Abstract
Human Tudor-SN is involved in the degradation of hyper-edited inosine-containing microRNA precursors, thus linking the pathways of RNA interference and editing. Tudor-SN contains four tandem repeats of staphylococcal nuclease-like domains (SN1–SN4) followed by a tudor and C-terminal SN domain (SN5). Here, we showed that Tudor-SN requires tandem repeats of SN domains for its RNA binding and cleavage activity. The crystal structure of a 64-kD truncated form of human Tudor-SN further shows that the four domains, SN3, SN4, tudor and SN5, assemble into a crescent-shaped structure. A concave basic surface formed jointly by SN3 and SN4 domains is likely involved in RNA binding, where citrate ions are bound at the putative RNase active sites. Additional modeling studies provide a structural basis for Tudor-SN's preference in cleaving RNA containing multiple I·U wobble-paired sequences. Collectively, these results suggest that tandem repeats of SN domains in Tudor-SN function as a clamp to capture RNA substrates.
INTRODUCTION
Posttranscriptional regulation, such as RNA interference, editing and decay, plays important roles in many cellular processes, including viral defense, chromatin remodeling, genome rearrangement and gene expression. A number of RNases are critically involved in these processes, such as the RNase III enzymes Dicer and Drosha, which digest primary double-stranded RNA transcripts to produce siRNA and miRNA in RNA silencing (1,2). Recently, a novel miRNase (ribonuclease for small interference micro RNA), Tudor-staphylococcal nuclease-like (SN), has been shown to be involved in the degradation of hyper-edited miRNA primary transcripts and intriguingly links the pathways between RNA editing and RNA interference (3).
Tudor-SN, also called p100 or SND1, has been identified in mammals, fishes, Drosophila, Caenorhabditis elegans, ciliates (tetrahymena) and fission yeast, but not in bacteria (4–8). It is an ubiquitous protein with similar mRNA levels in human pancreas, muscle, liver, lung, placenta, brain and heart (9). Tudor-SN was first characterized as a transcription coactivator, interacting with several specific transcription factors to activate their activities, including EBNA-2 (9), c-Myb (10), STAT6 (11–13) and STAT5 (14). Some of these transcription factors regulate important cellular signaling pathways. For example, c-Myb is involved in hematopoietic cell growth, differentiation and apoptosis, while STAT6 is a critical interleukin-4-induced transcription factor in immune and antiinflammatory responses. Tudor-SN was found to bridge between these specific transcription factors and component proteins of transcriptional machinery, for example, between c-Myb and Pim-1 (10), STAT6 and CBP (11), STAT6 and RNA HelicaseA (12) and STAT6 and PC1 (13). Activation of some of these transcription factors by Tudor-SN leads to cell proliferation and is linked to human diseases. For instance, Tudor-SN interacting with STAT6 and PC1 activates renal epithelial cell proliferation in autosomal-dominant polycystic kidney disease (ADPKD) (13). Up-regulation of Tudor-SN mRNA has also been observed in human colon cancer tissues and cell lines; however, in this case, posttranscriptional regulation of APC gene by Tudor-SN was suggested (15).
The involvement of Tudor-SN in posttranscriptional regulation was first hinted by the discovery that Tudor-SN is a component protein associated in RISC (RNA-induced silencing complex) (7). Human Tudor-SN was then shown to promote the cleavage of hyper-edited double-stranded RNA containing multiple I·U and U·I pairs (16,17). Subsequently, Tudor-SN was identified as a ribonuclease specific for inosine-containing primary transcripts of miRNA; it modulates miRNA processing and expression through RNA editing by ADAR (adenosine deaminase acting on RNA) (3,18). The primary transcript of human miRNA-142 is edited by ADAR enzymes, resulting in suppression of its processing by Drosha, and the edited transcripts are further degraded by Tudor-SN. These results demonstrate that Tudor-SN plays an important role in the regulation of the biogenesis and expression of some miRNAs. Moreover, Tudor-SN interacts with U5 snRNP (small nuclear ribonucleoproteins) and functions in spliceosome assembly and pre-mRNA splicing (19). Therefore, this interesting protein appears to play multiple roles in transcriptional regulation, RNA interference, RNA editing and RNA splicing.
Sequence analyses have shown that Tudor-SN contains four tandem repeats of SN domains (SN1 to SN4) followed by a tudor and C-terminal partial SN domain (SN5, see domain organization in Figure 1A) (20,21). The SN domains share ∼20% sequence identity with staphylococcal nuclease, which is a Ca2+-dependent extracellular nuclease produced by Staphylococcus aureus (22). The tudor domain, bearing a barrel-like fold, is a protein–protein interaction domain, usually interacting with methylated peptides, such as the ones identified in SMN (23), 53BP1 (24), SPF30 and TDRD (25). The crystal structure of a C-terminal fragment of Tudor-SN (residues 654–870) containing the tudor domain and the C-terminal SN domain (SN5) shows that a conserved aromatic cage in the tudor domain may bind the methylated peptides of its interaction partners in snRNPs (26). However, it remains uncertain how Tudor-SN binds RNA and participates in RNA interference, editing and degradation. Here, we address the role of Tudor-SN in RNA binding and cleavage by biochemical and X-ray crystal structure analysis. Our results demonstrate that Tudor-SN adopts an interesting means, using its tandem repeats of SN domains, to capture RNA substrates.
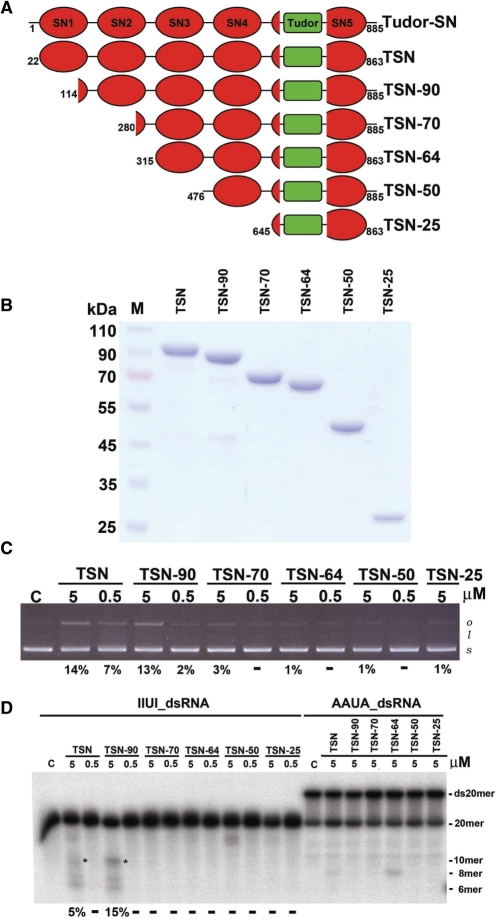
Figure 1.
