Abstract
Ribosomal RNAs (rRNAs), assisted by ribosomal proteins, form the basic structure of the ribosome, and play critical roles in protein synthesis. Compared to prokaryotic ribosomes, eukaryotic ribosomes contain elongated rRNAs with several expansion segments and larger numbers of ribosomal proteins. To investigate architectural evolution and functional capability of rRNAs, we employed a Tn5 transposon system to develop a systematic genetic insertion of an RNA segment 31 nt in length into Escherichia coli rRNAs. From the plasmid library harboring a single rRNA operon containing random insertions, we isolated surviving clones bearing rRNAs with functional insertions that enabled rescue of the E. coli strain (Δ7rrn) in which all chromosomal rRNA operons were depleted. We identified 51 sites with functional insertions, 16 sites in 16S rRNA and 35 sites in 23S rRNA, revealing the architecture of E. coli rRNAs to be substantially flexible. Most of the insertion sites show clear tendency to coincide with the regions of the expansion segments found in eukaryotic rRNAs, implying that eukaryotic rRNAs evolved from prokaryotic rRNAs suffering genetic insertions and selections.
INTRODUCTION
Ribosomes translate genetic information encoded in mRNAs into a corresponding sequence of amino acids to form a protein. Ribosomes consist of large and small subunits, each of which is a ribonucleoprotein complex formed by rRNAs and ribosomal proteins. The rRNAs form the basic structure of the ribosome, and play central roles in the fundamental processes of protein biosynthesis. Recent structural studies of each subunit and of the 70S ribosome revealed that the functional cores subserving mRNA decoding and peptide-bond formation consist entirely of rRNAs, thus implying that the ribosome is an RNA-based machine (1–7). Although the functional regions of rRNAs are highly conserved, the architecture of rRNAs diversifies amongst organisms and organelles. In mammalian mitochondria, the lengths of rRNAs are shortened to approximately half that of prokaryotic rRNAs. Many helices in rRNAs are shortened or missing, whereas all functional domains are conserved. Large regions of missing RNA segments are replaced by enlarged ribosomal proteins and other, mitochondria-specific proteins (8–13). In contrast, eukaryotic ribosomes contain elongated rRNAs and an increased number of ribosomal proteins (14–17). It is thought that the architecture of rRNAs might have coevolved with ribosomal proteins so as to preserve the fundamental structure and function of ribosomes in all domains of life. Variations in the RNA-to-protein ratio found in ribosomes from various organisms indicate that some degree of architectural flexibility of is permissible in the evolutionary refinement of ribosomal structure. Compared to Escherichia coli 23S rRNA, with 2904 nt, yeast (Saccharomyces cerevisiae) 26S rRNA consists of 3392 nt, while human 28S rRNA consists of 5025 nt (15). The additional residues in eukaryotic rRNAs are inserted at several specific sites in the secondary structures of prokaryotic rRNAs as expansion segments (ESs) (15). ESs vary in their size and sequence from species to species. ESs are categorized as 12 distinct segments (designated es1 to es12) in the small subunit rRNAs, and 41 distinct segments (designated ES1 to ES41) in the large subunit rRNAs (Figure 5A and B). As ESs can be found in nonconserved regions of rRNAs, it is thought that ES insertion does not disturb the fundamental function of rRNAs (15). ESs are known to contact with other ESs to form a large structural element of eukaryotic ribosomes (18–21). The structural diversity conferred upon eukaryotic ribosomes by ESs affects the complex regulatory mechanism of eukaryotic translation (14,22,23). Although the exact functions of ESs in rRNAs remain elusive, cryo-EM studies of eukaryotic ribosomes are providing clues revealing some of the functional aspects of ESs. ESs provide sites for eukaryote-specific intersubunit bridges, as well as scaffolds allowing additional proteins to bind to ribosomes (14). It has been revealed that ES24 near Helix 59 in the large subunit of the yeast 80S ribosome interacts directly with the Sec61 complex. This interaction suggests that ES24 plays an important role in the process of cotranslational protein translocation, by serving as an attachment site for the protein-conducting channel in endoplasmic reticulum (22). Upon binding of Sec61, ES27, an essential rod-like component, drastically changes its conformation, moving from the peptide exit site to a site close to L1 stalk. It has been proposed that movement of ES27 coordinates access of non-ribosomal protein factors to the peptide exit channel (22). In the protozoan Trypanosoma cruzi 80S ribosomes, es6 and es7 in the small subunit form a large domain which might assist in escorting mRNAs to the ribosome (19).
Figure 5.
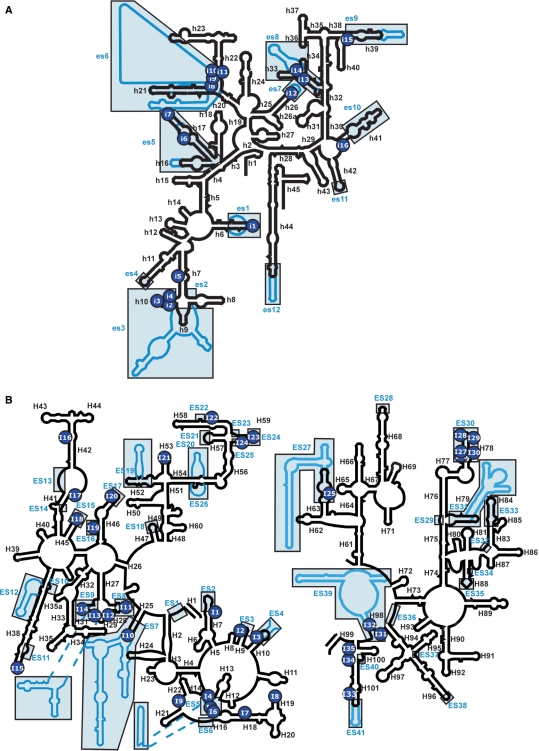
Secondary structure diagrams of E. coli rRNAs superimposed with S. cerevisiae rRNAs, indicating the sites of functional insertions. Escherichia coli 16S (A) and 23S (B) rRNAs are shown as black lines. Saccharomyces cerevisiae 18S (A) and 25S (B) rRNAs are shown as blue lines. Expansion segments are boxed and shaded in blue. The sites of functional insertions are mapped on the secondary structures of rRNAs using numbered blue circles.
