Abstract
STAT5A and STAT5B proteins belong to the family of signal transducers and activators of transcription. They are encoded by two separate genes with 91% identity in their amino acid sequences. Despite their high degree of conservation, STAT5A and STAT5B exert non-redundant functions, resulting at least in part from differences in target gene activation. To better characterize the differential contribution of STAT5A and STAT5B in gene regulation, we performed single or double knockdown of STAT5A and STAT5B using small interfering RNA. Subsequent gene expression profiling and RT-qPCR analyses of IL-3-stimulated Ba/F3-β cells led to the identification of putative novel STAT5 target genes. Chromatin immunoprecipitation assays analyzing the corresponding gene loci identified unusual STAT5 binding sites compared to conventional STAT5 responsive elements. Some of the STAT5 targets identified are upregulated in several human cancers, suggesting that they might represent potential oncogenes in STAT5-associated malignancies.
INTRODUCTION
The STAT (signal transducers and activators of transcription) family of transcription factors comprises seven members (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6), which are essential mediators of cytokine, growth factor and hormone responses. STAT proteins play important functions in a variety of pathways, from innate and acquired immunity to cell proliferation, differentiation and survival [reviewed in reference (1)]. Accordingly, inappropriate activation of STATs, in particular of STAT1, STAT3 and STAT5, is associated with a wide variety of human cancers and diseases, and constitutively activated STAT signaling directly contributes to oncogenesis (2,3).
As with other STAT family members, STAT5 is mostly present in the cytoplasm as latent monomers. Upon stimulation by a broad spectrum of cytokines (4), STAT5 is recruited to the cytokine receptor through an interaction between STAT5 SH2 domain and a specific receptor phosphotyrosine residue. STAT5 is then phosphorylated on a conserved carboxy-terminal tyrosine, leading to its dissociation from the receptor and dimer formation. Dimerization is mediated by a reciprocal SH2-phosphotyrosine interaction between two phospho-STAT5 molecules, and is a prerequisite for DNA binding activity (5). Although STAT5 monomers can shuttle between the cytoplasm and the nucleus, STAT5 dimers are actively translocated to and retained in the nucleus where they bind to specific DNA binding sites and activate transcription (1,6,7). Transcriptional activation by STAT5 involves recruitment of the coactivators CBP/p300 (8) and NCoA-1 (9), and requires a deacetylase activity (10,11).
STAT5A and STAT5B proteins are encoded by two distinct but chromosomally linked genes, and share 91% identity in their amino acid sequence. Interestingly, the most divergence is found within the carboxy-terminal phosphotyrosyl tail and transactivation domain (4). STAT5A and STAT5B exert redundant yet distinct functions, as revealed by single and double knockout mice studies (12,13). Thus, while STAT5A is essential for prolactin-dependent mammary gland development and lactogenesis (14,15), STAT5B mediates the sexual dimorphic effects of growth hormone (15–17). In the immune system, both STAT5A and STAT5B regulate IL-2 signaling, with a stronger contribution of STAT5B through regulation of both IL-2Rα and IL-2Rβ gene expression, while STAT5A only regulates IL-2-induced IL-2Rα expression (15,18,19).
Multiple factors contribute to the non-redundant functions of STAT5A and STAT5B. Differences in STAT5A/B mRNA levels (20–22), activation by tyrosine- and serine-phosphorylation (23–27), and nucleocytoplasmic shuttling (7,28,29), all regulating STAT5A and STAT5B availability in the nucleus have been reported.
Once in the nucleus, DNA binding as homodimers (STAT5A:STAT5A, STAT5B:STAT5B), heterodimers (STAT5A:STAT5B), as well as tetramers represent a major level of differential regulation of transcription by STAT5A and STAT5B. Although the DNA binding sequences of the various STAT proteins are very similar (TTC N3–4 GAA), clear differences in DNA binding specificities have been described (30,31). While both STAT5A and STAT5B are required for induction of the human Cis gene, they show distinct specificities in binding to the 4 STAT5 binding sites located in the proximal promoter in vitro (32). Subtle amino acid differences within the DNA binding domain of STAT5A and STAT5B have been shown to play a role in differential DNA binding specificities (33). STAT5 tetramerization represents another main level of transcriptional regulation. STAT5 binding sites are often found in tandem in gene regulatory regions, allowing cooperative binding through interactions involving the N-terminal domains of two neighboring STAT5 dimers (34–36). In vitro, STAT5A molecules bind DNA preferentially as tetramers, whereas STAT5B molecules form dimeric complexes (32,37). Interestingly, tetramerization is tightly coupled to tyrosine dephosphorylation and nuclear accumulation of STAT5 (38), suggesting that differential tetramerization of STAT5A and STAT5B may influence the downstream prolongation or attenuation of the STAT5 response of a particular target gene in vivo. In support of this hypothesis, STAT5A and STAT5B show distinct kinetics and dynamics of DNA binding in vivo (39).
Beside DNA binding specificity, differential transcriptional activation by STAT5A and STAT5B might possibly involve recruitment of specific cofactors through their divergent carboxy-terminal domain, although STAT5A- and STAT5B-specific cofactors have not been described thus far. Finally, differential attenuation mechanisms (dephosphorylation by phosphatases, degradation via the proteasome-dependent pathway, naturally occurring dominant negative truncated STAT5 variants, negative feedback loop by the Cis/SOCS proteins) might also participate in differential regulation by STAT5A and STAT5B (22,40–42).
As predicted from the non-redundancy of STAT5A and STAT5B functions in vivo, a differential contribution of STAT5A and STAT5B in the progression of various cancers has been reported. For example, while STAT5A activation has been associated with mammary cancer progression (43), STAT5B activation has been more specifically associated with squamous cell carcinoma of the head and neck (SCCHN) tumorigenesis (44,45). This implies that there are distinct downstream target genes acting as oncogenes that are specifically induced by either STAT5A or STAT5B.
Identification of novel STAT5-dependent oncogenes, a better characterization of the respective contribution of STAT5A and STAT5B in their regulation, and the identification of the level of differential regulation (activation, nuclear translocation, DNA binding, transcription) might offer novel possibilities for specific therapeutic intervention in STAT5-associated cancers.
With this in mind, we conducted a gene expression profiling study on IL-3-stimulated Ba/F3-β cells following small interfering RNA (siRNA)-mediated knockdown of both STAT5A and STAT5B expression. New STAT5-regulated genes were identified. The selective knockdown of STAT5A or STAT5B demonstrated that these proteins differentially influence the expression profiles of these genes in response to IL-3 stimulation. Specific STAT5 binding sites were identified within some of these genes using chromatin immunoprecipitation (ChIP), unveiling conventional as well as unusual binding site sequences and locations compared to STAT5 binding elements reported so far. Interestingly, some of the novel STAT5 targets are overexpressed in several cancers, suggesting that they might represent putative oncogenes.
MATERIALS AND METHODS
Cells
The IL-3-dependent murine proB cell line Ba/F3-β was grown in RPMI 1640 containing 10% FBS and 10 ng/ml IL-3 (10,46). For cytokine stimulation, cells were washed in RPMI 1640 and rested for 6 h in RPMI 1640 containing 10% FBS before addition of 10 ng/ml IL-3 for the indicated times.
Protein analysis
Cells were lysed and analyzed by western blot as previously described (10,46), using antibodies specific for STAT5A, STAT5B and STAT3 (sc-1081, sc-835 and sc-483 respectively, Santa Cruz Biotechnology, Santa Cruz, CA, USA).
mRNA analysis by RT-qPCR
RNA isolation, cDNAs synthesis (RT) and real-time quantitative PCR (qPCR) expression analysis were performed as previously described (10,46). Data were normalized to S9 ribosomal mRNAs and expressed as relative mRNA levels. Murine-specific forward and reverse real-time PCR primers used in this study are as follows. STAT5A: CGCTGGACTCCATGCTTCTC and GACGTGGGCTCCTTACACTGA; STAT5B: GGACTCCGTCCTTGATACCG and TCCATCGTGTCTTCCAGATCG; STAT3: AATGGAAATTGCCCGGATC and AGGCGAGACTCTTCCCACAG; Socs-3: CCAAGAACCTACGCATCCAGTG and CGTGGGTGGCAAAGAAAAGG; Pim-2: TACGCCTTCTTGACTGGTTCG and CATAGGCCGCTCAAGGACC; IL-2Rβ: AGAGAAGGGTTGGCGTAGGG and TTGTCAAAGGGATGGAAGTCG; Heparin binding EGF-like growth factor: CGTACTCCCTCTTGCAAATGC and GAGTCAGCCCATGACACCTGT; Socs-1: TTCTTGGTGCGCGACAGTC and AAGCCATCTTCACGCTGAGC; C3ar1: CTGACCTACACTCACGGCCTC and CCATTGCCTAGCAGTCCCAA; TNFRSF13b: CTGCTGTTTCTTGGTGGCCT and AGAGTTTGCTTGTGACCCACG; Slfn2: TGTACAAGTGAGCGCCGCT and TGCAGGCAACTCTGGTCTGA; Enah: TGTCAGACGCCACTCCTGATT and CTGCGTGTCTCAGTGGTGGT; Lama5: AGCGCTGTGGGAACTCTAACC and CCCTCCATGTCCACGAACTC. Primers specific for the following cDNAs have been already described: S9, Cis, c-Myc, Pim-1, Osm, Bcl-x, p21, Id, MKP-1, IL-4Rα, Stra13, Spi2.1, Ryk, TCRγ-V4 and IL-2Rα (10).
Small interfering RNA (siRNA)-mediated knockdown
STAT5A, STAT5B, STAT3 and ScI siRNA duplexes were synthesized using the Silencer siRNA Construction Kit (Ambion #1620, Austin, TX, USA), as formerly described (46). SiRNA sequences for STAT5A, STAT5B and ScI have been described (46). STAT3 siRNA sequence is (5′–3′): AAGCAGCAGCTGAACAACATG. SiRNA were transfected by electroporation as previously described (46). Briefly, 3.106 Ba/F3-β cells were transfected with a total amount of siRNA of either 10 μg [STAT5A/B double knockdown, using 2.5 μg each of a pool of four siRNAs (46)], 8 μg [STAT5A and STAT5B single knockdown, 4 μg each of STAT5A or STAT5B pool of two siRNAs (46)], or 5 μg (STAT3 knockdown), in the presence of 50 U of a FITC-labeled uptake control (Sequitur #3013, Invitrogen, Carlsbad, CA, USA). Four hours posttransfection, FITC-positive cells were sorted by FACS and cultured for 22 h in RPMI 1640 containing 10% FBS and 0.2 ng/ml IL-3, followed by 5 h resting (RPMI 1640 containing 10% FBS) and IL-3 stimulation (10 ng/ml) for 30 min and 2 h. Cells were harvested at time 0 of IL-3 stimulation for western blot analysis and at times 0, 0.5 and 2 h of IL-3 stimulation for RNA and microarray analysis. STAT5 western blot analysis following STAT5 knockdown has been published (46).
