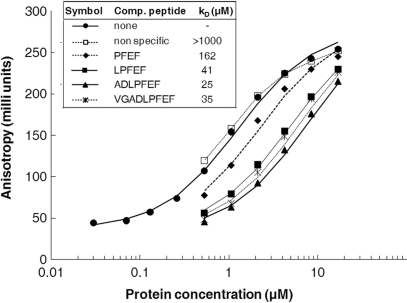
An initial prediction of the model was that the heptapeptide had a better affinity to PlcR than the shorter C-terminal PapR fragments, as it establishes more extensive contacts with PlcR. In contrast, longer C-terminal fragments should display a rather lower affinity due to the groove on PlcR being exactly the right size for the heptapeptide. We determined the binding affinity of PapR-derived peptides towards PlcR using competition experiments based on fluorescence anisotropy binding titrations. PlcR binding of the synthetic PapR5I (LPFEF) linked to a fluorescent label (fluorescein) was monitored by measuring fluorescence anisotropy changes in the presence of non-labelled peptides (Figure 3). Although fluorescein contributed to the free energy of binding, recognition of the fluorescent peptide probe was specific, as neither direct nor competitive binding was observed using control oligopeptides unrelated to the PapR C-terminal sequence. Assuming a single-site interaction model, a free energy change (▵G) of 7.9 ± 0.2 kcal/mol corresponding to a dissociation constant (Kd) of 1.3 µM was measured for the fluorescent peptide. Analysis of the competition profile yielded a Kd of 41 µM for the non-labelled pentapeptide LPFEF (▵G = 5.9 ± 0.2 kcal/mol). The heptapeptide (ADLPFEF) was a slightly better competitor than the 4, 5 or 9-residue PapR, displaying a Kd of 25 µM (Figure 3); this was consistent with our molecular model.
Figure 3.
Affinity of PlcR:PapR complexes. Anisotropy binding titrations of the fluorescein-labelled pentapeptide LPFEF (Fl- LPFEF) with PlcR, in the absence (closed circles) and in the presence of the competitor non-labelled peptides LPFEF (closed squares), PFEF (closed diamonds), ADLPFEF (closed triangles), VGADLPFEF (stars) and non-specific peptide (open squares). The concentration of Fl- LPFEF was 1.5 nM and that of the competitor peptides 121 µM. The Kd values shown correspond to analysis of the competition profiles using the model explained in Materials and methods section.
Penta- and hepta-peptides are comparable in their homologous activation potential
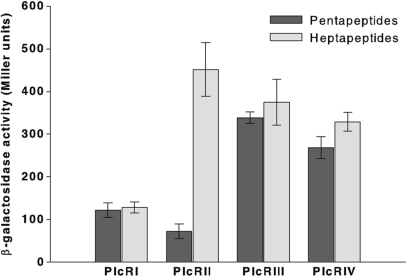
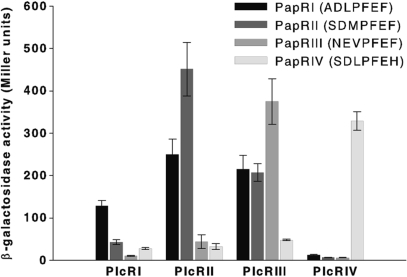
Slamti and Lereclus (2002) have shown that the PapR minimum size able to activate PlcR was the pentapeptide. To determine the effect of the two supplementary residues of the heptapeptide, we measured the activation level of each cognate PlcR–PapR complex using the 5- or the 7-amino-acid form of PapR. Plasmids containing plcR from one of the four pherotypes (PlcRI, PlcRII, PlcRIII or PlcRIV) (Supplementary Table S2) were introduced into the Bacillus reporter strain 407 (plcA′lacZ ΔplcR–papR) and synthetic PapRs were added to cultures at the onset of the stationary phase (T0). Thus, penta- or hepta- peptides of PapRI (LPFEF, ADLPFEF), PapRII (MPFEF, SDMPFEF), PapRIII (VPFEF, NEVPFEF) and PapRIV (LPFEH, SDLPFEH) were added to bacterial cultures expressing PlcRI, PlcRII, PlcRIII and PlcRIV, respectively. The activation of the plcA promoter was monitored using β-galactosidase assays 1 h after the peptides were added. The expression of plcA is controlled by PlcR (14), and thus, β-galactosidase production directly reflects PlcR activity in the bacterial cell. None of the four PlcRs was active in the absence of peptides (data not shown). Each PlcR in association with its specific PapR (5- or 7-amino-acids long) activated the expression the plcA′lacZ fusion (Figure 4). The results were consistent with our in vitro binding studies showing only a weak increase in affinity for the heptapeptide—PlcRI was similarly activated by its own specific 5- or 7-amino-acid PapRI. This was also the case for PlcRIII and PlcRIV and their cognate PapR penta- or hepta-peptides. However, the activity of PlcRII–PapRII was 6.5-fold greater with the heptapeptide added to the culture than the pentapeptide (Figure 4).