Different constructs of Tudor-SN, and their DNase and RNase activity assays. (A) Six constructs of Tudor-SN were prepared. (B) The purity of Tudor-SN proteins was assayed by 10% SDS–PAGE. (C) Plasmid digestion assays show that TSN, TSN-90 and TSN-70 had detectable DNase activity (14% to 3%), whereas TSN-64, TSN-50 and TSN-25 had residual activity (1%). (D) Tudor-SN truncated mutants were incubated with 20-bp RNAs (5′-end labeled on top strand) containing the wobble base-paired IIUI/UIUU sequence or Watson–Crick base-paired AAUA/UAUU sequence, in a reaction buffer for 8 h at 20°C. TSN and TSN-90 cleaved IIUI-dsRNA with the highest activities (5 and 15%), whereas TSN-70, TSN-64, TSN-50 and TSN-25 had no detectable activities (marked by -). The intensity of the cleavage product gel band of the 10-mer RNA (marked by *) was quantified and the substrate cleavage percentage was estimated, listed at the bottom of the gel. Undetectable cleavage was marked by -.
MATERIALS AND METHODS
Cloning, expression and protein purification
The full-length cDNA of human Tudor-SN (residues 1–885) was purchased from the Open Biosystems (Huntsville, AL, USA) (Clone ID: 3345037). The DNA fragments of TSN (residues 22–863), TSN-90 (residues 114–885), TSN-64 (residues 315–863), TSN-SN34 (residues 315–634) and TSN-25 (residue 645–863) were amplified by PCR and cloned into NdeI/EcoRI sites of expression vector pET-28c (Novagen, Madison, WI, USA) to generate N-terminal His-tagged constructs. The DNA fragments of TSN-70 (residues 280–885) and TSN-50 (residues 476–885) were cloned into NcoI/EcoRI sites of pET-28c to generate the C-terminal His-tagged constructs. Primers for the TSN fragments amplification were listed below: TSN: F:5′-CGGGGCGCATATGCGGCAGATCAACCTCAGC-3′ and R:5′-GAATTCTTAGGCGCTCTTGGCTGACTCTTG-3′; TSN-90: F: 5′-AGTTAACCATGGGCATGAGAGCTAATAATCCTGAG-3′ and R:5′-CCGAATTCTTGCGGCTGTAGCCAAATTCGTC-3′; TSN-70: F: 5′-AGTTAACCATGGGCACCCGGGGCGCAGAAAAGCG-3′ and R: 5′-CCGAATTCTTGCGGCTGTAGCCAAATTCGTC-3′; TSN-64: F: 5′-CGGGGCGCATATGGACAAGCAGTTTGTTGCCAAG-3′ and R: 5′-GAATTCTTAGGCGCTCTTGGCTGACTCTTG-3′; TSN-SN34: F: 5′-CGGGGCGCATATGGACAAGCAGTTTGTTGCCAAG-3′ and R: 5′-GAATTCTTAGGCCCAGACCTTCTCTTTCTT-3′; TSN-50: F: 5′-GGCAT GCCATGGGCATCCACCGTGTTGCAGATATA-3′ and R: 5′-CCGAATTCTTGCGGCTGTAGCCAAATTCGTC-3′; TSN-25: F: 5′-CGGGGCGCATATGCCAGTGCTGGAGGAG and R: 5′-GAATTCTTAGGCGCTCTTGGCTGACTCTTG-3′.
All the TSN constructs were expressed in BL21-CodonPlus(DE3)-RIPL cells (Stratagene, La Jolla, CA, USA) for the activity and filter-binding assays. Cells were grown in the LB medium containing appropriate antibiotics to a density of ∼0.6 OD600 and induced using 1 mM IPTG for 20 h at 20°C. After the induction, cells were harvested by centrifugation and resuspended in cold lysis buffer containing 20 mM Tris–HCl, pH 7.9, 500 mM NaCl, 5 mM imidazole, 10 mM β-mercaptoethanol and EDTA-free protease inhibitor cocktail (Roche, Mannheim, Germany). The cells were then lyzed by microfluidizer and the debris was removed by centrifugation (13 000 r.p.m., 30 min at 4°C). The cell extracts were then loaded onto the Ni-NTA affinity column (Qiagen Inc., Chatsworth, CA, USA) and washed extensively with wash buffer containing 20 mM Tris–HCl, pH 7.9, 500 mM NaCl, 40 mM imidazole and 10 mM β-mercaptoethanol. The purified proteins were eluted with wash buffer containing 250 mM imidazole. Peak fractions were pooled, diluted immediately into the buffer containing 50 mM HEPES pH 7.0, 10 mM EDTA, 10% glycerol and then loaded onto the HiTrap Heparin column (Amersham Pharmacia Biotech, Uppsala, Sweden) and eluted using the NaCl gradient. For the TSN-50 and TSN-25, diluted protein was loaded onto the HiTrap Q column (Pharmacia) and also eluted using the NaCl gradient. Peak fractions from Heparin (TSN, TSN-90, TSN-70, TSN-64, and TSN-SN34) or Q (TSN-50, TSN-25) were concentrated and further purified using the gel filtration chromatography column (Superdex 200, Phamacia) in the buffer containing 50 mM HEPES, pH 7.0, 250 mM NaCl, 10 mM β-mercaptoethanol and 10% glycerol. The purified protein samples were concentrated to ∼10 mg/ml and stored at −80°C.
