Abstract
Cellular proteins containing BTB and kelch domains have been shown to function as adapters for the recruitment of substrates to cullin-3-based ubiquitin ligases. Poxviruses are the only family of viruses known to encode multiple BTB/kelch proteins, suggesting that poxviruses may modulate the ubiquitin pathway through interaction with cullin-3. Ectromelia virus encodes four BTB/kelch proteins and one BTB-only protein. Here we demonstrate that two of the ectromelia virus encoded BTB/kelch proteins, EVM150 and EVM167, interacted with cullin-3. Similar to cellular BTB proteins, the BTB domain of EVM150 and EVM167 was necessary and sufficient for cullin-3 interaction. During infection, EVM150 and EVM167 localized to discrete cytoplasmic regions, which co-localized with cullin-3. Furthermore, EVM150 and EVM167 co-localized and interacted with conjugated ubiquitin, as demonstrated by confocal microscopy and co-immunoprecipitation. Our findings suggest that the ectromelia virus encoded BTB/kelch proteins, EVM150 and EVM167, interact with cullin-3 potentially functioning to recruit unidentified substrates for ubiquitination.
Keywords: poxvirus, ectromelia, cullin-3, ubiquitination, BTB/kelch
Introduction
In order to survive within the host, viruses have evolved an array of sophisticated mechanisms to evade the host anti-viral immune response (Finlay and McFadden, 2006; Tortorella et al., 2000). Typically, viral immune evasion mechanisms are directed towards the deregulation of anti-viral pathways such as apoptosis, antigen presentation, and cytokine activity (Finlay and McFadden, 2006; Tortorella et al., 2000). More recently, however, it has become clear that many viruses also regulate the ubiquitin protein modification pathway in order to ensure their survival (Banks et al., 2003; Shackelford and Pagano, 2005). Protein ubiquitination, which has emerged as a major mechanism for the control of protein degradation, regulates important cellular processes such as cell cycle, DNA repair, signal transduction, transcription, and antigen presentation (Pickart, 2001; Weissman, 2001). Ubiquitin is an evolutionarily conserved 76 amino acid protein that is covalently attached to proteins post-translationally (Pickart, 2001; Weissman, 2001). The addition of ubiquitin to target proteins results in either mono-ubiquitination, leading to the modification of protein function, or poly-ubiquitination, which typically targets proteins for degradation via the 26S proteasome (Pickart, 2001; Weissman, 2001). The specific addition of ubiquitin to protein substrates is a well-defined, multi-step process that requires a ubiquitin activation enzyme (E1), ubiquitin conjugating enzymes (E2), and ubiquitin ligases (E3) (Pickart, 2001; Weissman, 2001). Once activated, ubiquitin is transferred to a ubiquitin conjugating enzyme, and a ubiquitin ligase catalyses the transfer of ubiquitin from the conjugating enzyme to the substrate protein (Hatakeyama and Nakayama, 2003; Huibregtse et al., 1995; Joazeiro and Weissman, 2000). The human genome encodes an abundance of ubiquitin ligases, which function to provide substrate specificity through the recognition of target proteins and the subsequent conjugation of ubiquitin.
The multi-protein ubiquitin ligases include a family of conserved proteins referred to as cullins (Kipreos et al., 1996; Mathias et al., 1996). Cullin-based ubiquitin ligases contain one of seven cullin family members, cullin 1, 2, 3, 4a, 4b, 5 and 7. The cullin protein functions as a molecular scaffold for the RING-finger containing protein, Roc1, which facilitates the transfer of ubiquitin to a substrate protein (Ohta et al., 1999a). Substrate recruitment to cullin-based ubiquitin ligases occurs through specific linker and adapter proteins (Cardozo and Pagano, 2004; Petroski and Deshaies, 2005). Recently, proteins containing BTB domains, also known as Bric-a-Brac, Tramtrack, Broad complex or POZ domains, were shown to function as substrate specific adapters for the cullin-3 ubiquitin ligase complex (Furukawa et al., 2003; Geyer et al., 2003; Pintard et al., 2003b; Xu et al., 2003). Unlike other members of the cullin family which require separate linker and adapter proteins to recruit substrates to the ubiquitin ligase complex, cullin-3 substrates are recruited through a single protein containing a BTB domain as a linker to cullin-3 and a second protein:protein interaction domain, such as Kelch or MATH domain which interact with substrates (Furukawa et al., 2003; Geyer et al., 2003; Pintard et al., 2003b; Xu et al., 2003). The human genome encodes over 200 BTB domain-containing proteins, indicative of a wide range of potential substrate adapters for cullin-3-based ubiquitin ligases (http://www.sanger.ac.uk/Software/Pfam) (Bateman et al., 2000).
The sophisticated mechanisms utilized by viruses to counteract host anti-viral responses have become increasingly apparent, particularly when examining virus modulation of the ubiquitin pathway. Recent research has demonstrated that a number of viruses encode their own ubiquitin ligases. For example, herpes simplex virus encodes ICP0, a RING-finger containing protein that is required for lytic infection and reactivation from latency (Boutell et al., 2002; Everett, 2000). Members of the gamma-herpesvirus family regulate the levels of cell surface molecules, such as MHC class I, ICAM-1 and B7, through the expression of a membrane bound ubiquitin ligase (Coscoy and Ganem, 2001; Ishido et al., 2000; Lorenzo et al., 2002; Stevenson et al., 2000). In a similar fashion, M153R, encoded by myxoma virus, mediates the degradation of MHC class I and CD4 molecules (Guerin et al., 2002; Mansouri et al., 2003). Additionally, multiple members of the poxvirus family encode RING-finger containing proteins that are predicted to function as ubiquitin ligases (Afonso et al., 2000; Huang et al., 2004; Nerenberg et al., 2005). We recently demonstrated that the poxviral RING-finger containing protein p28 functions as a ubiquitin ligase mediating the formation of polyubiquitin conjugates (Huang et al., 2004; Nerenberg et al., 2005). In addition to encoding ubiquitin ligases, viruses have also evolved mechanisms to redirect specific substrates to cellular cullin-based ubiquitin ligases (Barry and Fruh, 2006). For example, the HIV protein Vif promotes APOBEC3G degradation through a cullin-5 based ubiquitin ligase (Yu et al., 2003). Similarly, paramyxovirus protein V prevents interferon signaling by degrading STAT3 through a cullin-4a-based ubiquitin ligase, while the adenovirus E4orf6 protein recruits p53 to a cullin-5-based ubiquitin ligase inducing p53 degradation (Querido et al., 2001; Ulane and Horvath, 2002). In addition, members of the poxvirus family encode multiple BTB/kelch proteins, suggesting that like their cellular counterparts the viral BTB/kelch proteins may function as cullin-3 substrate specific adapters (Shchelkunov et al., 2002; Totmenin et al., 2002).
