Abstract
An intricate interplay between DNA methylation and polycomb-mediated gene silencing has been highlighted recently. Here we provided evidence that Nervous System Polycomb 1 (NSPc1), a BMI1 homologous polycomb protein, plays important roles in promoting H2A ubiquitination and cooperates with DNA methylation in HOX gene silencing. We showed that NSPc1 stimulates H2A ubiquitination in vivo and in vitro through direct interaction with both RING2 and H2A. RT-PCR analysis revealed that loss of NSPc1, EZH2 or DNA methyltransferase 1 (Dnmt1), or inhibition of DNA methylation in HeLa cells de-represses the expression of HOXA7. Chromatin immunoprecipitation (ChIP) assays demonstrated that NSPc1, EZH2 and Dnmt1 bind to the promoter of HOXA7, which is frequently hypermethylated in tumors. Knockdown of NSPc1 results in significant reduction of H2A ubiquitination and DNA demethylation as well as Dnmt1 dissociation in the HOXA7 promoter. Meanwhile Dnmt1 deficiency affects NSPc1 recruitment and H2A ubiquitination, whereas on both cases EZH2-mediated H3K27 trimethylation remains unaffected. When EZH2 was depleted, however, NSPc1 and Dnmt1 enrichment was abolished concomitant with local reduction of H3K27 trimethylation, H2A ubiquitination and DNA methylation. Taken together, our findings indicated that NSPc1-mediated H2A ubiquitination and DNA methylation, both being directed by EZH2, are interdependent in long-term target gene silencing within cancer cells.
INTRODUCTION
Polycomb group (PcG) proteins were first discovered in Drosophila melanogaster and well known to be required to prevent inappropriate expression of homeotic (Hox) genes. They form multiprotein complexes (polycomb repressive complexes, PRCs) and possess intrinsic enzymatic activities towards histones through which they repress their target genes. The PRC2 (EZH2-EED) possesses H3K27 methyltransferase activity (1,2). And the PRC1 components were identified to ubiquitinate H2A (3), generally following the EZH2-mediaed H3K27 methylation (4). In these complexes, RING2 (/Ring1b) is the main catalytic subunit and its E3 ligase activity is known to be enhanced by BMI1 both in vivo and in vitro (5,6). A possible molecular mechanism of H2A ubiquitination in gene silencing has been recently suggested through preventing FACT recruitment and hence inhibiting transcriptional elongation (7).
DNA methylation is another type of repressive modification and its close relationship with PcG-mediated gene silencing has been widely studied in recent years (8–17). Abnormal DNA methylation is frequently recruited by PcG-mediated repressive mark to many tumor suppressor genes which are silenced in adult cancer cells (12,14,18). The finding that links EZH2 with DNMTs has established a role for this protein during the induction and targeting of DNA methylation (15). Recently PRC1 proteins also have been found to be associated with DNA methylation. Sakamoto et al. (10) found that Methylated DNA-binding domain proteins (MBD1) interact with Ring1b and hPc2 and they synergistically repress the transcription of HOXA genes. Furthermore, Dnmt1-associated protein 1 (Dmap1) known to associate with Dnmt1 was found to directly interact with Bmi1 and they were co-localized at their target genes (8). The regulator of DNA methylation Lsh brings Bmi1 and Dnmts together and controls Hox gene silencing (17). Yet the relationship between PRC1-mediated H2A ubiquitination and DNA methylation in mammals is barely investigated. The only indirect clue from research in Arabidopsis suggested that H2B deubiquitination by SUP32/UBP26 is required for heterochromatic histone H3 methylation and DNA methylation (19).
NSPc1 encodes a protein that is highly homologous to Mel18 and Bmi1 (20). We previously reported that NSPc1 promotes cell cycle progression through repressing the transcription of Cyclin-dependent kinases inhibitor (CKI) p21 (21). Then we tried to elucidate the mechanisms underlying its transcriptional repression. Tandem Affinity Purification (TAP) experiment identified NSPc1 as the core component of a new H2A ubiquitination complex (BCOR complex which contains RYBP, RING1, NSPc1 and RING2) (22). Here we provided evidence that NSPc1 promotes H2A ubiquitination in vivo and in vitro, during which the intact RING finger motif is required because it is necessary for its interaction with RING2. Analysis of target genes for NSPc1 in HeLa cells followed by ChIP assay revealed that NSPc1-mediated H2A ubiquitination is required for HOXA7 transcriptional repression. Moreover, manipulation of the H3K27 methylation, H2A ubiquitination and DNA methylation by knockdown EZH2, NSPc1 or Dnmt1, respectively, allowed us to address the relationship among these three epigenetic modifications. Thus, our work not only provided evidence that supports an essential role for NSPc1 in H2A ubiquitination, but also illustrated the importance of cooperation between H2A ubiquitination and DNA methylation in long-term gene silencing. It has important implications in understanding the molecular mechanisms by which polycomb proteins participate in stem cell self-renewal and cancer.
MATERIALS AND METHODS
Cell culture, cloning, antibodies and other reagents
HeLa, 293T and NT2/D1 cells (ATCC) were cultured in DMEM (GIBCO) supplemented with 10% FBS (Hyclone). NSPc1 was subcloned into the pcDEF-Flag vector (Shanghai Genomics) or pGEX6P-1 (Promega). H2A and RING2 [gifts from Zhang (3)] were subcloned into the pcDEF-Myc vector. NSPc1 RING finger mutant construct C56S was generated with the QuickChange Site-Directed Mutagenesis Kit (Stratagen) following the protocol recommended by the manufacturer. The primers used are listed in the Supplementary Table 1. Two siRNAs (sequences seen in the Supplementary Table 1) against human NSPc1 were used to knock down NSPc1 expression. And siRNA-2 was inserted into pSUPER-EGFP vector (pSE) as described previously (23). pGEX-MDM2 was a kind gift from Oren (24). The EED and EZH2 knockdown construct were nicely sent from Bernards (25). The target sequences against EED and EZH2 are listed in Supplementary Table 1. Lentivirus RNAi construct pLVTHM-Dnmt1 came from Jagodzinski, P. P. Polyclonal antibody against NSPc1 was raised in rabbit using synthetic peptides YRYDEQLNLCLERLSSGKDKNKSVL. Its specificity was confirmed by western blot and RNAi experiments (Supplementary Figure 1). Antibodies against EZH2 and Ring1B (specifically recognizes both mouse Ring1b and human RING2) were purchased from Aviva and MBL, respectively. Antibodies against H2A, ubiquitinated H2A (uH2A), trimethyl-H3 K27 (m3H3K27), Dnmt1 and HADC1 were from Upstate. Anti-β-tubulin was from Chemicon. DMSO and 5-aza-dC were purchased from Sigma. The recombinant E1 and a set of E2 proteins were from Calbiochem.