For crystallization experiments, TSN-64 was further overexpressed in BL21(DE3) harboring chaperone plasmid pG-KJE8 (Takara Bio Inc., Shiga, Japan) or methionine auxotrophic host B834 (Novagen). The wild-type TSN-64 protein were expressed and purified as described above, expect the final gel filtration buffer was changed to 50 mM HEPES, pH 7.0, 150 mM NaCl and 10 mM β-mercaptoethanol. The selenomethionine-labeled protein Se-TSN-64 was expressed from B834 in the minimal medium described in a previous study (27). The purification procedures of Se-TSN-64 were the same as wild-type TSN-64.
Crystallization, structural determination and refinement
Different truncated forms of Tudor-SN were screened for the initial crystallization conditions by the high-throughput robotic Honeybee (960 trials) system. The TSN-64 crystals appeared in Hampton Research Screen Index21 in 2 months. After minor modification, TSN-64 was crystallized to a suitable size in 3 weeks in a cold room in a solution containing 50 mM HEPES, pH 7.0, 150 mM NaCl, 10 mM β-mercaptoethanol, against a reservoir of 1.44 M tri-ammonium citrate pH 7.0 by the hanging-drop vapor diffusion method. Se-TSN-64 also crystallized under the same condition using the wild-type protein crystals as microseeds.
The Se-TSN-64 crystals were freshly soaked in the mother liquor containing 20% glycerol prior to the data collection at −150°C. Three multiwavelength anomalous diffraction (MAD) data sets were collected by a CCD detector at SPring-8 Taiwan beamline BL-12B2. Diffraction data were processed and scaled by HKL2000 (28). Eight selenium sites were located and the initial MAD phases of Se-TSN-64 were calculated by the CNS program (29). The Se-TSN-64 model, built by COOT (30), contained 537 residues (315–634 and 645–860) including a N-terminal vector encoded methinonine (Met314 in the model). The electron density maps at a loop between residues 634–645 and at the C-terminal tail (residues 861–863) were ill defined. The final Se-TSN-64 model had an R-factor of 17.46% for 51 544 reflections and an Rfree of 20.93% for 5780 reflections, in the resolution range of 50–1.9 Å. The data collection and refinement statistics are summarized in Table 1. All figures were generated with PyMOL (31) and the electric potential surfaces were calculated by APBS (32).
Table 1.
Data collection and refinement statistics
| Data collection and MAD phasing statistics | |||
| Crystal | Se-TSN-64 | ||
| Beamline | Spring-8 Taiwan beamline BL-12B2 | ||
| Wavelength (Å) | Peak 0.979389 | Inflection 0.979545 | Remote 0.964305 |
| Resolution (Å) | 1.85 | 1.9 | 1.9 |
| Space group | C2 | C2 | C2 |
| Cell constants (Å) | a = 97.17 | a = 97.54 | a = 97.41 |
| b = 91.78 | b = 92.03 | b = 91.93 | |
| c = 87.82 | c = 88.17 | c = 88.04 | |
| β = 91.27° | β = 91.25° | β = 91.26° | |
| Total reflections | 322 497 | 152 909 | 172 942 |
| Unique reflections | 64 175 | 60 203 | 68 052 |
| Completeness (%) | 97.9 | 97.9 | 97.8 |
| I/σ(I) – overall | 20.3 | 14.9 | 15 |
| I/σ(I) – last shell | 4.6 | 2.8 | 3.4 |
| Phasing power (centrics/acentrics) | 1.04/0.92 | ||
| Figure of merit (centrics/acentrics) | 0.67/0.45 | ||
| Refinement | |||
| Resolution (Å) | 50–1.9 | ||
| Reflections (working/test) | 51 544/5 780 | ||
| Rcryst/Rfree | 17.46/20.93 | ||
| R. M. S. bond length (Å) | 0.0053 | ||
| R. M. S. angles (°) | 1.23 | ||
| Ramachandran plot (most favored/additional allowed/generously allowed) | (90.2/9.4/0.4) | ||
| Number of atoms | |||
| Protein | 4242 | ||
| Water | 690 | ||
| Average B-factor (Å2) | |||
| Protein | 24.08 | ||
| Water | 37.48 | ||
Cleavage and filter-binding assays
The RNA oligonucleotides for RNase activity assays were purchased from Dharmacon (Lafayette, CO, USA) containing IIUI- or AAUA-sequence: 5′-ACUGGACAIIUICUCCGAGG-3′/5′-CCUCGGAGUIUUUGUCCAGU-3′ (or AAUA/UAUU in the center). The top strands of the double-stranded RNA were 5′end-labeled with [γ-32p]ATP by the T4 PNK (NEB, Beverly, MA, USA). The 1 pmole double-stranded RNA substrates were then incubated with different truncated forms of Tudor-SN in the reaction buffer containing 20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 2 mM MgCl2, 1 mM β-mercaptoethonal and 1% glycerol for 8 h at 20°C. After the incubation, 1 μl of Proteinase K (10 mg/ml) was added into the reaction mixture and incubated for 30 min at 37°C. An equal volume of 2× TBE–urea sample buffers (Bio-Rad, Richmond, CA, USA) were then added into the reaction mixtures and heated to 70°C for 2 min prior to 20% denaturing polyacrylamide gel electrophoresis. After the electrophoresis, the gels were exposed to the phosphorimaging plate (Fujifilm) and analyzed by the imaging system FLA-5000 (Fujifilm).
For filter-binding assays, double-stranded RNA were 5′-end labeled with [γ-32p]ATP in both strands by T4 PNK. The labeled dsRNA (25 fmol) were then incubated with the serial dilution of protein samples in the binding buffer containing 20 mM HEPES, pH 7.0, 20 mM NaCl, 10 mM EDTA, 5 mM β-mercaptoethanol and 10% glycerol for 30 min at 4°C. The reaction mixtures were then passed through the filter-binding assay apparatus (Bio-Dot SF microfiltration apparatus, Bio-Rad). After extensive washing, the protein–RNA complex-bound nitrocellulose membrane and free RNA bound nylon membrane were air dried and exposed to a phosphorimaging plate. The intensities of protein–RNA complex and free RNA were quantified by the program AlphaImager IS-2200 (Alpha Innotech, San Leandro, CA, USA). The binding percentages were calculated and normalized. The apparent Kd values were estimated by one-site binding curve fitting using GraphPad Prism 4. The atomic coordinates and structural factors of TSN-64 have been deposited in the Protein Data Bank with a PDB ID of 3BDL.
