Abstract
RNA polymerase (pol) III contains a dissociable subcomplex that is required for initiation, but not for elongation or termination of transcription. This subcomplex is composed of subunits RPC3, RPC6 and RPC7, and interacts with TFIIIB, a factor that is necessary and sufficient to support accurate pol III transcription in vitro. Direct binding of TFIIIB to RPC6 is believed to recruit pol III to its genetic templates. However, this has never been tested in vivo. Here we combine chromatin immunoprecipitation with RNA interference to demonstrate that the RPC3/6/7 subcomplex is required for pol III recruitment in mammalian cells. Specific knockdown of RPC6 by RNAi results in post-transcriptional depletion of the other components of the subcomplex, RPC3 and RPC7, without destabilizing core pol III subunits or TFIIIB. The resultant core enzyme is defective in associating with TFIIIB and target genes in vivo. Promoter occupancy by pol II is unaffected, despite sharing five subunits with the pol III core. These observations provide evidence for the validity in vivo of the model for pol III recruitment that was built on biochemical data.
INTRODUCTION
Pol III is the largest of the RNA polymerases, containing 17 subunits with an aggregate mass of around 700 kDa (1–5). All its products are short untranslated transcripts, which include tRNA and 5S rRNA, as well as the 7SL RNA component of the signal recognition particle. Other essential pol III products include the MRP, U6 and H1 RNAs, which are involved in processing of pre-rRNA, mRNA and tRNA respectively. Pol III has also been implicated recently in transcribing a subset of human microRNAs (6).
Pol III transcription is best characterized in Saccharomyces cerevisiae (1–5). Elegant biochemical analysis revealed that pol III from this yeast is recruited to its target genes through protein-protein interactions with the transcription initiation factor TFIIIB (7). Indeed, S. cerevisiae TFIIIB is necessary and sufficient to support multiple rounds of accurately-initiated transcription (7). In both yeast and mammals, TFIIIB is a complex containing the TATA-binding protein (TBP) and two associated factors, termed TFIIB-related factor 1 (Brf1) and B double prime 1 (Bdp1) (1,2,4). Of these, Brf1 is primarily responsible for contacting the polymerase (1,2,4). It can bind to the pol III-specific subunit RPC6 from yeast or humans, as shown by two-hybrid and GST pull-down assays with the isolated components (8–11). This is consistent with DNA photocrosslinking experiments with purified yeast pol III and TFIIIB, which indicate that RPC6 is located alongside Brf1 in the preinitiation complex (12). An interaction has also been detected between recombinant human RPC6 and TBP (11), but this was not observed in yeast.
Saccharomyces cerevisiae mutants of RPC6 are specifically defective in transcription initiation (13). In both budding yeast and human pol III, RPC6 forms a stable subcomplex with RPC3 and RPC7 (11,14,15) (Alternative names for subunits are listed in Table 1). Although it is retained in the presence of 2 M urea, the RPC3/6/7 subcomplex dissociates from the core of yeast or human pol III during native polyacrylamide gel electrophoresis or prolonged sucrose gradient sedimentation (11,14). The core human enzyme missing these three subunits is competent for transcript elongation and termination, but has lost the ability to initiate transcription in a promoter-directed manner (11). Accurate initiation can be restored by addition of the RPC3/6/7 subcomplex reconstituted from recombinant forms of its three components (11). A role in initiation is supported by electron microscopic analysis, which places the subcomplex at the DNA-binding cleft of yeast pol III (16). These observations resulted in a model in which the subcomplex provides the interface between TFIIIB and pol III core that is required to position the latter at the transcription start site.
Table 1.
Alternative names used for the pol III subunits investigated in this study
| Pol III subunit | Alternative names (H. sapiens) | Alternative names (S. cerevisiae) |
|---|---|---|
| RPC1 | RPC155 | C160 |
| RPC3 | RPC62 | C82 |
| RPC6 | RPC39 | C34 |
| RPC7 | RPC32 | C31 |
| RPB6 | RPB14.4 | ABC23 |
| RPB8 | RPB17 | ABC14.5 |
The model for pol III recruitment is based on extremely strong biochemical data, as well as yeast two hybrid evidence for interaction in vivo between overexpressed RPC6 and Brf1. However, it has yet to be confirmed in vivo under physiological conditions. We have attempted to do this in mammalian cells, using siRNAs directed against the RPC6 mRNA. We were interested to find that depleting endogenous RPC6 results in a specific post-transcriptional reduction of RPC7 and RPC3 protein levels, suggesting that subcomplex stability may depend on RPC6. As expected, this treatment compromises the expression of pol III products. Although occupancy of pol III templates by TFIIIB is unaffected, association of core polymerase subunits is compromised. Some of these core subunits are shared with pol II and their occupancy of pol II promoters remains normal. These data confirm that the RPC3/6/7 complex is necessary for specific recruitment of endogenous pol III to its target genes in vivo.