The availability of comparative and phylogenetic analyses of rRNA sequences with secondary structures provides us with many insights into the functional and structural evolution of rRNAs, whereas a purely experimental approach to investigating rRNA evolution is limited. Genetic insertion of short RNA segments into rRNAs is possible however, and allows us to probe ribosome architecture and function. Earlier experiments employing genetic insertion of RNA segments into rRNAs used cryo-electron microscopy to identify the placement of each rRNA helix within the ribosome structure (24,25). A 17-nt segment or a tRNA-like element was introduced at several positions of 23S rRNA by a conventional mutagenesis approach, and extra electron densities corresponding to the insertions were then observed. To examine the architectural evolution of rRNAs in an empirical and unbiased manner, it is necessary to design and adhere to a specific method for distinguishing functional insertions in rRNAs from a large number of random genetic insertions. Here, we describe a systematic genetic approach for selecting functional rRNA variants bearing short, inserted RNA segments. We previously developed a comprehensive genetic selection method which we named ‘systematic selection of functional sequences by enforced replacement’ (SSER) (26). This method allowed us to rapidly identify residues and sequences essential for ribosome function in E. coli cells, from randomized rRNA libraries. We employed this approach to analyze the peptidyl-transferase center (26), the conserved loop sequence of H69 (27) and the internal bulge sequence of H66 for the L2 binding site (28). For the current analysis, we constructed an rRNA library by randomly inserting a short RNA segment using a Tn5 transposon, and then subjected the library to SSER to isolate rRNA variants with functional insertions. To be identified as functional, the activity of the insertions needed to be high enough to rescue the Δ7rrn E. coli strain, in which all chromosomal rRNA operons are inactivated. The results of this study revealed that the architecture of E. coli rRNAs is particularly flexible. In addition, most of the sites receptive to insertion coincided with the ES regions found in eukaryotic rRNAs. Based on this study, we propose that eukaryotic rRNAs might have evolved from prokaryotic rRNAs through the process of genetic insertion and selection.
MATERIALS AND METHODS
Bacterial strains, plasmids and cultivation
The E. coli Δ7rrn strain TA542 (ΔrnEΔrrnBΔrrnAΔrrnHΔrrnG::catΔrrnC::catΔrrnD::catΔrecA56/pTRNA66 pHKrrnC) (29) was kindly provided by Dr Catherine L. Squires (Tufts Univ.). The rescue plasmid pRB101 (26) was constructed by introducing the SacB gene and rrnB operon into pMW118 (Ampr) (Nippon Gene). The plasmid pHKrrnC in strain TA542 was replaced with pRB101 to generate strain NT101, which was used as the host cell to construct variant strains in this study. Cells were grown at 37°C in 2× Luria-Bertani (2× LB, 2% Tryptone, 1% Yeast Extract and 1% NaCl) broth; for solid medium, 1.5% agar was added. Antibiotics were added at the following concentrations when required: 40 μg/ml spectinomycin (Spc), 100 μg/ml ampicillin (Amp), 50 μg/ml kanamycin (Km) and 100 μg/ml trimethoprim. To induce plasmid replacement in NT101, 5% sucrose was added to the LB broth. For the usage of Bsp1407I in the following experiment, A1394 in 16S rRNA of pRB102 (26) was mutated by U to erase the site for Bsp1407I, employing the Quik-Change site-directed mutagenesis kit (Stratagene) according to manufacturer's instruction, using the following primers: A1394U-F (5′cccgggccttgaacacaccgcccgtcacaccatggg3′) and A1394U-R (5′cgggcggtgtgaacaaggcccgggaacgtattcaccgtgg3′). NT102 is a series of E. coli strains in which pRB101 is replaced by pRB102 and its derivatives. To measure the growth rate of the NT102 variants, 2 μl of an overnight preculture was inoculated into 1 ml of 2× LB broth, then separated into five aliquots and plated in a flat-bottomed 96-well microplate (IWAKI). The microplate was incubated at 37°C with vigorous agitation in a Spectramax 190-plate reader (Molecular Device, Inc.), and absorbance at 600 nm (OD600) was monitored every 30 min.
Tn5 transposition of a short RNA segment in E. coli rRNAs
The nonautonomous Tn5 transposon, bearing a modified mosaic end (MME) was constructed by PCR amplification with primers RHI-F (5′ctgtctcttgtacacatcttgcggccgccaaccatcatcgatgaatt3′) and RHI-R (5′ctgtctcttgtacacatcttgcggccgccaaccctgaagcttgcatgcct3′), using EZ-Tn5™<DHFR-1>Transposon (Epcentre) as template DNA. For in vitro Tn5 transposition, 0.2 μl of EZ-Tn5™Transposase (Epcentre) was added to 500 ng of pRB102 (A1394U) and molar equivalent transposon in the 10-μl reaction buffer [50 mM Tris-acetate (pH 7.5), 150 mM potassium acetate, 10 mM magnesium acetate and 4 mM spermidine]. The reaction was incubated at 37°C for 2 h, then stopped by adding 1 μl of 1% SDS. The resulting mixture was heated at 70°C for 10 min. The reaction product was cleaned up using Mini-Elute columns (Qiagen, Germany), and eluted with 10 μl water. Two micro liters of the product was electroporated into E. coli DH5α Electro-Cells (Takara, Japan) in a 0.2-cm cuvette using Genepulser II (BioRad) and the following conditions: 1.8 kV, 100 Ω and 50 μF. The electrified cells were plated on LB plates containing Km (50 μg/ml) and trimethoprim (100 μg/ml). The plates were incubated at 37°C overnight, then all colonies (>10 000) were suspended in 3 ml LB broth. Plasmids were prepared from the suspension using a Mini-Elute column (Qiagen, Germany). The resulting plasmid library was digested with Bsp1407I (Takara, Japan) at 37°C overnight, then re-ligated using the Takara ligation kit version 2.4 (Takara, Japan) to construct the insertion library.
Selection of rRNA variants containing functional insertions
The detailed procedure for the SSER selection of rRNA has been described previously (26). The functionality of each insertion in pRB101 was tested using SacB-sucrose counter selection. The sacB gene from Bacillus subtilis is a counter-selectable marker that encodes levansucrase, which synthesizes fructan in the presence of sucrose. Fructan is toxic for a variety of bacteria, including E. coli. NT101 was transformed with the insertion library and spread onto LB plates containing Spc/Km. The Km-resistant cells transiently contain both pRB101 and pRB102 from the library. Transformant colonies were picked and suspended in LB broth and then spotted onto LB plates containing Spc/Km/sucrose to drive plasmid replacement. Sucrose-resistant colonies (600 colonies; NT102 variants) were then cultured in 2 × LB broth and plasmids purified by mini-prep, in preparation for the mapping. To check the plasmid replacement, each transformant was spotted onto two LB plates containing Spc/Amp and Spc/Km/sucrose. Zero growth of the spotted cells on the Amp-plate demonstrated complete plasmid replacement.