DNA microarray analysis
Ba/F3-β cells were washed in PBS, total RNA was isolated from unstimulated ScI siRNA-transfected Ba/F3-β cells, and IL-3-stimulated (30 min and 2 h) ScI siRNA- or STAT5A/B siRNA-transfected Ba/F3-β cells using the RNeasy Maxi kit, including an oncolumn DNase I treatment to eliminate genomic DNA contamination, according to the manufacturer's protocol (Qiagen, Valencia, CA, USA). DNase-treated total RNA (5 µg) was synthesized into biotinylated cRNA probe using one-cycle target labeling and IVT labeling (Affymetrix, Inc., Santa Clara, CA, USA) according to manufacturer's instructions. Fifteen microgram of biotinylated cRNA probe from each sample was hybridized onto murine U74A, U74B, U74C Affymetrix Microarray chips. The hybridized chips were then washed using the Affymetrix GeneChip Fluidics 400 station and scanned in the Affymetrix GeneChip Scanner 3000 (Affymetrix, Inc., Santa Clara, CA) according to manufacturer's instructions. The quality of cRNA probe synthesis and efficiency of hybridization was analyzed in the GeneChip Operating System for each Affymetrix chip once scanning was complete. Microarray data were normalized using MAS5 and then analyzed using the Genespring (Agilent, Palo Alto, CA, USA). Out of 36 767 genes, 14 553 passed our noise and P/M/A call filters. Genes were further selected for being stimulated at least 2-fold by IL-3 after 30 min (164 genes) and 2 h (553 genes) stimulation compared to the unstimulated cells (ScI-transfected cells). Then, IL-3 induced genes that were downregulated at least 2-fold in the STAT5A/B siRNA-transfected cells at 30 min and 2 h (12 and 29, respectively) were identified as putative STAT5 target genes and further investigated. The data discussed in this publication have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE10389.
ChIP
STAT5 ChIP was performed on unstimulated and IL-3-stimulated (45 min) Ba/F3-β cells, using STAT5A and STAT5B antibodies (sc-1081 and sc-835 respectively, Santa Cruz Biotechnology, Santa Cruz, CA, USA) or no antibody (protein A-sepharose control), and analyzed by real-time PCR as formerly described (10,46). Data are expressed as a percentage of input DNA. The genomic sequence of each gene (−6000 relative to the transcription start site up to the polyA signal) was searched for STAT5 binding sites using the Vector NTI software (Invitrogen, Carlsbad, CA, USA). Forward and reverse real-time PCR primers used for the mouse genomic DNA analysis are as follows. Primers specific for STAT5 binding sites-containing regions are underlined, the others being control primers. Cis (−184): GTCCAAAGCACTAGACGCCTG and TTCCCGGAAGCCTCATCTT; Cis (+4200): TACCCCTTCCAACTCTGACTGAGC and TTCCCTCCAGGATGTGACTGTG; IL-2Rα (−1270): GCATGATATGATGTGCAGTTTCTTC and TCAGGACTGGTGGTTGGTTG; IL-2Rα (−90): AAACACTGCCCACACCTCCT and TTTGCGGTAATTTTTCAAACCA; Spi2.1 (−1740): GCACAGTGGAAGGGAAAAGC and ACCCACCACATCCAACATCA; Spi2.1 (−127): AAATCACCCGGTCTGTCCAT and TGTTGATGCAGATAAGCTGTGC; Socs-1 (−2100): CAACATTGCTTGGCTAACGG and CTGGGAAAGCTCTGGGAAGTT; Socs-1 (−1200): GCCCACCAAGGGTTAGACTGA and TGAATTTGTGTGGCTGTGGTTG; Socs-1 (−480): GCCCCACAGGTCTCTAGGACTA and CTTGGCAATCTCGACCCTTC; Socs-3 (−1235): TTTCACCTCAATCACCTGCTCT and TCTGAACTTTCCCGATGGTG; Socs-3 (−345): CTGCCAGAAACCAGCCTTC and GCCCCCAACTTCTCATTCA; Socs-3 (−95): GCACAGCCTTTCAGTGCAGAG and GTATTTACCCGGCCAGTACGC; C3ar1 (−3000): ATGGCAGCTCACAACTGTCTGT and CGTGTCTGTGGGATGGTGTC; C3ar1 (−1850): ACACAGAGAAACCCTGCCTCA and CTTTGGGCGCTTATTGCTCT; C3ar1 (−1620): TTGAGAAAGTGCCTCCACCAGA and CCAAGAAAATGACCCACAGCC; C3ar1 (−480): TGCTTGGTAGCAATGCCTGT and TTCTCCTGCCTCCGTCTTCT; C3ar1 (+1380): GCCTTCAGCGAAGAGCTCAC and GAAGAGGCTTTGTCTTGGGTACA; TNFRSF13b (−600): TAAAGCCCTTGCCTGTGACC and GCTATCCCCGACAGAATGACA; TNFRSF13b (+1680): GGAACGGCAAGATGGAAGAT and CCATGGACTTCCCAGAAAGC; TNFRSF13b (+3780): GCAGCTGAGTGACAGCAGTGT and CCACAGTCTCCCTTGCTGTG; MKP-1 (−3450): TTCACGGAACACAGGCTCAC and GCCTTCTGGCTGATTTCCAC; MKP-1 (−1700): GGCAGCAGTTCCAGCTCTTT and GCTGTGGAGTTCTGCCCTCT; MKP-1 (+1680): TCCCCTGCTATCCTTTGCAT and AGGGTCTGCCTCTTGCAGTC; Osm (−226): CATCATCCTTGGGCGTGGGGC and CGCTCCTCCTCCCGTTTTCTTCG; Osm (+2875): CGGTCCACTACAACACCAGATGTC and TATCCCCAGAGAAAGCCACAGC; Pim-2 (−2740): TGATCCCAGCACTGAGGAGG and GCTGGCCTGCAACTCAAAAG; Pim-2 (−1515): TGAGTCTTGAGTCAACCAGTTTCTC and CGGAAACAGACACATCCACAC; Pim-2 (−1135): GCAGCACAAATATTTCTGTGAATGT and TCACAAGCACACAAGCACCA; Pim-2 (−545): AATTTTACCCTTCCCTGAAGAGTG and GATCAGGCAGCCTTTGCTC; Pim-2 (−270): CACCCCACCACACCAACA and TGCACGCTTCTGGGAATTC; IL-2Rβ (−5840): TGCCTTGGTCATGGTATCTGTT and AGGGACCCAGCTGAGATAAGG; IL-2Rβ (−840): GCCCCGATGAAGATGGTAATG and AGCTTCTGCCCTCTTGTGAAAAG; IL-2Rβ (+2725): GGCCTGAGCGATTCCTTTTC and AGGCGAGAAGCATCCACATC; Ryk (−4116): CCTTTTTCCAGTGGCTGTGA and GCTGCTGCTGTCTTTTGTCG; Ryk (−2190): AGGGGCCAATACCAGAATCAC and AGGAGAGGAACAGCTGGAATTG; Slfn2 (−400): TATCCCCTTTCCTGGTCCTG and GTGGATGGGAGAGGGAGTTG; Slfn2 (+4130): CGGCTCTGTCTGTTTTCCAGG and CCAGCAGCCTTTGCATTTTC; Slfn2 (+4680): CAAGCATAGCCTTCCCTGAAG and ATGCAGCAGCCAAATCCTC; Slfn2 (+4800): TACATTGCTTTCAGCGAAACG and GTTTGAGGGAGGAGCTCTTTGA; Slfn2 (+5040): GTACACGTGCAAATTCATCGAAG and CGCACACACATATGCACACAC; Id (−2600): CACAGTCTGTTCTTTGAAGTTGGTC and CCAATGCCTGAACCTCAGAAA; Id (−1000): CCTGGCGTCTAACGGTCTG and ACCCCCTCCCTTTCCTTTC; Id (+135): GCAGGCCCTAGCTGTTCG and GCTCCGACAGACCAAGTACCA; Id (+5358): GGACCAGGCGCTACTTTCC and TGGTTCCTCCTAGTCCTGGTTT; Id (+5690): GCTCTCAGGCATCCTGAGCT and CGATTTGACTTTGCAGGAACC.
Cancer panel
Paired human tumor (primary melanoma and renal tumors) and normal adjacent tissues (NAT) were obtained from patients undergoing routine therapeutic surgery from the following sources: Dr David Gotley of the Princess Alexandra Tumour Tissue Bank (Woolloongabba, Queensland Australia), Zoion Diagnostics (Hawthorne, New York, USA), National Disease Research Interchange (NDRI) (Philadelphia, PA, USA) and Zoion Diagnostics (Shrewsbury, MA, USA). Normal human tissue were obtained from patients undergoing routine surgery [NDRI, Ardais Corporation, Lexington, MA, USA and Cooperative Human Tissue Network (CHTN), Nashville, TN, USA], from short hour autopsy (≤5 h, Zoion Diagnostics), and from transplant donors (Anatomical Gift Foundation, Hanover, MD, USA). Tumor tissue was collected for each case, and when possible matching NAT was also collected. Each case was accompanied with a brief clinical history, pathology report and pathological diagnosis. All surgical samples were frozen as quickly as possible, typically within an hour of excision. All sample diagnoses and staging were confirmed internally by a pathologist. This information was used to select cases, 87 melanoma cases and 35 renal cell carcinoma cases to be studied for expression analysis. Cases excluded from the study had either other diagnoses or large areas of necrotic tissue, upon examination by an inhouse pathologist.
For real-time quantitative PCR analysis, total RNA was isolated using RNA STAT-60 (Tel-Test, Friendswood, TX, USA) according to manufacturer's instructions. After isopropanol precipitation, total RNA was re-extracted with phenol:chloroform:isoamyl alcohol (25:24:1) (Sigma Chemicals, Balcatta, Perth, Western Australia). Total RNA (5 μg) was subjected to treatment with DNase I (Roche Molecular Biochemicals, Indianapolis, IN, USA) according to manufacturer's instructions to eliminate possible genomic DNA contamination, and reverse transcribed using Superscript II (Invitrogen) according to manufacturer's instructions. Real-time quantitative PCR was conducted as described above and the data were normalized to ubiquitin mRNA levels. The human forward and reverse qPCR primers used were as follows. C3ar1: CTGGTGTCAGAATCATCGCAA and CATCCACAGATAGAGCAGGCC; c-Myc: AACAGGAACTATGACCTCG and AGCAGCTCGAATTTCTTC. The ubiquitin primers have been described (47). Statistical analysis of tumor expression was conducted using a Kruskal–Wallis non-parametric test.