RESULTS
Tandem repeats of SN domains are required for RNA binding and cleavage activity
Tudor-SN has been reported to contain both DNase and RNase activities (7). Interestingly, an amino acid sequence comparison between SN domains of Tudor-SN and staphylococcal nuclease showed that most of the critical active site residues have been mutated (21). To find out which SN domains are involved in substrate binding and cleavage, a number of deletion mutants of Tudor-SN were constructed (Figure 1A). We found that the full-length Tudor-SN was unstable and degraded into three major stable fragments after purification. According to the N-terminal sequencing analysis results of these stable degraded fragments, three truncated forms of Tudor-SN, TSN-90 (residues 114–885), TSN-70 (residues 280–885) and TSN-50 (residues 476–885), were constructed. Based on the secondary structure predictions, three more constructs, TSN (residues 22–863), TSN-64 (residues 315–863) and TSN-25 (residues 645–863) were prepared, in which the flexible loops at both the N- and C-terminal ends were deleted. All of these His-tagged Tudor-SN truncated proteins were then overexpressed in Escherichia coli and purified to homogeneity by chromatographic methods, using a Ni-NTA affinity column, followed by a heparin and a size exclusion column. The purified proteins were analyzed by 10% SDS–PAGE (Figure 1B) and had >98% purity with minor degraded fragments that were confirmed by the Tudor-SN-specific antibody (data not shown).
To test if the recombinant Tudor-SN proteins contained DNase activity, plasmid-nicking assays were carried out. The intensity of each substrate and product gel band was quantified and the estimated substrate cleavage percentages were listed at the bottom of the gel (Figure 1C). TSN, TSN-90 and TSN-70 had detectable DNase activity (14–3%), whereas TSN-64 TSN-50 and TSN-25 had residual activity (1%). This result suggests that the N-terminal SN domains, including SN1, and SN2, are required for efficient DNase activity. To test the RNase activity, 20-bp double-stranded RNAs, containing four wobble-paired IIUI/UIUU or four Watson–Crick base-paired AAUA/UAUU, were used as substrates for RNA digestion experiments. We found that TSN and TSN-90 cleaved IIUI-dsRNA with the highest activities (5 and 15%), whereas TSN-70, TSN-64, TSN-50 and TSN-25 had no detectable dsRNase activity (Figure 1D). Moreover, TSN and TSN-90 cleaved IIUI-dsRNA more efficiently than AAUA-dsRNA, demonstrating that Tudor-SN indeed prefers to cleave I·U-containing double-stranded RNA, consistent with previous studies (3,16).
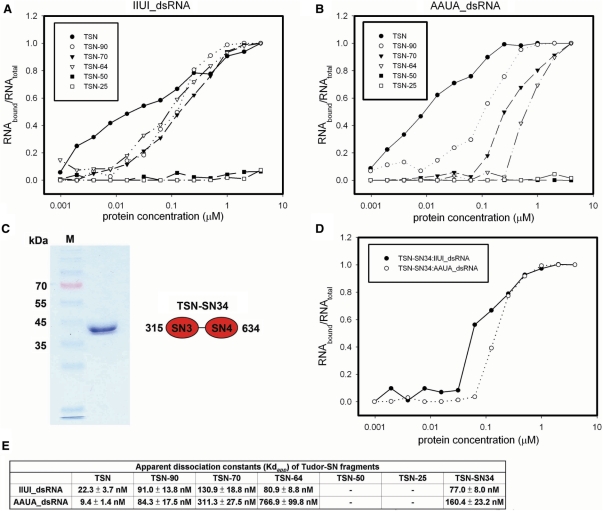
To find out why some of the truncated mutants do not cleave RNA efficiently, the dissociation constants between Tudor-SN truncated proteins and RNAs were measured by nitrocellulose filter-binding assays (Figure 2A and B), in the absence of magnesium ions. The results summarized in Figure 2E show firstly that the full-length TSN (residues 22–863) binds to IIUI- and AAUI-dsRNA with similar Kd values in the range of ∼10–20 nM. This result indicates that Tudor-SN does not bind site-specifically to the IIUI sequence, but it also binds well to Watson–Crick based-paired RNAs. Second, the truncated proteins containing four tandem SN (TSN), three tandem SN (TSN-90) and two tandem SN (TSN-70 and TSN-64) domains all bound RNA, with Kd values ranging from 20 to 700 nM. Nevertheless, the two truncated mutants, TSN-50 and TSN-25, which did not contain tandem SN domains, cannot bind either IIUI- or AAUA-dsRNA. These results suggest that the N-terminal SN domains are responsible for RNA binding and the C-terminal tudor and SN5 domains are not involved in RNA binding. To further investigate whether the C-terminal tudor and SN5 domains are required for RNA binding, we expressed and purified a C-terminal truncated mutant TSN-SN34 bearing only SN3 and SN4 domains. TSN-SN34 bound to IIUI- and AAUA-dsRNA with apparent Kd values in the range of ∼80–160 nM, comparable to those of TSN-64. This result confirmed that the C-terminal tudor and SN5 domains are dispensable in RNA binding, and a minimum of two tandem SN domains in Tudor-SN are required for efficient double-stranded RNA binding.
Figure 2.
Nitrocellulose filter-binding assays between truncated Tudor-SN mutants and RNA. (A) The binding assays show that TSN, TSN-90, TSN-70 and TSN64 all bind the 20-bp IIUI-dsRNA with comparable affinity, whereas TSN-50 and TSN-25 cannot bind RNA. (B) The filter-binding assays between Tudor-SN proteins and the 20-bp AAUA-dsRNA show that the truncated proteins containing more tandem repeats of SN domains bind AAUA-dsRNA better. TSN-50 and TSN-25 did not bind AAUA-dsRNA. (C) SDS–PAGE analysis of purified TSN-SN34. (D) The filter binding assays between TSN-SN34 and RNA. (E) The summary of the apparent dissociation constants (Kdapp) between Tudor-SN proteins and 20-bp IIUI- and AAUA-dsRNAs.