Mapping insertion sites in rRNAs
To elucidate the insertion site of the RNA fragment in pRB102, we amplified plasmids by means of multiply primed rolling circle amplification, employing phi29 DNA polymerase (30). Each colony on the Km-sucrose plate was picked and heated at 95°C for 3 min in 10-μl reaction buffer [20 mM Tris–HCl (pH 8.0), 40 mM NaCl, 1 mM EDTA, thiophosphate-modified random hexamer 5′-NpNpNpNpsNpsN-3′]. Next, 10 μl enzyme buffer [100 mM Tris–HCl (pH 8.0), 8 mM MgCl2, 100 mM ammonium sulfate, 0.6 mM dNTP, 400 μg/ml bovine serum albumin and 0.2 μl phi29 DNA polymerase] was added to the reaction mixture. Each plasmid was amplified by rolling circle amplification at 30°C for 48 h. As the insertion contains a Bsp1407I site, we mapped the site of insertion (Figure S2) using two combinations of restriction enzymes, Bsp1407I/BamHI and Bsp1407I/PvuI. The sites of insertion were determined by DNA sequencing using the ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
Translational fidelity measured by β-galactosidase activity
A series of Lac Z reporters were employed to measure the translational fidelity of the NT102 strains carrying the functional insertions in 16S rRNA, as described previously (27). The reporter plasmids pNT3-lacZ (+1), pNT3-lacZ (–1), pNT3-lacZ (UGA) and pNT3-lacZ (UAG) were used for measuring +1 or –1 frameshift, or UGA or UAG stop codon read through, respectively. As a positive control for UGA and UAG read through, we employed a NT102 strain with error prone mutations C912G and G885U in 16S rRNA. The NT102 strain carrying high-fidelity mutations C912G and G888U were employed as a control for +1 frameshift activity. The β-galactosidase assay was performed as described previously by Miller (31).
RESULTS
Random insertional mutagenesis of E. coli rRNAs by Tn5 transposition
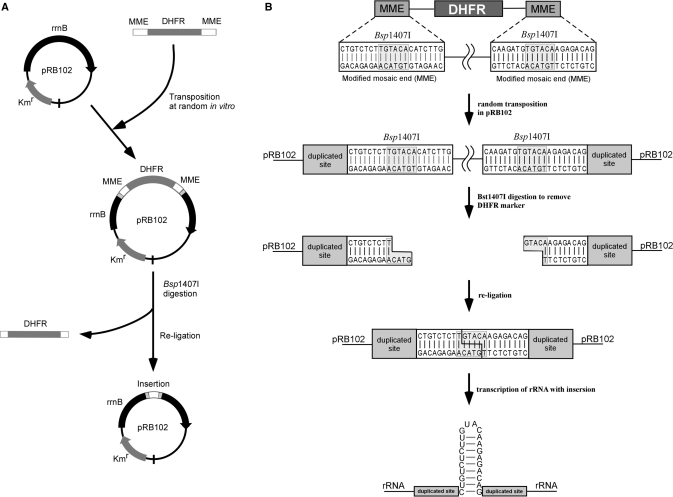
To search for functional insertions in rRNAs, we employed an in vitro Tn5 transposition system (32) to develop a procedure for the random genetic insertion of a short RNA segment into E. coli rRNAs (Figure 1A). We constructed a Tn5 transposon harboring dihydrofolatereductase (DHFR) gene as a selective marker, flanked by two modified mosaic end sequences (MMEs) bearing the cleavage site for restriction enzyme Bsp1407I (Figure 1B). The Tn5 transposon containing the DHFR marker was randomly inserted in vitro into a plasmid (pRB102) harboring the rrnB operon. At the sites of insertion, nine bases of the target sequences are duplicated. Next, E. coli DH5α was transformed with the resultant plasmid and selected using trimethoprim and Km. If the insertion has toxic effects, such as when it disrupts an antibiotic-resistant gene or the replication origin, no transformants are obtained. About 10 000 colonies were combined to make a library. The DHFR marker in each plasmid was removed by Bsp1407I digestion. After re-ligation, a 31-base insertion composed of the residual linker sequence (22 bases) and the duplicated sequence (nine bases) remained at the insertion site.
Figure 1.
Outline of the systematic genetic insertion of RNA segment in rRNAs. (A) A Tn5 transposon carrying a DHFR marker flanked by two MMEs was randomly inserted in vitro into a plasmid pRB102 harboring the rrnB operon, to construct the insertion library. Escherichia coli DH5α was transformed with the library and selected using trimethoprim and kanamycin. About 10 000 colonies were scraped together. To prepare the plasmid mixture, the DHFR marker in each plasmid was removed by Bsp1407I digestion. After re-ligation, the 31-base inserted RNA segment remained at the insertion site. (B) Detailed description of the site of insertion. MME contains a Bsp1407I site. At the sites of insertion, nine (or 10) bases of the target sequences are duplicated as a result of Tn5 transposition. It is just described as a ‘duplicated site’. The 31-nt inserted sequence, composed of residual linker sequence (22 nt) and duplicated sequence (9 or 10 nt), makes a short helix in the rRNA sequence.
Identification of functional insertions in rRNAs
We carried out a large-scale transformation of the plasmid library (containing the 31-base random insertion) into Δ7rrn strain NT101, and selected for Km-resistant transformants. The effect of this step was to exclude variants in which the incorporated plasmid had a dominant lethal insertion into a critical region of rRNA, such as the peptidyl-transferase center or the GTPase-associated region. The functionality of each insertion was tested using SacB-sucrose counter selection, as previously described (26) (Supplementary Figure S1). The Km-resistant cells transiently contain both pRB101 and pRB102 from the library. To promote plasmid replacement, we employed a method of counter selection with the sacB gene. Cells were picked and spotted onto selection plates containing Km and sucrose. If the incorporated pRB102 plasmid contained a functional sequence for ribosomal activity, it rapidly eliminated the pRB101 rescue plasmid, thus yielding sucrose-resistant cells (NT102 derivatives). Additionally, if the incorporated plasmid had a weak or nonfunctional ribosomal activity sequence, the rescue plasmid could not be replaced by the introduced plasmid, and the transformant would be sensitive to sucrose, due to the sacB gene. In this study, as a result of the selection, we obtained 600 functional variants. The position of RNA insertion was mapped in each variant (Supplementary Figure S2). Most insertions were found at the intergenic regions in the pRB102 plasmid, as expected. We obtained 51 unique insertions in the rRNAs, 16 insertions into 16S rRNA (designated i1 to i16) and 35 insertions into 23S rRNA (designated I1 to I35), respectively (Tables 1 and 2, Figures 2 and 3). At the insertion sites, nine or 10 bases of the target sequence were duplicated (Tables 1 and 2). A 22-base insertion was integrated between the duplicated sequences. When transcribed, the insertion sequence makes a short helix in rRNAs (Figure 1B).
Table 1.