RESULTS
Identification of novel STAT5-regulated genes in IL-3 stimulated Ba/F3-β cells
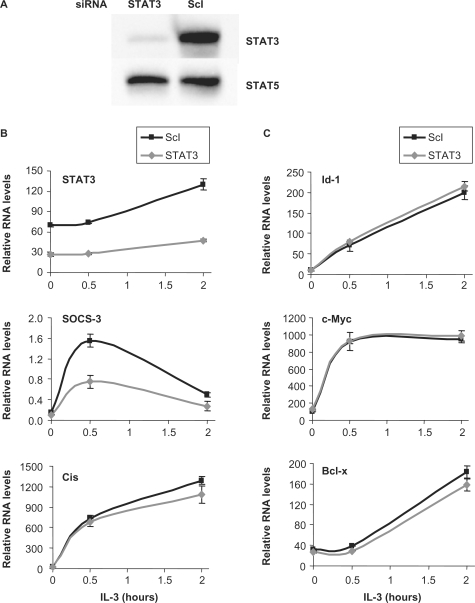
To identify novel IL-3-inducible STAT5 target genes in the proB cell line Ba/F3-β, we exploited an experimental system previously described by our group (46), in which expression of both STAT5A and STAT5B proteins (hereafter designated STAT5A/B) was knocked down using small interfering RNA (siRNA). Upon siRNA-mediated knockdown, STAT5A and STAT5B mRNA and protein levels were equally reduced by ∼60% [Figure 1A and reference (46)]. Following siRNA transfection, Ba/F3-β cells were starved from cytokine and stimulated with IL-3 for 30 min and 2 h. Isolated RNAs were hybridized on an Affymetrix DNA array covering the whole mouse genome to identify IL-3-induced genes that were downregulated in the STAT5A/B knockdown.
Figure 1.
STAT5 mRNA levels in Ba/F3-β cells upon STAT5A and STAT5B double (A) and single (B) knockdown. Ba/F3-β cells were transfected either with a non-specific siRNA (ScI), or with siRNAs specific for STAT5A (5A), STAT5B (5B) or both (5AB). Twenty-six hours later, cells were starved for 5 h and stimulated with IL-3. Cells were harvested at the indicated times, and mRNAs levels were monitored by RT-qPCR. A 60% reduction in STAT5 mRNA levels was detected in both the single and double knockdown, with the expected specificity. Reduction in STAT5 protein levels was verified by western blot and has been already published (46). STAT3 mRNA levels (this figure), as well as protein levels (46), remained unaffected, confirming the specificity of STAT5 targeting.
After 30 min of IL-3 stimulation, 164 probes were induced by IL-3 at least 2-fold, 12 of which were downregulated at least 2-fold in the STAT5A/B knockdown (corresponding to 11 different genes). Seven genes were validated by RT-qPCR (below and data not shown). Four are known genes (Cis, Socs-1, Heparin binding EGF-like growth factor and Stra13) and three are unknown genes. After 2 h of IL-3 stimulation, 553 genes were induced at least 2-fold by IL-3, 29 of which were downregulated at least 2-fold in the STAT5A/B knockdown (corresponding to 27 different genes). Twenty-one genes were validated by RT-qPCR (below and data not shown). Twelve are known genes (including the four already identified after 30 min stimulation) and nine are unknown genes. These known genes are shown in Table 1 with the corresponding Affymetrix data showing the fold induction by IL-3 and the fold inhibition following STAT5A/B knockdown.
Table 1.
IL-3 inducible genes downregulated upon STAT5A/B knockdown in Ba/F3-β cells (Affymetrix array data)
| Gene name | Accession no. | 30 min | 2 h | |||
|---|---|---|---|---|---|---|
| Fold up | Fold down | Fold up | Fold down | |||
| (IL-3) | (5AB) | (IL-3) | (5AB) | |||
| 1 | Cytokine inducible SH2-containing protein (Cis)a | NM_009895 | 10.12 | 2.82 | 14.43 | 2.43 |
| 2 | Cytokine inducible SH2-containing protein 1 (Socs-1)a | NM_009896 | 7.91 | 2.74 | 3.66 | 2.23 |
| 3 | Serine (or cysteine) proteinase inhibitor (Spi2.1)a | M64085 | 2.71 | 1.99 | 6.24 | 2.59 |
| 4 | Receptor-like tyrosine kinase (Ryk) | L21707 | 1.25 | 1.01 | 4.25 | 3.44 |
| 5 | Stimulated by retinoic acid 13 (stra13/clast5) | NM_011498 | 3.29 | 2.36 | 5.45 | 2.32 |
| 6 | Schlafen 2 (Sfln2) | NM_011408 | 2.72 | 1.35 | 14.17 | 5.92 |
| 7 | Serine/threonine kinase (Pim-2) | L41495 | 1.41 | 1.31 | 5.04 | 2.07 |
| 8 | Heparin binding epidermal growth factor-like growth factor | NM_010415 | 4.75 | 2.51 | 7.71 | 2.07 |
| 9 | G-protein coupled receptor; anaphylatoxin C3a receptor 1 (C3ar1) | NM_009779 | 1.76 | 2.92 | 2.75 | 3.34 |
| 10 | TNFRSF13b | NM_021349 | 1.41 | 1.46 | 2.19 | 3.25 |
| 11 | Enabled homolog (Drosophila) (enah); Mena; VASP; WASP | NM_010135 | 0.96 | 1.13 | 2.66 | 2.21 |
| 12 | Laminin, alpha 5 (Lama5) | U37501 | 1.08 | 1.22 | 2.60 | 2.01 |
aKnown STAT5 targets from the literature (see text).
The identification of known STAT5 target genes such as Cis, Spi2.1 and Socs-1 (48–50) demonstrated the validity of our experimental approach. Furthermore, Cis, Spi2.1, Ryk and Stra13 were identified by our group in a previous gene expression profiling study as inhibited by trichostatin A (TSA), a histone deacetylase inhibitor also acting as a potent repressor of STAT5-mediated transactivation (10). Our microarray analysis upon STAT5A/B knockdown thus confirmed several known STAT5 targets and revealed seven potential novel STAT5 target genes (Table 1).
STAT5 target genes are differentially regulated by STAT5A and STAT5B
We next examined the differential contribution of STAT5A and B proteins in the regulation of STAT5 target gene expression. Following the same strategy as above, expression of either STAT5A or STAT5B was specifically knocked down in Ba/F3-β cells by siRNA transfection. Reduction in STAT5 mRNA and protein levels upon siRNA-mediated knockdown in the single knockdown was similar (∼60%) to that observed in the double knockdown [Figure 1B and reference (46)]. In addition to the genes shown in Table 1, our study was broadened to additional putative STAT5 targets [derived from our formerly described gene profiling analysis of TSA sensitivity (10)] and to well-characterized STAT5 target genes (Table 2). mRNA levels upon IL-3 stimulation of a total of 24 genes were evaluated and compared in the double STAT5A/B and single STAT5A and STAT5B knockdown cells by RT-qPCR (Figure 2 and Table 2).
Table 2.
STAT5A/STAT5B relative contribution and STAT5 DNA binding characteristics on the known and novel STAT5 target genes evaluated in this study
| RT-qPCR | ChIP assay | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene name | Accession no. | % inhibition in 5AB KD | STAT5 binding | % of input DNAb | STAT5 binding sitec (positions relative to tss) | References | IL-3 induction profile | |
| STAT5A or STAT5B | ||||||||
| 1 | Cytokine inducible SH2-containing protein (Cis)a | NM_009895 | 59 | + | 6 | 4 c. in tandem (−230/−103) | (10,48), this study | Early, sustained |
| 2 | Cytokine inducible SH2-containing protein 3 (Socs-3)a | NM_007707 | 54 | + | 3.5 | 2 c. + 1 n.c. in tandem (−345/−64) | (53), this study | Early, transient |
| 3 | Oncostatin M (Osm)a | D31942 | 59 | + | 4 | 2 c. in tandem (−190/−95) | (93), this study | Early, transient |
| 4 | MAP kinase phosphatase 1 (MKP-1) | X61940 | 52 | + | 0.4 | 2 c. in tandem (−3450/−3414) | This study | Early, transient |
| 5 | Stimulated by retinoic acid 13 (stra13/clast5) | NM_011498 | 67 | − | − | [2 c. −680/−410] | This study | Early/late, sustained |
| 6 | Serine/threonine kinase (Pim-2) | L41495 | 56 | − | − | [4 c. −1520/−270] | This study | Late, sustained |
| 7 | Interleukin 2 receptor, beta (IL-2Rβ) | M28052 | 59 | − | − | [1 c. −5840; 1 c. +2725] | This study | Late, sustained |
| 8 | Heparin binding epidermal growth factor-like growth factor | NM_010415 | 71 | ND | ND | ND | This study | Late, sustained |
| 9 | Serine/threonine kinase (Pim-1)a | NM_008842 | 35 | ND | ND | ND | (10), this study | Early, sustained |
| 10 | T-cell receptor gamma, variable 4 (TCRγ-V4)a | NM_011558 | 36 | ND | ND | ND | (10), this study | Late, sustained |
| STAT5A and STAT5B | ||||||||
| (i) Equal contribution | ||||||||
| 11 | Cytokine inducible SH2-containing protein 1 (Socs-1)a | NM_009896 | 52 | + | 0.6 | 2 c. + 1 n.c. in tandem (−2095/−2061) | This study | Early, transient |
| 12 | G-protein coupled receptor; anaphylatoxin C3a receptor 1 (C3ar1) | NM_009779 | 91 | + | 1.1 | 7 n.c. (−2085/−1520) | This study | Early, sustained |
| 13 | TNFRSF13b | NM_021349 | 86 | + | 3 | 2 c. (+1662/+1735, intron 2) | This study | Late, sustained |
| 14 | Receptor-like tyrosine kinase (Ryk) | L21707 | 79 | − | − | [1 c. −2190; 7 n.c. −4150/+1] | This study | Late, sustained |
| 15 | Schlafen 2 (Sfln2) | NM_011408 | 90 | − | − | [no c. in promoter; 4 c. +4130/+5040] | This study | Late, sustained |
| 16 | p21 (WAF1/CIP1)a | NM_007669 | 66 | ND | ND | ND | (10), this study | Early, transient |
| (ii) A > B | ||||||||
| 17 | Serine (or cysteine) proteinase inhibitor (Spi2.1)a | M64085 | 85 | + | 0.9 | 1 c. + 1 n.c. in tandem (−127/−92) | (50), this study | Late, sustained |
| (iii) B > A | ||||||||
| 18 | Interleukin 2 receptor, alpha (IL-2Rα)a | NM_008367 | 90 | + | 2.5 | 1 c. + 1 n.c. in tandem (−1268/−1240) | (35), this study | Late, sustained |
| 19 | Interleukin 4 receptor, alpha (IL-4Rα) | M29854 | 62 | ND | ND | ND | (10), this study | Early, sustained |
| STAT5B specific | ||||||||
| 20 | Enabled homolog (Drosophila) (enah); Mena; VASP; WASP | NM_010135 | 79 | ND | ND | ND | This study | Late, sustained |
| 21 | Laminin, alpha 5 (Lama5) | U37501 | 82 | ND | ND | ND | This study | Late, sustained |
| ? | ||||||||
| 22 | Helix-loop-helix DNA binding protein regulator (Id-1)a | M31885 | 0 | + | 1.6 | 1 c. + 2 n.c. in tandem (+5350/+5710) | (11), this study | Early/late, sustained |
| 23 | Myelocytomastosis oncogene (c-Myc)a | X01023 | 0 | ND | ND | ND | (10), this study | Early, sustained |
| 24 | Bcl-xa | L35049 | 0 | ND | ND | [2c. in tandem, +386/+475, intron 1] | (10,39,85), this study | Late, sustained |
aKnown STAT5 target genes from the literature.
bNo antibody control generated background values of 0.01–0.1% of input DNA (see also Figure 4).
cValues in square bracket indicates putative binding motifs found in the genomic sequence that showed no STAT5 binding by ChIP.