MATERIALS AND METHODS
Cell culture
Immortalized mouse embryonic fibroblasts (MEF)/3T3 cells were cultured in DMEM supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml penicillin and 100 U/ml streptomycin.
RNAi
MEF/3T3 cells were transfected at 40% confluence to a final concentration of 100 nM siRNA oligonucleotides, delivered using Lipofectamine 2000 (Invitrogen). Medium was replaced after 5 h and cells were harvested after a further 48 h. The siRNAs used were RPC6 si1 5′-GGAAAAACUGGUCUAUCAA-3′, RPC6 si2 5′-GGAUUCUCAGAAUGCUGGU-3′ and GFP (Cycle 3), all from Ambion.
RT-PCR analysis
RT-PCR of ARPP P0 mRNA, 5S rRNA, 7SK, pre-rRNA and tRNA transcripts was performed as previously (17,18). RT-PCR of RPC6 mRNA used primers 5′-CACAAGAGGTGGAGGGAAAA-3′ and 5′-GGATCCTCTGTGGAGCTGAG-3′ to give a 213 bp product using the following cycling parameters: 95°C for 3 min, 22 cycles of (95°C for 1 min, 64°C for 30 s, 72°C for 1 min), 72°C for 5 min. RT-PCR of RPC7 mRNA used primers 5′-GTAGAATCTGCGGCTCCTTG-3′ and 5′-TACAGGAAACAGGGGTGGAG-3′ to give a 244 bp product using the following cycling parameters: 95°C for 3 min, 26 cycles of (95°C for 1 min, 55°C for 30 s, 72°C for 1 min), 72°C for 5 min. RT-PCR of RPC3 mRNA used primers 5′-ACACAAAAGCATCCCTGGAC-3′ and 5′-GGAGCTCCTCAACAATCAGC-3′ to give a 207 bp product using the following cycling parameters: 95°C for 3 min, 22 cycles of (95°C for 1 min, 59°C for 30 s, 72°C for 1 min), 72°C for 5 min.
Cell extracts and immunoblotting
Whole cell extracts for immunoblotting were prepared as described (19). Immunoblotting was performed as previously (20), using antibodies C11 against actin, N-15 against TFIIIC63, SI-1 against TBP (all Santa Cruz Biotechnologies); Ab26185 against RPC3 (Abcam); MAb c39 against RPC6, MAb c32 against RPC7, MAb B6-1 against RPB6, MAb B8-1 against RPB8 (21), 1900 against RPC1, Ab4 against TFIIIC220, 3208 against TFIIIC110, 3238 against TFIIIC102, 1898 against TFIIIC90, 128 against Brf1, 2663 against Bdp1 (22,23).
Transcription assays
Transcription in vitro was carried out as previously (20). The pLeu template has been described (24), the pol I pre-rRNA template was pMrWT (25) which was linearized using EcoR1.
