Abstract
The use of capillary electrophoresis with fluorescently labeled nucleic acids revolutionized DNA sequencing, effectively fueling the genomic revolution. We present an application of this technology for the high-throughput structural analysis of nucleic acids by chemical and enzymatic mapping (‘footprinting’). We achieve the throughput and data quality necessary for genomic-scale structural analysis by combining fluorophore labeling of nucleic acids with novel quantitation algorithms. We implemented these algorithms in the CAFA (capillary automated footprinting analysis) open-source software that is downloadable gratis from https://simtk.org/home/cafa. The accuracy, throughput and reproducibility of CAFA analysis are demonstrated using hydroxyl radical footprinting of RNA. The versatility of CAFA is illustrated by dimethyl sulfate mapping of RNA secondary structure and DNase I mapping of a protein binding to a specific sequence of DNA. Our experimental and computational approach facilitates the acquisition of high-throughput chemical probing data for solution structural analysis of nucleic acids.
INTRODUCTION
Nucleic acid structure contributes to cellular regulation (1–7). The ability to rapidly characterize the structure of nucleic acids with chemical and enzymatic probes is central to elucidating their functional roles (8,9). For example, the ENCODE project has identified critical sequences in the human genome which now require structural characterization (10,11). ‘Footprinting’ and chemical or enzymatic ‘mapping’ are synonymous terms for assays in which the accessibility of either the backbone or side-chains of a macromolecule is characterized by their reactivity to an exogenous probe (12–18). The key feature of chemical mapping is that it can report local changes in macromolecular structure with as fine as single-residue resolution (12,19–23). The products of DNA or RNA chemical mapping reactions have traditionally been separated by denaturing gel electrophoresis (GE) and imaged by autoradiography. Although advances in autoradiogram analysis have improved experimental throughput (23,24), further improvement is necessary for genomic-scale mapping analyses.
While capillary electrophoresis (CE)-based sequencers are ubiquitous at most institutions, their application to quantitative nucleic acid structural characterization requires special expertise (25–28). We therefore set out to harness the potential of CE for the structural characterization of nucleic acids by quantitative chemical mapping. A major limitation to their adoption for nucleic acid structural analysis is the absence of software that can quantitate the elution trace. The ‘base calling’ algorithms necessary for sequencing are not suitable for quantification of the chemical and enzymatic mapping data necessary for structural analysis (29–31). Rather, an algorithm capable of deconvoluting overlapping signal is necessary (23,24,32) along with software that transforms and manipulates the mapping data. To take advantage of high-throughput CE sequencers, we have developed the experimental protocols and the capillary automated footprinting analysis (CAFA) software described in this paper that builds upon tested GE analysis tools (23,24). The structural analyses derived from CAFA-based analysis will be a valuable addition to genome analyses (11,33,34).
MATERIALS AND METHODS
RNA, primers and size standard ladder
We prepared the L-21 ScaI Ribozyme by in vitro transcription and purification as described previously (35,36). HPLC-purified Cy5-labeled primers were obtained from Sigma Genosys and resuspended in TE buffer at 1 μM. The two primers we used in this study are, 5′ A CTC CAA AAC TAA TCA ATA TAC TTT C 3′ and 5′ GCA TCC ATA TCA ACA GAA GAT C 3′, and are complementary to nucleotides 409–384 and 255–234 of the Tetrahymena ribozyme, respectively. We purchased DNA size standard kits 400 and 600 from Beckman Coulter (PN 608098 and PN 608095).
Primer extension of cleaved/modified RNA
We used ∼5 pmol of RNA per mapping reaction and always ethanol precipitated the RNA prior to RT. We resuspended the precipitate in 9 μl of annealing buffer (50 mM Tris–Cl, pH 8.3, 60 mM NaCl, 10 mM DTT) and added 1 µl of fluorophore-labeled primer stock solution (1 μM) to each tube. We heated the samples to 85°C for 1 min, followed by slow cooling to 25°C for primer annealing, then added 9 µl of reverse transcription mix (4 μl of 5X RT buffer supplied with Superscript III-Invitrogen, 1 μl of 0.1 M DTT, 2 μl of RNase Inhibitor, 2 μl of 10 mM dNTP mix) in each tube. We incubated the solutions at 55°C for 5 min and then added 1 μl (200 U) of Superscript III (Invitrogen, Carlsbad, CA). The final reaction volume is 20 µl, which we incubated at 55°C for 15 min.
Upon completion of RT extension, we degraded the RNA by adding 2 µl of 2 N NaOH and incubating at 95°C for 3 min. To neutralize the solution, we added 2 µl of 2 N HCl followed by 3 μL of 3 M Na-acetate to facilitate cDNA precipitation and finally 80 µl of 100% ethanol. We centrifuged at 14 000 r.p.m. for 30 min to pellet the cDNA which we then dried and resuspended in 40 μl of the Sample Loading Solution® (Beckman, Fullerton, CA). We performed the dideoxy sequencing reactions (for markers) in the same way except that we added 0.25 mM of one of the ddNTPs.
Electrophoretic parameters
We separated the cDNAs by CE in a Beckman CEQ8000 Genetic Analysis System. The optimized parameters that produced peak traces at single nucleotide resolution for about 300 nt are: electro-kinetic injection voltage, 2 kV; electro-kinetic injection time, 7 s; denaturation, 95°C for 150 s; separation voltage, 3 kV; and capillary temperature, 60°C.