Figure 4.
Comparison of the activity of the four PlcRs in association with their cognate penta- and hepta-peptides. Synthetic penta- or hepta- peptides of PapRI (LPFEF, ADLPFEF), PapRII (MPFEF, SDMPFEF), PapRIII (VPFEF, NEVPFEF) and PapRIV (LPFEH, SDLPFEH) (2 µM) were added to a culture of 407 plcA′-lacZ ΔplcR-papR at stationary phase (OD600 3 ± 0.3), complemented with PlcRI (pHT304ΩplcRI), PlcRII (pHT304ΩplcRII), PlcRIII (pHT304ΩplcRIII) and PlcRIV (pHT304ΩplcRIV), respectively. β-Galactosidase assays were performed 1 h after peptide addition. Vertical bars: SEM.
Cross-talk between the four PlcR groups
Previous studies, restricted to the specific activation of PlcRI from Bacillus strain 407, revealed the existence of four PlcR groups (22). To determine the relationships between the four groups and the specificity of activation for the quorum-sensing system, we measured the activity of one PlcR from each group associated with one of the four heptapeptides using plcA-directed lacZ expression. This functional analysis showed that each specific pair (PlcRI:PapRI, PlcRII:PapRII, PlcRIII:PapRIII and PlcRIV:PapRIV) was more active than the heterologous pairs (Figure 5). However, specific activities ranged from 100 Miller units (PlcRI:PapRI) to 500 Miller units (PlcRII:PapRII). PlcRI:PapRI was also 3- to 4-fold less active than the non-cognate pairs, PlcRII:PapRI, PlcRIII:PapRI and PlcRIII:PapRII. PlcRIII was non-specifically activated by PapRI, PapRII, but not by PapRIV. PlcRIV was strongly and strictly activated by its corresponding heptapeptide (Figure 5). These results confirm the existence of four pherotypes. Furthermore, although activity of PlcRI and PlcRIV is dependent on their cognate peptides, PlcRII and III are less restrictive and can be activated by heterologous PapRs.
Figure 5.
Cross-talk between the four PlcR groups. Synthetic heptapeptides PapRI (ADLPFEF), PapRII (SDMPFEF), PapRIII (NEVPFEF) or PapRIV (SDLPFEH) (2 µM) were added to a stationary phase culture of 407 plcA′-lacZ ΔplcR-papR (OD600 3 ± 0.3) complemented with PlcRI, PlcRII, PlcRIII and PlcRIV, respectively. β-Galactosidase assays were performed 1 h after peptide addition. Vertical bars: SEM.