Co-immunoprecipitation
Cells were washed in ice-cold PBS and scraped into IP buffer (50 mM HEPES pH 7.5, 5 mM EDTA, 10 mM NaF, 150 mM NaCl, 25% glycerol, 0.5% Triton X-100, 0.5 mM PMSF, 0.5 µg/ml leupeptin, 0.7 µg/ml pepstatin, 0.5 µg/ml aprotinin, 40 µg/ml bestatin, 1 mM sodium vanadate and 50 mM β-glycerophosphate). After 15 min on ice, extracts were passed three times through a 26G needle and insoluble material was removed by centrifugation at 14 000 g for 15 min prior to immunoprecipitation. Extracts (500 µg) were pre-cleared on an orbital shaker with 30 µl of protein G–Sepharose beads and then incubated for 3 h at 4°C with 30 µl of protein G–Sepharose beads carrying 1 μg of pre-bound IgG. Supernatants were removed and beads washed three times with 500 µl of 20 mM HEPES pH 7.9, 12 mM MgCl2, 0.1 mM EDTA and 17% glycerol. Antibodies used for immunoprecipitations were normal mouse IgG (Santa Cruz Biotechnologies) and MAb B8-1 against RPB8 (31).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (26). Immunoprecipitated DNA was quantified by PCR using published primers and amplification procedures. Antibodies used were 1900 against RPC1 (RPC155) and 128 against Brf1 (22,23); Ab7 against TFIIIC220 (27); MAb c39 against RPC6 and MAb B6-1 against RPB8 (21); R-124 against Cyclin D1 and FL-109 against TFIIA (Santa Cruz Biotechnologies). Primers used for PCR reactions have been described elsewhere (17,28). Serial dilutions of input chromatin were used to establish that PCRs were in the linear range.
RESULTS
RNAi of RPC6 causes post-transcriptional depletion of RPC3 and RPC7
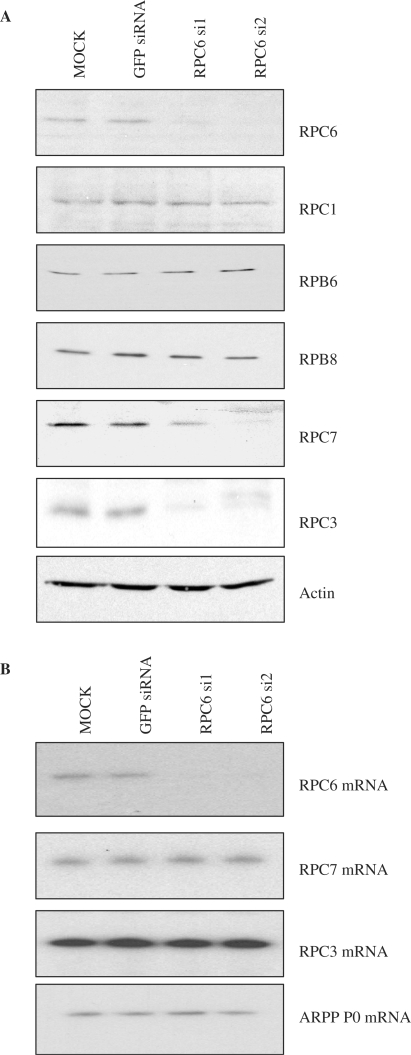
To examine the role of RPC6 in vivo, siRNAs were transiently transfected into immortalized mouse embryonic fibroblasts (MEFs). As a precaution against off-target effects, we used two alternative siRNAs, si1 and si2, that recognize distinct regions of the RPC6 mRNA (Figure 1A). Both were found by immunoblotting to reduce RPC6 expression, relative to controls that were either mock-transfected or transfected with an irrelevant siRNA against the mRNA encoding green fluorescent protein (GFP). This effect was specific, since none of the core subunits examined (RPC1, RPB6 and RPB8) showed any change. Expression of the actin loading control was also unaffected. However, levels of RPC3 and RPC7 were both diminished significantly. As described above, these two subunits form a subcomplex with RPC6 that can be dissociated from the core of pol III (11,14,15). Despite the clear decrease in their protein expression, the mRNAs that encode RPC3 and RPC7 were not affected by the RPC6 siRNAs (Figure 1B). This excludes the possibility that the siRNAs act directly in these cases and indicates that the selective depletion of RPC3 and RPC7 is a post-transcriptional effect. A plausible scenario is that the stability of these subunits is compromised in the absence of their partner RPC6. RPC7 is predicted to contain long stretches of disordered amino acid residues and to adopt a rather extended conformation (16). These features may contribute to its instability when deprived of its partners.
Figure 1.
Knockdown of RPC6 results in a post-transcriptional decrease in RPC3 and RPC7 protein levels. (A) Cell lysates were prepared from cells treated with GFP siRNA, RPC6 si1 siRNA, RPC6 si2 siRNA or mock-transfected with the siRNA dilutant (mock). These extracts were resolved on SDS-polyacrylamide gels and analysed by western immunoblotting with antibodies specific to the indicated proteins. (B) RT-PCR analysis of the indicated transcripts from cells treated as above.