Basis for choosing the fluorescent dyes
The CEQ fluorescence detection filter wheel has four filters with the following wave-length cutoffs: 1: 675 ± 2 nm; 2: 715 ± 2 nm; 3: 775 ± 2 nm; and 4: 820 ± 2 nm. The primers are labeled with the dye Cy5 whose emission max (668 nm) corresponds to the filter 1. The Beckman size standard fragments are labeled with the Beckman WellRed® D1 dye whose emission max (∼820 nm) corresponds to filter 4.
Direct end-labeling of DNA duplex and DNase I experiments
We used a singly end-labeled 110bp DNA fragment containing the AdMLP TATA Box sequence TATAAAAG. The DNA fragment was amplified by PCR from the plasmid pRW2 using one labeled and one unlabeled primer (37), followed by purification on a 6% nondenaturing polyacrylamide gel. The DNase I experiments were performed in the absence and presence of 195 nM TBP as described in (38) and references cited therein.
Direct radioactive end-labeling of RNA
End labeling of RNA with 32P was conducted as described at either the 5′ (36) or the 3′ ends (39,40). The labeled products of OH cleavage were visualized by GE and phosphor storage imaging and quantitated by the SAFA software (23).
CAFA software development
We developed CAFA within the Matlab version 7.4 development environment (The MathWorks, Inc., Natick, MA) and compiled on both the Windows XP and Apple OS 10 operating systems. The developed peak width model is based on the initial estimation of the distance between peaks,
| 1 |
where wi is the peak width, pi the peak position (in pixels) and A is a proportionality constant that is fit by bounded nonlinear least squares. We use a custom implementation of the bounded nonlinear least squares (41) for efficient and robust model fitting. Data normalization is a two-step procedure. Peak areas are first normalized to their mean values. The differences between the resulting profiles are then minimized, excluding data identified as having high-error based on the background lane (Figure 4B).
Figure 4.
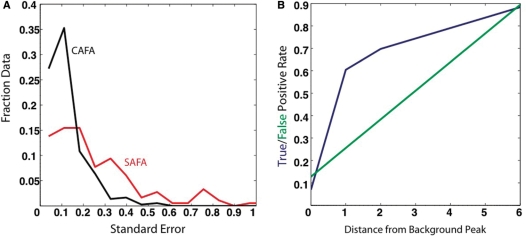
(A) Normalized histogram of the standard error for CAFA (black) and SAFA (red) analysis of the same RNA. The CAFA results have a smaller standard error than SAFA. (B) False-positive (green) and true-positive (blue) rates for the prediction of high-error data using the CAFA error prediction algorithm. CAFA identifies putative’ RT stop’ sites based on a background lane and flags this data for exclusion. We then analyze how well we are able to predict areas of low reproducibility in the data (as measured by repeating the experiments five times on independent samples) using the putative RT stops identified in the background lane. This figure demonstrates that if we exclude all data within one nucleotide of an RT stop, we are able to predict 62% of the high-error data. Therefore, this strategy can be used to identify high-error data without having to perform multiple repeats of the experiment. The user can adjust the number of nucleotides around the RT stop to exclude during the analysis.
RESULTS
Method development
Figure 1 illustrates the premise of CAFA for quantitation of nucleic acid structure probes such as the hydroxyl radical (·OH) radical, dimethyl sulfate (DMS), N-methylisatoic anhydride (NMIA), DNase I and base-specific nucleases (38,42,43). These probes cleave or modify a nucleic acid depending on the local chemical environment; modifications are transformed to cDNA fragments to facilitate electrophoretic separation for chemical mapping analysis. For example, if RNA is exposed to ·OH at conditions under which on average a molecule is cleaved once (Figure 1A) the extent of backbone cleavage at each residue is proportional to its solvent accessibility (8,43–45). The result of an ·OH mapping reaction is a population of RNA fragments that reflect the relative solvent accessible surface of each nucleotide (Figure 1B).
Figure 1.
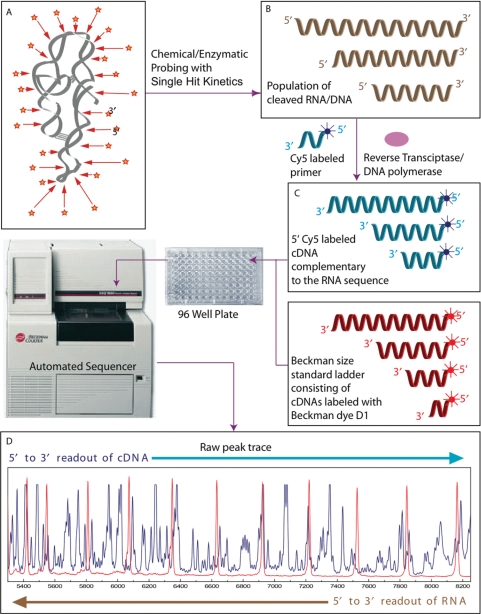
Basic premise of the CAFA approach. (A) DNA or RNA is exposed to a chemical or enzymatic probe that either cleaves or modifies it with single-hit kinetics such that the population of the reaction products (B) is related to the probe's reactivity. Extension of fluorescently labeled primers by RT or DNA polymerase generates a corresponding population of fluorescently labeled cDNA molecules (C). The cDNA samples are mixed with a Beckman size standard ladder (400 or 600). Each mixture is subject to CE yielding a trace of the size separated reaction products (D) to be analyzed by CAFA. The blue trace records the fluorescence emission of the Cy5 labeled cDNA fragments; the red peaks correspond to the Beckman WellRED® dye D1 present on the size standard fragments.