In vitro histone interaction assays
The core histones GST pull-down assay was done as previously described (24). In brief, about 5 μg GST fusion proteins, immobilized on glutathione sepharose 4B beads (Amersham), were incubated with about 10 μg of acid-extracted core histones in NP-40 buffer (120 mM NaCl, 1 mM EDTA, 20 mM Tris pH 8.0, 0.5% NP-40) supplemented with 0.5% BSA for 1 h at 4°C. After three washes in NP-40 buffer without BSA, the beads were boiled in sample loading buffer, and the released proteins were resolved by SDS–PAGE (12%). The gel was stained with Coomassie Brilliant Blue (CBB).
Co-immunoprecipitation analysis
293T cells were transfected with Myc-H2A, or co-transfected with Myc-RING2 and Flag-NSPc1 or RING finger mutant Myc-C56S using the Lipofectamin 2000 (Invitrogen). Twenty-four hour later, cells were harvested, washed twice with ice-cold PBS, re-suspended in NP-40 buffer and sonicated for three periods of 15 s on ice. For the endogenous CoIP assay for NSPc1 and H2A, wild-type HeLa cells were extracted similarly. The extracts were incubated with rabbit anti-Myc, anti-NSPc1 polyclonal antibody or IgG as a negative control for 3–4 h at 4°C in NP-40 buffer. And Protein A Sepharose beads (Roche) were added and incubated for another 1 h. Beads were then washed and boiled as mentioned earlier, followed by SDS–PAGE and western blot with respective antibodies.
In vivo ubiquitination analysis
Transfected 293T cells were harvested 48 h post transfection, lysed and extracts immunoprecipitated with anti-Myc antibody as described (24). Mouse anti-Myc and anti-HA antibodies were respectively used to detect the ubiquitinated H2A which was formed by the overexpressed Myc-H2A and HA-Ubiquitin.
In vitro ubiquitination assay
Nucleosomes were prepared from HeLa cells (Supplementary Data). In vitro ubiquitin ligase reaction was performed by incubating 0.1 μg ubiquitin activating enzyme E1 (Calbiochem), 0.6 μg ubiquitin conjugating enzyme UbcH5c (E2) (Calbiochem), 1 μg Flag-ubiquitin (Sigma), 5 μg prepared nucleosomes with indicated GST recombinant proteins in a 36 μl reaction containing 50 mM Tris–HCl (pH 7.9), 5 mM MgCl2, 2 mM NaF, 1 mM DTT, 2 mM ATP, 1 mM NaVO3. Purified GST-NSPc1 (3 mg/ml) was titrated to a series of concentration (0.2, 0.4, 1 mg/ml). After incubation at 37°C for 1 h, reaction was terminated by addition of sample loading buffer. The proteins were resolved in 12% SDS–PAGE and blotted with the anti-uH2A antibody.
Reverse transcription (RT)-PCR and quantitative real-time RT-PCR
Total RNA was isolated from HeLa or NT2/D1 cells using reagent Trizol (Invitrogen) according to the manufacturer's instructions and thereafter reverse-transcribed. RT-PCR exponential phase was determined on 25–35 cycles to allow semi-quantitative comparisons among cDNAs developed from identical reactions. Quantitative real-time PCR of the target cDNAs was performed by the SYBR Green method using Power SYBR Green PCR Master Mix (Takara). The fold relative enrichment was quantified together with normalization by the GAPDH level. The primers used are listed in Supplementary Table 1.
RNA interference and stable cell line establishment
Diluted siRNA (20 µM) or RNAi constructs were transfected by Lipofectamine 2000. Cells were cultured for 36 h and proceeded for the following experiment. An aliquot of cells were used for real-time PCR or western blot assay to confirm the RNAi efficiency.
For the convenience of collecting cells for the following ChIP experiment, we established the NSPc1 knockdown stable HeLa cell line. pSE-NSPc1 and control vector pSE were respectively transfected into HeLa cells. Two days later, G418 at 0.6 mg/ml was added to the medium until clones formed. Then individual GFP positive clones were amplified.
Chromatin immunoprecipitation
The ChIP assay was generally carried out as previously described with little modification (21). Diluted and cleared extracts corresponding to 2.5 × 106 HeLa cells were incubated with each of the following antibodies overnight at 4°C: 4 µg IgG control (Santa Cruz), 5 µg NSPc1 polyclonal antibody, 6 µg Ring1B monoclonal antibody (MBL), 4 µg EZH2 polycolonal anti-serum (Aviva), 4 µg uH2A (E6C5; Upstate), 4 µg m3H3K27, Dnmt1 and HDAC1 polycolonal antibody (Upstate) respectively. Protein A beads were then added to the antibody-chromatin mixture and incubated for an additional hour. Agarose beads were then washed and eluted with elution buffer containing 1% SDS and 0.1 M NaHCO3. Then cross-links were reversed in the presence of 200 mM NaCl at 65°C for 5 h, immunoprecipitated DNA was extracted by phenyl/chloroform, precipitated by ethanol, and analyzed by PCR using primer pairs listed in Supplementary Table 1.