Crystal structure of TSN-64
The different truncated forms of Tudor-SN were screened for crystallization conditions, and the construct of TSN-64 (residues 315–863) yielded diffraction-quality crystals. The crystal structure of TSN-64 was determined by MAD at a resolution of 1.9 Å based on the anomalous scattering from a Se-Met-labeled TSN-64 crystal, using diffraction data collected at Taiwan beamline BL-12B2 at SPring-8, Japan. The final structural model contained one protein molecule (residues 315–634 and 645–860), four citrate ions and 690 water molecules in an asymmetric unit of the C2 monoclinic cell (see diffraction and refinement statistics in Table 1). A long loop between SN4 and SN5 domains (residues 635–644) was disordered without visible electron density.
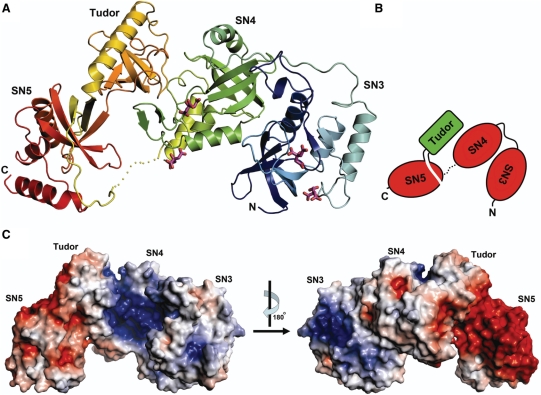
The four domains, SN3, SN4, tudor and SN5 in TSN-64 are arranged in a distorted crescent shape. SN5 is not a partial but a complete SN domain, but a tudor domain is inserted in SN5 and loops out to form a separated barrel-like domain (Figure 3A and B). SN4, though linked to SN5 in sequence, makes no contact to SN5, but is packed against the tudor domain. The four domains are well packed with reasonably buried interfaces to stabilize the structure (1541.6 Å2 between SN3 and SN4, 1339.1 Å2 between SN4 and tudor, and 628.9 Å2 between tudor and SN5). Therefore, the four domains should be assembled in a rigid way to maintain the overall crescent-shaped structure. Superimposition of the C-terminal tudor and SN5 domains in TSN-64 with the previously reported truncated Tudor-SN structure containing only the tudor and SN5 domains (26) (PDB accession code: 2O4X) gave an average RMSD of 0.77 Å for 195 Cα-atoms. This indicates that the interface between tudor and SN5 domains is well built to fix their relative domain orientations.
Figure 3.
Crystal structure of TSN-64. (A) The ribbon model of TSN-64 bound with four citrate ions. The four domains, SN3, SN4, Tudor and SN5, are rainbow colored from blue (N-terminus) to red (C-terminus). A loop between SN4 and SN5 (residues 635–644) is disordered and is displayed as a dotted line. (B) A schematic diagram representing the domain arrangement in TSN-64. The tudor domain is inserted in SN5 and packed between SN4 and SN5. (C) The electrostatic potential, mapped onto the solvent-accessible surfaces of TSN-64, calculated by APBS (32). The color scale was set from −5 kT/e (red) to +5 kT/e (blue). The molecular surfaces of tudor and SN5 are more acidic, whereas those of SN3 and SN4 are more basic.
The electrostatic potential mapping onto the solvent accessible surfaces of TSN-64 was calculated and shown in Figure 3C. The solvent accessible surfaces of SN5 domain are highly acidic, suggesting that the SN5 domain is likely not involved in nucleic acid interactions. On the other hand, the surfaces of SN3 and SN4 are more basic, especially in the concave surface of the crescent-shaped structure. Four citrate molecules, cocrystallized with TSN-64, are located at the basic surfaces, with two citrates bound near the putative active site region in the SN3 and SN4 domains, respectively. These citrate ion binding sites provide clues for the RNA phosphate binding sites (see the subsequent section). Based on the structural and electrostatic surface analysis results, we suggest that the nucleic acid substrates are bound to the SN3 and SN4 domains underneath the concave surface of the crescent-shaped structure.
Comparison of SN domains to staphylococcal nuclease
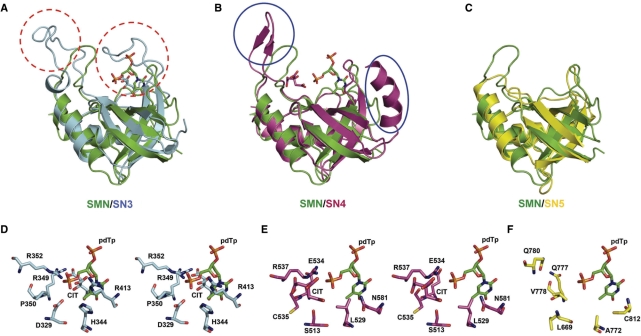
The three SN domains, SN3, SN4, and SN5, all bear an OB-fold of staphylococcal nuclease, containing mainly a five-stranded β-barrel packed against three α-helices (Figure 4) (33). Although the overall OB-fold structures are similar, minor differences are present in loop and flanking regions. Superimposition of respective SN domains with staphylococcal nuclease [PDB accession code:1EY0 (34)] shows that SN3 contains two extra long loops (Gly355-Ile371 and Ala400-Glu412), extending out from the side of the β-barrel; SN4 domain contains an extra α-helix (Thr486-Gly500) flanking the β-barrel and an extra pair of antiparallel β-strands (Arg540-Glu548) interacting with the tudor domain. SN5, on the other hand, more closely resembles the overall structure of staphylococcal nuclease, without the presence of extra long loops or inserted secondary structural elements.
Figure 4.
Structural comparison between staphylococcal nuclease (SMN) and SN domains in TSN-64. (A) Superposition of staphylococcal nuclease (PDB entry: 1EY0 (34)) and SN3 domain. SN3 contains extra long loops, marked by red dashed circles. (B) Superposition of SMN and SN4. SN4 contains an extra pair of β-strands and an α-helix, delineated by the blue circles. (C) Superposition of SMN and SN5. (D) The stereo view of the putative active site of SN3. A citrate ion (labeled as CIT) bound at the active site, overlapped with the pdTp bound at the active site of SMN [PDB accession code: 2ENB (35)]. (E) The stereo view of the putative active site of SN4. A citrate ion is bound next to the pdTp bound in SMN. (F) The putative active site in SN5. Most residues are hydrophobic, not appropriate for metal-ion binding or RNA hydrolysis.