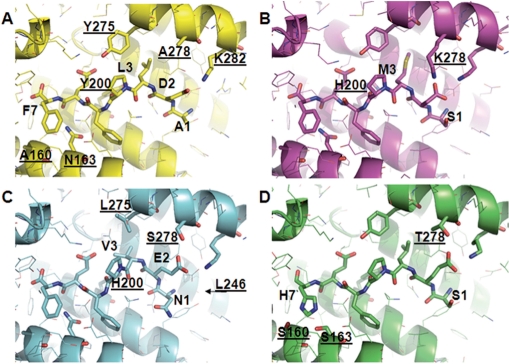
These observations are consistent with our structural models: PlcRI provides fewer interactions with PapR than PlcR of other groups (Figure 6A), explaining why PlcRI displays the lowest PapR-induced activity. In PlcRII, K278 is well positioned to form a salt-bridge with the heptapeptide aspartic acid (Figure 6B). The additional binding energy may, at least in part, account for the higher activation levels of PlcRII by PapR heptapeptides (Figure 5). The Y275L and Y200H substitutions in PlcRIII, combined with the PapR L3V substitution, are predicted to pull PapR closer to the C-terminal helix (Figure 6C). The hydrogen bond of H200 with the PapR P4 backbone, and because H200 is smaller than Y200, will result in the N-terminus of PapR being pulled a little closer towards H200. Thus, PapRIII E2 appears able to bind simultaneously to S278 and K282, and PlcRIII is able to accommodate the slightly bulkier N-terminal asparagine of PapRIII. PlcRIII has evolved to accommodate PapRIII with its bulkier N-terminal asparagine side chain, and thus should also be able to bind the smaller PapRI and PapRII peptides. However, the supplementary H-bond between PlcRIII S278 and PapRIII E2 appears inaccessible for D2 of PapRI and PapRII, explaining the lower activation rate for these heterologous interactions. PapRIV has the unique feature of a histidine in position 7. On the side of PlcRIV, this is accompanied by the changes A160S and N163S, which are positioned to form H-bonds with the nitrogens of the H7 side chain (Figure 6D). Together, these substitutions confer a polar, rather than a hydrophobic interaction between PapR position 7 and the underlying binding surface. As a result, PlcRIV will only be activated by its corresponding peptide.
Figure 6.
Model structures for the complexes formed by PlcRI–IV and their cognate PapR heptameric peptides. Underlined atom labels correspond to PlcR residues. (A) Molecular detail of PlcRI (ribbon representation) in a complex with PapRI (stick model). (B) PlcRII in a complex with PapRII. (C) PlcRIII in a complex with PapRIII. (D) PlcRIV in a complex with PapRIV. The arrow indicates the L246 residue.
TPR-mediated activation of the PlcR–PapR system
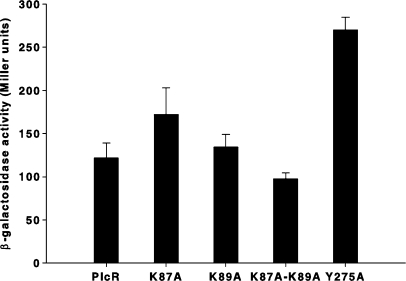
We next characterized the function of the interactions between PlcR and the conserved, non-specific, PapR hydrophobic core (FEF sequence in PapRI). Structural analysis of the PlcR–PapR interactions showed that the conserved PlcR K87 and K89 residues directly interact with PapR E6 and that the aromatic PlcR Y275 residue establishes a hydrogen bond with both PlcR K89 and PapR E6 [Figure 2B, (20)]. Substitution of these lysines by alanines (as a single mutation or in tandem) did not affect PlcR activity (Figure 7), suggesting that these interactions were not important for activation. Their interactions with PapR glutamic acids may function to selectively allow PapR, but not other similar autoinducers, to bind PlcR. The Y275A substitution results in a 2-fold increase of the PlcR response (Figure 7). The Y275 side chain stabilizes the position of the C-terminal capping helix of PlcR by intercalating with the preceding TPR repeat. In the PlcRI–PapRI complex, this C-terminal helix is quite mobile, as witnessed by above average B-factors (20). Possibly, a slight inward movement of this helix contributes to PlcR activation. This action may be facilitated in the Y275A mutant.
Figure 7.
Activation of modified PlcR proteins. Synthetic heptapeptides of PapRI (ADLPFEF) (2 µM) were added to a culture of 407 plcA′-lacZ ΔplcR–papR (OD600 3 ± 0.3) at stationary phase complemented with PlcRK87A, PlcRK89A, PlcRK87A-K89A and PlcRY275A, respectively. The PlcR–PapR complex activity was measured by β-galactosidase assays 1 h after peptide addition. Vertical bars: SEM.