Bisulfite genomic sequencing
Genomic DNA from differentially transfected or 5-aza-dC (Sigma) treated HeLa cells were extracted with the QIAamp mini purification kit (Qiagen) and treated with sodium bisulfite, followed by PCR amplification with specific primers as described (10). The resulting PCR products were purified and inserted into pGEM-T vector (Promega). The transformed 10 clones for each sample were sequenced.
RESULTS
NSPc1 enhances H2A ubiquitination in vivo and in vitro
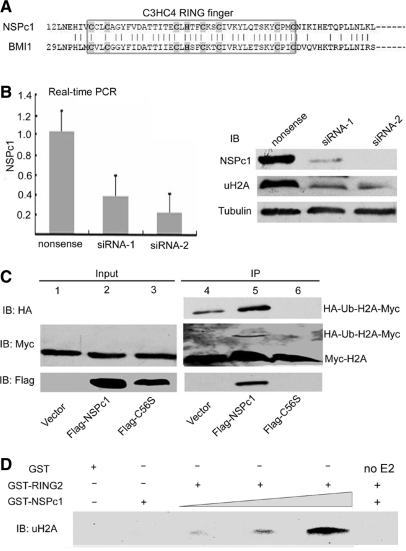
NSPc1 and BMI1 are highly identical in the N terminal C3HC4 type RING finger motif (Figure 1A). However, their roles in different H2A ubiquitination complexes remain elusive. First we verified that NSPc1 is required for keeping normal H2A ubiquitination. Western blot analysis with acid extracted histones (3) from HeLa cells demonstrated that the global level of H2A ubiquitination was reduced when NSPc1 was efficiently knocked down by two specific siRNAs (Figure 1B). The reduction of H2A ubiquitination was not caused by unspecific knockdown of BMI1 as the expression level of BMI1 was even slightly up-regulated (Supplementary Figure 2A).
Figure 1.
NSPc1 enhances H2A ubiquitination in vivo and in vitro. (A) Protein sequence alignment of NSPc1 and BMI1. The protein sequence similarity between NSPc1 and BMI1 is about 63%, with high identity (69%) in the C3HC4 type RING finger motif. (B) Real-time PCR and western blot assays showed the knockdown efficiency of two specific siRNAs against NSPc1, GAPDH and tubulin were used as control. Ubiquitinated H2A level was examined by western blot, tubulin was used as loading control. (C) 293T cells in 10 cm dish were transfected with the indicated combinations of plasmids: 2 μg Myc-H2A, 2 μg HA-Ubiquitin, 6 μg Flag-NSPc1 or Flag-C56S. Cells were harvested 48 h later, and extracts were immunoprecipitated with anti-Myc (Rabbit) antibody, followed by SDS–PAGE and western blot analysis with anti-Myc (mouse) to visualize H2A-Myc and HA-Ub-H2A-Myc, or anti-Flag (mouse) to visualize input or immunoprecipitated NSPc1- Flag or C56S- Flag, or anti-HA (mouse) antibody to visualize HA-Ub-H2A-Myc. (D) NSPc1 enhances E3 ligase activity of GST-RING2 in vitro. Different concentrations of GST-NSPc1 and/or GST-RING2 were incubated in ligase buffer and samples were analyzed for uH2A by western blot as described in Materials and Methods. E2 was omitted in the reaction in lane 6.
To further determine whether NSPc1 promotes H2A ubiquitination in vivo, 293T cells were co-transfected with Myc-H2A and HA-Ubiquitin, with control vector or Flag-NSPc1. Cell extracts were subjected to immunoprecipitation of Myc-H2A, and western blot analysis with anti-Myc antibody (Figure 1C, right upper panel) to detect all forms (unmodified and ubiquitinated) of Myc-H2A in the precipitates, whereas ubiquitin conjugates were also determined by anti-HA antibody (Figure 1C, right middle panel). As shown in Figure 1C, cells transfected with control vector displayed low level or even undetectable monoubiquitinated Myc-H2A (lane 4 upper and middle panels). However, inclusion of NSPc1 led to a pronounced increase in ubiquitinated Myc-H2A (lane 5 upper and middle panels). Since RING finger motif is important for the protein interaction, we then asked whether this domain is necessary for H2A ubiquitination. When the RING finger mutant Flag-C56S was co-transfected instead, the ubiquitinated Myc-H2A was barely detectable.
The in vivo results are further supported by the in vitro ubiquitination assay. Our unpublished data suggested NSPc1 recombinant protein alone does not show ubiquitin E3 ligase activity. Consistent with this, GST-NSPc1 failed to ubiquitinate H2A, however it enhanced the E3 ligase activity of GST-RING2 in a dose-dependent manner (Figure 1D). The reaction was abolished when absent of UbcH5c (Lane 6). Together, these data suggest NSPc1 contributes to H2A ubiquitination in vivo and in vitro, which is highly dependent on its RING finger motif. Among the E3 ligase complex NSPc1 might help RING2 to anchor to the nucleosome and stabilize the E3 ligase activity.