Poxviruses are currently the only viruses known to encode BTB/kelch proteins. Although, the presence of BTB/kelch domains were first documented in poxviruses more than 10 years ago, the specific function of the poxvirus BTB/kelch proteins is currently undefined (Koonin et al., 1992; Senkevich et al., 1993). Multiple BTB/kelch proteins are present in a wide range of poxviruses and genome analysis has revealed the presence of over 100 BTB/kelch proteins, which are subdivided into six BTB/kelch protein families (Ehlers et al., 2002; Shchelkunov et al., 2002). Proteins representing four of the BTB/kelch families are encoded within the genome of ectromelia virus strain Moscow, the causative agent of mousepox (Chen et al., 2003). Our studies have shown that at least two of the ectromelia virus encoded BTB/kelch proteins, EVM150 and EVM167, interact with cullin-3 and that the BTB domain is both necessary and sufficient for cullin-3 interaction. EVM150 and EVM167 co-localize with both cullin-3 and conjugated ubiquitin to discrete punctate regions within the cytoplasm of infected cells and EVM150 and EVM167 co-precipitate with conjugated ubiquitin. Our observations, which describe the first function for EVM150 and EVM167, suggest that the BTB/kelch proteins function by modulating the ubiquitin pathway through an interaction with cullin-3-based ubiquitin ligases.
Results
Ectromelia virus encodes four BTB/kelch proteins and one BTB-only protein
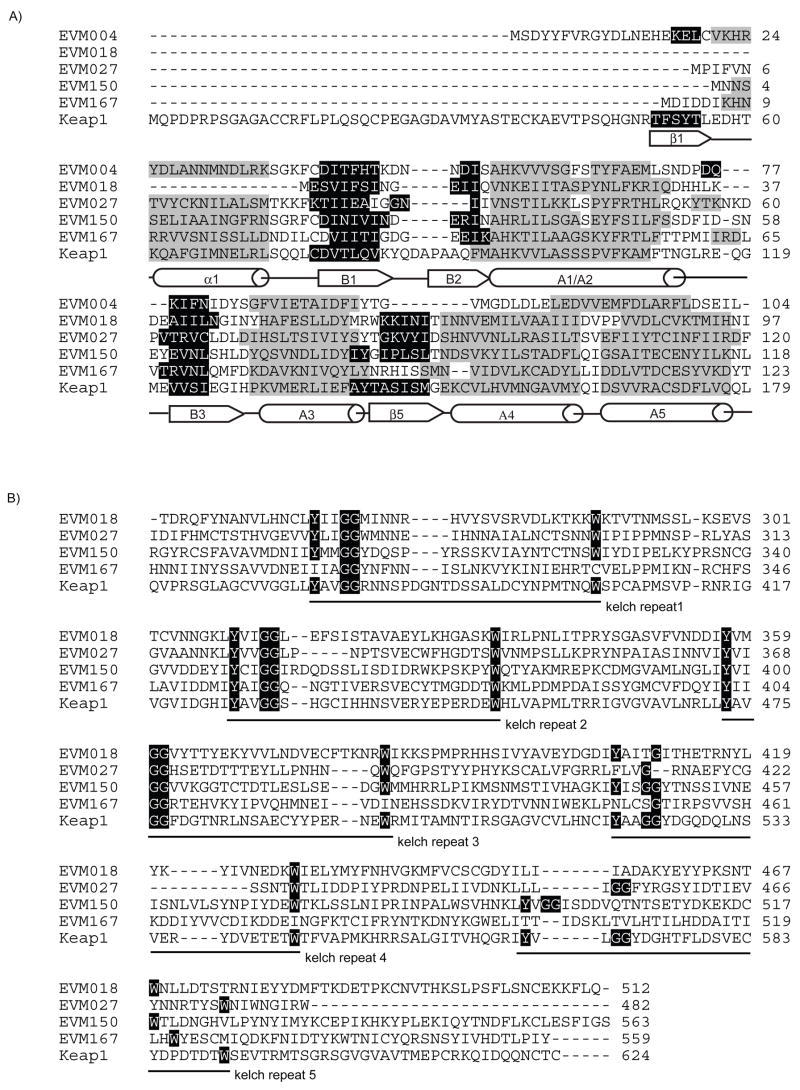
Members of the poxvirus family contain multiple genes encoding proteins with both BTB and kelch domains (Shchelkunov et al., 2002; Totmenin et al., 2002). Although the poxvirus BTB/kelch genes were identified more than 10 years ago, the specific function of these proteins during infection remains unknown (Koonin et al., 1992; Senkevich et al., 1993). Ectromelia virus, the causative agent of mousepox, is predicted to express four BTB/kelch proteins, encoded by EVM018, EVM027, EVM150, and EVM167, and one BTB-only containing protein, EVM004 (Chen et al., 2003; Esteban and Buller, 2005). All the ectromelia virus BTB-containing genes are located within regions of the genome that typically encode proteins important for modulating the host immune response and cellular signaling (Chen et al., 2003). Although the overall sequence similarity is typically low between BTB family members, BTB domains share a conserved secondary structure composed of a series of alpha helices and beta sheets (Stogios et al., 2005). Figure 1A shows the five BTB containing proteins encoded by ectromelia virus and the BTB domain of Keap1, a cellular BTB/kelch-containing protein (Stogios et al., 2005). The predicted secondary structure was overlayed onto the sequences and similar to Keap1, EVM004, EVM018, EVM027, EVM150, and EVM167 are predicted to form a core BTB fold composed of alpha helices and beta sheets (Stogios et al., 2005) (Fig. 1A). Additionally, EVM018, EVM027, EVM150 and EVM167 also contain a series of evolutionarily conserved C-terminal kelch repeats of 44 to 56 amino acids in size that are characterized by conserved double glycine residues and aromatic residues (Adams et al., 2000). Sequence alignments indicate that EVM018 and EVM167 contain three kelch repeats, while EVM027 and EVM150 contain four and five kelch repeats, respectively (Fig. 1B and Fig. 2A).