The C-terminal helix of PlcR is required but not sufficient for specific PapR-mediated activation
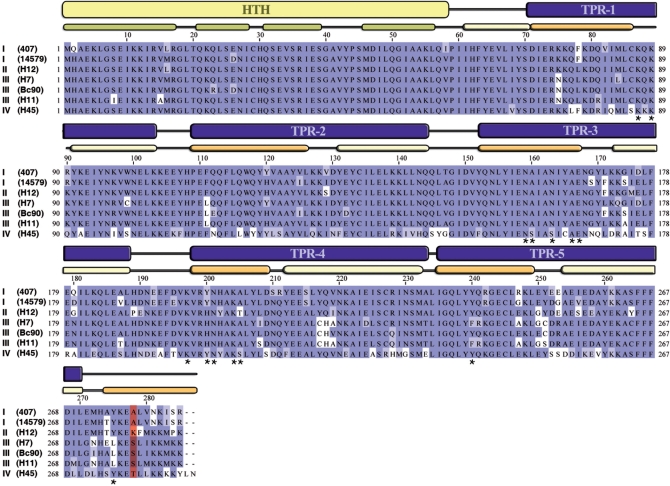
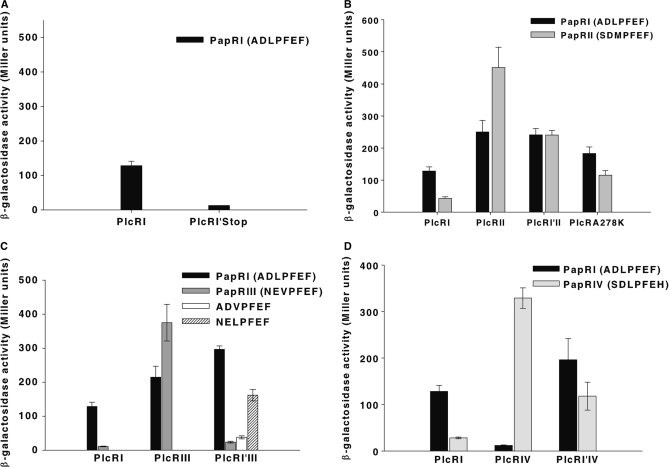
To investigate the regions of PlcR conferring specificity towards PapR, 29 PlcR sequences were compared (22). This analysis revealed seven characteristic PlcRs, which differed only in their TPR domains (Figure 8). Furthermore, only position 278 characterizes the four specific PlcR groups: at position 278, group I PlcRs contained an alanine, group II a lysine, group III a serine and group IV a threonine (Figure 8). Inspection of the 3D PlcRI model showed that A278 is located on the C-terminal helix, oriented towards the bound PapRI heptapeptide (Figure 2). To investigate the role of the C-terminal helix of PlcR and specifically of its residue 278, the following constructions have been done: (i) a PlcR molecule was truncated at position 278, (ii) three chimeric molecules were constructed by exchanging the C-terminal ends starting from the position 278 of PlcRI with that of the three other groups and (iii) PlcRI A278 was substituted by K278 according to PlcRII sequence. Chimeras between PlcRI and PlcRII, PlcRI and PlcRIII, PlcRI and PlcRIV were called PlcRI'II, PlcRI'III and PlcRI'IV, respectively. PlcRI A278K substitution was called PlcRA278K (Supplementary Table S2). The PlcR constructs were introduced into the plcR–papR-null mutant 407 strain. The constructs were then functionally analysed by testing their ability to activate a lacZ reporter fused to the plcA promoter, in response to various PapRs. None of the chimeras, PlcRI'II, PlcRI'III and PlcRI'IV, was active in the absence of peptide (data not shown). Truncated PlcR could not activate lacZ expression in response to PapR addition (Figure 9A), suggesting that the C-terminal helix is required for PlcR activation or protein stability. Indeed, we cannot assure the stability of the mutant form, or its correct folding because immunodetection of cytosolic PlcR (either truncated or wild type) was unsuccessful.
Figure 8.
Sequence alignment of seven representative PlcR sequences. Sequence alignment of seven representative PlcRs was performed with JalView, using the Blosum62 colouring scheme. The strain names are indicated in parentheses and the roman numbers refer to the PlcR groups. Based on 3D-structure, HTH domain and TRP domains are indicated in yellow and in blue, respectively. The α-helices from the HTH domain are in green. The first and the last α-helices from TPR domains are in light yellow and in orange, respectively. Asterisks represent PlcR residues implicated in PapR binding. Position 278 is coloured in red.
Figure 9.
Activation of chimeric PlcR proteins. Synthetic heptapeptides of PapRI (ADLPFEF), PapRII (SDMPFEF), PapRIII (NEVPFEF) or PapRIV (SDLPFEH) (2 µM) were added to a culture of 407 plcA′-lacZ ΔplcR–papR (OD600 3 ± 0.3) at stationary phase complemented with PlcRI, PlcRI′stop (A), PlcRI′II and PlcRA278K (B), PlcRI′III (C) and PlcRI′IV (D), respectively. The PlcR–PapR complex activity was measured by β-galactosidase assays 1 h after peptide addition. Vertical bars: SEM.