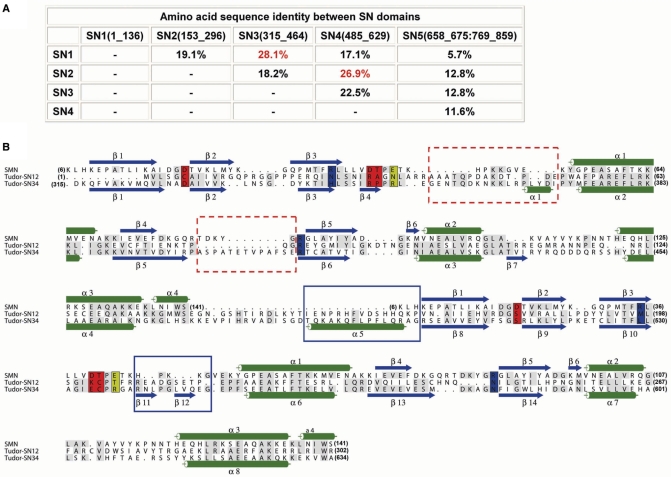
Interestingly, the structural-based sequence alignment suggests that SN1 domain is more similar to SN3 with a sequence identity of 28.1% (Figure 5A). Moreover, SN1 contains the sequences that may align with the two extra long loops in SN3. On the other hand, the sequence of SN2 is more similar to that of SN4 (26.9% identity), because only SN2 bears the inserted sequences that may align with the extra N-terminal α-helix in SN4 (from Thr486-Gly500, marked in boxes in Figure 5B). In the crystal structure of TSN-64, SN3 and SN4 jointly form a structural module with a basic surface ideal for nucleic acid binding. Taken together, these results suggest that SN1 and SN2 likely also form a similar structural module for nucleic acid substrate recognition. The sequence alignment of SN1-SN2 to SN3-SN4 is shown in Figure 5B.
Figure 5.
Sequence comparison of SN domains and structural-based sequence alignment between SMN and SN domains in Tudor-SN. (A) The sequence identities between different SN domains in Tudor-SN show that SN1 shares the highest sequence identity with SN3 (28.1%), while SN2 shares the highest sequence identity with SN4 (26.9%). (B) Sequence alignment between SMN and Tudor-SN. The secondary structures of SMN and TSN-64 are displayed at the top and bottom of the sequences, respectively. As SN1 shares higher sequence identity with SN3, and SN2 shares higher sequence identity with SN4, SN1-SN2 domains are aligned with SN3–SN4.
Two citrate ions are located at the basic surface near the putative active sites in the SN3 and SN4 domains, respectively. Superimposition of SN3 and SN4 separately with the staphylococcal nuclease–pdTp complex [PDB accession code: 2ENB (35)] shows that a citrate ion bound in SN3 overlaps with pdTp, and a citrate ion bound in SN4 is located right next to pdTp. This result suggests that the citrate ions bound in TSN-64 likely mimic the phosphate-binding sites of nucleic acid molecules. Several residues surrounding the citrate ions, such as Asp329, His344, Arg349, Arg352 and Arg413 in SN3, and Ser513, Cys535, Arg537, Glu554 and Asn581 in SN4 (Figure 4D and E) are candidates for catalytic residues involved in metal-ion binding, phosphate binding or phosphodiester bond hydrolysis. In contrast, the residues surrounding the corresponding putative active site in SN5 (Figure 4F) are mostly hydrophobic, such as Leu669, Ala772, Val778, Gln777, Gln780 and Cys812, consistent with the hypothesis that this domain may not participate in nucleic acid binding or hydrolysis.
DISCUSSION
SN domains in RNA binding and cleavage
Our deletion experiments in Tudor-SN show that a minimum of two tandem repeats of SN domains are required for sufficient RNA binding. The recombinant Tudor-SN proteins indeed prefer to cleave a 20-bp I·U-containing RNA over an A·U-containing RNA. The crystal structure of TSN-64 further shows that SN3 and SN4 domains form a compact structural module and jointly generate a basic surface suitable for RNA recognition. The question is how these two SN domains work together to recognize a double-stranded RNA.
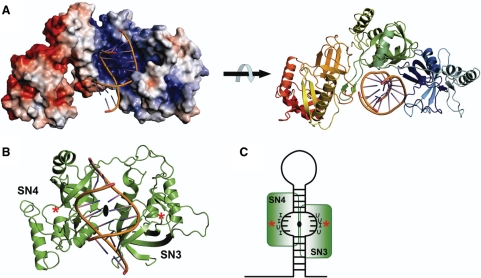
We noticed that the two SN domains in TSN-64 are related to each other by a pseudo 2-fold symmetry (Figure 6B). This 2-fold symmetry might be linked to the protein function of RNA recognition since the two sugar-phosphate backbones of a double-helical RNA are also related approximately by a 2-fold symmetry. Therefore, the structural module of SN3–SN4 seems ideal for the interactions with a double-stranded RNA. We thus built a complex model by superimposition of the dyad axis between SN3 and SN4 in TSN-64 to the dyad axis in a 16-bp RNA [PDB accession code: 1DI2 (36)]. In this way, one of the rotational axes of the RNA molecule in the complex was fixed by the superposition. Another rotational axis was further constrained by the shape of the cleft, which allows the RNA helix to pass through in only one direction. The final model, displayed in Figure 6A, has a double-stranded RNA snugly bound at the concave side of the crescent-shaped structure, with one strand of phosphate backbone interacting mainly with one SN domain. It is possible that the SN1 and SN2 domains assemble together with SN3 and SN4 to create another concave surface for RNA binding.
Figure 6.
Structural model of TSN-64 bound with a double-stranded RNA. (A) Two different views of the model show that a dsRNA may bind snugly at the concave basic surface in TSN-64 to SN3 and SN4 domains. (B) A pseudo 2-fold symmetry axis is identified between SN3 and SN4 domains. The structural model was built by aligning the dyad axis between a 16-bp RNA [PDB accession code: 1DI2 (36)] to the dyad axis between SN3 and SN4. Two stars mark the likely location of putative active sites in SN3 and SN4, based on the pdTp-binding site in staphylococcal nuclease. (C) The part of double-stranded RNA containing mis-paired I·U or U·I sequences may have an open conformation. The phosphate backbones may displace from a canonical A-form conformation so that they can be bound near the active sites in the SN3 and SN4 domains.