Depletion of RPC3/6/7 compromises pol III transcription in vitro and in vivo
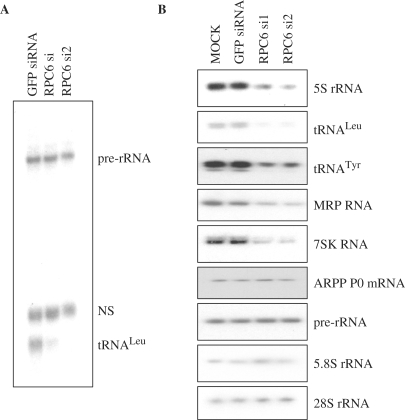
Transcription assays were carried out in vitro using whole cell extracts prepared from MEFs after RNAi. The ability to transcribe a tRNALeu template was severely compromised by transfection with si1 or si2 against RPC6, relative to the GFP control siRNA (Figure 2A). In contrast, little or no effect was seen in parallel on pol I transcription from an rRNA promoter (Figure 2A). This observation supports the evidence published by Wang et al. (11) that the RPC3/6/7 subcomplex is required for promoter-directed pol III transcription in vitro.
Figure 2.
RNAi of RPC6 specifically compromises pol III transcription in vitro and in vivo. (A) Whole-cell extracts (20 μg) from cells transfected with GFP siRNA, RPC6 si1 siRNA or RPC6 si2 siRNA were used to transcribe tRNALeu, and pre-rRNA templates (250 ng). NS = template-independent, non-specific band. (B) RT-PCR analysis of the indicated transcripts from cells treated as in Figure 1.
To investigate if this effect is replicated in vivo, RT-PCR reactions were carried out over a linear range with RNA extracted from the transfected cells. Expression of every pol III product examined was lower in the cells treated with the RPC6 siRNAs compared to controls that were mock-transfected or treated with GFP siRNA (Figure 2B). This was the case for 5S rRNA, tRNALeu, tRNATyr, MRP and 7SK RNAs, which provide representatives of each of the three main promoter types that are utilized by pol III (2,3). In contrast, little or no change was detected in levels of the pol II-dependent mRNA encoding ARPP P0 or the pol I-dependent pre-rRNA, 5.8S rRNA and 28S rRNA (Figure 2B). We conclude that depletion of the RPC3/6/7 subcomplex causes a specific defect in pol III transcription, both in vitro and in vivo.
Interaction between pol III and TFIIIB is disrupted by RPC6 RNAi
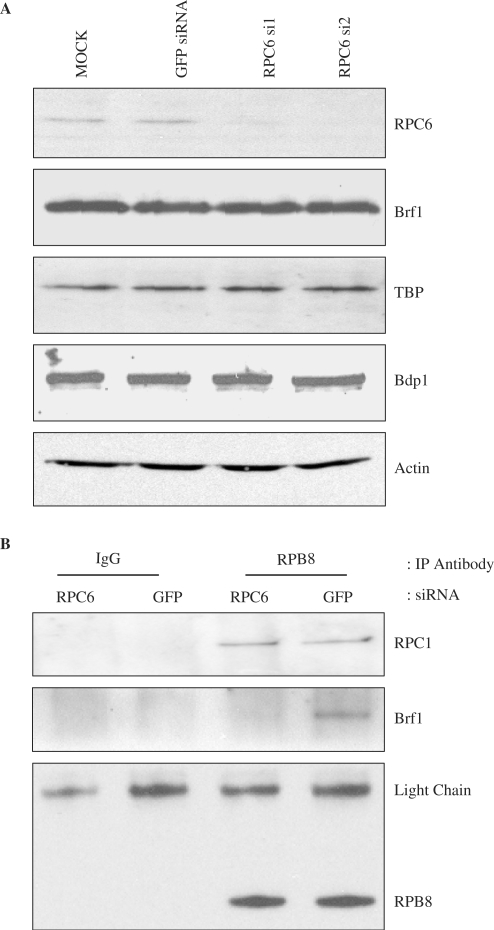
Pull-down assays using recombinant proteins indicated that RPC6 binds directly to the TFIIIB components TBP and Brf1 (29). We used co-immunoprecipitation assays to investigate whether the interaction between endogenous pol III and TFIIIB is disrupted by loss of RPC6. Although expression of the TFIIIB subunits (Brf1, TBP and Bdp1) is not affected by RPC6 siRNAs (Figure 3A), co-immunoprecipitation of Brf1 with pol III was greatly diminished, as determined using an antibody against RPB8 (Figure 3B). In contrast, interaction of the core pol III subunit RPC1 with RPB8 was not compromised by the RPC6 siRNA (Figure 3B). These data provide evidence that the RPC3/6/7 subcomplex is important for the stable association of endogenous pol III with TFIIIB.