Abbreviations: c., consensus; n.c., non-consensus; ND, not determined; tss, transcription start site.
Figure 2.
Expression of known and putative STAT5 target genes in STAT5A and STAT5B double (A) and single (B) knockdown. RNAs were isolated from Ba/F3-β cells treated as described in Figure 1, and analyzed by RT-qPCR. Six distinct expression profiles could be recognized, represented here by selected genes (see also Table 2), revealing a clear differential contribution of STAT5A and STAT5B in regulating expression of STAT5 target genes.
Interestingly, four distinct expression patterns were observed. First, a group of genes including Cis (Figure 2), Socs-3, Osm, Pim-1, Pim-2, MKP-1, Stra13, IL-2Rβ, TCRγ-V4 and Heparin binding EGF-like growth factor, which were downregulated in the double knockdown were weakly or even not affected in the single knockdown (Figure 2 and Table 2), indicating functional redundancy between STAT5A and STAT5B proteins. A second pattern was STAT5-regulated genes affected in both single knockdowns, although at various degrees. Genes such as C3ar1 (Figure 2), Socs-1, TNFRSF13b, Ryk, Slfn2 and p21, revealed an equal requirement for both STAT5A and STAT5B (Figure 2 and Table 2), whilst other genes showed a preferential contribution of either STAT5A (Spi2.1) or STAT5B (IL-2Rα, IL-4Rα) (Figure 2 and Table 2). Third, Enah (Figure 2) and Lama5 genes revealed a clear specificity toward STAT5B, while none of the evaluated genes showed an exclusive specificity toward STAT5A (Figure 2 and Table 2). Finally and unexpectedly, three known STAT5 targets (Id-1, c-Myc and Bcl-x) remained unaffected in the STAT5A/B double knockdown, and hence in the single knockdown (Figure 2 and Table 2). To rule out that this absence of effect was due to compensatory mechanisms mediated by STAT3, the other main STAT molecule activated in Ba/F3-β in response to IL-3 (51,52), expression of Id-1, c-Myc and Bcl-x was evaluated in IL-3-stimulated Ba/F3-β cells following siRNA-mediated knockdown of STAT3 (Figure 3A and B). While IL-3 induction of Socs-3, a well-characterized STAT3 target (53) was repressed following STAT3 knockdown (Figure 3B), IL-3 induction of Id-1, c-Myc and Bcl-x remained unaffected (Figure 3C) as was IL-3 induction of Cis, another STAT5 target (Figure 3B). This suggests that Id-1, c-Myc and Bcl-x are not STAT3 targets and therefore that the lack of effect in the STAT5A/B knockdown cannot be attributable to functional redundancy between the two STAT molecules.
Figure 3.
STAT3 and STAT5 do not have redundant functions in regulating STAT5 target gene expression in IL-3-stimulated Ba/F3-β cells. Ba/F3-β cells were transfected with a siRNA specific for STAT3 or with a control siRNA (ScI), as described in Figure 1. (A) At the time of IL-3 stimulation, a specific reduction in STAT3 protein levels upon STAT3 knockdown was verified by western blot using antibodies directed against STAT3 or STAT5 as a control. (B and C) mRNA levels for STAT3, SOCS-3—a STAT3 target—and Cis, Id-1, c-Myc and Bcl-x were analyzed as described in Figures 1 and 2.
This experimental approach thus confirmed that STAT5A and STAT5B exert overlapping as well as distinct functions in vivo at the level of transcriptional regulation, and led to the recognition of distinct categories of STAT5 target genes based on the differential contribution of STAT5A and STAT5B in their regulation.
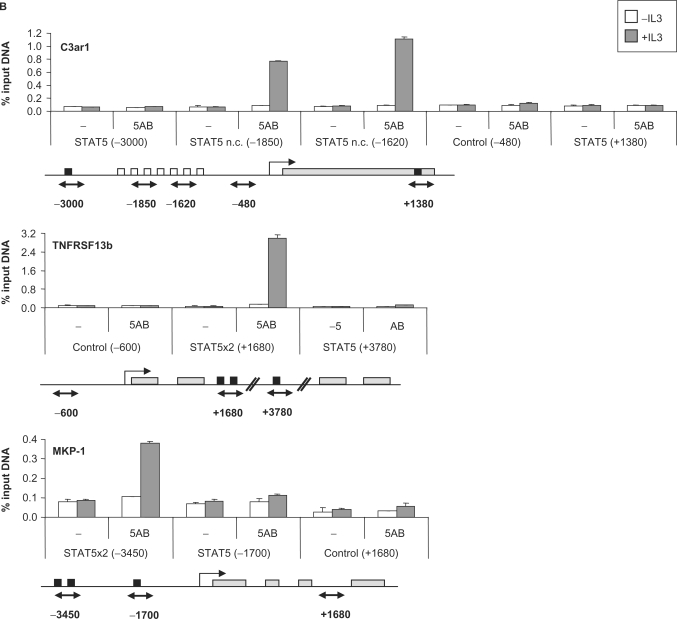
STAT5 is recruited at unusual sites to the C3ar1, MKP-1 and TNFRSF13b gene loci
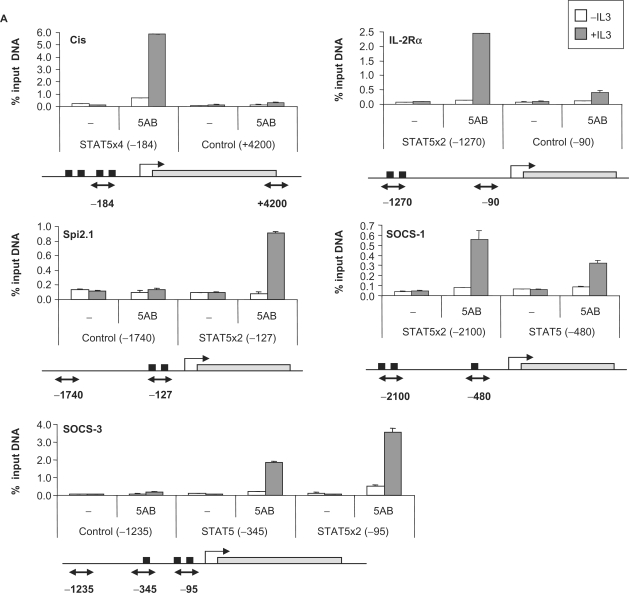
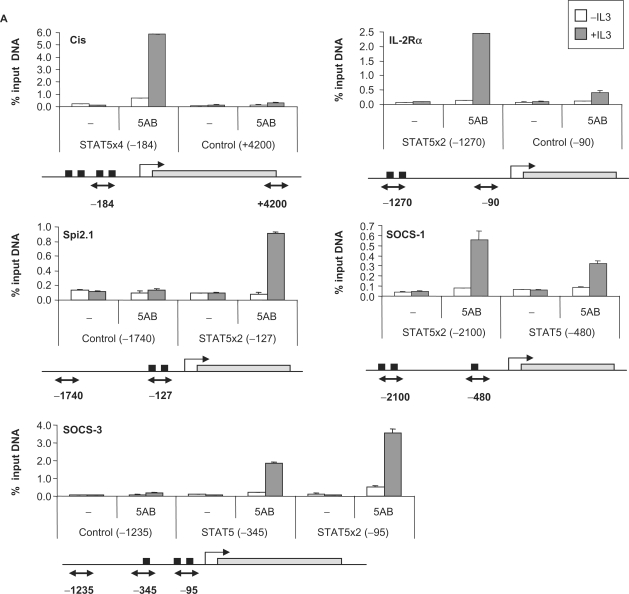
To address how these differences in gene expression relate to STAT5 DNA binding in vivo, and to separate direct STAT5 targets from genes indirectly regulated by STAT5, ChIP was performed on a subset of the above genes. Known STAT5 target genes with characterized (Cis, Osm, Socs-3, Spi2.1, IL-2Rα and Id-1) or uncharacterized (Socs-1) STAT5 binding sites and putative STAT5 targets (MKP-1, Stra13, Pim-2, C3ar1, TNFRSF13b, Ryk, Slfn2 and IL-2Rβ) were investigated. ChIP was performed on unstimulated and IL-3-stimulated (45 min) Ba/F3-β cells using antibodies directed against STAT5A and STAT5B, and the immunoprecipitated genomic DNA was analyzed by real-time PCR. First, the genomic sequence (from −6000 relative to the transcription start site up to the polyA signal) of each gene was scanned for the presence of putative consensus STAT5 binding motifs (TTCNNNGAA) as well as non-consensus motifs, and real-time PCR primer pairs were designed to examine these regions as well as remote control regions (10,46).
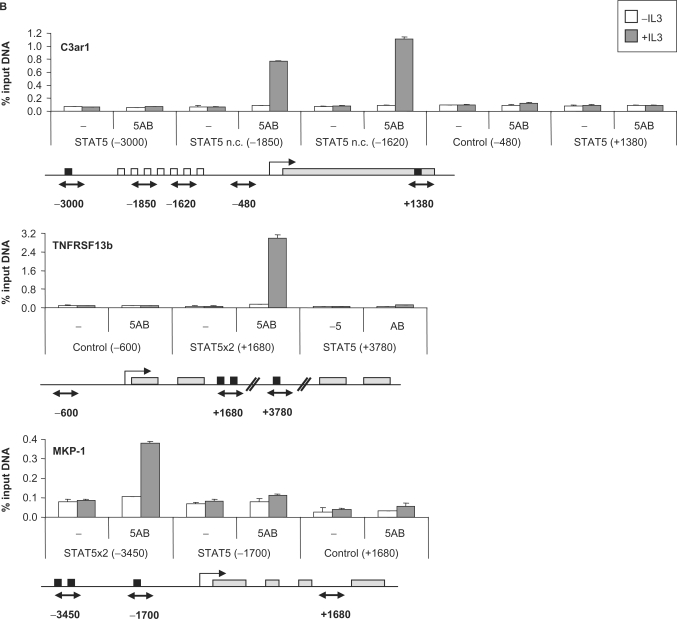
STAT5 recruitment to the known STAT5 target genes Cis, Osm, Socs-3, Spi2.1, IL-2Rα and Id-1 was detected at the expected position (Figure 4A and Table 2). Additionally, we were able to identify the so far uncharacterized STAT5 binding site of the Socs-1 gene at position −2100 relative to the transcription start site (Figure 4A). No STAT5 binding could be detected by ChIP on the Stra13, Pim-2, Ryk, Slfn2 and IL-2Rβ putative STAT5 target genes, even though consensus STAT5 binding sites were found within the investigated regions (Table 2). STAT5 recruitment was detected, however, within the C3ar1, TNFRSF13b and MKP-1 genes (Figure 4B).
Figure 4.
STAT5 recruitment in vivo on known (A) and novel (B) STAT5 target genes. Ba/F3-β cells were stimulated with IL-3 for 45 min and ChIP was performed as described in Materials and Methods section, using antibodies directed against STAT5A and STAT5B (5AB) or no antibody as a control (−). The isolated genomic DNA was analyzed by real-time PCR using primers specific for STAT5 binding sites or control regions, as indicated under each graph. Black squares indicate consensus STAT5 binding sites, open squares non-consensus (n.c.) STAT5 binding sites and gray rectangles represent open reading frames (A) or exons (B). Exons/introns were not represented in (A) for convenience. Positions are relative to the transcription start site.