The PlcRI'II chimera was similarly activated by both PapRI and PapRII. This activation was about 2-fold greater than that of wild-type PlcRI by PapRI, and 2-fold less than that of PlcRII by PapRII (Figure 9B). The PlcRI'III chimera was activated about 2-fold better by PapRI than PlcRI. However, PlcRI'III was not activated by PapRIII (Figure 9C). According to the structural models (Figure 6C), the PapRIII valine is highly specific to PlcRIII, requiring a leucine in position 275 of PlcR. Indeed, a V3 to L3 modified PapRIII (NELPFEF) partly restores the PlcRI'III-activating potential, but it is still less than that observed with PapRI (ADLPFEF). The PlcRI'IV chimera also showed an intermediate phenotype in relation to the PapRI and PapRIV peptides (Figure 9D). The PlcRA278K mutant was activated by both PapRI and PapRII. This activation was similar to PlcRI'II response (Figure 9B). Altogether, these studies suggest a role for the C-terminal PlcR helix in modulating PapR response. In addition, it appears that this role is in part due to the 278 residue. All chimeric proteins were more responsive to PapRI than wild-type PlcRI. We investigated structural and sequence data highlighting positions 278 and 281 to understand better why all chimeric proteins were more responsive to PapRI than PlcRI. In groups II, III and IV, these positions provide side chains capable of forming ionic or polar bonds with E2 or D2 of PapR. The increase in chimera reactivity to PapRI can therefore be, at least partly, explained by greater affinities for PapRI. In turn, the PlcRI portion of the chimera is less optimal for PapRII, III and IV, in relation to their affinity and thus, activation capacity is diminished. Additionally, position 283 might also play a role: PlcRII, III and IV, but not PlcRI have an amino acid with a long aliphatic side chain in this location (M, M and K, respectively, as opposed to I in PlcRI). These side chains point towards the preceding TPR motif, as observed as a result of the Y275A mutation, but these side chains are on the other face of the C-terminal helix. We cannot exclude that these residues contribute partly to activation by modulating the stability and position of the C-terminal helix.
DISCUSSION
Dissecting specificity and function in the PapR/PlcR quorum sensing
Quorum-sensing peptides that bind to a membrane receptor, such as the competence stimulating factor from B. subtilis and Streptococcus species and the virulence Agr signal in Staphylococcus, have been characterized and purified in the culture medium (34–37). In contrast, signalling peptides that bind directly to the effector (PlcR, Rap phosphatases) have more often been identified on the basis of genetic studies (4,16,38). Nevertheless, Solomon et al. (39) have identified an extra-cellular peptide that controls, via Rap phosphatase, the competence/sporulation pathways in B. subtilis. In this study, we purified a processed active PapR in the culture supernatant from a group I strain (Bacillus strain 407) and showed that the detectable form of secreted PapR is the heptapeptide ADLPFEF. In addition, this heptapeptide was also found in the cytoplasm, suggesting that PapR does not undergo further processing during or after uptake. However, the isolation of the heptapeptide does not preclude the presence of other minor undetectable active forms.
The PlcR-related Rap-Phr competence and sporulation system in B. subtilis involves the cleavage of the signalling peptide in the extra-cellular medium by one or several peptidases such as subtilisin, Vpr and Epr (40). The cleavage of Phr requires an alanine residue just before the N-terminus of the cleavable peptide bond (41). In B. subtilis strain 168, introduction of plcR and papR genes activate the expression of PlcR-regulated genes (12,14), indicating that PapR processing also occurs in this heterologous host. The heptapeptide PapR sequence is also preceded by an alanine. Apart from suggesting that PapR is 7-amino-acid-long, these results also suggest that Phr and PapR might be processed via a similar mechanism.