Selected functional insertions in 16S rRNA from the random insertion library
| Insertion variants | Helix numbers | Duplicated sequences with nt numbers | Duplicated sites | Doubling time | Overlapping or close expansion segments |
|---|---|---|---|---|---|
| WT | 51.54 ± 2.27 | ||||
| i1 | h6 | AAGCUUGCU (9) | 80–88 | 49.08 ± 7.78 | es1 |
| i2 | h10 | AGGGGGACC (9) | 199–207 | 48.37 ± 1.04 | es3 |
| i3 | h10 | CCUUGGGGC (9) | 206–214 | 49.57 ± 5.42 | es3 |
| i4 | h7,h10 | GCCUCUUGC (9) | 213–221 | 99.22 ± 10.10 | es3 |
| i5 | h7 | GAUGUGCCC (9) | 227–235 | 175.49 ± 23.76 | – |
| i6 | h17 | GGGAGGAAG (9) | 445–453 | 84.65 ± 7.83 | es5 |
| i7 | h17 | AGUUAAUAC (9) | 461–469 | 48.90 ± 5.75 | es5 |
| i8 | h21,h22 | AGCUUGAGU (9) | 649–657 | 74.54 ± 9.39 | es6 |
| i9 | h21,h22 | GCUUGAGUC (9) | 650–658 | 81.27 ± 2.17 | es6 |
| i10 | h22 | GUCUCGUAG (9) | 656–664 | 48.19 ± 3.37 | es6 |
| i11 | h22 | GACGAAGAC (9) | 742–750 | 52.95 ± 3.58 | es6 |
| i12 | h26 | CCCUUGAGG (9) | 839–847 | 69.08 ± 9.82 | es7 |
| i13 | h33 | CCACGGAAG (9) | 998–1006 | 59.80 ± 4.38 | es8 |
| i14 | h33 | GAAUGUGCC (9) | 1020–1028 | 74.17 ± 4.39 | es8 |
| i15 | h39,h40 | GGAGACUGCC (10) | 1153–1162 | 45.70 ± 13.65 | es9 |
| i16 | h41 | GUGCGUCGU (9) | 1290–1298 | 52.69 ± 5.77 | es10 |
Table 2.
Selected functional insertions in 23S rRNA from the random insertion library
| Insertion variants | Helix numbers | Duplicated sequences with nt numbers | Duplicated sites | Doubling time | Overlapping or close expansion segments |
|---|---|---|---|---|---|
| WT | 51.54 ± 2.27 | ||||
| I1 | H7 | GUUAUAACC (9) | 98–106 | 54.52 ± 2.58 | ES2(l) |
| I2 | H9 | GUGUGUUUC (9) | 132–140 | 52.91 ± 0.40 | ES3(l) |
| I3 | H10 | AUCAUUAAC (9) | 149–157 | 55.48 ± 3.78 | ES4(l) |
| I4 | H13,H14 | GAGCAGCCC (9) | 261–269 | 52.39 ± 0.99 | ES5(l) |
| I5 | H14,H16 | GCCCAGAGC (9) | 266–274 | 77.81 ± 5.09 | ES5(l) |
| I6 | H14,H16 | GCCCAGAGCC (10) | 266–275 | 52.94 ± 1.47 | ES5(l) |
| I7 | H18 | GUGUGUGUG (9) | 283–291 | 46.70 ± 3.57 | – |
| I8 | H19 | CGUCUGGAA (9) | 301–310 | 59.90 ± 3.64 | – |
| I9 | H22 | GAAUAUGGG (9) | 400–408 | 74.28 ± 1.21 | – |
| I10 | H25 | CGCUUAGGC (9) | 542–550 | 53.75 ± 1.09 | ES7(l) |
| I11 | H28 | GAAUAGGGG (9) | 612–620 | 87.39 ± 11.77 | ES8(l) |
| I12 | H28,H29 | GCCGAAGGG (9) | 622–630 | 99.15 ± 12.58 | ES8(l) |
| I13 | H29 | GGGAAACCG (9) | 628–636 | 54.73 ± 0.66 | ES9(l) |
| I14 | H31 | GUCUUAACU (9) | 638–646 | 75.93 ± 5.89 | ES9(l) |
| I15 | H38 | GGGUCAUCC (9) | 881–889 | 41.20 ± 1.19 | ES11(l) |
| I16 | H42 | GGCCCAGAC (9) | 1041–1049 | 91.28 ± 8.23 | – |
| I17 | H41 | GGGCUAAAC (9) | 1137–1145 | 50.84 ± 0.43 | – |
| I18 | H45 | CGCUUAUGC (9) | 1170–1178 | 47.32 ± 2.00 | ES15(l) |
| I19 | H46 | CUGUAAGCC (9) | 1200–1208 | 58.10 ± 1.22 | ES16(l) |
| I20 | H46 | GGUGUGCUG (9) | 1215–1223 | 42.19 ± 0.79 | ES17(l) |
| I21 | H53 | GGUUAAUAU (9) | 1388–1396 | 52.65 ± 0.93 | – |
| I22 | H58 | GCUGAGGCG (9) | 1511–1519 | 87.74 ± 10.07 | ES22(l) |
| I23 | H59 | GCACUACGG (9) | 1530–1538 | 54.29 ± 0.25 | ES24(l) |
| I24 | H59 | GUGCUGAAG (9) | 1538–1546 | 56.95 ± 3.45 | ES25(l) |
| I25 | H63 | GCUGAAAUC (9) | 1740–1748 | 60.27 ± 0.00 | ES27(l) |
| I26 | H78 | GGCUUUGAA (9) | 2127–2135 | 57.93 ± 0.74 | ES30(l) |
| I27 | H78 | GCUUUGAAG (9) | 2128–2136 | 49.36 ± 3.77 | ES30(l) |
| I28 | H78 | GUGGACGCC (9) | 2138–2146 | 53.32 ± 0.41 | ES30(l) |
| I29 | H78 | AGUCUGCAU (9) | 2147–2155 | 52.92 ± 0.81 | ES30(l) |
| I30 | H78 | GCAUGGAGC (9) | 2152–2160 | 66.53 ± 7.29 | ES30(l) |
| I31 | H94,H98 | CCCUGACCC (9) | 2787–2795 | 75.22 ± 4.15 | ES39(l) |
| I32 | H98 | CUUUAAGGG (9) | 2795–2803 | 51.86 ± 0.80 | ES39(l) |
| I33 | H101 | GCGUUGAGC (9) | 2862–2870 | 77.48 ± 4.53 | ES41(l) |
| I34 | H101 | GUACUAAUG (9) | 2877–2885 | 86.51 ± 5.50 | – |
| I35 | H99,H101 | CUAAUGAAC (9) | 2880–2888 | 80.98 ± 2.86 | – |
Figure 2.