NSPc1 directly interacts with H2A and RING2
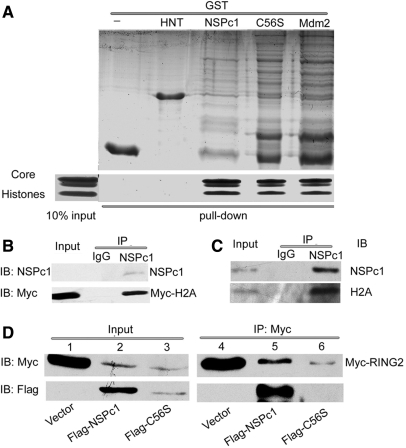
As polycomb proteins generally act as transcriptional co-repressors and NSPc1 promotes H2A ubiquitination, we therefore employed a pull-down assay to test whether NSPc1 interacts directly with core histones. As shown in Figure 2A, core histones bound to GST-NSPc1, GST-C56S and GST-MDM2 [positive control (24)], but not to GST or GST-HNT (a cell membrane protein). Thus, NSPc1 interacts directly and specifically with core histones in vitro, independent on the RING finger motif. Next we examined NSPc1-H2A interaction in vivo. First we transfected Myc-H2A into 293T cells, and then carried out the co-immunoprecipitation assay with anti-Myc antibody. As a result, endogenous NSPc1 was co-immunoprecipitated with Myc-H2A (Figure 2B, upper panel, the third lane) even when it was as low as undetectable in 2% input extract (the first lane), but was not precipitated with control IgG antibody (the second lane). Furthermore, interactions at physiological levels were suggested by the finding that endogenous NSPc1 specifically brought down endogenous H2A from HeLa cells in co-immunoprecipitation assays (Figure 2C). Together, these observations implied that NSPc1 interacts with H2A, presumably also with other histones, both in vitro and in living cells.
Figure 2.
NSPc1 interacts with H2A and RING finger dependently with RING2. (A) NSPc1 interacts with core histones in vitro. The indicated recombinant GST fusion proteins, immobilized on beads, were incubated with core histones. Bound proteins were boiled and directly resolved by SDS–PAGE, followed by CBB staining. Lower panel: The first lane is 10% of the input core histones mix employed for the in vitro binding assay. Upper panel: GST fusion proteins used for the pull-down assay. (B) Endogenous NSPc1 associates with transfected H2A in vivo. 293T cells were transfected with 7 μg Myc-H2A. Cell extracts was immunoprecipitated with anti-NSPc1 antibody and blotted with anti-NSPc1 antibody (upper panel) and anti-Myc antibody (lower panel). Two percent of the input was loaded. (C) Endogenous NSPc1 associates with endogenous H2A in vivo. HeLa cell extracts was incubated with IgG or anti-NSPc1 antibody, and immunoprecipitated proteins were analyzed by anti-H2A antibody. Two percent of the input was loaded. (D) NSPc1 interacts with RING2, dependent on intact RING finger motif of NSPc1. 293T cells in 6 cm dish were co-transfected with 3 μg Myc-RING2 and 5 μg Flag-pcDEF (control vector), Flag-NSPc1 or Flag-C56S. Twenty-four hour later cell extracts were immunoprecipitated with anti-Myc antibody (lanes 4–6) and blotted with anti-Myc to visualize Myc-RING2 (upper panel), or anti-FLAG to visualize FLAG-NSPc1 or Flag-C56S (lower panel). Lanes 1–3 depicts 2% of the input cell extract.
The RING finger motif of NSPc1 is indispensable for its H2A ubiquitination activity but its mutation does not affect the interaction with histones, is it required to interact with RING2? To answer this question, we co-transfected Myc-RING2 and Flag-NSPc1 or Flag-C56S and detected the interaction using co-immunoprecipitation assay. As shown in Figure 2D, Myc-RING2 interacted with Flag-NSPc1 (lower panel, lane 5) but not with Flag-C56S (lower panel, lane 6). Hence it suggests that the RING finger dependent interaction between NSPc1 and RING2 is required for their synergistic E3 ligase activity towards H2A. Further characterization of the phenotype of NSPc1 RING finger mutant may be interesting research issues.
NSPc1 knockdown results in alterations in HOX genes expression
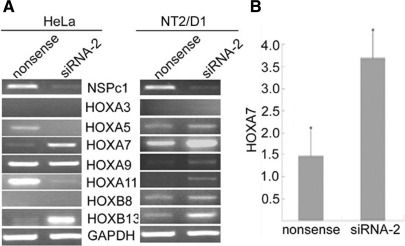
Having demonstrated that the knockdown of NSPc1 results in a significant decrease in the global H2A ubiquitination level, we investigated their role in HOX gene expression. Using RT-PCR screening of HOX gene expression in NSPc1 knockdown HeLa and an embryonal carcinoma cell line NT2/D1, we found that several HOXgenes were de-repressed, among which HOXA7 and HOXB13 expression were significantly up-regulated (Figure 3A). HOXA7 gene up-regulation was also found in Ring2 and Bmi1 knockout mouse (4,26), so it was chosen for further study in HeLa cells. The HOXA7 depression in NSPc1 knockdown HeLa cells was confirmed by real-time PCR (Figure 3B). It is well known that the regulation of HOX genes expression is distinct and highly dynamic in a cell-type-, signaling pathway- and gene-dependent manner. So it is reasonable that BMI1 shares the same target HOX genes with NSPc1 or acts separately, despite of their high homology (Supplementary Figure 3).
Figure 3.
Effects of NSPc1 knockdown on HOX genes expression. (A) HOX genes expression was analyzed by RT-PCR using total RNA isolated from NSPc1 knockdown HeLa and NT2/D1 cells and compared to control cells. GAPDH was used as control for equal RNA input. (B) HOXA7 de-repression in HeLa cells by NSPc1 knockdown was confirmed by real-time PCR.