This model of Tudor-SN–RNA complex is consistent with our biochemical and structural data, except that the phosphate backbones are far away from the putative active sites in SN3 and SN4 domains (marked as stars in Figure 6B). Previous biochemical studies showed that Tudor-SN efficiently cleave only the dsRNAs containing multiple I·U and U·I wobble base pairs, but not the RNAs containing the isosteric G·U and U·G wobble base pairs or Watson–Crick A·U base pairs (3,16,17,37). It has been shown that the RNA containing tandem I·U pairs are significantly less stable than the ones containing G·U or A·U base pairs; the melting points of an 8-bp RNA containing two or three tandem I·U base pairs are >20°C lower than those of G·U or A·U base-paired RNAs (38). Therefore, the dsRNA containing multiple I·U pairs might have a specific structure that is less stable and deviates considerably from a classical A-form structure. It is likely that the I·U-containing RNA bound at Tudor-SN has a distorted open conformation so that the RNA backbones are displaced from a well-annealed structure and thus are bound at the active sites (schematic diagram in Figure 6C). Only the hyper-edited double-stranded RNA may adopt this specific conformation and therefore only the RNA substrates containing multiple I·U and U·I pairs can be cleaved efficiently by Tudor-SN. This explains previous observations showing that the in vitro-edited pri-miRNA-142 can be degraded sensitively by Tudor-SN in proportion to the number of A→I modifications (3). Moreover, our hypothesis that two SN domains form a structural module for dsRNA binding is further supported by sequence data showing that Tudor-SN from various species contains even numbers of tandem repeats of SN domains, that is, either four for human, mouse, fish, drosophila, C. elegans and fission yeast or two for tetrahymena.
Biological roles of Tudor-SN and its substrates
Our biochemical and structural data support the previous finding that Tudor-SN functions as a miRNase (RNase for microRNA), which is more specific for I·U-containing double-stranded RNA. To date, the ADAR deaminase-edited primary transcript of miRNA-142 is the only identified natural substrate for Tudor-SN (3). Mature miRNA-142 expression levels increased substantially in ADAR1 null or ADAR2 null mice since Drosha processed only the unedited primary miRNA transcripts but not the hyper-edited ones. Tudor-SN plays an opposite role in that it degrades the hyper-edited miRNA more efficiently than the unedited ones. Over the decades, considerable lines of evidence have accumulated showing that miRNAs play key roles in the regulation of gene expression and affect mRNA degradation and translation (39). Therefore, besides functioning as a transcription coactivator and regulating gene expression at the transcriptional level, Tudor-SN may likely degrade negative regulators of hyper-edited miRNA and thus upregulate gene expression at the posttranscriptional level.
Apart from hyper-edited primary transcripts of miRNA, are there other RNA types that may be good candidate substrates for Tudor-SN? Firstly, bioinformatics screening for A→I RNA editing sites based on the comparison of cDNA or EST sequences with the corresponding genome sequences predicted that >85% of pre-mRNAs are edited, with the majority in introns (∼90%) and UTRs containing Alu repeats or LINE repeats in human transcriptome (40–43). These A→I edited pre-mRNAs, containing inverted repeats that fold into double-stranded RNA, are candidate substrates for Tudor-SN, which in turn, may control gene expression of mRNAs bearing repeat sequences (44). Second, the cytoplasmic ADAR1 is induced by interferon (45), implying that this enzyme is involved in antiviral defense. Combined with the finding demonstrating that a number of viral RNAs are hyper-edited, Tudor-SN might target these hyper-edited viral RNAs and work together with ADAR1 to combat viral infections (17). Third, up- or down-regulation of a number of miRNA levels has been shown to play a role in antiviral defense during viral infection not only in plants and invertebrates (46), but also in mammals, through the interferon system (47). Therefore, it will be intriguing to find out whether Tudor-SN is involved in the regulation of these host miRNA levels. As there is no direct evidence for the role of Tudor-SN in the degradation of hyper-edited pre-mRNA, viral RNA or host miRNA, more studies are needed to further reveal Tudor-SN's natural substrates.
How Tudor-SN's RNase activity is regulated is another important issue that has not yet been addressed. Tudor-SN is an ubiquitous protein present in the nucleus (4,9), cytoplasm (5,16), or both (7). It has been shown that a recombinant Tudor-SN, supplemented with a limited amount of Xenopus laevis oocyte extract, cleaved a IIUI-containing dsRNA much more efficiently than the unsupplemented one (16), indicating that some other factors may be involved in promoting its RNase activity. The recombinant Tudor-SN purified from yeast enhanced the cleavage of IIUI-dsRNA (20-mer) but no RNase activity was detected when the RNA substrates were incubated with 5 μM Tudor-SN alone (37). In contrast, the recombinant flag tagged Tudor-SN from HeLa cell digested inosine hyper-edited miRNA efficiently (3). In this study, the recombinant Tudor-SNs were purified from E. coli, and they had weak DNase and RNase activities. It is likely that Tudor-SN needs other cofactors to enhance its nuclease activity, or Tudor-SN prefers to digest a longer native hyper-edited substrate. The tudor domain in Tudor-SN is known to be a protein–protein interaction domain and it could interact with other cofactors for the regulation of Tudor-SN's activities. The SN5 domain, appearing not to be involved in nucleic acid interactions, may also be a good candidate for the screening of protein–protein interaction partners. Our structure analysis thus paves the way for future functional analyses of this versatile protein in RNA editing, interference and splicing.