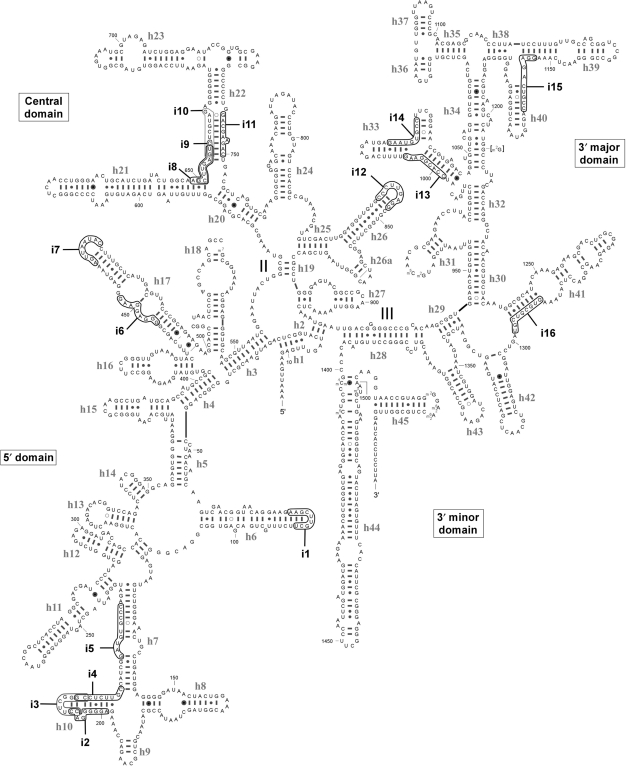
Sites of functional insertions on the secondary structure of E. coli 16S rRNA. The duplicated sequences at the sites of the 16 functional insertions (i1–i16) are shown by open ellipses. Numbering of the sites is described in Table 1. Helices (h1–h45) in 16S rRNA are numbered.
Figure 3.
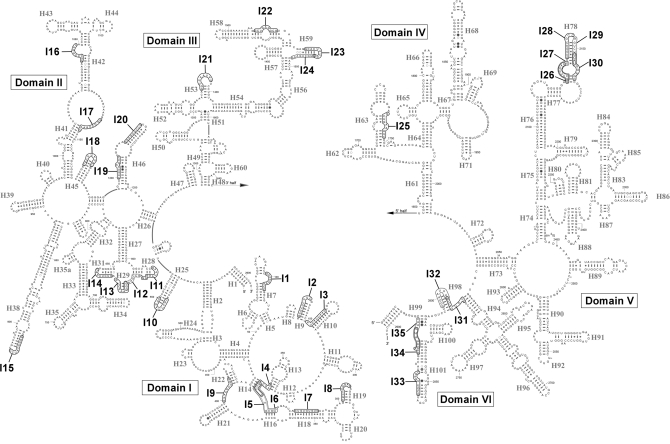
Sites of functional insertions on the secondary structure of E. coli 23S rRNA. The duplicated sequences at the sites of the 35 functional insertions (I1–I35) are shown by open ellipses. Numbering of the sites is described in Table 2. Helices (H1–H101) in 23S rRNA are numbered.
Insertion sites shape structural clusters on the surface of ribosomes
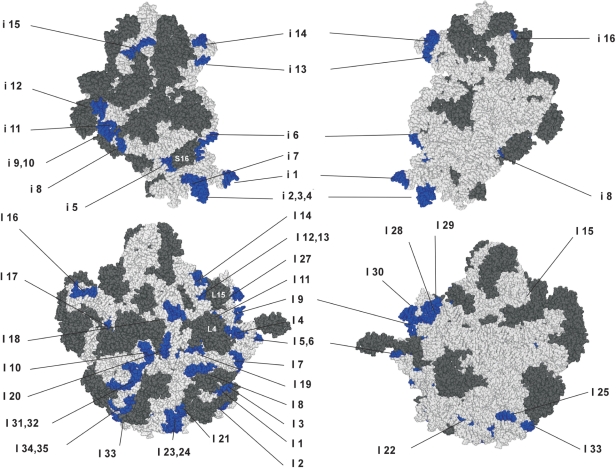
In 16S rRNA (Figure 2), the 16 functional insertions were widely distributed. Seven insertions (i1–7) were found in the 5′ domain, five insertions (i8–12) in the central domain, and four insertions (i13–16) in the 3′ major domain. No insertions were found in the 3′ minor domain, which contains the decoding center and a critical region for subunit association (5). There were two areas within 16S rRNA where we observed clusters of functional insertions. The first was in helix 10, where three insertions, i2, i3 and i4, were located. The second was at the branch point of helices 21 and 22, where i8, i9 and i10 were so densely mapped that the duplicated sites of i8 and i9 nearly overlapped. We suggest that it is not an accidental event that multiple insertions occurred at specific regions in rRNAs. Since the variants carrying these insertions were the clones that had survived selection following generation of the rRNA random insertion library, it is likely that the sites for multiple insertions are structurally and functionally tolerated in ribosomal architecture. When the insertion-receptive sites were mapped onto the tertiary structure of the 30S ribosome (Figure 4), most could be found in the solvent side and peripheral regions of the ribosome, avoiding any highly conserved functional domains or protein-binding sites. No insertions were found at the interface surface. Within the tertiary structure of the ribosome, i1 and i7 are located close to the i2–4 cluster at the bottom of 30S ribosome, despite their distance from each other within the secondary structure of 16S rRNA (Figure 2). Similarly, i11 and i12 reside close to the i8–10 cluster. These observations indicate that functional insertions tend to form a large cluster on the surface of ribosome.
Figure 4.
Sites of functional insertions mapped on the tertiary structure of E. coli ribosomes. The sites of insertions are marked in blue on the E. coli 30S (upper panels) and 50S (lower panels) subunits, showing numbering of insertions. The left and right figures represent the solvent side and the interface, respectively. RNAs and proteins are shown in light gray and dark gray, respectively. Ribosomal proteins S16, L4 and L15 are indicated. The atomic coordinates of each subunit were obtained from the Protein Data Bank (code 2AW4 and 2AVY) (5). The three-dimensional structures were displayed using Ras Top version 2.0.3.
In 23S rRNA (Figure 3), the 35 functional insertions we identified were also widely distributed among the six domains; nine insertions (I1–9) in domain I, 11 insertions (I10–20) in domain II, four insertions (I21–24) in domain III, one insertion (I25) in domain IV, five insertions (I26–30) in domain V and five insertions (I31–35) in domain VI. As observed in 16S rRNA (Figure 2), we also observed clustering of insertions in the secondary structure of 23S rRNA, with I4–6, I11–14 and I26–30 clustered together. We observed only one insertion (I25) in domain IV, and one cluster (I26–30) in domain V, likely because these domains shape the inter-subunit surface, and contain numerous critical regions for 50S function, such as the peptidyl-transferase center (1,33), intersubunit bridges and factor binding sites (3,5). When the observed insertion-receptive sites were mapped onto the 50S ribosome (Figure 4), most were seen on the solvent side and peripheral regions of 50S, avoiding any protein-binding sites. As in 16S rRNA, no insertions were found at the interface surface. In the 50S tertiary structure, the characteristic I26–30 cluster in Helix 78 of domain V is located in the L1 stalk. We found that I21–I24 in domain III shape a structural cluster at the base of 50S. Finally, I10 in domain II was located close to I31 and I32 in domain VI, forming an interdomain cluster in the lower part of the solvent side.