Figure 3.
RNAi of RPC6 specifically compromises association of Brf1 with polIII. (A) Cell lysates were prepared from cells treated with GFP siRNA, RPC6 si1 siRNA, RPC6 si2 siRNA or mock-transfected with the siRNA dilutant (mock). These extracts were resolved on SDS-polyacrylamide gels and analysed by western immunoblotting with antibodies specific to the indicated proteins. (B) Extracts (500 μg) prepared from cells treated with GFP siRNA or RPC6 si2 siRNA were immunoprecipitated with normal mouse IgG (lanes 1 and 2) or anti-RPB8 (lanes 3 and 4). Precipitates were resolved by SDS-PAGE and then analysed by western blotting with antibodies to Brf1, RPC1 and RPB8 antibodies.
RNAi of RPC6 selectively reduces crosslinking of pol III to its target genes in vivo
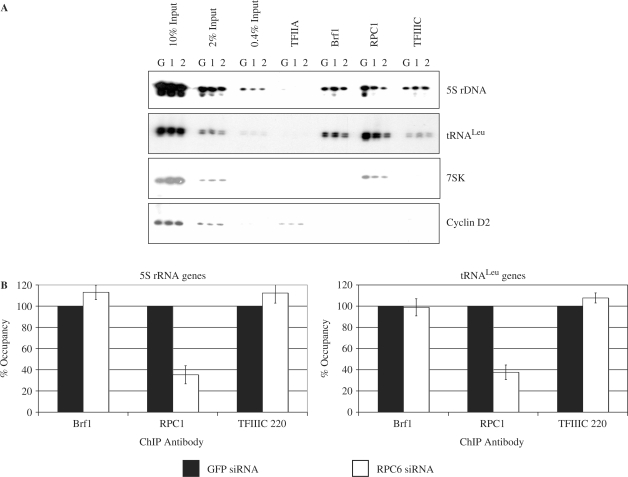
ChIP assays were used to investigate gene occupancy in vivo by pol III, TFIIIB and TFIIIC (Figure 4A). The primers employed detect crosslinking throughout the promoter and transcribed regions. As expected, the core pol III subunit RPC1 was clearly detected at 5S rRNA, tRNA and 7SK genes, but not at the cyclin D2 promoter, which directs pol II transcription. In contrast, the pol II-specific factor TFIIA is crosslinked to the cyclin D2 promoter, but not to any of the pol III templates, as reported previously (30). Brf1 and TFIIIC were detected at 5S rRNA and tRNA genes, but not at the 7SK gene, which has a type III promoter that does not utilize Brf1 or TFIIIC (2). These controls confirm the specificity of the assays. Although expression of RPC1 was unaffected (Figure 1A), its crosslinking to pol III templates was diminished in cells transfected with either siRNA against RPC6, compared with those receiving the negative control siRNA against GFP (Figure 4A and B). In contrast, no consistent change was found for Brf1 or TFIIIC. These data suggest that depletion of RPC3/6/7 compromises gene occupancy by pol III despite the presence of the transcription factors that are required for its recruitment.
Figure 4.
RNAi of RPC6 specifically compromises crosslinking of pol III to target genes. (A) ChIP assay showing crosslinking of TFIIA, Brf1, RPC1 and TFIIIC220 to tRNALeu, 5S rRNA and 7SK genes and the cyclin D2 promoter in cells transfected with GFP siRNA (G), RPC6 si1 siRNA (1) and RPC6 si2 siRNA (2). (B) PCR products from (A) and an additional independent experiment were quantified by densitometry and normalized to the appropriate input. The average fold change in Brf1, TFIIIC220 and RPC1 crosslinking relative to GFP siRNA is shown for 5S rRNA and tRNALeu genes after treatment with siRNAs against RPC6 (results for si1 and si2 are combined). Error bars indicate the standard deviation from the mean.