Our results confirmed that the PapR pentapeptides retained the ability to activate PlcR of all groups. However, pentapeptides displayed a slightly lower affinity for their cognate PlcR molecule than heptapeptides. Furthermore, pentapeptides were less group-selective. Indeed, the PlcRII activity is lower with its cognate pentapeptide PapRII than with the heterologous pentapeptide PapRI (Supplementary Figure S1). We have previously reported that the N-terminal amino acid of the PapR pentapeptide is implicated in selection; thus, we conclude that the three N-terminal residues of PapR direct group specificity. Our anisotropy assays and structural analysis support that the impact of these residues lies mainly in modulating the affinity of a given PapR towards various PlcR groups. The K87A and K89A mutants did not abolish PlcR activation by PapR, suggesting that activation of PlcR by PapR is mainly triggered by the hydrophobic interactions of PapR residues 3, 5 and 7 (L, F and F in PapRI) with helices 5 and 7 of the TPR-containing domain of PlcR. We speculate that lysines 87 and 89 function as gatekeepers, selecting PapR from other oligopeptides by ionic interactions with the glutamic acid of the FEF PapR core motif. Similarly, the central proline residue may be required for the PapR peptides to fit into the binding groove on PlcR, thus helping discriminate against other cell–cell signalling peptides.
Our results also highlight a role for the residue 278 within the PlcR C-terminal capping helix in specific activation. PapR may enhance the TPR modifications necessary for activation by pulling the PlcR C-terminal helix a little towards the centre of the molecule. Hydrophobic interactions between PlcR Y275 and PapR L3, and polar interactions between PlcR Y275 and PapR E4 appear important for this effect; PlcR group-specific residues between position 278 and the C-terminus also appear important. The enhanced activity of the PlcR Y275A mutant indicates that a slight rearrangement of the C-terminal helix accompanies PlcR activation. We can, however, not exclude a role of this helix in recruiting additional components.
Evolution of the specific PlcR–PapR activation
The plcR and papR genes are adjacent on the bacterial chromosome. Structural and phylogenetic analyses (20) and the observation that a fused PlcR–PapR protein is able to activate the PlcR regulon (42) support that the plcR and papR gene ancestors were initially one single gene. Our results now infer that following their separation, selective pressure on plcR and papR genes has led to their co-evolution, producing distinct PlcR/PapR groups. The mutation rate of PlcR is higher than that of other chromosomal genes, whereas inactivation of PlcR by a nonsense mutation in the B. anthracis strains leads to a low mutation rate similar to other chromosomal genes (43). The plcR sequence analysis showed that mutations are not clustered, both at the nucleotide and amino acid levels (43); however, the PlcR residues that interact with the non-selective core motif of PapR are conserved, except for the F7 to H7 mutation seen in the highly specific PlcRIV (Figure 8). Together, these observations support that efficient bacterial survival requires PlcR to associate with PapR, but also to allow variability and adaptability of this interaction, achieving strain-specificity among Bacilli.
Ji et al. (44) suggested that group-specific differences in the expression of virulence factors or other extra-cellular factors in Staphylococcus could be related to differences in disease patterns. However, current data in relation to PlcR-regulated virulence do not appear to corroborate a link between virulence characteristics and PlcR/PapR groups: the pathogenic B. cereus strains 391–98 [responsible for the death of three people (45)] and G9241 [possessing the anthrax toxin genes and a functional PlcR regulon (10)] belonged both to PlcR group IV, whereas the virulence of other group IV strains has not been highlighted (i.e. B. thuringiensis serotype 45 type strain, B. thuringiensis Bt51 and B. weihenstephanensis KBAB4). In contrast, several mammalian pathogen strains belong to the PlcRIII group: B. thuringiensis 97-27 subsp. konkukian, isolated from a necrotic human wound (46); B. cereus E33L, isolated from a carcass swab (47); and the major producer of commercially used biopesticides, the B. thuringiensis strain kurstaki HD1 Dipel (48).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank N. Daou, C. Nielsen-LeRoux and N. Ramarao for the critical reading of the manuscript and G. André-Leroux for helpful discussions. We also acknowledge S. Leppla for the gift of antibodies and A. Guillot for the mass spectrometry technical help, A. Chavanieu and J.