Figure 1.
Proteins encoded by ectromelia virus contain BTB and kelch domains. (A) Amino acid sequence of BTB domains of EVM004, EVM018, EVM027, EVM150, EVM167 and the BTB domain of Keap1. The ectromelia virus-encoded BTB proteins retain the predicted BTB fold containing a conserved series of alpha helices (grey) and beta sheets (black). Secondary structures were predicted using 3D-PSSM and PYRE. (B) Alignment of the kelch domain of EVM018, EVM027, EVM150 and EVM167 with the kelch domain of Keap1. Alignments were generated using ClustralW. EVM018 and EVM167 are predicted to contain 3 complete kelch repeats while EVM027 and EVM150 are predicted to contain 4 and 5 kelch repeats, respectively.
Figure 2.
EVM004, EVM018, EVM027, EVM150 and EVM167 are transcribed early during ectromelia virus infection. (A) Schematic representation of the ectromelia virus BTB/kelch proteins EVM018, EVM027, EVM150, EVM167 and the BTB-only containing protein EVM004. (B) CV-1 cells were infected with ectromelia virus at an MOI 5 in the presence and absence of cytosine arabinoside (AraC) and RNA isolated at the indicated times. RNA was subjected to RT-PCR followed by PCR using gene specific primers to detect transcripts for EVM004, EVM018, EVM027, EVM150, EVM167, EVM058 and cellular β-actin.
To determine if the ectromelia virus BTB-containing genes were transcribed during infection we examined mRNA levels using RT-PCR. After infecting CV-1 cells with ectromelia virus strain Moscow, RNA was extracted at the indicated time points and subjected to RT-PCR using primers specific for each BTB domain. In all cases, transcripts were detected at 4, 12 and 24 hours post infection (Fig. 2B). In the presence of cytosine arabinoside, an inhibitor of DNA synthesis and, therefore, an inhibitor of poxvirus late gene expression, transcripts were still detected for EVM004, EVM018, EVM027, EVM150, and EVM167, indicating that the BTB-containing genes were transcribed early during infection (Fig. 2B). In contrast, transcripts for EVM058, which encodes a homologue to the vaccinia virus I5L gene, a predicted late gene, were not detected in presence of cytosine arabinoside (Fig. 2B).
EVM150 and EVM167 interact with cullin-3
Cellular proteins containing both BTB and kelch domains have been shown to function as adapter proteins for cullin-3-based ubiquitin ligases (Furukawa et al., 2003; Geyer et al., 2003; Pintard et al., 2003b; Xu et al., 2003). The BTB domain interacts with cullin-3, while the kelch domain specifically recruits proteins to the cullin-3 complex for ubiquitination (Fig. 3A). EVM018, EVM027, EVM150, and EVM167 all contain N-terminal BTB domains followed by a series of conserved kelch repeats, suggesting that the ectromelia virus BTB/kelch proteins potentially function as substrate specific adapters for cullin-3-based ubiquitin ligase complexes.
Figure 3.

EVM150 and EVM167 interact with cullin-3. (A) A schematic representation of cullin-3 based ubiquitin ligase. A single protein containing both BTB and kelch domains serves as the substrate specific adaptor for cullin-3 based ubiquitin ligases. The BTB domain interacts with cullin-3 and the kelch domain recruits substrates for ubiquitination. (B) HEK293T cells were co-transfected with constructs to express EGFP-tagged versions of FPV039BH1, EVM018, EVM027, EVM150, EVM167 and Flag-cullin-3. Lysates were western blotted with anti-Flag to indicate expression of Flag-cullin-3. Immunoprecipitation and western blotting with an antibody specific for EGFP demonstrated expression of FPV039BH1, EVM018, EVM150 and EVM167. Interaction between EVM150 and EVM167 and cullin-3 was demonstrated by western blotting the immunoprecipitations with anti-Flag. (C) Reciprocal immunoprecipitations were performed with anti-Flag to confirm the interaction between EVM150, EVM167 and cullin-3. Western blotting of lysates with anti-EGFP demonstrated expression of FPV039BH1, EVM150 and EVM167. Immunoprecpitation with anti-Flag demonstrated expression of cullin-3 in the co-transfected samples. Interaction between EVM150, EVM167 and cullin-3 was demonstrated by subjecting the immunoprecipitations to western blotting with anti-EGFP.
To determine whether the ectromelia virus BTB/kelch proteins interact with cullin-3, we generated N-terminal EGFP-tagged versions of each full-length gene. HEK293T cells were co-transfected with pcDNA-Flag-cullin-3 and either pEGFP-EVM018, pEGFP-EVM027, pEGFP-EVM150, pEGFP-EVM167, or pEGFP-FPV039BH1, a cytoplasmic version of the fowlpox virus Bcl-2 protein, as a negative control. Interactions between cullin-3 and the virus encoded BTB/kelch proteins were then assessed by co-immunoprecipitation. Western blotting of the cellular lysates with an anti-Flag antibody indicated that Flag-cullin-3 was expressed in all co-transfected cells (Fig. 3B). Immunoprecipitation and western blotting with an antibody specific for EGFP indicated expression of EGFP-FPV039BH1, EGFP-EVM018, EGFP-EVM027, EGFP-EVM150, and EGFP-EVM167 (Fig. 3B). The subsequent western blotting of the immunoprecipitations with an anti-Flag antibody demonstrated a clear interaction between Flag-cullin-3 and EVM150 and EVM167 (Fig. 3B). However, no interaction was observed between Flag-cullin-3 and EVM018, EVM027 or FPV039BH1 (Fig. 3B). To confirm the interactions between cullin-3 and EVM150 and EVM167, we performed reciprocal immunoprecipitations with the anti-Flag antibody (Fig. 3C). Expression of EGFP-EVM150, EGFP-EVM167, and EGFP-FPV039BH1 was detected in the lysates by blotting with anti-EGFP (Fig. 3C). Immunoprecipitation of cell lysates with anti-Flag and western blotting with anti-Flag or anti-EGFP demonstrated precipitation of Flag-tagged cullin-3 and the co-immunoprecipitation of EVM150 and EVM167, confirming the previous observation that EVM150 and EVM167 interact with cullin-3 (Fig. 3C).