The sequence and position of the functional STAT5 binding elements evaluated in this study are presented in Table 3. In all cases, STAT5 binding sites are present in tandem of two or more consensus or/and non-consensus motifs. While 4 out of the 10 elements analyzed were located in the proximal region of the promoter (<400 bp upstream of the transcription start site; Cis, Osm, Socs-3 and Spi2.1), as typically described for STAT5 target genes, four were located in the distal promoter (−1200 to −3450; MKP-1, Socs-1, C3ar1 and IL-2Rα), one within an intron (+1660; TNFRSF13b) and one several kilobase downstream of the gene (+5350; Id-1) (Table 3). The most unforeseen finding was STAT5 recruitment at the C3ar1 distal promoter (−2085/−1520) that lacks consensus STAT5 motifs but comprises seven non-consensus elements (Figure 4B and Table 3). The resolution of the ChIP experiment does not, however, allow for the discrimination of the relative involvement of each motif over this 565-bp long region.
Table 3.
Nucleotide sequences of known and newly identified functional STAT5 binding sites (ChIP assay)
| Gene name | Positionb | STAT5 binding site (bold; 5′-3′) (consensus: TTC NNN GAA) | References | |
|---|---|---|---|---|
| 1 | Cisa | (−230/−103) | TTC CTG GAA AG TTC TTG GAA (82) TTC TAG GAA GATGAGGC TTC CGG GAA | (10,48), this study |
| 2 | Socs-3a | (−345/−64) | TTC TTA GAA (241) TTA CAA GAA GACCGGCCGGGCAG TTC CAG GAA | (53), this study |
| 3 | Osma | (−190/−95) | TTC GAA GAA (76) TTC CCA GAA | (93), this study |
| 4 | Spi2.1a | (−127/−92) | TTC CCA GAA ATCACCCGGTCTGTCCAT TTC TCA TTA | (50), this study |
| 5 | IL-2Rαa | (−1268/−1240) | TTC TGA GAA GTACCAGACAT TTC TGA TAA | (35), this study |
| 6 | Id-1a | (+5350/+5710, 4.2 kb downstream gene) | TTC CTG GAA (320) TTC TAT CAA CTGCTCTGAGG TTC CTG CAA | (11), this study |
| 7 | Socs-1a | (−2095/−2061) | TTC AGG GAA CTGAAC TTC CCA GAG CT TTC CCA GAA | This study |
| 8 | MKP-1 | (−3450/−3414) | TTC ACG GAA CACAGGCTCACGGGGTT TTC TTG GAA | This study |
| 9 | TNFRSF13b | (+1662/+1735, intron 2) | TTC TTG GAA (56) TTC TGG GAA | This study |
| 10 | C3ar1 | (−2085/−1520) | TTA GCA GAA (150) TTT CAG GAA (80) TTG GAA AAA AAA TTG GAT GAA (120) … | This study |
| … TTC TTG TAA (20) TTT CTG GAA (130) TTT GAA GAA |
aKnown STAT5 target genes.
bRelative to the transcription start site (except for TNFRSF13b: relative to the translation start site).
Interestingly, the potency of STAT5 DNA binding in vivo to the various elements varied significantly from 0.4 to 6% of input DNA (Figure 4 and Table 2). Although these differences might reflect variability in antibody accessibility, they might also reveal differences in DNA binding affinities on the various target genes in vivo.
The newly identified STAT5 targets C3ar1, MKP-1 and TNFRSF13b are potential oncogenes
Constitutive activation of STAT5A and STAT5B has been associated with a wide variety of human cancers, especially hematologic malignancies (leukemia, lymphoma and myeloma), as well as solid tumors such as breast cancer, SCCHN or melanoma (2,3, 45,54–58).
Our gene expression profiling study combined with ChIP analysis identified three novel direct STAT5 target genes, namely, TNFRSF13b, MKP-1 and C3ar1 raising the possibility that overexpression of these genes in STAT5-associated malignancies plays a role in tumorigenesis. Interestingly, tumor necrosis factor receptor superfamily, member 13B (TNFRSF13b) has been associated to lymphoid and non-lymphoid malignancies and to immunologic disorders (59–64), while MKP-1 (MAP kinase phosphatase-1) has been associated to a variety of human cancers (65–67).
C3ar1 encodes a G-protein coupled seven-transmembrane receptor for the anaphylatoxin and chemoattractant C3a, an important inflammatory mediator (68). C3ar1 is implicated in Th2 development and is linked to the pathogenesis of asthma (69,70). It is expressed in normal kidney (71) and its upregulation is associated to the development of lupus nephritis (72). Beside its implication in inflammatory diseases, C3ar1 overexpression has been recently associated to FMS-like tyrosine kinase 3 (FLT3) D835/I836 mutants-mediated cytogenetically normal acute myeloid leukemia (CN-AML) (73), which is also associated to constitutive STAT5 signaling (74–77).
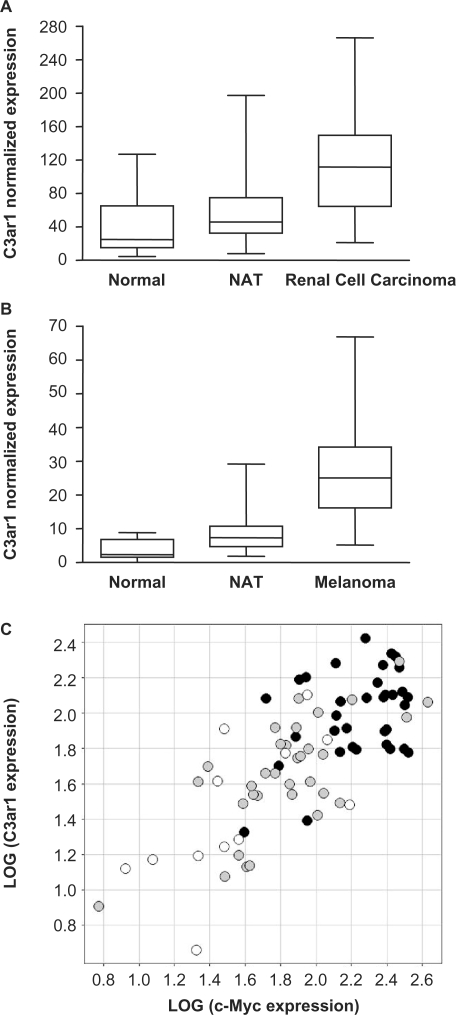
To further correlate expression of C3ar1 with the development of cancer, we monitored C3ar1 mRNA levels on various cancer panels (Figure 5). Interestingly, C3ar1 expression was found to be significantly upregulated in renal clear cell carcinoma (Figure 5A) and melanoma (Figure 5B). Although activation of STAT5 signaling was shown to directly contribute to the progression of malignant melanoma (55,56,58,78), an implication of STAT5 in renal cell carcinoma has not yet been clearly established (2,57,79). Expression of c-Myc, an oncogene regulated by numerous transcription factors including STAT5, was also upregulated in renal cancer (Figure 5C). Upregulation of c-Myc and C3ar1 was observed in 24 out of a panel of 30 pairs (80%) of ‘normal adjacent tissue’ and ‘renal cell carcinoma’ (not shown), and their expression correlated well in both normal and malignant tissues (Figure 5C). The observation that the newly identified STAT5 target gene C3ar1 is upregulated in several cancers (AML, renal cell carcinoma and melanoma) some of which being clearly associated to constitutive STAT5 signaling (AML and melanoma), therefore, raises the possibility that C3ar1 upregulation might play a role in tumorigenesis as a STAT5 target, and opens the door to further investigation.
Figure 5.
C3ar1, a novel STAT5 target gene, is overexpressed in (A, C) renal cell carcinoma and (B) melanoma. Expression of C3ar1 and c-Myc was evaluated by RT-qPCR on a panel of cDNAs samples from various human tumors and the corresponding NAT, as well as from normal tissues, as described in Materials and Methods section. mRNA levels were normalized to ubiquitin mRNA levels. C3ar1 is overexpressed in renal cell carcinomas (n = 35) (A) and melanoma tumors (n = 94) (B) compared to normal tissues [renal (n = 12) and skin (n = 15), respectively] and NAT [NAT renal (n = 33) and NAT skin (n = 91), respectively]. P-values for the difference in medians between tumors, normal tissues and NAT are <0.0001 by a two-tailed non-parametric Mann-Whitney test (Graph Pad Prism). The box represents 25–75th percentile of the data, the line represents the median and whiskers are the minimum and maximum values. (C) Expression of C3ar1 and c-Myc is upregulated in renal cell carcinoma (black dots) compared to NAT (gray dots) and normal tissues (white dots). Additionally, expression of C3ar1 and c-Myc is correlated in renal tumor and normal tissue. The squared correlation value, R2 is 55%.
DISCUSSION
Using a gene expression profiling approach followed by a ChIP analysis we were able to identify novel IL-3-dependent STAT5 target genes in the proB-derived Ba/F3-β cell line. Furthermore, a siRNA-mediated knockdown of STAT5A and STAT5B proteins demonstrated a clear functional difference between both proteins in regulating expression of these genes in vivo.
Our microarray analysis showed that 5–7% of the IL-3 inducible genes were (directly or indirectly) regulated by STAT5 in Ba/F3-β cells. Although this approach does not allow us to identify all STAT5 target genes, as revealed by the absence of effect of STAT5 knockdown on some known STAT5 targets such as c-Myc, Id-1 or Bcl-x (Table 2), it suggests that most of the IL-3 inducible genes are likely regulated through the MAP kinase and PI3-Kinase pathways which are also activated in response to IL-3 in these cells (80). Reciprocally, out of the genes downregulated in the STAT5A/B knockdown, only 20% were IL-3 inducible (not shown), suggesting that STAT5 may exert additional functions in regulating cytokine-independent genes. This proposition corroborates a recent ChIP-based study that identified STAT5 binding sites in the vicinity of IL-3 independent genes in Ba/F3 cells (39).
Although differential transcriptional regulation by STAT5A and STAT5B on a particular target gene has been already described (31), our study is the first to evaluate the relative contribution of STAT5A and STAT5B in the regulation of several STAT5 target genes in vivo. Since STAT5A and STAT5B are activated in a similar fashion in Ba/F3-β cells in response to IL-3 (39), the differential contribution of STAT5A and STAT5B is very likely attributable to differences in the regulatory potential of the two proteins, based in part on the DNA binding activity of the various STAT5 complexes. In support of this, STAT5 ChIP efficiency varied from 0.4 to 6% of input DNA, although these values should be taken with caution since differences in epitope availability might significantly contribute to the ChIP efficiency. These differences might also reflect the distinct kinetics of STAT5A and STAT5B DNA binding observed in vivo (39).