ACKNOWLEDGEMENTS
This work was supported by research grants from Academia Sinica and the National Science Council, Taiwan, ROC. Portions of this research were carried out at the National Synchrotron Radiation Research Center (BL-13B1 and BL-13C1), a national user facility supported by the National Science Council of Taiwan, ROC. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Research Program for Genomic Medicine. C.-L.L. is an Academia Sinica Postdoctoral Fellow, and H.S.Y. is an Academia Sinica Investigator Awardee. Funding to pay the Open Access publication charges for this article was provided by Academia Sinica.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ding S-W, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mello CC, Conte DJ. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 3.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broadhurst MK, Wheeler TT. The p100 coactivator is present in the nuclei of mammary epithelial cells and its abundance is increased in response to prolactin in culture and in mammary tissue during lactation. J. Endocrinol. 2001;171:329–337. doi: 10.1677/joe.0.1710329. [DOI] [PubMed] [Google Scholar]
- 5.Zhao CT, Shi KH, Liang LY, Yan Y, Postlethwait J, Meng AM. Two variants of zebrafish p100 are expressed during embryogenesis and regulated by nodal signaling. FEBS Lett. 2003;543:190–195. doi: 10.1016/s0014-5793(03)00445-9. [DOI] [PubMed] [Google Scholar]
- 6.Abe S, Wang P.-L, Takahashi F, Sasaki E. Structural analysis of cDNAs coding for 4SNc-Tudor domain protein from fish and their expression in yellowtail organs. Marine Biotechnol. 2005;7:677–686. doi: 10.1007/s10126-004-5137-z. [DOI] [PubMed] [Google Scholar]
- 7.Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- 8.Howard-Till RA, Yao M.-C. Tudor nuclease genes and programmed DNA rearrangements in Tetrahymena thermophila. Eukaryot. Cell. 2007;6:1795–1804. doi: 10.1128/EC.00192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leverson J, Koskinen PJ, Orrico F, Rainio E.-M, Jalkanen KJ, Dash AB, Eisenman RN, Ness SA. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell. 1998;2:417–425. doi: 10.1016/s1097-2765(00)80141-0. [DOI] [PubMed] [Google Scholar]
- 11.Valineva T, Yang J, Palovuori R, Silvennoinen O. The transcriptional ac-activator protein p100 recruits histone acetyltransferase activity to STAT6 and mediates interaction between the CREB-binding protein and STAT6. J. Biol. Chem. 2005;280:14989–14996. doi: 10.1074/jbc.M410465200. [DOI] [PubMed] [Google Scholar]
- 12.Valineva T, Yang J, Sivernnoinen O. Characterization of RNA helicase A as component of STAT6-dependent enhancement. Nucleic Acid Res. 2006;34:3938–3946. doi: 10.1093/nar/gkl539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Low SH, Vasanth S, Larson CH, Mukherjee S, Sharma N, Kinter MT, Kane ME, Obara T, Weimbs T. Polycystin-1, STAT6, and p100 function in a pathway that transduces ciliary mechanosensation and it activated in polycystic kidney disease. Develop. Cell. 2006;10:57–69. doi: 10.1016/j.devcel.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Paukku K, Yang J, Silvennoinen O. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcriptional 5. Mol. Endocrinol. 2007;17:1805–1814. doi: 10.1210/me.2002-0256. [DOI] [PubMed] [Google Scholar]
- 15.Tsuchiya N, Ochiai M, Nakashima K, Ubagai T, Sugimura T, Nakagama H. SND1, a component of RNA-induced silencing complex, is up-regulated in human colon cancers and implicated in early stage colon carcinogenesis. Cancer Res. 2007;67:9568–9576. doi: 10.1158/0008-5472.CAN-06-2707. [DOI] [PubMed] [Google Scholar]
- 16.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nature Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 17.Scadden AD, Smith CWJ. Specific cleavage of hyper-edited dsRNAs. EMBO J. 2001;20:4243–4252. doi: 10.1093/emboj/20.15.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precusor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Valineva T, Hong J, Bu T, Yao Z, Jensen ON, Frilander MJ, Silvennoinen O. Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acid Res. 2007;35:4485–4494. doi: 10.1093/nar/gkm470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callebaut I, Mornon JP. The human EBNA-2 coactivator p100: multidomain organization and relationship to the staphylococcal nuclease fold and t the tudor protein involved in Drosophila melanogaster development. Biochem. J. 1997;321:125–132. doi: 10.1042/bj3210125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponting CP. P100, a transcriptional coactivator, is a human homologue of staphylococcal nuclease. Protein Sci. 1997;6:459–463. doi: 10.1002/pro.5560060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cotton FA, Hazen EEJ, Legg MJ. Staphylococcal nuclease: proposed mechanism of action based on structure of enzyme-thymidine 3′,5′-bisphosphate-calcium ion complex at 1.5-Angstrom resolution. Proc. Natl Acad. Sci. USA. 1979;76:2551–2555. doi: 10.1073/pnas.76.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selenko P, Sprangers R, Stier G, Buhler D, Fisher U, Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 24.Huyen Y, Zgheib O, DiTullio RAJ, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 25.Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 26.Shaw N, Zhao M, Cheng C, Xu H, Saarikettu J, Li Y, Da Y, Yao Z, Silvennoinen O, Yang J, et al. The multifunctional human p100 protein ‘hooks’ methylated ligands. Nat. Struct. Mol. Biol. 2007;14:779–784. doi: 10.1038/nsmb1269. [DOI] [PubMed] [Google Scholar]
- 27.Li CL, Hor LI, Chang ZF, Tsai LC, Yang WZ, Yuan HS. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J. 2003;22:4014–4025. doi: 10.1093/emboj/cdg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Delano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: Delano Scientific; 2002. [Google Scholar]
- 32.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theobald DT, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Lu Z, Sakon J, Stites WE. Increasing the thermostability of staphylococcal nuclease: implications for the origin of protein thermostability. J. Mol. Biol. 2000;303:125–130. doi: 10.1006/jmbi.2000.4140. [DOI] [PubMed] [Google Scholar]
- 35.Libson AM, Gittis AG, Lattman EE. Crystal structures of the binary Ca2+ and pdTp complexes and the ternary complex of the Asp21–>Glu mutant of staphylococcal nuclease. Implications for catalysis and ligand binding. Biochemistry. 1994;33:8007–8016. [PubMed] [Google Scholar]
- 36.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA- protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scadden ADJ, O’Connell MA. Cleavage of dsRNAs hyper-edited by ADARs occurs at preferred editing sites. Nucleic Acid Res. 2005;33:5954–5964. doi: 10.1093/nar/gki909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serra MJ, Smolter PE, Westhof E. Pronouced instability of tandem IU base pairs in RNA. Nucleic Acid Res. 2004;32:1824–1828. doi: 10.1093/nar/gkh501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2007;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 40.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptosome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 44.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell. Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cullen BR. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat. Immunol. 2006;7:563–567. doi: 10.1038/ni1352. [DOI] [PubMed] [Google Scholar]
- 47.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–923. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]