The BTB domain of EVM150 and EVM167 is required for interaction with cullin-3
Cellular BTB-containing proteins interact with cullin-3 through the BTB domain (Furukawa et al., 2003; Geyer et al., 2003; Pintard et al., 2003b; Xu et al., 2003). Therefore, to determine if the BTB domains of EVM150 and EVM167 are responsible for binding cullin-3, we assessed the ability of the BTB domains of EVM150 and EVM167 to interact with cullin-3. We generated EGFP-tagged versions of the BTB domain that included the first 118 amino acids of EVM150 and the first 123 amino acids of EVM167. HEK293T cells were co-transfected with pEGFP-EVM150BTB, pEGFP-EVM167BTB, or pEGFP-FPV039BH1 and pcDNA-Flag-cullin-3. Western blotting of the cellular lysates with anti-Flag antibody indicated that Flag-cullin-3 was expressed (Fig. 4A). Immunoprecipitation with anti-EGFP precipitated approximately equal levels of EGFP-FPV039BH1, EGFP-EVM150BTB, and EGFP-EVM167BTB (Fig. 4A). Cell lysates were subjected to immunoprecipitation with an anti-EGFP antibody and western blotted with anti-Flag to detect Flag-cullin-3. A clear interaction between EGFP-EVM150BTB, EGFP-EVM167BTB and Flag-cullin-3, but not EGFP-FPV039BH1, was detected, establishing that the BTB domains were sufficient for interaction with cullin-3 (Fig. 4A). These observations were confirmed by performing reciprocal immunoprecipitations using an anti-Flag antibody, again demonstrating EVM150BTB and EVM167BTB interaction with cullin-3 (Fig. 4B). Furthermore, co-transfection with the EGFP-tagged versions of the kelch domains of EVM150 and EVM167 with Flag-cullin-3 demonstrated that EGFP-EVM150kelch and EGFP-EVM167kelch were unable to interact with cullin-3 (Fig. 4C). These studies established that the kelch domains alone are unable to interact with cullin-3 and that the BTB domains are both necessary and sufficient for interaction with cullin-3 (Fig. 4C).
Figure 4.
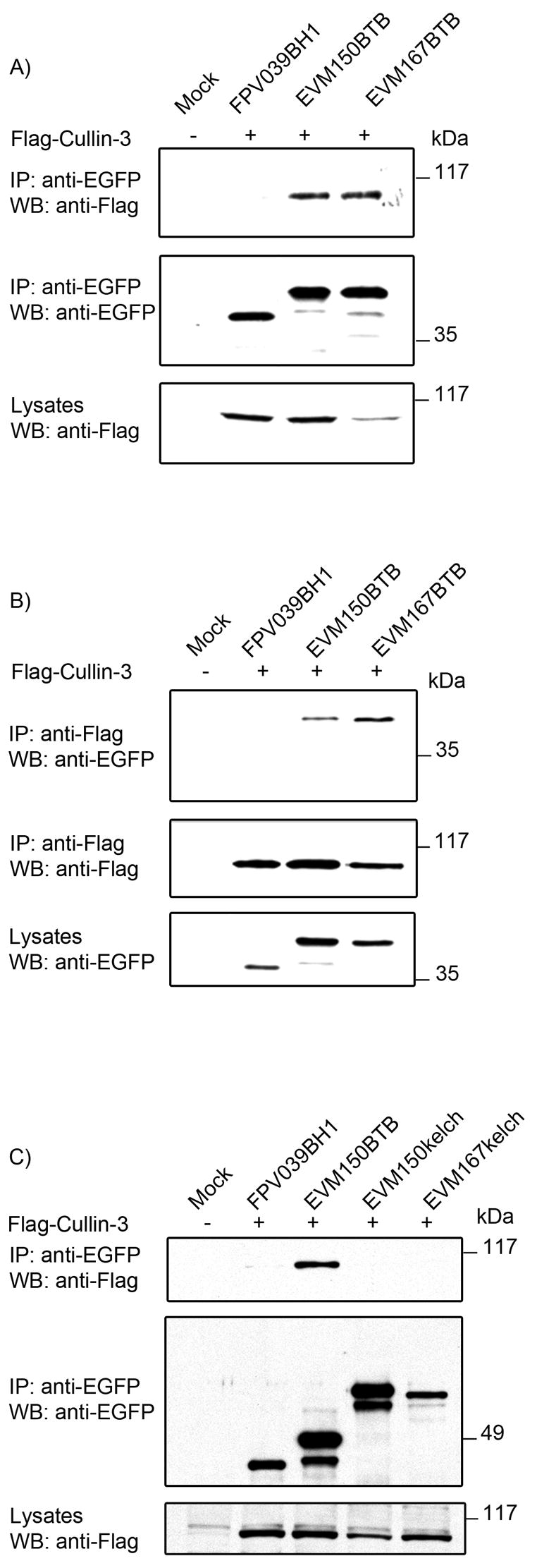
The BTB domain of EVM150 and EVM167 is required for interaction with cullin-3. (A) HEK293T cells were co-transfected with constructs to express EGFP-tagged BTB-only domains of EVM150 and EVM167 and co-transfected with Flag-cullin-3 and interaction assessed by co-immunoprecipitation. Westen blotting of lysates with anti-Flag indicated expression of Flag-cullin-3. Immunoprecipitation and western blotting with an antibody specific for EGFP demonstrated expression of FPV039BH1, and the BTB-only domains of EVM150 and EVM167. Interaction between the BTB domains of EVM150 and EVM167 and cullin-3 was demonstrated by subjecting the immunoprecipitations to western blotting with anti-Flag. (B) To confirm interaction between the BTB domains of EVM150 and EVM167 and cullin-3 reciprocal immunoprecipitations were performed with anti-Flag. Expression of FPV039BH1, EVM150BTB and EVM167BTB was assessed by western blotting lysates with anti-EGFP. Immunoprecpitation with anti-Flag demonstrated expression of cullin-3 in the co-transfected samples. Interaction between the BTB domains of EVM150 and EVM167 with cullin-3 was demonstrated by subjecting the immunoprecipitations to western blotting with anti-EGFP. (C) The kelch domains of EVM 150 and EVM167 do not interact with cullin-3. HEK293T cells were transfected with constructs to express EGFP-tagged kelch-only domains of EVM150, EVM167 and as a positive control the BTB-only domain of EVM150. Cells were co-transfected with Flag-cullin-3 and interaction assessed by co-immunoprecipitation. Lysates were subjected to western blotting with anti-Flag to indicate expression of Flag-cullin-3. Immunoprecipitation and western blotting with an antibody specific for EGFP demonstrated expression of FPV039BH1, BTB-only domain of EVM150 and the kelch-only domains of EVM150 and EVM167. Lack of interaction between the kelch domains of EVM150 and EVM167 and cullin-3 was demonstrated by western blotting the immunoprecipitations with anti-Flag.