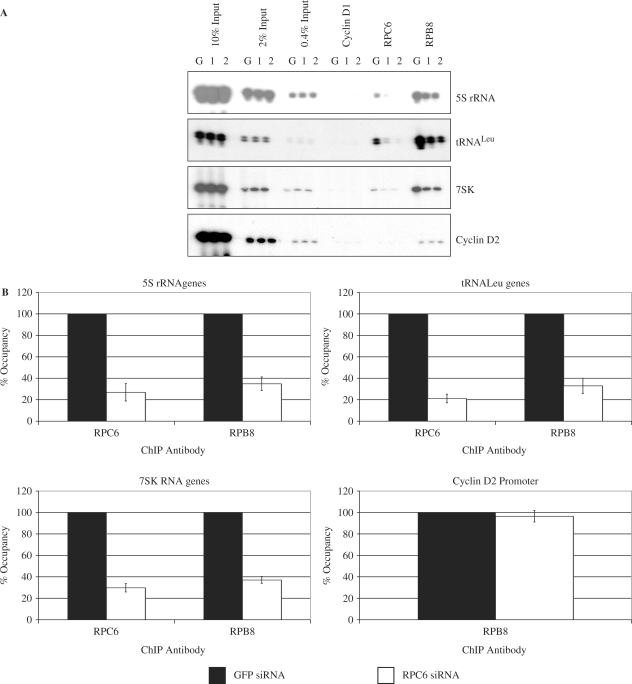
Further ChIP assays were performed with antibodies that recognize additional polymerase subunits. As expected, RPC6 was detected at pol III templates in cells transfected with control siRNA against GFP, but was not detected at the cyclin D2 promoter (Figure 5A). In contrast, the RPB8 subunit, which is shared between pols II and III, was detected at all the genes examined. Transfection with either RPC6 siRNA resulted in a clear decrease in occupancy of this subunit on tRNA, 5S rRNA and 7SK genes, compared to the GFP negative control (Figure 5A). This is consistent with the reduced RPC6 protein levels. In addition, crosslinking of RPB8 was reduced on the pol III-transcribed genes, despite the unchanged levels of this subunit (Figure 1A). On average, crosslinking of this subunit to pol III templates was reduced by RPC6 siRNA to approximately 35% of control (Figure 5B). This change is specific, as it was not seen on the cyclin D2 promoter, where RPB8 is recruited as part of pol II. These data demonstrate that depletion of the RPC3/6/7 subcomplex compromises crosslinking of core polymerase subunits to pol III- but not pol II-transcribed genes.
Figure 5.
RNAi of RPC6 compromises crosslinking of pol III but not pol II to target genes in vivo. (A) ChIP assay showing crosslinking of cyclin D1, RPC6, and RPB8 to tRNALeu, 5S rRNA and 7SK genes and the cyclin D2 promoter in cells transfected with GFP siRNA (G), RPC6 si1 siRNA (1) or RPC6 si2 siRNA (2). (B) PCR products from (A) and an additional independent experiment were quantified by densitometry and normalized to the appropriate input. The average decrease in polymerase crosslinking relative to GFP siRNA is shown for 5S rRNA, tRNALeu and 7SK genes and the cyclin D2 gene promoter after treatment with siRNAs against RPC6 (results for si1 and si2 are combined). Error bars indicate the standard deviation from the mean.
DISCUSSION
These experiments have tested directly the model that the subcomplex comprising RPC3, RPC6 and RPC7 mediates specific recruitment of pol III to its genetic templates. Our data provide evidence that this is indeed the case in vivo, thereby providing support for a key aspect of our current understanding of the mechanistic basis of pol III transcription. RPC6 was depleted using two independent siRNAs in order to avoid off-target effects. This resulted in a post-transcriptional decrease in the levels of RPC3 and RPC7 proteins, which suggests that binding to RPC6 is required for the stability of the other subcomplex components in vivo. A specific decrease was observed in the expression of pol III products. ChIP assays demonstrated that gene occupancy by TFIIIB and TFIIIC was undiminished, but crosslinking of pol III subunits was compromised severely. These data can be explained by a specific defect in pol III recruitment.