The analysis of the relative contribution of STAT5A and STAT5B to the expression of a number of STAT5-regulated genes revealed that approximately a third of these genes showed a strict requirement for both proteins, while a similar proportion showed redundancy between both STAT5 proteins (Table 2). A requirement for both STAT5A and STAT5B may suggest that transcription is dependent on STAT5A:STAT5B heterodimers. An alternative explanation is that STAT5A and STAT5B homodimers bind selectively to either one of tandemly arranged binding sites, consistent with observations in vitro (32,37). Redundancy between both STAT5 proteins suggests that STAT5A and STAT5B homodimers are equally efficient in DNA binding and transcriptional activation of the corresponding genes. It is likely that STAT5A:STAT5B heterodimers play little function in that context. Unexpectedly, IL-3-mediated induction of the Cis gene was not preferentially affected upon STAT5A knockdown in vivo. This contradicts the in vitro data showing a clear preference for STAT5A tetramers over STAT5B homodimers (32). This demonstrates that in vivo DNA binding activity involves other components, such as chromatin configuration, accessory regulatory elements and/or cofactors, which are lacking in in vitro experiments. Interestingly, some genes (Enah and Lama5) were specifically affected by the absence of one particular STAT5 protein in vivo (Table 2), in accordance with the non-redundancy of STAT5A and STAT5B functions in vivo (12,13,31).
STAT5A/B gene targeting studies showed that IL-2 signaling is impaired in STAT5A/B knockout mice, due to a diminished expression of both IL-2Rα and IL-2Rβ in T and NK cells (15,18,19). These studies suggested that while IL-2 induction of IL-2Rα requires both STAT5A and STAT5B, IL-2 induction of IL-2Rβ preferentially involves STAT5B (18,19). In the IL-3-stimulated proB Ba/F3-β cell line, expression of IL-2Rα was also dependent on both STAT5A and STAT5B, with a stronger participation of STAT5B (Figure 2 and Table 2). However, IL-3 induction of IL-2Rβ was redundantly regulated by STAT5A and STAT5B, thus revealing a functional difference between IL-2 and IL-3 signaling in regulating transcription of the IL-2Rβ gene. On the other hand, since no functional STAT5 binding site was found within the IL-2Rβ gene (Table 2), a direct regulation by STAT5 remains hypothetical.
The serine/threonine protein kinases Pim-1 and Pim-2 belong to the same family of proto-oncogenes, with partially redundant functions in IL-2 and IL-3 signaling in vivo (81–83). While Pim-1 is a known STAT5 target (10,41,84), regulation of Pim-2 expression by STAT5 remains elusive. We show here that IL-3 induction of Pim-2 is diminished in STAT5A/B siRNA-transfected Ba/F3-β cells. However, expression of both genes appears to be differentially regulated by IL-3 in these cells. First, while Pim-1 is induced as an early response gene (10) (Table 2), Pim2 is a late response gene (Table 2 and data not shown). Second, while IL-3 induction of Pim-1 is repressed by the deacetylase inhibitor—and inhibitor of STAT5-mediated transcription—TSA (10), induction of Pim-2 is not (data not shown). In addition, no STAT5 binding was detected within the Pim-2 gene in vivo, although four consensus binding sites were found between positions −1520 and −270 of the Pim-2 promoter. It remains, thus, unclear whether Pim-2 is a direct target of STAT5, in contrast to Pim-1. Collectively, although they belong to the same family of proto-oncogenes, Pim-1 and Pim-2 regulation by STAT5 appears to involve distinct mechanisms.
Surprisingly, IL-3 induction of the known STAT5 targets Id-1, c-Myc and Bcl-x was not diminished upon siRNA-mediated STAT5A/B knockdown (Figure 2 and Table 2), although all three genes were upregulated in Ba/F3 cells expressing a constitutively active form of STAT5A (designated 1*6) (10,84), and IL-3-mediated induction was inhibited by TSA (10). The fact that STAT5A/B knockdown does not affect expression of these genes cannot be explained by compensatory effects of STAT3 (Figure 3), and thus suggests either that robust IL-3-mediated STAT5-independent regulation of these genes does not lead to reduced expression upon STAT5 knockdown, or that these genes are subject to an alternative mechanism of transcriptional regulation by STAT5. Interestingly, Id-1, c-Myc and Bcl-x bear unconventional STAT5 binding site locations. STAT5 binds within the first intron of the Bcl-x gene and >4 kb downstream of the Id-1 gene (11,39,85). A preliminary attempt to identify STAT5 binding within the proximal c-Myc promoter in vivo was not successful (A.R., unpublished data), suggesting that the STAT5 responsive element might be located elsewhere. It is therefore likely that transcriptional regulation by STAT5 through remote regulatory elements involves an alternative mechanism, possibly involving chromatin components and/or additional factors. In an attempt to explain how a 60% reduction in STAT5 protein levels upon siRNA knockdown does not influence expression of these genes, we can already rule out that STAT5 is constitutively bound to DNA even in the absence of IL-3 (7,39), since STAT5 binding on the Id-1 gene in vivo in unstimulated cells was at background levels and was clearly induced in response to IL-3 (Table 2 and data not shown). Alternatively, stable STAT5-containing complexes might persist in the nucleus of unstimulated cells, which could be recruited to DNA upon IL-3 stimulation. Further investigations would be necessary to address that complex question. In particular, it would be interesting to explore STAT5 recruitment by ChIP upon siRNA-mediated STAT5 knockdown.
The analysis of the newly identified and previously characterized STAT5 DNA binding sites (Table 3) shows that all elements consist of a tandem arrangement of at least two consensus motifs [TTCNNNGAA or more precisely TTCYNUGAA, in agreement with published data (30)] or of one consensus plus at least one non-consensus motifs (i.e. containing one mismatch), suggesting that cooperation through STAT5 tetramerization might be essential for efficient transcriptional activation in vivo (32,34–37). One exception was the C3ar1 gene, lacking consensus binding sites but encompassing a cluster of seven non-consensus motifs that efficiently bound STAT5 in vivo. Interestingly, Nelson et al. [see supplementary materials (39)] recently identified novel STAT5 target genes presenting a similar configuration. This observation suggests that the lack of consensus sequence can be compensated by the presence of multiple non-optimal binding sites which might allow cooperative binding of STAT5. The identification of the TNFRSF13b STAT5 binding sites within the second intron (Table 3) confirms a recent ChIP-based genome-wide survey showing that STAT5 binding sites are often located within introns (39), a characteristic also shared by the STAT5 target gene Bcl-x (85). Further studies (EMSA, reporter assays) will be needed to investigate the functionality of these newly identified STAT5 binding elements.
Out of the STAT5-regulated genes tested by ChIP, five showed no detectable binding within the region investigated (Table 2). An absence of binding could signify either that the binding site is located outside the investigated region (−6000/polyA), as such elements can be located up to 200 kb downstream the transcription start site (11,86), or that it is mediated through interaction with other transcription factors (87) or more likely that regulation of gene expression by STAT5 is indirect. It is more likely that the late response genes, for which no STAT5 binding sites were identified, such as Pim-2, IL-2Rβ, Ryk and Slfn2, are indirectly regulated by STAT5. In support to that proposition, these genes did not show the expected expression pattern either in response to TSA (Pim-2, see above) or in the Ba/F3-1*6 cell line expressing a constitutively active form of STAT5A (IL-2Rβ, Ryk and Slfn2, data not shown). Alternatively, we cannot exclude that the time point chosen to perform the ChIP assay (45 min of IL-3 stimulation) did not allow significant detection of STAT5 recruitment.
Table 2 summarizes the results of our study. It did not reveal a clear correlation between position of the binding site, potency of DNA binding, specificity toward STAT5A or STAT5B, or expression profile (early versus late response genes). This demonstrates that transcriptional regulation by STAT5 in vivo is complex, probably involving local chromatin structure and additional factors, likely responsible for the specificity of gene regulation. On the other hand, and as predicted, a correlation was found between the degree of inhibition in the STAT5A/B knockdown and the specific requirement for ‘STAT5A or STAT5B’ versus ‘STAT5A and STAT5B’. While the expression of genes that were redundantly regulated by STAT5A and STAT5B (Table 2, genes numbered 1–10) was inhibited to 55% in average in the double knockdown, the genes that were dependent on the presence of only one or of both STAT5 proteins (Table 2, genes numbered 11–21) were more dramatically affected in the double knockdown (78% inhibition in average). Again, exceptions were observed, emphasizing the existence of additional levels of regulation.
We identified three novel STAT5 target genes, namely TNFRSF13b, MKP-1 and C3ar1, that show distinct expression profiles in response to IL-3, distinct requirements for STAT5A and STAT5B, and distinct STAT5 DNA binding characteristics (Tables 2 and 3). All the three genes have been previously associated with tumorigenesis (63–66,73). TNFRSF13b encodes the transmembrane activator and CAML interactor (TACI) protein and is a receptor for TNFSF13/APRIL and TNFSF13B/TALL1/BAFF/BLYS ligands. It is involved in the stimulation of B and T cell function and the regulation of humoral immunity (60,62,64,88–90). Defect in TNFRSF13b is associated with lymphoid and non-lymphoid malignancies, autoimmune disorders and is a cause of immunodeficiencies, such as common variable immunodeficiency (CVID), and immunoglobulin A deficiency (IgAD) (59–64). MKP-1 is a dual specificity phosphatase essential for the dephosphorylation of MAP kinases p38 and JNK (91). MKP-1 acts as a negative regulator of the synthesis of proinflammatory cytokines in vivo, hence playing a pivotal role in the feedback control of the innate immune response (91), a function also shared by the STAT5 target gene SOCS-1 (92). MKP-1 was found overexpressed in a number of human cancer tissues or cell lines (65–67) and to contribute to tumorigenesis in pancreatic cancer and non-small-cell lung cancer (NSCLC) (65,66). We showed here that C3ar1 is upregulated in renal cancer and melanoma. Whether this newly described cancer association is the consequence of STAT5 constitutive activation remains to be established, but, together with its previous association with mutant FLT3-mediated CN-AML (73), it raises the possibility that C3ar1 overexpression is involved in tumorigenesis in STAT5-associated malignancies. Our data demonstrate the efficiency of an approach combining gene expression profiling, ChIP and cancer panel expression analysis to identify potential STAT5-dependent oncogenes.