-F. Guichou for the synthesis of PapR peptides. S.Z. is the recipient of a Marie-Curie European International Fellowship (MCEIF-CT-2004-007320). L.B. received a PhD grant from the Institut National de la Recherche Agronomique (MICA) and the région Ile de France. This work was supported by the Agence Nationale de la Recherche (PNRA 013-04). Funding to pay the Open Access publication charges for this article was provided by INRA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dunny GM, Leonard BA. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 2.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 3.Shi K, Brown CK, Gu ZY, Kozlowicz BK, Dunny GM, Ohlendorf DH, Earhart CA. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc. Natl Acad. Sci. USA. 2005;102:18596–18601. doi: 10.1073/pnas.0506163102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perego M, Brannigan JA. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides. 2001;22:1541–1547. doi: 10.1016/s0196-9781(01)00490-9. [DOI] [PubMed] [Google Scholar]
- 5.Granum PE, Lund T. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 1997;157:223–228. doi: 10.1111/j.1574-6968.1997.tb12776.x. [DOI] [PubMed] [Google Scholar]
- 6.Kotiranta A, Lounatmaa K, Haapasalo M. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2000;2:189–198. doi: 10.1016/s1286-4579(00)00269-0. [DOI] [PubMed] [Google Scholar]
- 7.Miller JM, Hair JG, Hebert M, Hebert L, Roberts F.J., Jr., Weyant RS. Fulminating bacteremia and pneumonia due to Bacillus cereus. J. Clin. Microbiol. 1997;35:504–507. doi: 10.1128/jcm.35.2.504-507.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callegan MC, Booth MC, Jett BD, Gilmore MS. Pathogenesis of gram-positive bacterial endophthalmitis. Infect. Immun. 1999;67:3348–3356. doi: 10.1128/iai.67.7.3348-3356.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilliard NJ, Schelonka RL, Waites KB. Bacillus cereus bacteremia in a preterm neonate. J. Clin. Microbiol. 2003;41:3441–3444. doi: 10.1128/JCM.41.7.3441-3444.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, et al. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl Acad. Sci. USA. 2004;101:8449–8454. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klee SR, Ozel M, Appel B, Boesch C, Ellerbrok H, Jacob D, Holland G, Leendertz FH, Pauli G, Grunow R, et al. Characterization of Bacillus anthracis-like bacteria isolated from wild great apes from Cote d’Ivoire and Cameroon. J. Bacteriol. 2006;188:5333–5344. doi: 10.1128/JB.00303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agaisse H, Gominet M, Okstad OA, Kolsto AB, Lereclus D. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 1999;32:1043–1053. doi: 10.1046/j.1365-2958.1999.01419.x. [DOI] [PubMed] [Google Scholar]
- 13.Okstad OA, Gominet M, Purnelle B, Rose M, Lereclus D, Kolsto AB. Sequence analysis of three Bacillus cereus loci carrying PIcR-regulated genes encoding degradative enzymes and enterotoxin. Microbiology. 1999;145:3129–3138. doi: 10.1099/00221287-145-11-3129. [DOI] [PubMed] [Google Scholar]
- 14.Lereclus D, Agaisse H, Gominet M, Salamitou S, Sanchis V. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J. Bacteriol. 1996;178:2749–2756. doi: 10.1128/jb.178.10.2749-2756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gominet M, Slamti L, Gilois N, Rose M, Lereclus D. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 2001;40:963–975. doi: 10.1046/j.1365-2958.2001.02440.x. [DOI] [PubMed] [Google Scholar]
- 16.Slamti L, Lereclus D. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 2002;21:4550–4559. doi: 10.1093/emboj/cdf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature. 2003;423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- 18.Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, et al. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 19.Rasko DA, Ravel J, Okstad OA, Helgason E, Cer RZ, Jiang L, Shores KA, Fouts DE, Tourasse NJ, Angiuoli SV, et al. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 2004;32:977–988. doi: 10.1093/nar/gkh258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Declerck N, Bouillaut L, Chaix D, Rugani N, Slamti L, Hoh F, Lereclus D, Arold ST. Structure of PlcR: insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. Proc. Natl Acad. Sci. USA. 2007;104:18490–18495. doi: 10.1073/pnas.0704501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 22.Slamti L, Lereclus D. Specificity and polymorphism of the PlcR-PapR quorum-sensing system in the Bacillus cereus group. J. Bacteriol. 2005;187:1182–1187. doi: 10.1128/JB.187.3.1182-1187.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lereclus D, Arantes O, Chaufaux J, Lecadet M. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 1989;51:211–217. doi: 10.1016/0378-1097(89)90511-9. [DOI] [PubMed] [Google Scholar]