Although it has not been reported in mammals, an interaction has been detected in S. cerevisiae between Brf1 and RPC9 (also called C17) (31). This is consistent with the presence of RPC9 in a stalk that is presented on the surface of pol III alongside the RPC3/6/7 complex, providing the potential for simultaneous binding of RPC6 and RPC9 to Brf1 (16). Our data suggest that if such an interaction is conserved in mammals, it is insufficient to allow robust recruitment of pol III to promoters in the absence of RPC3/6/7.
7SK genes have TATA-containing type III promoters, similar to those of some mammalian U6 genes (1–3). A paradigm U6 promoter was shown not to utilize Brf1 (32) and we found evidence that 7SK genes are not occupied by Brf1 in vivo (Figure 4). Instead, these promoters require a form of TFIIIB in which Brf1 is replaced by a shorter family member called Brf2, with which it has <18% identity (32,33). Despite this limited homology, it was anticipated that Brf1 and Brf2 perform similar functions. Our data provide support for this assumption, by showing that pol III recruitment to 7SK genes requires the same RPC3/6/7 subcomplex that binds to Brf1 (Figures 4 and 5). As far as we are aware, this has not been shown previously. It is likely that Brf1 and Brf2 present a similar surface to pol III that can be recognized by RPC3/6/7.
The original model of pol III recruitment was based on biochemical assays with fractionated components, which showed that TFIIIB and TFIIIC are both assembled prior to pol III on a tRNA or 5S rRNA gene. However, evidence has also been presented for a holoenzyme, in which pol III is pre-assembled with TFIIIB (29,34,35). Our data imply that holoenzyme formation is not required for TFIIIB recruitment in vivo, as ChIP assays provide no evidence for decreased gene occupancy by TFIIIB when its interaction with pol III is compromised (Figure 4). This by no means invalidates the holoenzyme model, but does suggest that efficient interaction with pol III is not a pre-requisite for TFIIIB to occupy its target promoters.
Much of what we currently know about the mechanics of pol III transcription was gleaned through biochemical analyses. There is therefore value in testing the extent to which current models can correctly predict molecular behaviour under physiological conditions in vivo. A combination of RNAi with ChIP has allowed us to test the prediction that RPC6 is crucial for recruitment of pol III to its chromosomal templates. Our data provide strong support for this model of one of the key steps in gene transcription.
ACKNOWLEDGEMENTS
We thank Peter Cook for antibodies against RPC6, RPC7, RPB8 and RPB6 and Yuhong Shen and Arnie Berk for Ab4. This work was supported by the Biotechnology and Biological Sciences Research Council and Cancer Research UK. Funding to pay the Open Access publication charges for this article was provided by Cancer Research UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J. Mol. Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 2.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 3.White RJ. 2002. RNA Polymerase III Transcription. (Landes Bioscience, Austin, TX) [Google Scholar]
- 4.Kassavetis GA, Geiduschek EP. Transcription factor TFIIIB and transcription by RNA polymerase III. Biochem. Soc. Trans. 2006;34:1082–1087. doi: 10.1042/BST0341082. [DOI] [PubMed] [Google Scholar]
- 5.Cramer P. Recent structural studies of RNA polymerases II and III. Biochem. Soc. Trans. 2006;34:1058–1061. doi: 10.1042/BST0341058. [DOI] [PubMed] [Google Scholar]
- 6.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 7.Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EP. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 8.Werner M, Chaussivert N, Willis IM, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 9.Khoo B, Brophy B, Jackson SP. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 10.Andrau JC, Sentenac A, Werner M. Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol. 1999;288:511–520. doi: 10.1006/jmbi.1999.2724. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Roeder RG. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 12.Bartholomew B, Durkovich D, Kassavetis GA, Geiduschek EP. Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brun I, Sentenac A, Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997;16:5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valenzuela P, Hager GL, Weinberg F, Rutter WJ. Molecular structure of yeast RNA polymerase III: demonstration of the tripartite transcription system in lower eukaryotes. Proc. Natl Acad. Sci. USA. 1976;73:1024–1028. doi: 10.1073/pnas.73.4.