ACKNOWLEDGEMENTS
We want to thank Jeanine Mattson for her assistance with the microarray analysis and Joachim Griesenbeck for critically reading the manuscript. Schering-Plough Biopharma was fully supported by Schering-Plough Corporation. Funding to pay the Open Access publication charges for this article was provided by Schering-Plough Corporation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ivashkiv LB, Hu X. Signaling by STATs. Arthritis Res. Ther. 2004;6:159–168. doi: 10.1186/ar1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 3.Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J. Cell. Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 4.Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine Growth Factor Rev. 1999;10:131–157. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 5.Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer T, Vinkemeier U. Nucleocytoplasmic shuttling of STAT transcription factors. Eur. J. Biochem. 2004;271:4606–4612. doi: 10.1111/j.1432-1033.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- 7.Zeng R, Aoki Y, Yoshida M, Arai K, Watanabe S. Stat5B shuttles between cytoplasm and nucleus in a cytokine-dependent and -independent manner. J. Immunol. 2002;168:4567–4575. doi: 10.4049/jimmunol.168.9.4567. [DOI] [PubMed] [Google Scholar]
- 8.Pfitzner E, Jahne R, Wissler M, Stoecklin E, Groner B. p300/CREB-binding protein enhances the prolactin-mediated transcriptional induction through direct interaction with the transactivation domain of Stat5, but does not participate in the Stat5-mediated suppression of the glucocorticoid response. Mol. Endocrinol. 1998;12:1582–1593. doi: 10.1210/mend.12.10.0180. [DOI] [PubMed] [Google Scholar]
- 9.Litterst CM, Kliem S, Marilley D, Pfitzner E. NCoA-1/SRC-1 is an essential coactivator of STAT5 that binds to the FDL motif in the alpha-helical region of the STAT5 transactivation domain. J. Biol. Chem. 2003;278:45340–45351. doi: 10.1074/jbc.M303644200. [DOI] [PubMed] [Google Scholar]
- 10.Rascle A, Johnston JA, Amati B. Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5. Mol. Cell. Biol. 2003;23:4162–4173. doi: 10.1128/MCB.23.12.4162-4173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu M, Nie L, Kim SH, Sun XH. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta. EMBO J. 2003;22:893–904. doi: 10.1093/emboj/cdg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akira S. Functional roles of STAT family proteins: lessons from knockout mice. Stem Cells. 1999;17:138–146. doi: 10.1002/stem.170138. [DOI] [PubMed] [Google Scholar]
- 13.Lin JX, Leonard WJ. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19:2566–2576. doi: 10.1038/sj.onc.1203523. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 15.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 16.Park SH, Liu X, Hennighausen L, Davey HW, Waxman DJ. Distinctive roles of STAT5a and STAT5b in sexual dimorphism of hepatic P450 gene expression. Impact of STAT5a gene disruption. J. Biol. Chem. 1999;274:7421–7430. doi: 10.1074/jbc.274.11.7421. [DOI] [PubMed] [Google Scholar]
- 17.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc. Natl Acad. Sci. USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imada K, Bloom ET, Nakajima H, Horvath-Arcidiacono JA, Udy GB, Davey HW, Leonard WJ. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J. Exp. Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakajima H, Liu XW, Wynshaw-Boris A, Rosenthal LA, Imada K, Finbloom DS, Hennighausen L, Leonard WJ. An indirect effect of Stat5a in IL-2-induced proliferation: a critical role for Stat5a in IL-2-mediated IL-2 receptor alpha chain induction. Immunity. 1997;7:691–701. doi: 10.1016/s1074-7613(00)80389-1. [DOI] [PubMed] [Google Scholar]
- 20.Ambrosio R, Fimiani G, Monfregola J, Sanzari E, De Felice N, Salerno MC, Pignata C, D’Urso M, Ursini MV. The structure of human STAT5A and B genes reveals two regions of nearly identical sequence and an alternative tissue specific STAT5B promoter. Gene. 2002;285:311–318. doi: 10.1016/s0378-1119(02)00421-3. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc. Natl Acad. Sci. USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen H, Haldosen LA. EGF modulates expression of STAT5 in mammary epithelial cells. Exp. Cell Res. 1998;243:347–358. doi: 10.1006/excr.1998.4160. [DOI] [PubMed] [Google Scholar]
- 23.Kirken RA, Malabarba MG, Xu J, Liu X, Farrar WL, Hennighausen L, Larner AC, Grimley PM, Rui H. Prolactin stimulates serine/tyrosine phosphorylation and formation of heterocomplexes of multiple Stat5 isoforms in Nb2 lymphocytes. J. Biol. Chem. 1997;272:14098–14103. doi: 10.1074/jbc.272.22.14098. [DOI] [PubMed] [Google Scholar]
- 24.Meinke A, Barahmand-Pour F, Wohrl S, Stoiber D, Decker T. Activation of different Stat5 isoforms contributes to cell-type-restricted signaling in response to interferons. Mol. Cell. Biol. 1996;16:6937–6944. doi: 10.1128/mcb.16.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moucadel V, Constantinescu SN. Differential STAT5 signaling by ligand-dependent and constitutively active cytokine receptors. J. Biol. Chem. 2005;280:13364–13373. doi: 10.1074/jbc.M407326200. [DOI] [PubMed] [Google Scholar]
- 26.Storz P, Doppler H, Pfizenmaier K, Muller G. Insulin selectively activates STAT5b, but not STAT5a, via a JAK2-independent signalling pathway in Kym-1 rhabdomyosarcoma cells. FEBS Lett. 1999;464:159–163. doi: 10.1016/s0014-5793(99)01689-0. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita H, Xu J, Erwin RA, Farrar WL, Kirken RA, Rui H. Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. J. Biol. Chem. 1998;273:30218–30224. doi: 10.1074/jbc.273.46.30218. [DOI] [PubMed] [Google Scholar]
- 28.Kazansky AV, Rosen JM. Signal transducers and activators of transcription 5B potentiates v-Src-mediated transformation of NIH-3T3 cells. Cell Growth Differ. 2001;12:1–7. [PubMed] [Google Scholar]
- 29.Phung-Koskas T, Pilon A, Pous C, Betzina C, Sturm M, Bourguet-Kondracki ML, Durand G, Drechou A. STAT5B-mediated growth hormone signaling is organized by highly dynamic microtubules in hepatic cells. J. Biol. Chem. 2005;280:1123–1131. doi: 10.1074/jbc.M409918200. [DOI] [PubMed] [Google Scholar]
- 30.Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J. Biol. Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 31.Frasor J, Park K, Byers M, Telleria C, Kitamura T, Yu-Lee LY, Djiane J, Park-Sarge OK, Gibori G. Differential roles for signal transducers and activators of transcription 5a and 5b in PRL stimulation of ERalpha and ERbeta transcription. Mol. Endocrinol. 2001;15:2172–2181. doi: 10.1210/mend.15.12.0745. [DOI] [PubMed] [Google Scholar]
- 32.Verdier F, Rabionet R, Gouilleux F, Beisenherz-Huss C, Varlet P, Muller O, Mayeux P, Lacombe C, Gisselbrecht S, Chretien S. A sequence of the CIS gene promoter interacts preferentially with two associated STAT5A dimers: a distinct biochemical difference between STAT5A and STAT5B. Mol. Cell. Biol. 1998;18:5852–5860. doi: 10.1128/mcb.18.10.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boucheron C, Dumon S, Santos SC, Moriggl R, Hennighausen L, Gisselbrecht S, Gouilleux F. A single amino acid in the DNA binding regions of STAT5A and STAT5B confers distinct DNA binding specificities. J. Biol. Chem. 1998;273:33936–33941. doi: 10.1074/jbc.273.51.33936. [DOI] [PubMed] [Google Scholar]
- 34.John S, Vinkemeier U, Soldaini E, Darnell J.E., Jr, Leonard WJ. The significance of tetramerization in promoter recruitment by Stat5. Mol. Cell. Biol. 1999;19:1910–1918. doi: 10.1128/mcb.19.3.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer WK, Reichenbach P, Schindler U, Soldaini E, Nabholz M. Interaction of STAT5 dimers on two low affinity binding sites mediates interleukin 2 (IL-2) stimulation of IL-2 receptor alpha gene transcription. J. Biol. Chem. 1997;272:31821–31828. doi: 10.1074/jbc.272.50.31821. [DOI] [PubMed] [Google Scholar]
- 36.Xu X, Sun YL, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 37.Soldaini E, John S, Moro S, Bollenbacher J, Schindler U, Leonard WJ. DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol. Cell. Biol. 2000;20:389–401. doi: 10.1128/mcb.20.1.389-401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer T, Hendry L, Begitt A, John S, Vinkemeier U. A single residue modulates tyrosine dephosphorylation, oligomerization, and nuclear accumulation of stat transcription factors. J. Biol. Chem. 2004;279:18998–19007. doi: 10.1074/jbc.M400766200. [DOI] [PubMed] [Google Scholar]
- 39.Nelson EA, Walker SR, Alvarez JV, Frank DA. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J. Biol. Chem. 2004;279:54724–54730. doi: 10.1074/jbc.M408464200. [DOI] [PubMed] [Google Scholar]
- 40.Brierley MM, Fish EN. Stats: multifaceted regulators of transcription. J. Interferon Cytokine Res. 2005;25:733–744. doi: 10.1089/jir.2005.25.733. [DOI] [PubMed] [Google Scholar]
- 41.Mui AL, Wakao H, Kinoshita T, Kitamura T, Miyajima A. Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: role of Stat5 in proliferation. EMBO J. 1996;15:2425–2433. [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle JN. Naturally occurring dominant negative variants of Stat5. Mol. Cell. Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren S, Cai HR, Li M, Furth PA. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene. 2002;21:4335–4339. doi: 10.1038/sj.onc.1205484. [DOI] [PubMed] [Google Scholar]
- 44.Leong PL, Xi S, Drenning SD, Dyer KF, Wentzel AL, Lerner EC, Smithgall TE, Grandis JR. Differential function of STAT5 isoforms in head and neck cancer growth control. Oncogene. 2002;21:2846–2853. doi: 10.1038/sj.onc.1205385. [DOI] [PubMed] [Google Scholar]
- 45.Xi S, Zhang Q, Gooding WE, Smithgall TE, Grandis JR. Constitutive activation of Stat5b contributes to carcinogenesis in vivo. Cancer Res. 2003;63:6763–6771. [PubMed] [Google Scholar]
- 46.Rascle A, Lees E. Chromatin acetylation and remodeling at the Cis promoter during STAT5-induced transcription. Nucleic Acids Res. 2003;31:6882–6890. doi: 10.1093/nar/gkg907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol. Cell. Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- 49.Monni R, Santos SC, Mauchauffe M, Berger R, Ghysdael J, Gouilleux F, Gisselbrecht S, Bernard O, Penard-Lacronique V. The TEL-Jak2 oncoprotein induces Socs1 expression and altered cytokine response in Ba/F3 cells. Oncogene. 2001;20:849–858. doi: 10.1038/sj.onc.1204201. [DOI] [PubMed] [Google Scholar]