- 24.Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lecadet MM, Blondel MO, Ribier J. Generalized transduction in Bacillus thuringiensis var. berliner 1715 using bacteriophage CP-54Ber. J. Gen. Microbiol. 1980;121:203–212. doi: 10.1099/00221287-121-1-203. [DOI] [PubMed] [Google Scholar]
- 26.Agaisse H, Lereclus D. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 1994;13:97–107. doi: 10.1111/j.1365-2958.1994.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 27.Guerout-Fleury AM, Shazand K, Frandsen N, Stragier P. Antibiotic resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 28.Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lereclus D, Agaisse H, Gominet M, Chaufaux J. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spo0A mutant. Bio/Technology (NY) 1995;13:67–71. doi: 10.1038/nbt0195-67. [DOI] [PubMed] [Google Scholar]
- 30.Bouillaut L, Ramarao N, Buisson C, Gilois N, Gohar M, Lereclus D, Nielsen-Leroux C. FlhA influences Bacillus thuringiensis PlcR-regulated gene transcription, protein production, and virulence. Appl. Environ. Microbiol. 2005;71:8903–8910. doi: 10.1128/AEM.71.12.8903-8910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lereclus D, Agaisse H, Grandvalet C, Salamitou S, Gominet M. Regulation of toxin and virulence gene transcription in Bacillus thuringiensis. Int. J. Med. Microbiol. 2000;290:295–299. doi: 10.1016/S1438-4221(00)80024-7. [DOI] [PubMed] [Google Scholar]
- 32.Royer CA. Improvements in the numerical analysis of thermodynamic data from biomolecular complexes. Anal. Biochem. 1993;210:91–97. doi: 10.1006/abio.1993.1155. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 35.Havarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl Acad. Sci. USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen FC, Fimland G, Scheie AA. Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol. Microbiol. 2006;61:1322–1334. doi: 10.1111/j.1365-2958.2006.05312.x. [DOI] [PubMed] [Google Scholar]
- 37.Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl Acad. Sci. USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Core L, Perego M. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol. Microbiol. 2003;49:1509–1522. doi: 10.1046/j.1365-2958.2003.03659.x. [DOI] [PubMed] [Google Scholar]
- 39.Solomon JM, Lazazzera BA, Grossman AD. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 40.Lanigan-Gerdes S, Dooley AN, Faull KF, Lazazzera BA. Identification of subtilisin, Epr and Vpr as enzymes that produce CSF, an extracellular signalling peptide of Bacillus subtilis. Mol. Microbiol. 2007;65:1321–1333. doi: 10.1111/j.1365-2958.2007.05869.x. [DOI] [PubMed] [Google Scholar]
- 41.Stephenson S, Mueller C, Jiang M, Perego M. Molecular analysis of Phr peptide processing in Bacillus subtilis. J. Bacteriol. 2003;185:4861–4871. doi: 10.1128/JB.185.16.4861-4871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pomerantsev AP, Pomerantseva OM, Leppla SH. A spontaneous translational fusion of Bacillus cereus PlcR and PapR activates transcription of PlcR-dependent genes in Bacillus anthracis via binding with a specific palindromic sequence. Infect. Immun. 2004;72:5814–5823. doi: 10.1128/IAI.72.10.5814-5823.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ko KS, Kim JW, Kim JM, Kim W, Chung SI, Kim IJ, Kook YH. Population structure of the Bacillus cereus group as determined by sequence analysis of six housekeeping genes and the plcR Gene. Infect. Immun. 2004;72:5253–5261. doi: 10.1128/IAI.72.9.5253-5261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 45.Lund T, De Buyser ML, Granum PE. A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 2000;38:254–261. doi: 10.1046/j.1365-2958.2000.02147.x. [DOI] [PubMed] [Google Scholar]
- 46.Hernandez E, Ramisse F, Ducoureau JP, Cruel T, Cavallo JD. Bacillus thuringiensis subsp. konkukian (serotype H34) superinfection: case report and experimental evidence of pathogenicity in immunosuppressed mice. J. Clin. Microbiol. 1998;36:2138–2139. doi: 10.1128/jcm.36.7.2138-2139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han CS, Xie G, Challacombe JF, Altherr MR, Bhotika SS, Brown N, Bruce D, Campbell CS, Campbell ML, Chen J, et al. Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J. Bacteriol. 2006;188:3382–3390. doi: 10.1128/JB.188.9.3382-3390.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.