The growth phenotypes and translational fidelity of the insertion variants
As the selected variants contain homogeneous ribosomes bearing mutant rRNAs with functional insertions, the functionality of each insertion variant is reflected by its growth phenotype. To examine the growth phenotype of the insertion variants carefully, we generated new constructs of each variant, to avoid the emergence of secondary mutations at unexpected positions, or compensatory mutations during selection and subculturing. The growth rate of each variant was measured to estimate its ribosomal activity (Tables 1 and 2). Most of variants were healthier than expected, despite having the 31-base insertion. With the exception of i5, all variants had doubling times no more than twice those of wild-type controls. It is known that mutations in 16S rRNA can affect translational fidelity. We therefore examined whether ribosomes bearing insertions in 16S rRNA would affect the accuracy of mRNA decoding, by measuring the ability of ribosome variants to translate engineered +1 and –1 frameshift sites or stop codons in β-galactosidase mRNA (Table 3).
Table 3.
Translational fidelity of the insertion variants
| TGA readthrough | TAG readthrough | +1 frameshift | −1 frame shift | |
|---|---|---|---|---|
| WT | 1554.51 ± 56.67 | 478.24 ± 6.67 | 1417.82 ± 19.04 | 627.10 ± 25.25 |
| i1 | 2372.28 ± 255.48 | 460.37 ± 7.54 | 1487.78 ± 41.77 | 565.57 ± 12.31 |
| i2 | 2710.00 ± 242.18 | 506.31 ± 14.62 | 1620.09 ± 75.47 | 718.86 ± 22.97 |
| i3 | 2024.91 ± 154.42 | 503.43 ± 13.60 | 1664.08 ± 61.25 | 563.24 ± 35.90 |
| i4 | 1662.30 ± 30.34 | 431.84 ± 14.35 | 2478.71 ± 103.74 | 716.23 ± 10.59 |
| i5 | 1592.13 ± 35.39 | 515.12 ± 12.90 | 2191.23 ± 101.81 | 980.59 ± 45.59 |
| i6 | 2031.94 ± 57.51 | 479.67 ± 11.95 | 1530.43 ± 28.66 | 758.71 ± 89.74 |
| i7 | 2033.93 ± 33.65 | 450.46 ± 93.47 | 1987.88 ± 92.36 | 666.91 ± 108.23 |
| i8 | 3092.56 ± 254.82 | 720.81 ± 24.92 | 1872.57 ± 116.67 | 753.92 ± 24.62 |
| i9 | 2974.65 ± 118.29 | 507.09 ± 23.58 | 1263.33 ± 20.92 | 624.27 ± 26.69 |
| i10 | 2425.90 ± 98.83 | 615.20 ± 23.19 | 2091.87 ± 10.91 | 767.14 ± 18.66 |
| i11 | 2110.55 ± 89.58 | 448.28 ± 10.32 | 1580.58 ± 227.63 | 635.59 ± 20.91 |
| i12 | 1301.44 ± 22.44 | 471.18 ± 15.85 | 2364.53 ± 82.43 | 690.35 ± 24.04 |
| i13 | 2468.02 ± 108.39 | 468.26 ± 7.93 | 1878.99 ± 66.33 | 606.35 ± 32.77 |
| i14 | 1817.86 ± 53.79 | 386.89 ± 13.15 | 1590.20 ± 27.20 | 569.86 ± 68.35 |
| i15 | 2235.41 ± 71.80 | 454.46 ± 8.06 | 1810.91 ± 107.38 | 751.10 ± 73.97 |
| i16 | 2052.02 ± 68.13 | 372.22 ± 5.89 | 1882.35 ± 39.22 | 741.94 ± 18.00 |
| HF | 1182.61 ± 32.54 | 326.03 ± 1.02 | 4176.75 ± 145.47 | 783.93 ± 35.45 |
| EP | 11510.89 ± 510.24 | 1101.03 ± 10.97 | 3456.16 ± 123.92 | 477.12 ± 15.25 |
Mean β-galactosidase activities (±SD values) were measured in each insertion variant bearing 4 different lacZ reporter constructs; pNT3-lacZ (+1), pNT3-lacZ (−1), pNT3-lacZ (UGA) and pNT3-lacZ (UAG) which is responsible for +1 frameshift, −1 frameshift, UAG read through and UGA read through, respectively.
The above-mentioned i5 variant, in which the insertion site is located on helix 7 of 16S rRNA, exhibited doubling time 3-fold longer than that of wild-type control (i5, 175.49 min; wild type, 51.54 min). Variant i5 showed strong +1 as well as –1 frameshift activities, but did not show any read through activities (Table 3). The result suggested that the growth defect carried by i5 is partly explained by the enhanced frameshift activity due to the insertion at helix 7. Within the tertiary structure (Figure 4), i5 overlaps with the binding site for S16 (34). In addition, it is located within the interior of the 30S particle. Thus, insertion of the RNA segment at i5 might perturb correct folding of the adjacent regions. Variant i4 also exhibited a growth defect (doubling time of 99.22 min). Like i5, i4 showed enhanced frameshift activities (Table 3). As i4 is close to i5, a similar mechanism of ribosomal dysfunction might be involved. The insertion site i12 was located at the tip of helix 26. Enhanced frameshift activity was also observed in i12, however this variant exhibited only mild growth retardation. Variants i8 and i9, in which the insertion sites are localized at the branch point of helices 21 and 22, showed a slight reduction of growth rate, and enhanced readthrough activities.
In the clustered insertion I11–14 in domain II, variants I11 and I12 showed intermediate reductions in the growth rate (87.39 min and 99.15 min, respectively). We noticed on the 50S structure that I11 and I12 are localized behind L4 and L15, respectively (5,35,36). Insertions at these positions might influence protein binding during ribosome assembly, or configuration of these proteins. I16 exhibited a mild reduction of growth rate (91.28 min). I16 overlaps with one of binding sites of L10 in the GTPase-associated region (37), and as such its phenotypic expression might affect factor binding and/or GTPase activity.
DISCUSSION
Using SSER, we have described here a method for the systematic genetic insertion of short RNA segment into rRNAs, and have demonstrated that rRNAs are substantially tolerant toward short insertions. The main advantage of this method is that it can be used to test a large number of random genetic insertions in vivo, and that it permits neutral genetic selection of functional insertions, eliminating experimental bias. Although we were able to identify 51 sites with functional insertions in 16S and 23S rRNAs, the selection was unlikely to have been saturated, since most of the sequenced variants did not overlap with each other. However, judging from the density of the mapped insertion sites at the intergenic regions in pRB102 (Supplementary Figure S2), the randomness of the insertion library was sufficient for neutral genetic selection of functional insertions to occur. Due to experimental limitations imposed by the Tn5 transposon system, only a short, 22-base helix could be inserted into rRNAs in this study. Use of a different system would allow the insertion of longer and different sequences, and would likely produce different results.