The peak profiles analyzed by CAFA are fluorescently labeled DNA separated and detected by the sequencer. For DNA singly end-labeled with a fluorescent dye prior to probing (direct labeling), the mapping reaction products are themselves subjected to electrophoretic separation. However, when fluorescent dyes are susceptible to degradation by a footprinting probe (e.g. ·OH) or the mapping reaction yields chemical modification that must be converted to nucleic acid fragments (e.g. DMS), proportionate postlabeling of the reaction products (indirect labeling) is advantageous. Indirect labeling methods include extension of a fluorescently labeled primer to yield fluorescent cDNA complements of the chemically mapped nucleic acid. This strategy can also be used with whole-cell extracts of RNA, as the primer will selectively anneal to the RNA of interest (8). An advantage of indirect labeling for RNA analysis is the resistance to nuclease degradation of cDNA transcripts. This advantage is critical if the sequencer used is maintained by a facility that does not conduct RNase free operations. Therefore, the CAFA approach does not require a dedicated or modified capillary sequencer.
CAFA can be used both with direct and indirect labeling strategies of nucleic acids. Incorporation of a fluorophore-labeled nucleotide at the 5′ or 3′ end of a nucleic-acid prior to cleavage/modification (also referred to as direct labeling) is a routine technique summarized in the Materials and methods section. Our protocol for indirect labeling of RNA (post cleavage/modification), tailored to CAFA builds upon the advances of others (8,26,28) and is illustrated for the ·OH mapping analysis of RNA folding. Unlabeled RNA is cleaved by ·OH as described below and the resultant fragments converted to cDNA for analysis. Cy5-labeled primers complementary to the 3′ end of the RNA template are annealed and extended by reverse transcription (RT) to generate labeled cDNA strands for samples analyzed by the Beckman CEQ8000 (CEQ) sequencer (Figure 1B). Elongation of the transcript terminates at sites of backbone cleavage producing a population of Cy5-cDNA molecules proportional to the population of unlabeled chemical mapping reaction products (Figure 1C). The CEQ size separates the products and records fluorescent intensity as a function of time in an ‘elution trace’ that is readily exported from the instrument for analysis (Figure 1D). Each peak in the trace represents cDNA molecules of n, n + 1, n + 2 … lengths. The area of each peak is proportional to the amount of cDNA present (Figure 1D).
CAFA automatically quantifies the area of the individual peaks of a trace. We adjust the sample injection time to maximize the usable fluorescence signal without saturating the detector. We also optimized the CE run parameters to maximize peak separation by systematically analyzing the denaturation and separation temperatures and the separation voltage (Supplementary Figure 5 and Table 1).
Implementation of CAFA
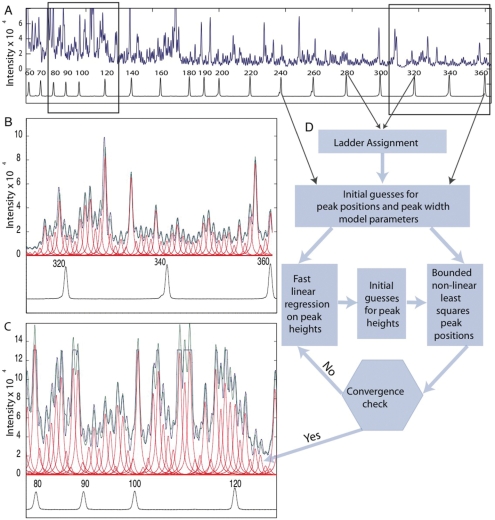
In a typical RNA experiment, CEQ analysis of the Cy5-cDNA products of ·OH mapping yield a trace such as that shown in Figure 2A for the Mg2+-folded L-21 Tetrahymena thermophila group I intron. The observed (blue) trace is characteristic of the Mg2+-folded ribozyme (13,19,20,46). Three hundred nucleotides are easily read in this experiment. Panels B and C illustrate the ‘best’ and ‘worst’ data by zooming in on 45 nt in regions of high and low peak separation, respectively. The Beckman size markers labeled with the D1 WellRED® dye are co-electrophoresed with the samples to reference the position of the sample peaks (Figure 2B and C; black traces below the peak profiles).
Figure 2.
CE traces (blue), CAFA models estimating peak area (green), individual peaks (red) and size ladder (black) with automated peak assignments. (A) The complete trace of the L-21 T. Thermophila group I intron with a CAFA model fit to the data. (B) and (C) Enlargements of right and left ends, respectively, of the CE trace. (D) A flow chart summarizing CAFA fitting showing the data inputs discrete steps [data (square boxes), and binary decisions (hexagons)]. The peaks present in the size ladder trace are initially assigned using a threshold peak picking algorithm (manually adjusted with a ‘sensitivity’ slider). These initial guesses of the peak positions are refined using bounded nonlinear least squares. The fast linear regression is used to update peak heights if convergence has not been achieved resulting in a peak model that fits the data optimally and reproducibly.