De-repression of HOXA7 correlates with decreased H2A ubiquitination in the promoter
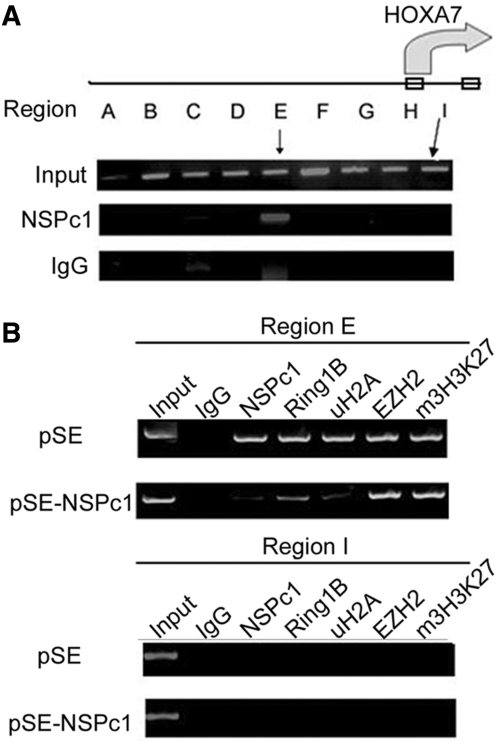
To understand the relationship between NSPc1, H2A ubiquitination and HOXA7 de-repression, we first examined NSPc1 association status across the large range of HOXA7 gene locus (∼5 kb) by ChIP assays. Results shown in Figure 4A indicate that NSPc1 binding was restrictedly enriched in Region E, about 2 kb upstream of transcription start site. The observed binding is specific, as the enrichment is barely observed in a parallel ChIP experiment with the NSPc1 stable knockdown HeLa cells (Figure 4B, upper and lower panels, the slight band might be due to NSPc1 incomplete knockdown). Consistent with the interaction between NSPc1 and RING2, ChIP with anti-Ring1B (/RING2) showed that RING2 is also enriched in the same region. And the deficiency of NSPc1 resulted in reduced association with RING2 and significant decrease of H2A ubiquitination in the region (Figure 4B, lower panel). The polycomb binding and H2A ubiquitination were restricted to the promoter region, as no similar result was observed in a parallel analysis of Region I spanning the intron between exon1 and exon2. All these results indicate that NSPc1 represses HOXA7 transcription through promoting H2A ubiquitination in the promoter regions.
Figure 4.
De-repression of HOXA7 in NSPc1 knockdown HeLa cells correlates with a decreased level of promoter H2A ubiquitination. (A) ChIP analysis across the upstream regions of HOXA7 gene with an antibody against NSPc1 or control IgG in wild-type HeLa cells to identify binding regions. The HOXA7 upstream regulatory regions (A–H, about 4 kb) and a 5′-UTR region (Region I) were indicated in the diagram on top of the panel. Each region spans about 500 bp. The first two exons are indicated by two open boxes. Results were revealed by EtBr staining of agarose gel containing PCR amplified ChIP DNA. (B) ChIP analysis in the control HeLa cells (upper panel) and NSPc1 stable knockdown HeLa cells (lower panel) with indicated antibodies. Regions E and I were examined, respectively.
NSPc1 recruitment, H2A ubiquitination and DNA methylation are all affected by EZH2/EED knockdown
Most recent studies support the notion that PRC1 recruitment and H2A ubiquitination are downstream events of EZH2-mediated H3K27 methylation (4). We further tried to verify whether NSPc1 follows this rule. First we confirmed by real-time PCR assay that EZH2 is also required for HOXA7 transcriptional repression because HOXA7 expression was significantly up-regulated (Figure 5A) after EZH2 was efficiently knocked down (Supplementary Figure 2A and B). As shown in Figure 4B, loss of NSPc1 binding and decreased H2A ubiquitination did not affect the binding of EZH2 (the expression level of EZH2 is up-regulated as seen in Supplementary Figure 2A) and the H3K27 methylation level was unaltered, either. As expected, EZH2 depletion resulted in reduced EZH2 binding, concomitant with reduced H3K27 trimethylation. More importantly, the decrease in EZH2 and me3H3K27 occupancy at the promoter coincided with a significant reduction of NSPc1 binding and H2A ubiquitination (Figure 5B) despite that the expression of NSPc1 remained constant (Supplementary Figure 2). This mode of action of polycomb proteins is consistent with the categorizing PcG proteins into two distinct subsets: initiation complex and maintenance complex (27).
Figure 5.
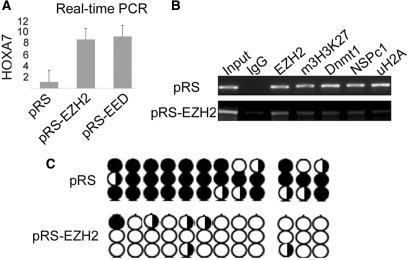
EZH2 knockdown affects Dnmt1 recruitment and reduces H2A ubiquitination concomitant with HOXA7 de-repression. (A) HOXA7 expression level was detected by real-time PCR assay in pRS-EZH2 and control vector transfected HeLa cells. (B) ChIP analysis in the binding regions of HOXA7 with indicated antibodies. The experiment was performed in pRS-EZH2 (lower panel) and control vector (upper panel) transfected HeLa cells. (C) Genomic DNA derived from EZH2 knockdown and control HeLa cells was subjected to bisulfite sequencing to examine the HOXA7 proximal promoter regions rich with CpG islands (across Regions G and H). Methylated CpGs are presented by black circles and unmethylated sites by open circles. Circles of half open and half black represents variation of methylation status.
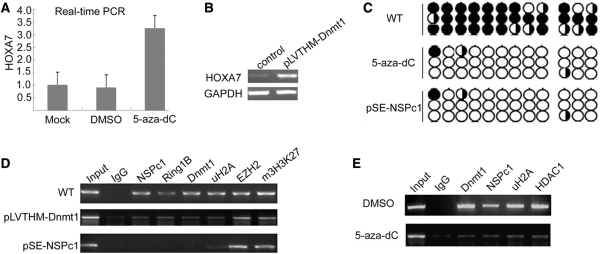
HOXA gene cluster was frequently found to be hypermethylated in tumors (10,18,28). But when EZH2 was knocked down, there was a pronounced loss of CpG methylation compared with control HeLa cells (Figure 5C). This is consistent with the finding that DNA methylation is directly controlled by EZH2 (15). Then we treated HeLa cell with Dnmts inhibitor 5-aza-2-deoxycytidine (5-aza-dC; 5 μM) for 3 days. Under this hypomethylation condition, real-time PCR analysis showed that HOXA7 gene expression was significantly activated compared with the untreated or DMSO-treated cell (Figure 6A). The HOXA7 de-repression was also observed by knocking down Dnmt1, the only DNA methyltransferase responsible for the maintenance of DNA methylation (Figure 6B). Furthermore, Dnmt1 was unable to be recruited to the HOXA7 promoter region after EZH2 was knocked down (Figure 5B); while Dnmt1 deficiency did not affect EZH2 recruitment and H3K27 trimethylation (Figure 6C, the middle panel). The dissociation of NSPc1 and Dnmt1 in EZH2 knockdown cells (Figure 5B) was specific because the binding of Sp1 in the promoter region was unaltered (Supplementary Figure 4). Collectively, NSPc1 and Dnmt1 recruitment, H2A ubiquitination and DNA methylation are downstream events of EZH2-mediated H3K27 methylation.