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werner M, Hermann S, Treich I, Sentenac A, Thuriaux P. Effect of mutations in a zinc-binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol. Cell. Biol. 1992;12:1087–1095. doi: 10.1128/mcb.12.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Torneo C, Bottcher B, Riva M, Carles C, Steuerwald U, Ruigrok R.WH, Sentenac A, Muller CW, Schoehn G. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol. Cell. 2007;25:813–823. doi: 10.1016/j.molcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Winter AG, Sourvinos G, Allison SJ, Tosh K, Scott PH, Spandidos DA, White RJ. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumours. Proc. Natl Acad. Sci. USA. 2000;97:12619–12624. doi: 10.1073/pnas.230224097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daly NL, Arvanitis DA, Fairley JA, Gomez-Roman N, Morton JP, Graham SV, Spandidos DA, White RJ. Deregulation of RNA polymerase III transcription in cervical epithelium in response to high-risk human papillomavirus. Oncogene. 2005;24:880–888. doi: 10.1038/sj.onc.1208031. [DOI] [PubMed] [Google Scholar]
- 19.Innes F, Ramsbottom B, White RJ. A test of the model that RNA polymerase III transcription is regulated by selective induction of the 110 kDa subunit of TFIIIC. Nucleic Acids Res. 2006;34:3399–3407. doi: 10.1093/nar/gkl432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White RJ, Gottlieb TM, Downes CS, Jackson SP. Mitotic regulation of a TATA-binding-protein-containing complex. Mol. Cell. Biol. 1995;15:1983–1992. doi: 10.1128/mcb.15.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones E, Kimura H, Vigneron M, Wang Z, Roeder RG, Cook PR. Isolation and characterization of monoclonal antibodies directed against subunits of human RNA polymerase I, II and III. Exp. Cell Res. 2000;254:163–172. doi: 10.1006/excr.1999.4739. [DOI] [PubMed] [Google Scholar]
- 22.Sutcliffe JE, Brown T.RP, Allison SJ, Scott PH, White RJ. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol. Cell. Biol. 2000;20:9192–9202. doi: 10.1128/mcb.20.24.9192-9202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairley JA, Scott PH, White RJ. TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J. 2003;22:5841–5850. doi: 10.1093/emboj/cdg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White RJ, Stott D, Rigby P.WJ. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989;59:1081–1092. doi: 10.1016/0092-8674(89)90764-2. [DOI] [PubMed] [Google Scholar]
- 25.Rudloff U, Eberhard D, Tora L, Stunnenberg H, Grummt I. TBP-associated factors interact with DNA and govern species specificity of RNA polymerase I transcription. EMBO J. 1994;13:2611–2616. doi: 10.1002/j.1460-2075.1994.tb06551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 27.Shen Y, Igo M, Yalamanchili P, Berk AJ, Dasgupta A. DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol. Cell. Biol. 1996;16:4163–4171. doi: 10.1128/mcb.16.8.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, et al. Identification of a Wnt/Dvl/β-catenin -> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Luo T, Roeder RG. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fairley JA, Kantidakis T, Kenneth NS, Intine RV, Maraia R, White RJ. Human La is found at RNA polymerase III-transcribed genes in vivo. Proc. Natl Acad. Sci. USA. 2005;102:18350–18355. doi: 10.1073/pnas.0506415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferri ML, Peyroche G, Siaut M, Lefebvre O, Carles C, Conesa C, Sentenac A. A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol. Cell. Biol. 2000;20:488–495. doi: 10.1128/mcb.20.2.488-495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schramm L, Pendergrast PS, Sun Y, Hernandez N. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 2000;14:2650–2663. doi: 10.1101/gad.836400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabart P, Murphy S. BRFU, a TFIIB-like factor, is directly recruited to the TATA-box of polymerase III small nuclear RNA gene promoters through its interaction with TATA-binding protein. J. Biol. Chem. 2001;276:43056–43064. doi: 10.1074/jbc.M108515200. [DOI] [PubMed] [Google Scholar]
- 34.Burke DJ, Soll D. Functional analysis of fractionated Drosophila Kc cell tRNA gene transcription components. J. Biol. Chem. 1985;260:816–823. [PubMed] [Google Scholar]
- 35.Wingender E, Jahn D, Seifart KH. Association of RNA polymerase III with transcription factors in the absence of DNA. J. Biol. Chem. 1986;261:1409–1413. [PubMed] [Google Scholar]