CAFA fits the blue elution trace to a model consisting of a series of Lorentzian line shapes (Figure 2; red lines) by assigning peak positions based on the size ladder and then refining the global peak model using bounded nonlinear least-squares (41). Figure 2D schematizes CAFA's peak-fitting algorithm. Our novel iterative approach to peak fitting adjusts the three parameters of each Lorentzian (height, width and position) in succession. The initial peak position guesses are generated by linear interpolation between the size standard peaks. We assume that the width of individual peaks is a function of elution volume; we call this a ‘two-parameter peak-width model’ [Equation (1), Materials and methods section). Fast linear regression adjusts the peak heights to yield a refined set of initial guesses for the height, width and position of each peak. Bounded nonlinear least-squares iteratively adjusts the peak positions to no more than 30% of their width. We use a relative tolerance of 10–6 on the gradients of the objective function (difference between data and model) to determine convergence (diamond box, Figure 2D). CAFA flags data that do not converge. The peak areas are calculated from the best-fit peak positions and the width and height values derived from the peak model. Each experiment includes a ‘background’ trace, which refers to a primer extension reaction run on the unmodified RNA. Certain positions in the RNA cause the RT to stop, and we refer to these sites as ‘RT stops’. The background trace is also fit to identify such ‘RT stops’ for exclusion in the final analysis. We can fit 600 peaks in several seconds on a 2 GHz single-core processor with 1 Gb of RAM.
Validation of CAFA
Four assumptions underlie nucleic acid structural analysis by CAFA: (i) Peak position is unambiguously assigned against a concomitantly electrophoresed set of size standards; (ii) peak area is proportional to nucleic acid concentration; (iii) peak area is reproducible; and (iv) the peak profiles discerned from mapping experiments correlate with the structural features of the analyzed nucleic acid.
We addressed point 1 by separately analyzing the cDNA products produced by primer extension on the full-length unfolded Tetrahymena RNA in the presence of ddCTP and the cDNA derived from primer extension on the RNase T1 cleaved fragments of the same molecule. We analyzed these peak traces by CAFA and verified correspondence to the positions of guanosine (G) nucleotides in the RNA (Supplementary Figure 1). The excellent alignment of the cleavage product traces (blue) with the ladder (black) demonstrates that comparison with the ladder accurately assigns the initial peak positions for CAFA peak fitting. Analysis of corresponding peaks between the ladder and digest traces (for example G110 and position 300 on the ladder, Supplementary Figure 1) revealed that differences in peak position are ≤30% of the peak width. We use this 30% bound during CAFA peak fitting.
We demonstrate point 2 by analyzing a set of serially diluted samples of cDNA molecules (Supplementary Figure 2). The relationship between cDNA concentration and resolved peak area is linear within the dynamic range used for our analyses (R2 = 0.97, χ2 = 10–14) and is sufficient for accurate quantitative chemical mapping analysis. We confirmed linear fluorescence response by also establishing that the Mg2+ folding isotherms, derived from OH footprinting experiments, generated by CAFA recapitulate those obtained by GE and SAFA (Supplementary Figure 3) (23,39). We used 1 pmol of fluorophore-labeled primers (final concentration 0.05 μM) for our RT reactions to maintain the total amount of fluorophore-labeled cDNA within the estimated dynamic range.
To assess point 3, we compared the error in CAFA quantified data for a single ·OH mapping experiment that was divided into five aliquots with five ‘independent’ identical reactions each extended from two different primers. The aliquots of a single experiment run in separate capillaries either along a row or a column of the 96-well plate and therefore analyzed separately, deviate by <1% standard error compared with 12% error among the experimental replicates. Thus, CEQ separation and detection are not a major source of error in CAFA quantified data. We collected data on all eight capillaries of our CEQ-8000 sequencer. Our observation of a 12% error therefore encompasses any error that is the result of the experimental and analysis protocols. It is thus an upper estimate of the error of the technique.
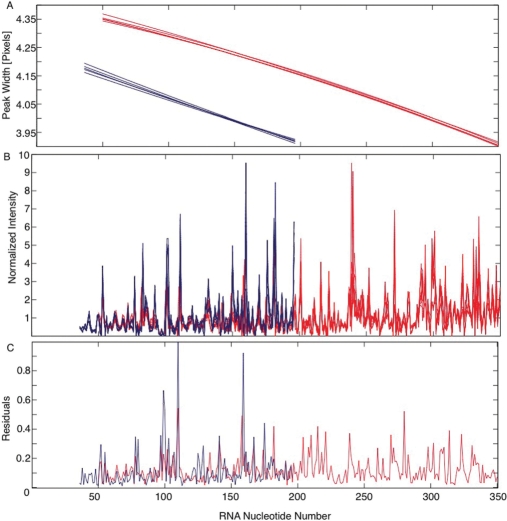
As seen for the independent reactions analyzed by different primers, longer cDNA fragments have narrower and better-separated peaks (Figure 3A). This behavior is opposite to that observed in GE. High reproducibility is observed in well-resolved regions (Figure 2B), while more variability is present, as expected, in peaks fit to poorly separated traces (Figure 2C). We analyzed the effect of peak resolution by comparing the peak fits to nucleotides obtained from the two different primers, which have different peak widths (Figure 3A; red and blue, respectively). Quantitative comparison of these data required the development of a novel normalization of the peak areas of each data set to compensate for variation in RT efficiency, fluorophore concentration and sample uptake (Figure 3B). The normalization approach incorporated into CAFA initially divides the peak areas by their mean and then performs an optimization that minimizes the pair wise differences between the traces. CAFA analyzed data are highly reproducible with the normalized standard deviations ≤12%. Although standard error is slightly higher in less well-separated regions of the trace the regions of abnormally large standard error (≥20%) correlate with strong sequence-dependent RT stops present in the cDNA.
Figure 3.