Nevertheless, the secondary structures of bacterial 16S and 23S rRNAs are highly conserved, we obtained 51 rRNA variants with the 31-base insertion. Most of the viable functional insertions were found in the solvent side and peripheral regions of both subunits, but not in the highly conserved functional regions, indicating that toxic insertions resulting in lethal phenotypes were successfully excluded during selection. In one sense, the procedure comprised a functional probing of rRNA structure. Especially noteworthy is our finding that most insertions can be found on the surface of each subunit, and avoid protein-binding sites, although I11 and I12 might affect the L4 and L15-binding sites (5,35,36), respectively. It seems that sites on the surface of the ribosome more easily accommodate the additional helix. Our examination of the growth phenotype and translational fidelity of each variant allows us to speculate upon the result of inserting a short RNA segment. However, for a more complete understanding, we require information on the actual ribosomal structure following insertion, since it is likely that the insertion will induce structural alteration of the target region. Moreover, such alteration might be compensated for by structural rearrangement of adjacent regions, which might in turn affect other, more distant regions. Structural characterization by cryo-electron microscopy will be necessary to understand the phenotypic behavior of each ribosome.
The structural diversity of eukaryotic ribosomes is illustrated by the acquisition of ESs at several specific sites in the secondary structures of prokaryotic rRNAs (14,15). If eukaryotic rRNAs evolved from prokaryotic rRNAs suffering random genetic insertions and selection, sites of functional insertions could be expected to coincide with the regions of the ESs found in eukaryotic rRNAs. In 16S rRNA, 16 functional insertions can be superimposed onto ESs in eukaryotic 18S rRNAs (Figure 5A). Among 16 insertions, 15 show a clear tendency to coincide with the regions of the ESs found in yeast 18S rRNAs (Figure 5A, Table 1). The insertion i1 overlaps well with es1 which is localized at the tip of helix 6 (in the spur region). Insertions i6 and i7 occur in the region corresponding to that of es5; i15 and i16 reside near the regions es9 and es10, respectively; i13 and i14 overlap with es8 on helices 33 and 34, in a region termed the ‘beak’ in prokaryotic ribosomes. In yeast, helix 33 is slightly longer than the corresponding region in 16S rRNA, but helix 33a is replaced by eukaryote-specific protein rpS0 (14). The clustered insertion i2–4 is densely mapped on helix 10, where the large expansion segment es3 is localized. es3 forms an eukaryote-specific subdivision named ‘left foot’. It is known that human es3 is 45-bases longer than yeast es3, suggesting that es3 has been elongated by genetic insertion during the process of eukaryotic evolution. Another clustered insertion, i8–11 at the branch point of helices 21 and 22, coincides with es6, which is the largest segment, averaging 250-nt in length (18). By interacting with es3, es6 forms an extra density in the lower left part of the body of the 40S subunit (14). On the same side of the 40S subunit, es7 overlaps with i12 at the tip of helix 26. A striking feature involving es6 and es7 can be seen in the 40S subunit from protozoan Trypanosoma cruzi (19). Trypanosoma cruzi 18S rRNA contains large es6 and es7 sequences of 504 nt and 147 nt, respectively. Cryo-EM revealed that es6 together with es7 makes up a large helical structure termed the ‘turret’ which is located at the left side of 40S body (19). As the turret structure only exists in Tryponosomas, it was speculated that acquisition of these large expansion segments is involved in unique mechanism of Tryponosoma-specific translation initiation. The high frequency of functional insertion at these positions of E. coli 16S rRNA strongly suggests that eukaryotic rRNAs evolved through a process of genetic insertion, to acquire a unique module that facilitates their characteristic functions in protein synthesis. Concerning es12 at the tip of helix 44, we did not observe any functional insertions around this region, possibly because the genetic selection was not saturated. In fact, we were able to elongate helix 44 by artificial insertion of a short RNA segment of 19 nt (data not shown). Accordingly, we found that most of the ES sites in 16S rRNA were tolerant of genetic insertions. Meanwhile, only one variant, i5, did not overlap with any ESs. Variant i5 was the least robust strain selected in this study, exhibiting a defect in reading frame maintenance. As i5 is the site for S16 binding (34), and is localized inside the 30S particle, i5 should not be an appropriate insertion for eukaryotic rRNAs.
In 23S rRNA, 35 functional insertions are superimposed with ESs in eukaryotic 28S rRNAs (Figure 5B). As in 16S rRNA, the functional insertions showed a tendency to coincide with the regions of the ESs found in yeast 25S rRNAs. However, nine insertions did not overlap with the ESs (Figure 5B, Table 2). In domain I, among six ESs (ES1–6), four ESs overlapped with functional insertions (I1–6). Notably, I4–I6 were clustered and mapped onto ES5. Domain II contains 11 ESs (ES7–17). Seven were covered by functional insertions. I10 coincides with ES7 at the tip of helix 25. ES7 is the largest expansion segment in domain II. In S. cerevisiae, ES7 is broadly fused with ES39, another major expansion segment in domain VI (14). Near region ES11, I15 is localized at the tip of helix 38, which is known as ‘A-site finger (ASF)’. ASF is a conserved motif, known to form the intersubunit bridge B1a with protein S13 in the small ribosomal subunit (3). We previously reported that ASF acts as a functional attenuator for translocation (38), nevertheless ASF could be genetically shortened. These observations indicate that ASF is tolerant of genetic insertion and deletion. Domain II also contains the GTPase-associated region, which is one of the most important functional domains in rRNAs (39). Notably, I16 is located in the GTPase-associated region. According to the three-dimensional structure of the 50S subunit, I16 overlaps with the binding site of L10 (37), which is a stalk base protein to interact with L7/12 proteins. The slow growth phenotype (91.28 min) of the I16 variant might arise from altered assembly of the L10·(L7/12)6 complex, affecting the GTPase activity of translational factors. I17 is found in the connecting region between helices 41 and 42. In the 50S structure, I17 coincides with one of the interacting sites of L13 (35). However, the I17 variant has a normal growth phenotype. Among the nine ESs (ES18–26) in domain III, only three feature insertions. I23 and I24 map to helix 59, which coincides with ES24. As helix 59 is localized near the exit site of the peptide tunnel, it provides an attachment site for protein complexes that associate with nascent polypeptide chains, such as signal recognition particles (40) and protein-conducting channels (41). In eukaryotic ribosomes, ES24 contains the site for Sec61 interaction during protein translocation (22). Although it was thought that insertions at helix 59 might affect the interactions of these factors, both I23 and I24 show healthy phenotypes (Table 2). Domain IV has only two expansion segments (ES27 and ES28), likely since it has numerous contacts with domain V, and occupies the intersubunit region of the large subunit. We found I25 in the area of ES27. In S. cerevisiae, ES27, which is termed the ‘yeast spine’, is a large, rod-shaped domain which undergoes a significant conformational change when the ribosome binds to Sec61 (22). In domain V, all of the five insertions (I26–30) identified were densely located within helix 78, which is the site for ES30 in eukaryotic rRNAs. In domain VI, two insertions, I31 and I32, overlapped with a large expansion segment ES39, which forms a huge domain with ES7, as described above.