Figure 6.
NSPc1-mediated H2A ubiquitination and DNA methylation are cooperative in HOXA7 silencing. (A) HOXA7 expression level was examined with Real-time PCR assay in differentially treated HeLa cells. (B) HOXA7 expression level was examined by RT-PCR assay in Dnmt1 knockdown and control HeLa cells. (C) ChIP analysis was performed in untreated, Dnmt1 and NSPc1 knockdown HeLa cells with the indicated antibodies to detect H2A ubiquitination or association of specific proteins in the HOXA7 promoter regions. (D) Genomic DNA derived from Dnmt1 and NSPc1 knockdown HeLa cells as well as the control HeLa cells was subjected to bisulfite sequencing to examine the HOXA7 proximal promoter regions rich with CpG islands. (E) ChIP analysis was performed in control and 5-aza-dC-treated HeLa cells with the indicated antibodies.
NSPc1 complex and DNA methylation cooperate in HOXA7 silencing
All above results suggested that DNA methylation and H2A ubiquitination are both required for HOXA7 gene silencing, which prompted us to ask whether these two epigenetic modifications were connected. ChIP analysis in Dnmt1 knockdown HeLa cells demonstrated that not only Dnmt1 recruitment was lost at the HOXA7 promoter, but also NSPc1 and RING2 recruitment was affected, while EZH2 recruitment and H3K27 trimethylation remained unaffected (Figure 6C, the middle panel).
Interestingly, when we turned back to perform ChIP assay in the Regions C–H in NSPc1 knockdown HeLa cells, we found Dnmt1 binding was undetectable in the whole promoter regions (Figure 6C, the bottom panel) although its expression appeared up-regulated (Supplementary Figure 2A). This result was further supported by the bisulfite sequencing. In the same amplified CpG region (across Regions G and H), at most three sites kept the methylated state in NSPc1 knockdown HeLa cells (Figure 6D, lower panel) consistent with the sequencing result in 5-aza-dC-treated cells (Figure 6D, middle panel) while at least 26 sites were methylated in wild-type cells (Figure 6D, upper panel). Similar results were also seen in glioblastoma cell T98G (data not shown). So NSPc1 binding is required for Dnmt1 recruitment and maintenance of the DNA methylation level.
In order to further elucidate whether Dnmt1 activity accounts for the synergistic effect with NSPc1 complex on transcriptional repression, we compared their association status in 5-aza-dC and DMSO-treated HeLa cells, respectively. Consistent with previous results in colon and renal cancer cells (29,30), 5-aza-dC treatment significantly disassociate Dnmt1, NSPc1 and HDAC1 from HOXA7 promoter regions (Figure 6E), suggesting that 5-aza-dC disrupts the formation of both H2A ubiquitination complex and HDAC complex. Hence we can speculate that proper expression level and activity of Dnmt1 responsible for the maintenance of DNA methylation are required for the normal function of NSPc1.
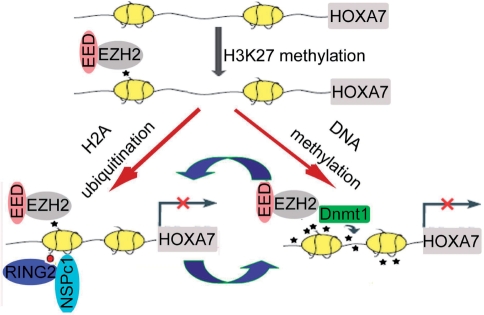
An integration of these results suggests a model illustrated in Figure 7 which has partly shed light on the transcriptional repression mechanism for novel polycomb gene NSPc1. EZH2-mediated H3K27 methylation, present in stem/precursor cells as well as in tumor cells, alone is insufficient to maintain gene silencing. Recruitment of NSPc1 complex and Dnmt1 leads to the formation of more repressive chromatin structure. Our present evidence supports that NSPc1-mediated H2A ubiquitination and DNA methylation are interdependent and mutually reinforce each other, repressing the transcription of HOXA7 synergistically. And HOXB13, another target gene of NSPc1 (as seen in Figure 3), was also found being silenced in a similar way (Supplementary Figure 5).Therefore this study added a new example of cooperation between PcG-mediated silencing and DNA methylation.
Figure 7.
Model for interdependent and self-reinforcing role of NSPc1-mediated H2A ubiquitination and DNA methylation in HOX gene silencing. EZH2-mediated H3K27 methylation presets at some genes but does not necessarily repress their transcription. During tumorigenesis, EZH2 recruits NSPc1 complex and Dnmt1, leading to further modifications such as H2A ubiquitination and maintenance of DNA methylation. And these two latter epigenetic modifications are not separate but interdependent and contribute to the formation of more repressive complexes. Disturbance of either of them will affect the other modification and de-repress the target genes. Methylated CpGs and H3K27 are presented by small black stars. The purple ball stands for attached ubiquitin on H2AK119.