A highly conserved region in the N-terminus of cullin-1 is important for the interaction with the linker protein SKP1 (Zheng et al., 2002). Significantly, a similar conserved region in cullin-3 has recently been shown to be necessary for interacting with cellular BTB domains (Furukawa et al., 2003). Therefore, to determine if the BTB domains of EVM150 and EVM167 interact with the N-terminal region of cullin-3 similar to cellular BTB domains, we transfected HEK293T cells with either pEGFP-EVM150, pEGFP-EVM167, or pEGFP-FPV039BH1 and co-transfected the cells with either pcDNA-Myc-cullin-3 or pcDNA-Myc-cullin-3ΔN41. Myc-cullin-3ΔN41 is missing 41 amino acids in the N-terminal region of cullin-3 which are required to mediate the interaction with cellular BTB-domain containing proteins (Furukawa et al., 2003). Western blotting of cell lysates with anti-Myc established that both Myc-cullin-3 and Myc-cullin-3ΔN41 were expressed in all the co-transfected cells (Fig. 5A), and immunoprecipitation indicated that all EGFP-tagged proteins were expressed (Fig. 5A). Subsequent western blotting of the immunoprecpitations with an anti-Myc antibody demonstrated an interaction between Myc-tagged cullin-3 and EGFP-tagged EVM150 and EVM167, supporting our previous observations. No interaction, however, was detected between EVM150 or EVM167 and Myc-cullin3ΔN41 (Fig. 5A). We also performed co-immunoprecipitations with the BTB domains of EVM150 and EVM167 and no interaction between Myc-cullin-3ΔN41 and the BTB domain-only constructs of EVM150 and EVM167 were detected (Fig. 5B). Reciprocal immunoprecipitations confirmed these results (data not shown), indicating that amino acids 34 to 74 in the N-terminus of cullin-3 are required for interaction with EVM150 and EVM167 (Fig. 5B). To further illustrate that the BTB-cullin-3 interaction was specific for the N-terminal region of cullin-3, we used a second deletion construct of cullin-3, Myc-cullin-3ΔRoc1, which is missing amino acids 597 to 615 and unable to interact with the RING-finger protein Roc1 (Furukawa et al., 2003). Co-expression of EGFP-EVM150 or EGFP-EVM167 with Myc-cullin-3ΔRoc1 and co-immunoprecipitation demonstrated an interaction between cullin-3 and EVM150 and EVM167 irrespective of the presence of the Roc1 binding site (Furukawa et al., 2003) (Fig. 5C). These data illustrate that EVM150 and EVM167 interact with the N-terminal region of cullin-3 through the BTB domain.
Figure 5.
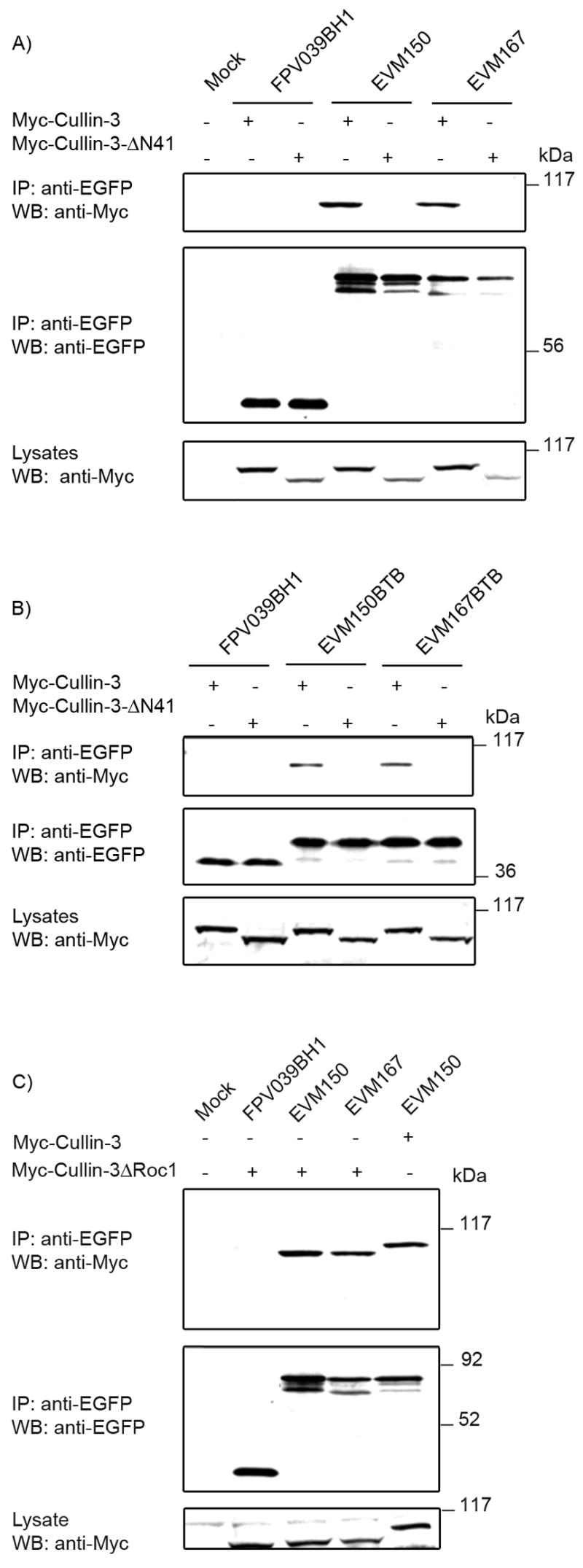
A conserved region in the N-terminus of cullin-3 is necessary for EVM150 and EVM167 interaction with cullin-3. (A) HEK293T cells were co-transfected with constructs to express EGFP-tagged full-length versions of EVM150, EVM167 and co-transfected with Myc-cullin-3 or Myc-cullin-3ΔN41. Interaction was assessed by co-immunoprecipitation with anti-EGFP. Western blotting of lysates with anti-Myc showed expression of Myc-cullin-3 and Myc-cullin-3ΔN41. Immunoprecipitation and western blotting with an antibody specific for EGFP demonstrated expression of FPV039BH1, EVM150 and EVM167. Interaction between EVM150 and EVM167 and cullin-3 but not Myc-cullin-3ΔN41 was determined by western blotting the immunoprecipitations with anti-Myc. (B) Expression of the BTB-only domains of EVM150 and EVM167 retain the ability to interact with Myc-cullin-3 but not Myc-cullin-3ΔN41. (C) EVM150 and EVM167 interact with both cullin-3 and cullin-3ΔRoc1. HEK293T cells were co-transfected with constructs to express EGFP-tagged full-length versions of EVM150, EVM167 and co-transfected with Myc-cullin-3ΔRoc1. As a positive control, EVM150 was co-transfected with Myc-cullin-3. Lysates were subjected to western blotting with anti-Myc to indicate expression of Myc-cullin-3 and Myc-cullin-3ΔRoc1. Immunoprecipitation and western blotting with an antibody specific for EGFP demonstrated expression of FPV039BH1, EVM150 and EVM167. Interaction between EVM150 and EVM167 and cullin-3 and Myc-cullin-3ΔRoc1 was demonstrated by subjecting the immunoprecipitations to western blotting with anti-Myc.