An assessment of CAFA reproducibility by analyzing five independent experiments with primers (red and blue) that anneal to the 3′ and middle of the RNA, respectively. (A) Peak widths (in pixels) as a function of RNA nucleotide number for each experimental replicate. (B) Normalized peak areas as a function of RNA nucleotide number for the five experimental repeats. (C) The standard error of each nucleotide's peak is calculated as the standard deviation divided by the mean.
Figure 4A compares histograms summarizing the distribution of the standard error of resolved peaks obtained using CAFA [excluding intrinsic RT stops but including data obtained for both primers (Figure 2)] and the identical sequence analyzed using 32P direct labeling, GE and SAFA (23). The mean error is much lower for the CAFA analysis of indirectly labeled RNA. It is noteworthy that in the CAFA analysis there is almost a complete absence of peaks with standard errors ≥0.3. Thus, CAFA analysis of CE separations is clearly superior to GE-based analysis.
Automatic exclusion of unreliable peaks
The ∼11% of the CAFA data with a standard error ≥20% is predominantly comprised of peaks at or near RT intrinsic stops. An automated algorithm identifies and marks for exclusion these unreliable data. Our method requires that a ‘background’ trace of cDNA transcribed from sample that is not chemically modified accompany the experimental samples. The algorithm flags peaks in the background trace whose area is 3-fold greater than the mean background. These peaks correspond to intrinsic stops in cDNA or degradation products in the sample traces.
We document the performance of our method for identifying high-error data by computing false and true-positive rates with respect to high-error peak identification (Figure 4B). We identify 9% of the high-error data if only background peaks are used (distance 0 on the ordinate of Figure 4B). The true-positive and false-positive rates increase to 62% and 16%, respectively if peaks within 1 nt of the background peaks are included. Both error rates increase as the distance from each background peak increases. Since a distance of one offers the best improvement of true-positives versus false positives (Figure 4B), CAFA automatically flags RT stops and excludes all peaks within one nucleotide when provided with a background cDNA trace. This procedure identifies and excludes a majority of the unreliable data without the need to carry out multiple [experimental] repeats.
Applications of CAFA
To illustrate the generality of CAFA and its value for structural characterization, we used it to structurally analyze several nucleic acids previously studied using 32P labeling, GE and SAFA (38,39,47,48). First, we compared the solvent accessible surface of the Tetrahymena group I intron derived from the ·OH reactivity profile to the values calculated from the crystal structure (Figure 5A) (49,50). We measured a correlation coefficient of 0.75 between the calculated (using the mean accessibility of the five sugar-carbon atoms) and experimentally determined accessibility profiles; the agreement between experiment and a prediction allows us to use ·OH mapping data as a filter for structural modeling (Jonikas, Radmer, Laederach, Altman, submitted for publication). The Mg2+ midpoint of several sites of protection for folding of the L-21 T. Thermophila group I intron are identical to those previously determined (Figure 5B and Supplementary Material Figure 3).
Figure 5.
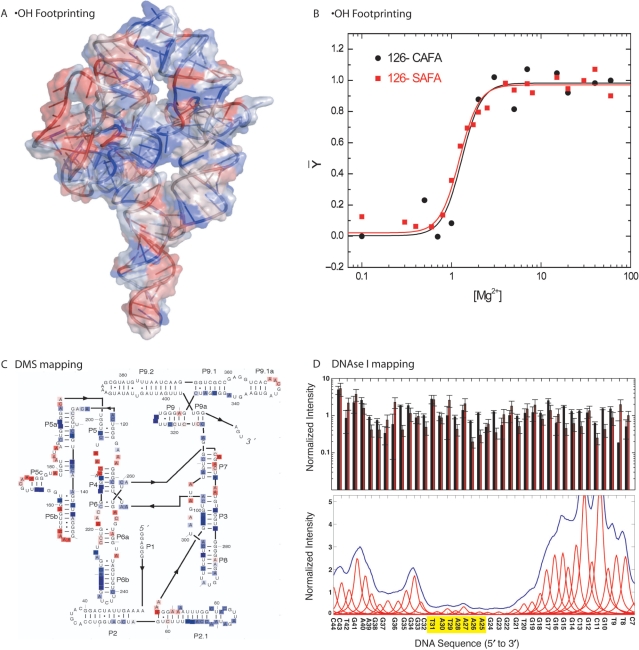
Different applications of CAFA to nucleic acid structural mapping. (A) Mapping of ·OH reactivity onto the crystal structure of the L-21 T. thermophila group I intron (45,46). Note that suppression (blue) and enhancement (red) of ·OH reactivity closely map the solvent accessibility of the structure. (B) Comparison of Mg2+-dependent folding isotherms of nucleotide 126 in the T. thermophila group I intron analyzed by SAFA (red) and CAFA (black) analysis. (C) DMS mapping of L-21 T. thermophila group I intron to determine its secondary structure performed in the presence of 100 mM KCl and no Mg2+. Nucleotides protected from DMS modification are boxed in blue. Those that are reactive are boxed in red. Raw data for the DMS experiments is plotted in Supplementary Material Figure 7. (D) Mapping by DNase I of the binding of TBP to a TATA Box present on a DNA duplex. In the top panel we illustrate the reproducibility of CAFA (black) relative to SAFA (red, 38) determined peak areas for the same DNA molecule without protein bound. In the bottom panel, we illustrate automated CAFA analysis of the bound DNA clearly identifying the TATA binding site (yellow).