Most of the functional insertions obtained in this study overlapped with the regions in which ESs reside in eukaryotic rRNAs. Considering that eukaryotic ribosomes evolved from prokaryotic ribosomes, elongation of rRNAs might have coevolved with ribosomal proteins. In this study, we clearly showed that genetic elongation of rRNAs is possible without any effects upon ribosomal proteins. Thus, it can be speculated that RNA elongation by genetic insertions together with participation of new proteins might have occurred in the early evolution of eukaryotic ribosomes. In contrast, several insertions did not overlap with ESs, such as i5, I7, I8, I9, I16, I17, I21, I34 and I35 (Tables 1 and 2, Figure 5A and B). Novel expansion segments might be found in these regions in organisms which have not yet been analyzed. Otherwise, if RNA segments had been inserted at these sites in the process of evolution, such variants might have been excluded as a result of poor survival. In this study, we obtained 51 functional insertions in E. coli rRNAs. These variants are available as useful resources for functional and architectural analyses of rRNAs. Moreover, this study represents a further step forward in the development of a system to better investigate the function of ribosomal insertions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to the Suzuki lab members for fruitful discussion and advice, and especially to Drs Neuza S. Sato and Kei Kitahara for technical support. We would also like to thank Dr Catherine L. Squires (Tufts Univ.) for providing us with valuable strains and plasmids. This work was supported by grants-in-aid for scientific research on priority areas from the Ministry of Education, Science, Sports, and Culture of Japan (to T.S.), the Human Frontier Science Program (RGY23/2003) (to T.S.), and the Global COE Program for Chemistry Innovation (to T.Y.). Funding to pay the Open Access publication charges for this article was provided by the Japan Ministry of Education, Science, Sports and Culture. Funding to pay the Open Access publication charges for this article was provided by the Japan Ministry of Education, Science, Sports and Culture.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 2.Wimberly BT, Brodersen DE, Clemons W.M., Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 3.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 4.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 5.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 6.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 7.Selmer M, Dunham CM, Murphy FVT, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J. Biol. Chem. 2001;276:33181–33195. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J. Biol. Chem. 2001;276:21724–21736. doi: 10.1074/jbc.M100432200. [DOI] [PubMed] [Google Scholar]
- 10.Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 11.Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J. Biol. Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 12.O’Brien TW. Properties of human mitochondrial ribosomes. IUBMB Life. 2003;55:505–513. doi: 10.1080/15216540310001626610. [DOI] [PubMed] [Google Scholar]
- 13.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 14.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 15.Gerbi SA. Expansion Segments: Regions of Variable Size that Interrupt the Universal Core Secondary Structure of Ribosomal RNA. New York, pp. 71–87: CRC Press; 1996. [Google Scholar]
- 16.Planta RJ, Mager WH. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Wittmann-Liebold B. Structure, Function, and Genetics of Ribosome. In: Hardesty B, Kramer G, editors. Ribosomal Proteins: Their Structure and Evolution. New York, pp. 326–361: Springer; 1986. [Google Scholar]
- 18.Alkemar G, Nygard O. Secondary structure of two regions in expansion segments ES3 and ES6 with the potential of forming a tertiary interaction in eukaryotic 40S ribosomal subunits. RNA. 2004;10:403–411. doi: 10.1261/rna.5135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao H, Ayub MJ, Levin MJ, Frank J. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc. Natl Acad. Sci. USA. 2005;102:10206–10211. doi: 10.1073/pnas.0500926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson J, Sengupta J, Gursky R, Nissen P, Frank J. Comparison of fungal 80 S ribosomes by cryo-EM reveals diversity in structure and conformation of rRNA expansion segments. J. Mol. Biol. 2007;369:429–438. doi: 10.1016/j.jmb.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 23.Wilson DN, Nierhaus KH. The ribosome through the looking glass. Angew. Chem. Int. Ed. Engl. 2003;42:3464–3486. doi: 10.1002/anie.200200544. [DOI] [PubMed] [Google Scholar]
- 24.Spahn CM, Grassucci RA, Penczek P, Frank J. Direct three-dimensional localization and positive identification of RNA helices within the ribosome by means of genetic tagging and cryo-electron microscopy. Structure. 1999;7:1567–1573. doi: 10.1016/s0969-2126(00)88347-1. [DOI] [PubMed] [Google Scholar]
- 25.Matadeen R, Sergiev P, Leonov A, Pape T, van der Sluis E, Mueller F, Osswald M, von Knoblauch K, Brimacombe R, Bogdanov A, et al. Direct localization by cryo-electron microscopy of secondary structural elements in Escherichia coli 23 S rRNA which differ from the corresponding regions in Haloarcula marismortui. J. Mol. Biol. 2001;307:1341–1349. doi: 10.1006/jmbi.2001.4547. [DOI] [PubMed] [Google Scholar]
- 26.Sato NS, Hirabayashi N, Agmon I, Yonath A, Suzuki T. Comprehensive genetic selection revealed essential bases in the peptidyl-transferase center. Proc. Natl Acad. Sci. USA. 2006;103:15386–15391. doi: 10.1073/pnas.0605970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirabayashi N, Sato NS, Suzuki T. Conserved loop sequence of helix 69 in Escherichia coli 23 S rRNA is involved in A-site tRNA binding and translational fidelity. J. Biol. Chem. 2006;281:17203–17211. doi: 10.1074/jbc.M511728200. [DOI] [PubMed] [Google Scholar]
- 28.Kitahara K, Kajiura A, Sato NS, Suzuki T. Functional genetic selection of Helix 66 in Escherichia coli 23S rRNA identified the eukaryotic-binding sequence for ribosomal protein L2. Nucleic Acids Res. 2007;35:4018–4029. doi: 10.1093/nar/gkm356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires CL. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 1999;181:3803–3809. doi: 10.1128/jb.181.12.3803-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller JH. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor, New York, pp. 72–74: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 32.Goryshin IY, Reznikoff WS. Tn5 in vitro transposition. J. Biol. Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 33.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 34.Brodersen DE, Clemons W.M., Jr., Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 35.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 36.Stelzl U, Spahn CM, Nierhaus KH. Selecting rRNA binding sites for the ribosomal proteins L4 and L6 from randomly fragmented rRNA: application of a method called SERF. Proc. Natl Acad. Sci. USA. 2000;97:4597–4602. doi: 10.1073/pnas.090009297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egebjerg J, Douthwaite SR, Liljas A, Garrett RA. Characterization of the binding sites of protein L11 and the L10.(L12)4 pentameric complex in the GTPase domain of 23 S ribosomal RNA from Escherichia coli. J. Mol. Biol. 1990;213:275–288. doi: 10.1016/S0022-2836(05)80190-1. [DOI] [PubMed] [Google Scholar]
- 38.Komoda T, Sato NS, Phelps SS, Namba N, Joseph S, Suzuki T. The A-site finger in 23 S rRNA acts as a functional attenuator for translocation. J. Biol. Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 39.Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 40.Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 41.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks C.L., III, Ban N, Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.