EVM150 and EVM167 co-localize with cullin-3 during virus infection
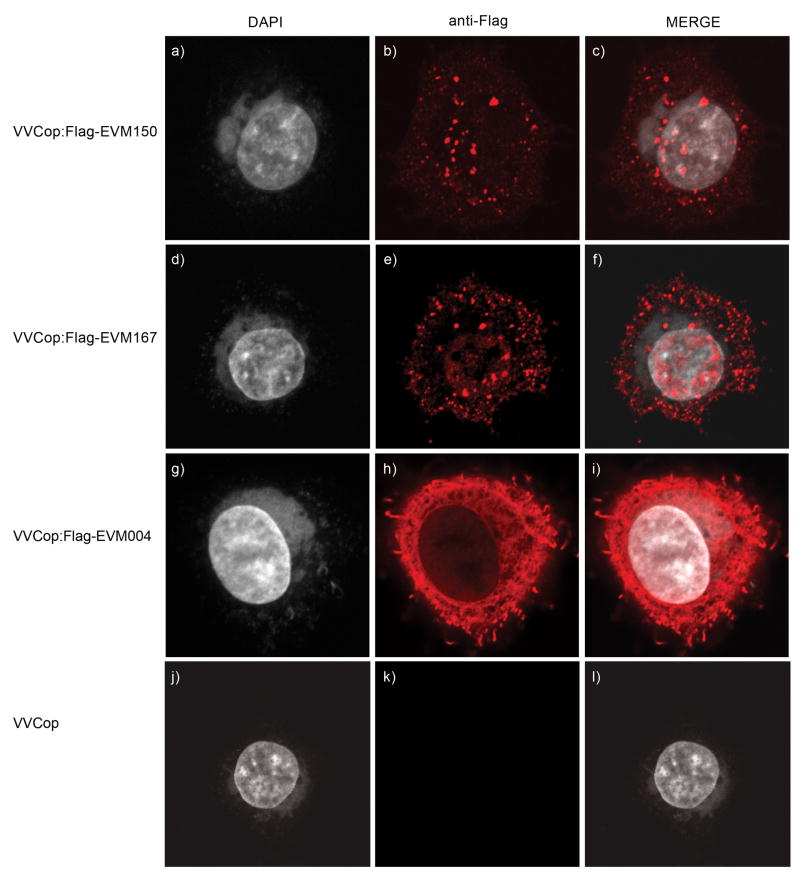
To further investigate the function of EVM150 and EVM167 we generated recombinant vaccinia viruses that expressed Flag-tagged versions of EVM004, EVM150 and EVM167. The subcellular localization of EVM004, the BTB-only containing protein, and EVM150 and EVM167 was examined in HeLa cells during infection. Both Flag-EVM150 and Flag-EVM167 showed a subcellular distribution during infection; appearing as discrete punctate regions within the cytoplasm (Fig. 6 a–f). In some cells, low levels of Flag-EVM150 and Flag-EVM167 were detected in the nucleus (Fig. 6 a–f). In contrast, expression of Flag-EVM004 during infection, which naturally encodes only a BTB domain, resulted in cytoplasmic staining pattern distinct from Flag-EVM150 or Flag-EVM167 (Fig. 6 g–i), while no anti-Flag staining was detected in VV(Cop) infected cells as expected (Fig. 6 j–l). To demonstrate cells were infected and to determine if the punctate cytoplasmic expression of EVM150 and EVM167 co-localized with cytoplasmic viral replication factories generated during poxvirus infection, cells were co-stained with DAPI, which stains both nuclear DNA and DNA-containing cytoplasmic virus factories (Fig. 6). This approach demonstrated no obvious accumulation of EVM150 or EVM167 at the virus factories (Fig. 6 a–f).
Figure 6.
EVM150 and EVM167 localize to cytoplasmic regions during poxvirus infection. HeLa cells were infected with VVCop:Flag-EVM150 (a–c), VVCop:Flag-EVM167 (d–f), VVCop:Flag-EVM004 (g–i) or VV(Cop) (j–l) at an MOI of 5. Twelve hours post infection cells were fixed and stained with DAPI and anti-Flag to visualize the viral factories and EVM150, EVM167 and EVM004.