We used CAFA to map the secondary structure of the Mg2+-free (10 M sodium cacodylate buffer with an additional 100 mM KCl) Tetrahymena group I intron. Secondary but not tertiary structure is present under these experimental conditions. DMS reactivity was used to probe for base pairing. Unpaired adenines and cytosines are methylated during a short incubation with DMS; the modifications terminate RT extension. Normalization of the data from a single experiment allows precise identification of the methylation-induced RT stops over the full length of the ribozyme (except for the sequence annealed to the primer). Ninety percent of the crystallographically determined base-paired nucleotides are identified with a 5% false-positive rate (Figure 5C, colored blue and red, respectively). We identify the base-paired residues by considering any residue base-paired that is 30% or more protected relative to the mean protection observed for the entire molecule. These data show that the pseudo-knot P3 helix is formed (51,52).
DNase I is often used to map the binding of proteins to specific sequences of duplex DNA. We used CAFA to analyze the interaction of the Saccharomyces cerevisiae TATA-binding protein (TBP) to a TATA Box sequence to which it specifically binds (38,53,54). DNA directly labeled by PCR was analyzed in the absence and presence of a saturating concentration of TBP (Figure 5C). CAFA analysis yields a DNase I footprint of TBP comparable to the one obtained with 32P-DNA and GE (Figure 5D and Supplementary Material Figure 6). The DNase I footprinting traces fit well despite the large disparity among the peak heights due to the nuclease sequence preferences. The utilization of the size ladder to provide initial peak positions, together with the peak width constraint allow CAFA to rapidly and accurately quantitate the DNase I traces. The examples shown in Figure 5 highlight the generality of CAFA with regard to the nature of the mapping probe and the phenomenon being investigated.
DISCUSSION
Quantitative analysis of CE separations of the products of chemical and enzymatic mapping (footprinting) by CAFA yields more results, more quickly with better precision compared to GE based methods. The quality and rapidity of data analysis enabled by CAFA reduces the problem of characterizing RNA solution structure to common practice. We release CAFA as free open-source software with the hope that it will stimulate quantitative study of nucleic acid structure and function as has our GE-based SAFA software (23,25,28,55). Furthermore, the fact that this analysis can be run on a standard instrument could lead to core facilities offering CAFA analysis in addition to sequencing.
The experimental versatility and simplicity of quantitative solution mapping enabled by CAFA, melded with indirect labeling, allows very long lengths of nucleic acids to be efficiently interrogated with single nucleotide resolution. Combined with in vivo chemical mapping protocols (8), CAFA provides an excellent platform for structural characterization of nucleic acids as well as nucleic-acid protein interactions inside the cell. The combination of automated analysis and the high-throughput achieved with multicapillary machines enables genomic scale studies now to be undertaken. Furthermore, the throughput of the technique could be further increased by simultaneously running samples with different colored dyes within a single capillary. The fundamental fitting algorithms implemented in CAFA are compatible with this approach, although the costs of synthesizing additional colored primers may not always justify extending the approach in this way.
CAFA accommodates the biggest technical hurdle to indirect labeling by RT primer extension; sequence-specific pauses and stops. While our experimental protocols minimize the frequency of these stops, their predictability from a ‘background trace’, makes it possible to flag these sites of low reliability data for exclusion in subsequent analysis (Figure 4B). The ability to accurately exclude erroneous data early in the analysis procedure is critical to high-throughput data analysis. Our approach is conservative in that we choose to exclude more data (16% false positives, Figure 4B) to ensure that the remaining data are accurate.
CAFA is a standalone application with a graphical user interface (Supplementary Figure 4) that accommodates a variety of experimental protocols. The software takes a raw CE-trace, fits a peak model to it and thus quantitates the relative amount of each mapping reaction product (Figure 1). The output peak areas are associated with nucleotide numbers corresponding to the DNA reference peaks of the size standard ladder; these numbers are then related to either the source from which the cDNA was transcribed (indirect labeling) or the directly labeled sample. CAFA and its documentation show how data can be associated with the sample sequence based on concomitant analysis of the appropriate sequence reference ladders. Postprocessing tools are provided to facilitate this task. We observed excellent agreement between the Beckman size ladder and a T1 digest and are confident in the accurate assignment of sequence to peaks (Supplementary Figure 1). However, it is possible that systemic shifts in sequence assignment could occur for molecules with extreme GC content. For this reason, we recommend calibration of the ladder against the RNA upon initiation of a new study to identify systematic bias and allow for its correction.