- 50.Wood TJ, Sliva D, Lobie PE, Goullieux F, Mui AL, Groner B, Norstedt G, Haldosen LA. Specificity of transcription enhancement via the STAT responsive element in the serine protease inhibitor 2.1 promoter. Mol. Cell. Endocrinol. 1997;130:69–81. doi: 10.1016/s0303-7207(97)00075-0. [DOI] [PubMed] [Google Scholar]
- 51.Lacronique V, Boureux A, Monni R, Dumon S, Mauchauffe M, Mayeux P, Gouilleux F, Berger R, Gisselbrecht S, Ghysdael J, et al. Transforming properties of chimeric TEL-JAK proteins in Ba/F3 cells. Blood. 2000;95:2076–2083. [PubMed] [Google Scholar]
- 52.Nagata Y, Todokoro K. Interleukin 3 activates not only JAK2 and STAT5, but also Tyk2, STAT1, and STAT3. Biochem. Biophys. Res. Commun. 1996;221:785–789. doi: 10.1006/bbrc.1996.0674. [DOI] [PubMed] [Google Scholar]
- 53.Auernhammer CJ, Bousquet C, Melmed S. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc. Natl Acad. Sci. USA. 1999;96:6964–6969. doi: 10.1073/pnas.96.12.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barash I. Stat5 in the mammary gland: controlling normal development and cancer. J. Cell. Physiol. 2006;209:305–313. doi: 10.1002/jcp.20771. [DOI] [PubMed] [Google Scholar]
- 55.Mirmohammadsadegh A, Hassan M, Bardenheuer W, Marini A, Gustrau A, Nambiar S, Tannapfel A, Bojar H, Ruzicka T, Hengge UR. STAT5 phosphorylation in malignant melanoma is important for survival and is mediated through SRC and JAK1 kinases. J. Invest. Dermatol. 2006;126:2272–2280. doi: 10.1038/sj.jid.5700385. [DOI] [PubMed] [Google Scholar]
- 56.Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23:8017–8023. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 57.Turkson J, Jove R. STAT proteins: novel molecular targets for cancer drug discovery. Oncogene. 2000;19:6613–6626. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- 58.Wellbrock C, Weisser C, Hassel JC, Fischer P, Becker J, Vetter CS, Behrmann I, Kortylewski M, Heinrich PC, Schartl M. STAT5 contributes to interferon resistance of melanoma cells. Curr. Biol. 2005;15:1629–1639. doi: 10.1016/j.cub.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 59.Castigli E, Wilson SA, Garibyan L, Rachid R, Bonilla F, Schneider L, Geha RS. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat. Genet. 2005;37:829–834. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- 60.Gross JA, Johnston J, Mudri S, Enselman R, Dillon SR, Madden K, Xu W, Parrish-Novak J, Foster D, Lofton-Day C, et al. TACI and BCMA are receptors for a TNF homologue implicated in B-cell autoimmune disease. Nature. 2000;404:995–999. doi: 10.1038/35010115. [DOI] [PubMed] [Google Scholar]
- 61.Salzer U, Chapel HM, Webster AD, Pan-Hammarstrom Q, Schmitt-Graeff A, Schlesier M, Peter HH, Rockstroh JK, Schneider P, Schaffer AA, et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat. Genet. 2005;37:820–828. doi: 10.1038/ng1600. [DOI] [PubMed] [Google Scholar]
- 62.Salzer U, Jennings S, Grimbacher B. To switch or not to switch—the opposing roles of TACI in terminal B cell differentiation. Eur. J. Immunol. 2007;37:17–20. doi: 10.1002/eji.200636914. [DOI] [PubMed] [Google Scholar]
- 63.Seshasayee D, Valdez P, Yan M, Dixit VM, Tumas D, Grewal IS. Loss of TACI causes fatal lymphoproliferation and autoimmunity, establishing TACI as an inhibitory BLyS receptor. Immunity. 2003;18:279–288. doi: 10.1016/s1074-7613(03)00025-6. [DOI] [PubMed] [Google Scholar]
- 64.Yan M, Wang H, Chan B, Roose-Girma M, Erickson S, Baker T, Tumas D, Grewal IS, Dixit VM. Activation and accumulation of B cells in TACI-deficient mice. Nat. Immunol. 2001;2:638–643. doi: 10.1038/89790. [DOI] [PubMed] [Google Scholar]
- 65.Chattopadhyay S, Machado-Pinilla R, Manguan-Garcia C, Belda-Iniesta C, Moratilla C, Cejas P, Fresno-Vara JA, de Castro-Carpeno J, Casado E, Nistal M, et al. MKP1/CL100 controls tumor growth and sensitivity to cisplatin in non-small-cell lung cancer. Oncogene. 2006;25:3335–3345. doi: 10.1038/sj.onc.1209364. [DOI] [PubMed] [Google Scholar]
- 66.Liao Q, Guo J, Kleeff J, Zimmermann A, Buchler MW, Korc M, Friess H. Down-regulation of the dual-specificity phosphatase MKP-1 suppresses tumorigenicity of pancreatic cancer cells. Gastroenterology. 2003;124:1830–1845. doi: 10.1016/s0016-5085(03)00398-6. [DOI] [PubMed] [Google Scholar]
- 67.Wang HY, Cheng Z, Malbon CC. Overexpression of mitogen-activated protein kinase phosphatases MKP1, MKP2 in human breast cancer. Cancer Lett. 2003;191:229–237. doi: 10.1016/s0304-3835(02)00612-2. [DOI] [PubMed] [Google Scholar]
- 68.Kohl J. Anaphylatoxins and infectious and non-infectious inflammatory diseases. Mol. Immunol. 2001;38:175–187. doi: 10.1016/s0161-5890(01)00041-4. [DOI] [PubMed] [Google Scholar]
- 69.Ali H, Panettieri R.A., Jr Anaphylatoxin C3a receptors in asthma. Respir. Res. 2005;6:19. doi: 10.1186/1465-9921-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drouin SM, Corry DB, Hollman TJ, Kildsgaard J, Wetsel RA. Absence of the complement anaphylatoxin C3a receptor suppresses Th2 effector functions in a murine model of pulmonary allergy. J. Immunol. 2002;169:5926–5933. doi: 10.4049/jimmunol.169.10.5926. [DOI] [PubMed] [Google Scholar]
- 71.Braun MC, Reins RY, Li TB, Hollmann TJ, Dutta R, Rick WA, Teng BB, Ke B. Renal expression of the C3a receptor and functional responses of primary human proximal tubular epithelial cells. J. Immunol. 2004;173:4190–4196. doi: 10.4049/jimmunol.173.6.4190. [DOI] [PubMed] [Google Scholar]
- 72.Bao L, Osawe I, Haas M, Quigg RJ. Signaling through up-regulated C3a receptor is key to the development of experimental lupus nephritis. J. Immunol. 2005;175:1947–1955. doi: 10.4049/jimmunol.175.3.1947. [DOI] [PubMed] [Google Scholar]
- 73.Whitman SP, Ruppert AS, Radmacher MD, Mrozek K, Paschka P, Langer C, Baldus CD, Wen J, Racke F, Powell BL, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111:1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilliland DG, Griffin JD. Role of FLT3 in leukemia. Curr. Opin. Hematol. 2002;9:274–281. doi: 10.1097/00062752-200207000-00003. [DOI] [PubMed] [Google Scholar]
- 75.Mizuki M, Fenski R, Halfter H, Matsumura I, Schmidt R, Muller C, Gruning W, Kratz-Albers K, Serve S, Steur C, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96:3907–3914. [PubMed] [Google Scholar]
- 76.Spiekermann K, Bagrintseva K, Schwab R, Schmieja K, Hiddemann W. Overexpression and constitutive activation of FLT3 induces STAT5 activation in primary acute myeloid leukemia blast cells. Clin. Cancer Res. 2003;9:2140–2150. [PubMed] [Google Scholar]
- 77.Spiekermann K, Dirschinger RJ, Schwab R, Bagrintseva K, Faber F, Buske C, Schnittger S, Kelly LM, Gilliland DG, Hiddemann W. The protein tyrosine kinase inhibitor SU5614 inhibits FLT3 and induces growth arrest and apoptosis in AML-derived cell lines expressing a constitutively activated FLT3. Blood. 2003;101:1494–1504. doi: 10.1182/blood-2002-04-1045. [DOI] [PubMed] [Google Scholar]
- 78.Morcinek JC, Weisser C, Geissinger E, Schartl M, Wellbrock C. Activation of STAT5 triggers proliferation and contributes to anti-apoptotic signalling mediated by the oncogenic Xmrk kinase. Oncogene. 2002;21:1668–1678. doi: 10.1038/sj.onc.1205148. [DOI] [PubMed] [Google Scholar]
- 79.Song C, Jun SY, Hong JH, Ahn H. Transforming growth factor-beta downregulates interleukin-2-induced phosphorylation of signal transducer and activator of transcription 5 in human renal cell carcinoma. J. Cancer Res. Clin. Oncol. 2007;133:487–492. doi: 10.1007/s00432-007-0192-2. [DOI] [PubMed] [Google Scholar]
- 80.Martinez-Moczygemba M, Huston DP. Biology of common beta receptor-signaling cytokines: IL-3, IL-5, and GM-CSF. J. Allergy Clin. Immunol. 2003;112:653–665. doi: 10.1016/S0091. [DOI] [PubMed] [Google Scholar]
- 81.Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J, Berns A. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol. Cell. Biol. 2004;24:6104–6115. doi: 10.1128/MCB.24.13.6104-6115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Z, Bhattacharya N, Weaver M, Petersen K, Meyer M, Gapter L, Magnuson NS. Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J. Vet. Sci. 2001;2:167–179. [PubMed] [Google Scholar]
- 83.White E. The pims and outs of survival signaling: role for the Pim-2 protein kinase in the suppression of apoptosis by cytokines. Genes Dev. 2003;17:1813–1816. doi: 10.1101/gad.1123103. [DOI] [PubMed] [Google Scholar]
- 84.Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui AL, Kitamura T. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 1999;18:4754–4765. doi: 10.1093/emboj/18.17.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Silva M, Benito A, Sanz C, Prosper F, Ekhterae D, Nunez G, Fernandez-Luna JL. Erythropoietin can induce the expression of bcl-x(L) through Stat5 in erythropoietin-dependent progenitor cell lines. J. Biol. Chem. 1999;274:22165–22169. doi: 10.1074/jbc.274.32.22165. [DOI] [PubMed] [Google Scholar]
- 86.Nelson EA, Walker SR, Li W, Liu XS, Frank DA. Identification of human STAT5-dependent gene regulatory elements based on interspecies homology. J. Biol. Chem. 2006;281:26216–26224. doi: 10.1074/jbc.M605001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19:2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- 88.von Bulow GU, Bram RJ. NF-AT activation induced by a CAML-interacting member of the tumor necrosis factor receptor superfamily. Science. 1997;278:138–141. doi: 10.1126/science.278.5335.138. [DOI] [PubMed] [Google Scholar]
- 89.Wang H, Marsters SA, Baker T, Chan B, Lee WP, Fu L, Tumas D, Yan M, Dixit VM, Ashkenazi A, et al. TACI-ligand interactions are required for T cell activation and collagen-induced arthritis in mice. Nat. Immunol. 2001;2:632–637. doi: 10.1038/89782. [DOI] [PubMed] [Google Scholar]
- 90.Yan M, Marsters SA, Grewal IS, Wang H, Ashkenazi A, Dixit VM. Identification of a receptor for BLyS demonstrates a crucial role in humoral immunity. Nat. Immunol. 2000;1:37–41. doi: 10.1038/76889. [DOI] [PubMed] [Google Scholar]
- 91.Wang X, Liu Y. Regulation of innate immune response by MAP kinase phosphatase-1. Cell. Signal. 2007;19:1372–1382. doi: 10.1016/j.cellsig.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakagawa R, Naka T, Tsutsui H, Fujimoto M, Kimura A, Abe T, Seki E, Sato S, Takeuchi O, Takeda K, et al. SOCS-1 participates in negative regulation of LPS responses. Immunity. 2002;17:677–687. doi: 10.1016/s1074-7613(02)00449-1. [DOI] [PubMed] [Google Scholar]
- 93.Yoshimura A, Ichihara M, Kinjyo I, Moriyama M, Copeland NG, Gilbert DJ, Jenkins NA, Hara T, Miyajima A. Mouse oncostatin M: an immediate early gene induced by multiple cytokines through the JAK-STAT5 pathway. EMBO J. 1996;15:1055–1063. [PMC free article] [PubMed] [Google Scholar]