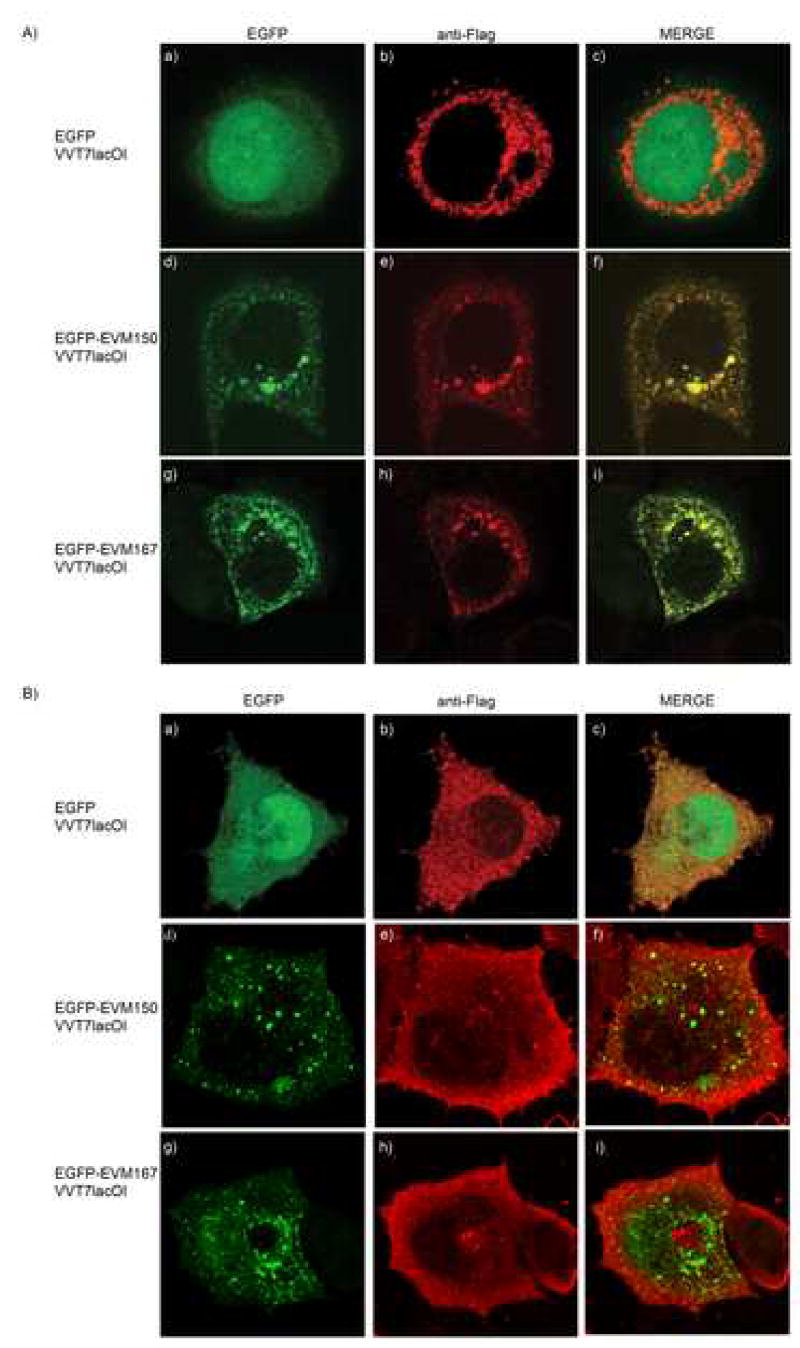
Prompted by our previous observations establishing that EVM150 and EVM167 interact with ectopically expressed cullin-3 (Fig. 3, 4 and 5), we tested the possibility that ectopically expressed cullin-3 co-localized with EVM150 and EVM167 during infection. HeLa cells were infected with the T7 polymerase-expressing vaccinia virus, VVT7lacOI and co-transfected with pcDNA-Flag-cullin-3 and either pEGFP, pEGFP-EVM150, or pEGFP-EVM167 all under control of the T7 promoter (Alexander et al., 1992; Fuerst et al., 1986). Cullin-3 expression was detected by staining cells with an anti-Flag antibody, and EGFP fluorescence indicated the localization of EVM150 and EVM167. HeLa cells infected and expressing EGFP demonstrated nuclear and cytoplasmic EGFP expression (Fig. 7A a). Expression of Flag-cullin-3, however, resulted in a punctate cytoplasmic distribution that did not co-localize with EGFP (Fig. 7A b and c). HeLa cells infected and co-transfected with pEGFP-EVM150 and pcDNA-Flag-cullin-3 displayed a punctate cytoplasmic staining pattern for EGFP-EVM150 (Fig. 7A d), as previously observed for cells infected with VVCop:Flag-EVM150 (Fig. 6). Significantly, EGFP-EVM150 and cullin-3 demonstrated clear co-localization (Fig. 7 d–f), and a similar co-localization between EGFP-EVM167 and cullin-3 was also observed (Fig. 7 g–i). As a control, we performed a similar co-localization experiment using an N-terminal cullin-3 deletion mutant, cullin-3(200–768), that lacked the BTB interaction domain (Fig. 7B) (Wilkins et al., 2004). HeLa cells were infected with VVT7lacOI and co-transfected with pEGFP, pEGFP-EVM150 or pEGFP-EVM167 and pGEMT-Flag-cullin-3(200–768). In contrast to the punctate localization pattern observed for cullin-3 (Fig. 7A), localization of Flag-cullin-3(200–768) was diffuse throughout the cytoplasm (Fig. 7B b) and did not co-localize with EGFP (Fig. 7B a–c), EGFP-EVM150 (Fig 7B d–f) or EGFP-EVM167 (Fig. 7B g–i) supporting our previous co-immunoprecipitation data and confirming that the interaction between cullin-3 and EVM150 and EVM167 requires the N-terminus of cullin-3.
Figure 7.
EVM150 and EVM167 co-localize with Flag-cullin-3 during virus infection. A) HeLa cells were infected with VVT7lacOI (MOI 5) and co-tranfected with pEGFP (a–c), pEGFP-EVM150 (d–f) or pEGFP-EVM167 (g–i) and Flag-cullin-3. Expression was induced by the addition of IPTG. Twelve hours post-infection, cells were fixed and cullin-3 localization visualized by staining with anti-Flag. EGFP fluorescence indicated the localization of EVM150 and EVM167. B) HeLa cells were infected with VVT7lacOI (MOI 5) and co-transfected with pEGFP (a–c), pEGFP-EVM150 (d–f) or pEGFP-EVM167 (g–i) and pGEMT-Flag-cullin-3(200–768).