The three applications of CAFA we demonstrated are: determination of nucleotide solvent accessibility with ·OH footprinting, secondary structure mapping using DMS, and protein-binding site identification on DNA. Common to each problem is the need to accurately determine the peak areas corresponding to each nucleotide, which is at the heart of the CAFA algorithm. Given the experimental versatility of RT-based indirect labeling and the availability of the CAFA software, CE appears poised to replace GE as the method of choice for the high-throughput analysis of nucleic acid structure as it already has for DNA sequencing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We wish to thank Kevin Wilkinson, Rick Russell, Dan Herschlag and Kevin Weeks for advice and assistance with experiments. We also wish to thank Joerg Schlatterer and Elizabeth Jamison for help with the experimental protocols. This work was funded by National Institutes of Health Grants P01-GM66275, U54-GM072970 (National Centers for Biomedical Computation), P41-EB0001979, and K99/R00 (GM079953) award to A.L. and the NSF 0443508 for the RNA Ontology Consortium. Funding to pay the Open Access publication charges for this article was provided by the Wadsworth Center, Albany, NY.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laederach A, Chan JM, Schwartzman A, Willgohs E, Altman RB. Coplanar and coaxial orientations of RNA bases and helices. RNA. 2007;13:643–650. doi: 10.1261/rna.381407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flam F. Hints of a language in junk DNA. Science. 1994;266:1320. doi: 10.1126/science.7973718. [DOI] [PubMed] [Google Scholar]
- 5.Duarte CM, Wadley LM, Pyle AM. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. Nucleic Acids Res. 2003;31:4755–4761. doi: 10.1093/nar/gkg682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culver GM. Assembly of the 30S ribosomal subunit. Biopolymers. 2003;68:234–249. doi: 10.1002/bip.10221. [DOI] [PubMed] [Google Scholar]
- 7.Ashraf SI, Kunes S. A trace of silence: memory and microRNA at the synapse. Curr. Opin. Neurobiol. 2006;16:535–539. doi: 10.1016/j.conb.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Adilakshmi T, Lease RA, Woodson SA. Hydroxyl radical footprinting in vivo: mapping macromolecular structures with synchrotron radiation. Nucleic Acids Res. 2006;34:e64. doi: 10.1093/nar/gkl291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lease RA, Adilakshmi T, Heilman-Miller S, Woodson SA. Communication between RNA folding domains revealed by folding of circularly permuted ribozymes. J. Mol. Biol. 2007;373:197–210. doi: 10.1016/j.jmb.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elnitski LL, Shah P, Moreland RT, Umayam L, Wolfsberg TG, Baxevanis AD. The ENCODEdb portal: simplified access to ENCODE Consortium data. Genome Res. 2007;17:954–959. doi: 10.1101/gr.5582207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinstock GM. ENCODE: more genomic empowerment. Genome Res. 2007;17:667–668. doi: 10.1101/gr.6534207. [DOI] [PubMed] [Google Scholar]
- 12.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5′ domain. J. Mol. Biol. 2005;351:508–519. doi: 10.1016/j.jmb.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Brenowitz M, Senear DF, Shea MA, Ackers GK. ‘Footprint’ titrations yield valid thermodynamic isotherms. Proc. Natl Acad. Sci. USA. 1986;83:8462–8466. doi: 10.1073/pnas.83.22.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenowitz M, Senear DF, Shea MA, Ackers GK. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 15.Das R, Kwok LW, Millett IS, Bai Y, Mills TT, Jacob J, Maskel GS, Seifert S, Mochrie SG, Thiyagarajan P, et al. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J. Mol. Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- 16.Gross P, Arrowsmith CH, Macgregor R.B., Jr. Hydroxyl radical footprinting of DNA complexes of the ets domain of PU.1 and its comparison to the crystal structure. Biochemistry. 1998;37:5129–5135. doi: 10.1021/bi972591k. [DOI] [PubMed] [Google Scholar]
- 17.Loizos N. Mapping protein-ligand interactions by hydroxyl-radical protein footprinting. Methods Mol. Biol. 2004;261:199–210. doi: 10.1385/1-59259-762-9:199. [DOI] [PubMed] [Google Scholar]
- 18.Silverman JA, Harbury PB. Rapid mapping of protein structure, interactions, and ligand binding by misincorporation proton-alkyl exchange. J. Biol. Chem. 2002;277:30968–30975. doi: 10.1074/jbc.M203172200. [DOI] [PubMed] [Google Scholar]
- 19.Brenowitz M, Chance MR, Dhavan G, Takamoto K. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical ‘footprinting’. Curr. Opin. Struct. Biol. 2002;12:648–653. doi: 10.1016/s0959-440x(02)00366-4. [DOI] [PubMed] [Google Scholar]
- 20.Sclavi B, Woodson S, Sullivan M, Chance M, Brenowitz M. Following the folding of RNA with time-resolved synchrotron X-ray footprinting. Methods Enzymol. 1998;295:379–402. doi: 10.1016/s0076-6879(98)95050-9. [DOI] [PubMed] [Google Scholar]
- 21.Strahs D, Brenowitz M. DNA conformational changes associated with the cooperative binding of cI-repressor of bacteriophage lambda to OR. J. Mol. Biol. 1994;244:494–510. doi: 10.1006/jmbi.1994.1748. [DOI] [PubMed] [Google Scholar]
- 22.Gross P, Yee AA, Arrowsmith CH, Macgregor R.B., Jr. Quantitative hydroxyl radical footprinting reveals cooperative interactions between DNA-binding subdomains of PU.1 and IRF4. Biochemistry. 1998;37:9802–9811. doi: 10.1021/bi9731448. [DOI] [PubMed] [Google Scholar]