DISCUSSION
Here, we provided evidence that PRC1 component NSPc1, recruited by H3K27 methylation, regulates HOX genes expression through promoting H2A ubiquitination in their promoter regions. The sequential modifications are also supported by the recent study of histone demethylases (31). Furthermore, we found that either loss of NSPc1 or disruption of DNA methylation was associated with gene de-repression. And these two epigenetic players are mutually required and enforce each other. It is interesting that NSPc1-BCOR complex contains FBXL10 (22), a JmjC-domain-containing protein that has recently been identified as histone H3K36 demethylase (32). Therefore a combination of H2A ubiquitination and histone demethylation could be utilized by NSPc1 to direct gene repression. In addition, either knockdown of EZH2, NSPc1 or disruption of Dnmt1 affects HDAC1 recruitment (Figure 6E, Supplementary Figure 5C). So there could be a complex network of epigenetic modifications involved in HOX genes silencing. As Dnmt1 fails to immunoprecipitate NSPc1 in our experiment (data not shown), they may exist in a distinct functional complex. It is speculative that some unknown proteins associated with Dnmt1 (like MeCP2, MBDs or HDACs) could possibly coordinate this crosstalk in the regulation of transcriptional repression of the target genes.
In the past 2 years, the combination of ChIP with hybridization of precipitated DNA to genomic tiling arrays have begun to give a genome wide picture of PcG targets in both mammals and D. melanogaster (33–37). In embryonic stem (ES) cells and adult stem cells, a high proportion of target genes represent those associated with differentiation and development, and silencing of these genes contributes to maintaining the pluripotency or multipotency. In a screen for genes that contribute to ES cells self-renewal, Nspc1 was identified as one of the candidates that maintain the pluripotent phenotype of mouse ES cells even in the absence of LIF. It is interesting that Bmi1 is incapable of rescuing the property of undifferentiated ES cell under the differentiation-inducing conditions (38). Therefore it is worth further exploring how Nspc1 keeps differentiation specific gene silenced in ES cells. In our work, we also confirmed that NSPc1-mediated H2A ubiquitination was required for p21 transcriptional repression (data not shown). Since CKIs play active roles in controlling stem cell proliferation (39), it is tempting to speculate that the role of Nspc1 in ES cell self-renewal is at least partially linked to its function in H2A ubiquitination. And identification of distinct repressive complexes associated with Nspc1 and Bmi1 may help to disclose their functional differences in ES cell self-renewal.
It is now widely accepted that cancer is originated from cancer stem cells (14,40,41). In normal stem/precusor cells, unique bivalent chromatin pattern keeps key developmental genes in a repressed but activatable state (42,43). EZH2-mediated trimethylation is mainly responsible for the repressive modification. The stem cell PcG targets are more vulnerable than non-targets to DNA hypermethylation (43), turning the reversible state to permanent silencing. The loss of function of these genes, in turn, locks stem/precursor cells into abnormal clonal expansion which begins a process of neoplastic initiation and progression (12,14,18,44). This sequential cascade seems to have established a functional link of different epigenetic factors in tumorigenesis. Yet DNA methylation is far from the only and final contributor for stable silencing. As this study and others have revealed (8,17), methylated DNA could further recruit polycomb repressive complex and HDAC complex. How all these different complexes are integrated to silence specific genes awaits further clarification.
During embryogenesis and organogenesis, HOX genes exert their function by regulating cell proliferation and differentiation. And many HOX genes display altered expression patterns in a variety of tumors (45–47). For example, HOXB13 is DNA hypermethylated and silenced in melanoma, renal cell carcinoma and prostate cancer (48–50). Its overexpression suppresses the growth of prostate cancer cells (51). We and other research groups have found that NSPc1 is highly expressed in a variety of tumors (52,53). It could be possible that an increase of NSPc1 promotes the binding of DNMTs, specific CpG islands undergoing de novo methylation, causing HOX genes stably silenced and thereby predisposing to cancer. However HOX genes can so far be defined as only tumor modulators rather than as oncogenes or tumor suppressors (54). Therefore, we could not conclude that its activity to enhance proliferation is exactly due to silencing HOX genes. To really understand the role for NSPc1 in HOX gene silencing, in vivo experiments in knockout mouse and tumor xenograft model might be required.
In summary, our study established a mutually reinforcing loop between NSPc1-mediated H2A ubiquitination and DNA methylation in gene silencing in adult cancer cells. Disturbance of any level of these events could de-repress the target genes followed by the cell fate change. Considered that epigenetic regulation is generally reversible, further identification and functional study of histone demethylases, histone deubiquitinases and DNA demethylases could bring us new strategies for cancer treatment and prevention.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Y. Zhang, R. Bernards, G. Xu, M. Oren and Jagodzinski, P. P. for plasmids; H. Wang for technical assistance in nucleosome preparation. This work was supported by grants from National Sciences Foundation of China No. 30430200, 30600166, 30721063 and 90612019; ‘863’ No. 2006AA02Z13 and 2006AA02A304; ‘973’ No. 2004CB518604 and 2005CB522507, 2006CB504100; Program for New Century Excellent Talents in University No. NCET-07-0505. And we apologize for not citing all relevant references, owing to space constraints. Funding to pay the Open Access publication charges for this article was provided by National Sciences Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 4.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei J, Zhai L, Xu J, Wang H. Role of Bmi1 in H2A ubiquitylation and Hox gene silencing. J. Biol. Chem. 2006;281:22537–22544. doi: 10.1074/jbc.M600826200. [DOI] [PubMed] [Google Scholar]
- 7.Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Negishi M, Saraya A, Miyagi S, Nagao K, Inagaki Y, Nishikawa M, Tajima S, Koseki H, Tsuda H, Takasaki Y, et al. Bmi1 cooperates with Dnmt1-associated protein 1 in gene silencing. Biochem. Biophys. Res. Commun. 2007;353:992–998. doi: 10.1016/j.bbrc.2006.12.166. [DOI] [PubMed] [Google Scholar]
- 9.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Sakamoto Y, Watanabe S, Ichimura T, Kawasuji M, Koseki H, Baba H, Nakao M. Overlapping roles of the methylated DNA binding protein MBD1 and polycomb group proteins in transcriptional repression of HOXA genes and heterochromatin foci formation. J. Biol. Chem. 2007;282:16391–16400. doi: 10.1074/jbc.M700011200. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez-Munoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol. Cell. Biol. 2005;25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohm J, McGarvey K, Yu X, Cheng L, Schuebel K, Cope L, Mohammad H, Chen W, Daniel V, Yu W, et al. A stem cell -like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Gen. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Gen. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 14.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat. Gen. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 15.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 16.Tanay A, O'Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc. Natl Acad. Sci. USA. 2007;104:5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K. Lsh controls Hox gene silencing during development. Proc. Natl Acad. Sci.USA. 2007;104:14366–14371. doi: 10.1073/pnas.0703669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc. Natl Acad. Sci. USA. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, Bressan RA, Zhu JK. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447:735–738. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 20.Nunes M, Blanc I, Maes J, Fellous M, Robert B, McElreavey K. NSPc1, a novel mammalian Polycomb gene, is expressed in neural crest-derived structures of the peripheral nervous system. Mech. Dev. 2001;102:219–222. doi: 10.1016/s0925-4773(01)00288-x. [DOI] [PubMed] [Google Scholar]
- 21.Gong Y, Yue J, Wu X, Wang X, Wen J, Lu L, Peng X, Qiang B, Yuan J. NSPc1 is a cell growth regulator that acts as a transcriptional repressor of p21Waf1/Cip1 via the RARE element. Nucleic Acids Res. 2006;34:6158–6169. doi: 10.1093/nar/gkl834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 24.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 25.Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Jorgensen HF, Giadrossi S, Casanova M, Endoh M, Koseki H, Brockdorff N, Fisher AG. Stem cells primed for action: polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle. 2006;5:1411–1414. doi: 10.4161/cc.5.13.2927. [DOI] [PubMed] [Google Scholar]
- 27.Gil J, Bernard D, Peters G. Role of polycomb group proteins in stem cell self-renewal and cancer. DNA Cell Biol. 2005;24:117–125. doi: 10.1089/dna.2005.24.117. [DOI] [PubMed] [Google Scholar]
- 28.Novak P, Jensen T, Oshiro MM, Wozniak RJ, Nouzova M, Watts GS, Klimecki WT, Kim C, Futscher BW. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66:10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- 29.Sharma D, Blum J, Yang X, Beaulieu N, Macleod AR, Davidson NE. Release of methyl CpG binding proteins and histone deacetylase 1 from the Estrogen receptor alpha (ER) promoter upon reactivation in ER-negative human breast cancer cells. Mol. Endocrinol. 2005;19:1740–1751. doi: 10.1210/me.2004-0011. [DOI] [PubMed] [Google Scholar]
- 30.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Gen. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 31.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 32.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 33.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Gen. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 36.Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Gen. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 37.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 38.Pritsker M, Ford NR, Jenq HT, Lemischka IR. Genomewide gain-of-function genetic screen identifies functionally active genes in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 2006;103:6946–6951. doi: 10.1073/pnas.0509861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruggeman SW, van Lohuizen M. Controlling stem cell proliferation: CKIs at work. Cell Cycle. 2006;5:1281–1285. doi: 10.4161/cc.5.12.2806. [DOI] [PubMed] [Google Scholar]
- 40.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat. Rev. Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 42.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 43.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nature Rev. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 44.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 45.Cillo C, Cantile M, Faiella A, Boncinelli E. Homeobox genes in normal and malignant cells. J. Cell. Physiol. 2001;188:161–169. doi: 10.1002/jcp.1115. [DOI] [PubMed] [Google Scholar]
- 46.Drabkin HA, Parsy C, Ferguson K, Guilhot F, Lacotte L, Roy L, Zeng C, Baron A, Hunger SP, Varella-Garcia M, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16:186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 47.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 48.Muthusamy V, Duraisamy S, Bradbury CM, Hobbs C, Curley DP, Nelson B, Bosenberg M. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res. 2006;66:11187–11193. doi: 10.1158/0008-5472.CAN-06-1274. [DOI] [PubMed] [Google Scholar]
- 49.Okuda H, Toyota M, Ishida W, Furihata M, Tsuchiya M, Kamada M, Tokino T, Shuin T. Epigenetic inactivation of the candidate tumor suppressor gene HOXB13 in human renal cell carcinoma. Oncogene. 2006;25:1733–1742. doi: 10.1038/sj.onc.1209200. [DOI] [PubMed] [Google Scholar]
- 50.Jung C, Kim RS, Lee SJ, Wang C, Jeng MH. HOXB13 homeodomain protein suppresses the growth of prostate cancer cells by the negative regulation of T-cell factor 4. Cancer Res. 2004;64:3046–3051. doi: 10.1158/0008-5472.can-03-2614. [DOI] [PubMed] [Google Scholar]
- 51.Jung C, Kim RS, Zhang HJ, Lee SJ, Jeng MH. HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res. 2004;64:9185–9192. doi: 10.1158/0008-5472.CAN-04-1330. [DOI] [PubMed] [Google Scholar]
- 52.Gieseg MA, Man MZ, Gorski NA, Madore SJ, Kaldjian EP, Leopold WR. The influence of tumor size and environment on gene expression in commonly used human tumor lines. BMC Cancer. 2004;4 doi: 10.1186/1471-2407-4-35. Paper 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sultmann H, von Heydebreck A, Huber W, Kuner R, Buness A, Vogt M, Gunawan B, Vingron M, Fuzesi L, Poustka A. Gene expression in kidney cancer is associated with cytogenetic abnormalities, metastasis formation, and patient survival. Clin. Cancer Res. 2005;11:646–655. [PubMed] [Google Scholar]
- 54.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat. Rev. Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.