- 23.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takamoto K, Chance MR, Brenowitz M. Semi-automated, single-band peak-fitting analysis of hydroxyl radical nucleic acid footprint autoradiograms for the quantitative analysis of transitions. Nucleic Acids Res. 2004;32:E119. doi: 10.1093/nar/gnh117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badorrek CS, Weeks KM. RNA flexibility in the dimerization domain of a gamma retrovirus. Nat. Chem. Biol. 2005;1:104–111. doi: 10.1038/nchembio712. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 27.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNA(Asp) transcripts. J. Am. Chem. Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 29.Dolnik V. DNA sequencing by capillary electrophoresis (review) J. Biochem. Biophys. Methods. 1999;41:103–119. doi: 10.1016/s0165-022x(99)00041-x. [DOI] [PubMed] [Google Scholar]
- 30.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 31.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 32.Shadle SE, Allen DF, Guo H, Pogozelski WK, Bashkin JS, Tullius D. Quantitative analysis of electrophoresis data: novel curve fitting methodology and its application to the determination of a protein-DNA binding constant. Nucleic Acids Res. 1997;25:850–860. doi: 10.1093/nar/25.4.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Dewey TG. Cluster analysis of dynamic parameters of gene expression. J. Bioinform. Comput. Biol. 2003;1:447–458. doi: 10.1142/s0219720003000307. [DOI] [PubMed] [Google Scholar]
- 34.Masseroli M, Martucci D, Pinciroli F. GFINDer: Genome Function INtegrated Discoverer through dynamic annotation, statistical analysis, and mining. Nucleic Acids Res. 2004;32:300. doi: 10.1093/nar/gkh432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Celander DW, Cech TR. Iron(II)-ethylenediaminetetraacetic acid catalyzed cleavage of RNA and DNA oligonucleotides: similar reactivity toward single- and double-stranded forms. Biochemistry. 1990;29:1355–1361. doi: 10.1021/bi00458a001. [DOI] [PubMed] [Google Scholar]
- 36.Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 37.Henegariu O, Bray-Ward P, Ward DC. Custom fluorescent-nucleotide synthesis as an alternative method for nucleic acid labeling. Nat. Biotechnol. 2000;18:345–348. doi: 10.1038/73815. [DOI] [PubMed] [Google Scholar]
- 38.Sprouse RO, Brenowitz M, Auble DT. Snf2/Swi2-related ATPase Mot1 drives displacement of TATA-binding protein by gripping DNA. EMBO J. 2006;25:1492–1504. doi: 10.1038/sj.emboj.7601050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shcherbakova I, Gupta S, Chance MR, Brenowitz M. Monovalent ion-mediated folding of the Tetrahymena thermophila ribozyme. J. Mol. Biol. 2004;342:1431–1442. doi: 10.1016/j.jmb.2004.07.092. [DOI] [PubMed] [Google Scholar]
- 40.Huang Z, Szostak JW. A simple method for 3′-labeling of RNA. Nucleic Acids Res. 1996;24:4360–4361. doi: 10.1093/nar/24.21.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coleman TF, Li Y. An interior, trust region approach for nonlinear minimization subject to bounds. SIAM J. Optim. 1996;6:418–445. [Google Scholar]
- 42.Lempereur L, Nicoloso M, Riehl N, Ehresmann C, Ehresmann B, Bachellerie JP. Conformation of yeast 18S rRNA. Direct chemical probing of the 5′ domain in ribosomal subunits and in deproteinized RNA by reverse transcriptase mapping of dimethyl sulfate-accessible. Nucleic Acids Res. 1985;13:8339–8357. doi: 10.1093/nar/13.23.8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 44.Sclavi B, Woodson S, Sullivan M, Chance MR, Brenowitz M. Time-resolved synchrotron X-ray ‘footprinting’, a new approach to the study of nucleic acid structure and function: application to protein-DNA interactions and RNA folding. J. Mol. Biol. 1997;266:144–159. doi: 10.1006/jmbi.1996.0775. [DOI] [PubMed] [Google Scholar]
- 45.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 46.Shcherbakova I, Mitra S, Beer RH, Brenowitz M. Fast Fenton footprinting: a laboratory-based method for the time-resolved analysis of DNA, RNA and proteins. Nucleic Acids Res. 2006;34:e48. doi: 10.1093/nar/gkl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laederach A, Shcherbakova I, Liang M, Brenowitz M, Altman RB. Local kinetic measures of macromolecular structure reveal partitioning among multiple parallel pathways from the earliest steps in the folding of a large RNA molecule. J. Mol. Biol. 2006;358:1179–1190. doi: 10.1016/j.jmb.2006.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laederach A, Shcherbakova I, Jonikas MA, Altman RB, Brenowitz M. Distinct contribution of electrostatics, initial conformational ensemble, and macromolecular stability in RNA folding. Proc. Natl Acad. Sci. USA. 2007;104:7045–7050. doi: 10.1073/pnas.0608765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo F, Gooding AR, Cech TR. Comparison of crystal structure interactions and thermodynamics for stabilizing mutations in the Tetrahymena ribozyme. RNA. 2006;12:387–395. doi: 10.1261/rna.2198206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol. Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Rook MS, Treiber DK, Williamson JR. Fast folding mutants of the Tetrahymena group I ribozyme reveal a rugged folding energy landscape. J. Mol. Biol. 1998;281:609–620. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- 52.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J. Mol. Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 53.Gupta S, Cheng, Mollah AK, Jamison E, Morris S, Chance MR, Khrapunov S, Brenowitz M. DNA and protein footprinting analysis of the modulation of DNA binding by the N-terminal domain of the Saccharomyces cerevisiae TATA binding protein. Biochemistry. 2007;46:9886–9898. doi: 10.1021/bi7003608. [DOI] [PubMed] [Google Scholar]
- 54.Khrapunov S, Brenowitz M. Influence of the N-terminal domain and divalent cations on self-association and DNA binding by the Saccharomyces cerevisiae TATA binding protein. Biochemistry. 2007;46:4876–4887. doi: 10.1021/bi061651w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vicens Q, Gooding AR, Laederach A, Cech TR. Local RNA structural changes induced by crystallization are revealed by SHAPE. RNA. 2007;13:536–548. doi: 10.1261/rna.400207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.