EVM150 and EVM167 interact with endogenous cullin-3 and Roc1 during infection
Since cullin-3 is typically expressed at low levels within the cell we sought to confirm the interaction between EVM150 and EVM167 with endogenous levels of cullin-3 during virus infection. HEK293T cells were infected with VV(Cop) or recombinant VVCop:Flag-EVM004, VVCop:Flag-EVM150, or VVCop:Flag-EVM167. Western blotting of cellular lysates with anti-cullin-3 showed the presence of cullin-3 in all samples and the expression of Flag-EVM004, Flag-EVM150 and Flag-EVM167 was detected by blotting with anti-Flag lysates (Fig. 8A). Immunoprecipitation using a cullin-3 specific antibody precipitated cullin-3, as expected and also precipitated Flag-EVM150 and Flag-EVM167 (Fig. 8A). In contrast, no co-immuoprecipitation of Flag-EVM004 with endogenous cullin-3 was detected, indicating that EVM150 and EVM167 interacted with endogenous levels of cullin-3 during infection (Fig. 8A). This interaction was further supported by the ability to detect EVM150 and EVM167 interaction with cullin-3 by a reciprocal immunoprecipitation (Fig. 8B) clearly indicating that EVM150 and EVM167 associated with cullin-3 based ubiquitin ligases.
Figure 8.
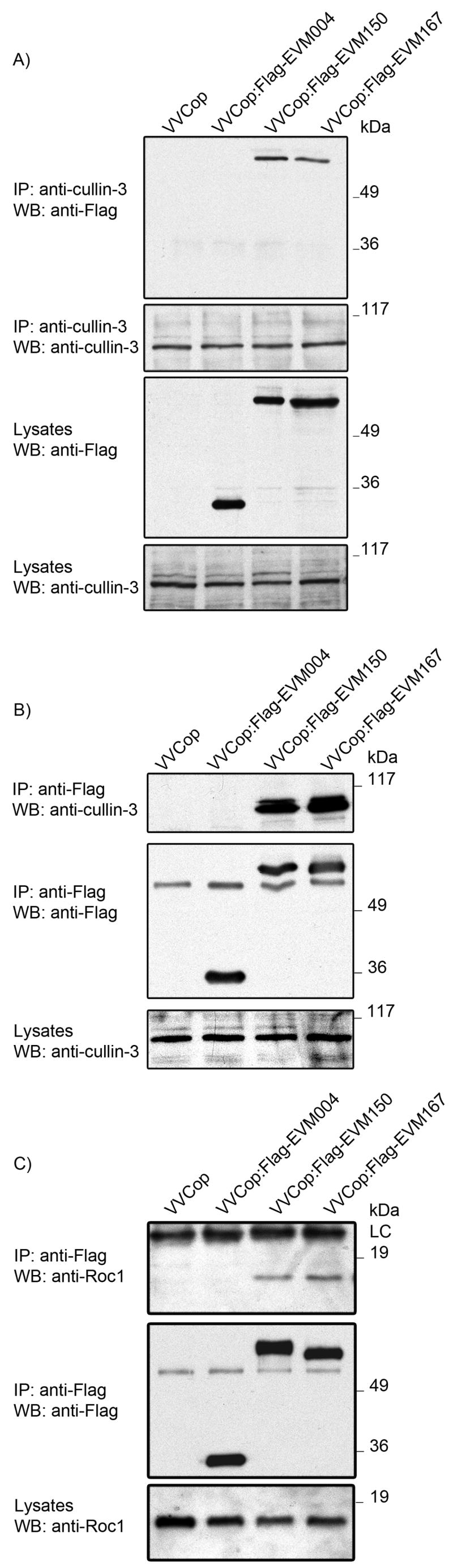
EVM150 and EVM167 interact with endogenous cullin-3 and Roc1 during infection. HEK293T cells were infected with VV(Cop), VVCop:FlagEVM004, VVCop:FlagEVM150 or VVCop:Flag167 at an MOI 5. Twelve hours post-infection cell lysates were generated and EVM150 and EVM167 interaction with endogenous cullin-3 assessed by co-immunoprecipitation. (A) Endogneous cullin-3 expression was detected by immunoprecipitation and western blotting with a cullin-3 specific antibody. Interaction between EVM150 and EVM167 and cullin-3 was demonstrated by western blotting with anti-Flag. (B) Interaction between cullin 3 and EVM150 and EVM167 was assessed by reciprocal immunoprecipitation using anti-Flag followed by western blotting with anti-cullin 3. (C) Roc1 co-immunoprecipitates with EVM150 and EVM167. Cellular lysates were analyzed by immunoprecipitaiton with anti-Flag followed by western blotting with anti-Roc1.
To determine if EVM150 and EVM167 interacted with a potentially functional cullin-3 based ubiquitin ligase we analyzed immunoprecipitations for the presence of Roc1, the RING finger protein that functions as the ubiquitin ligase and is therefore an essential component of cullin-3 based ubiquitin ligases (Furukawa et al., 2002). Cells infected with VV(Cop), VVCop:Flag-EVM004, VVCop:Flag-EVM150 and VVCop:Flag-EVM167 showed no change in the expression level of Roc1 (Fig. 8C) and significantly, Roc1 co-immunoprecipitated with both EVM150 and EVM167 indicative of interaction with a functional cullin-3 based ubiquitin ligase complex.
EVM150 and EVM167 associate with conjugated ubiquitin
The observation that EVM150 and EVM167 interacted with endogenous cullin-3 and Roc1 in infected cells suggested that the poxviral BTB/kelch proteins may form functional cullin-3 based ubiquitin ligases resulting in ubiquitination. Given that potential substrates for EVM150 and EVM167 are currently unknown we instead sought to determine if EVM150 and EVM167 co-immunoprecipitated with conjugated ubiquitin. Cells were infected with VV(Cop) or recombinant vaccinia viruses expressing Flag-EVM150, Flag-EVM167 or Flag-EVM004, and the Flag-tagged proteins were immunoprecipitated with anti-Flag antibody and the immunocomplexes were subjected to western blotting with an antibody (FK2) that recognizes conjugated ubiquitin but not free ubiquitin (Fujimuro et al., 1994) (Fig. 9). This approach detected the immunoprecipitation of Flag-tagged EVM004, EVM150 and EVM167 and demonstrated the presence of conjugated ubiquitin in EVM150 and EVM167 imunoprecipitations indicating that both EVM150 and EVM167 associated with ubiquitinated proteins. Notably, VV(Cop) and VV-Flag-EVM004 infected cells showed little co-immunoprecipitation of conjugated ubiquitin (Fig. 9).
Figure 9.
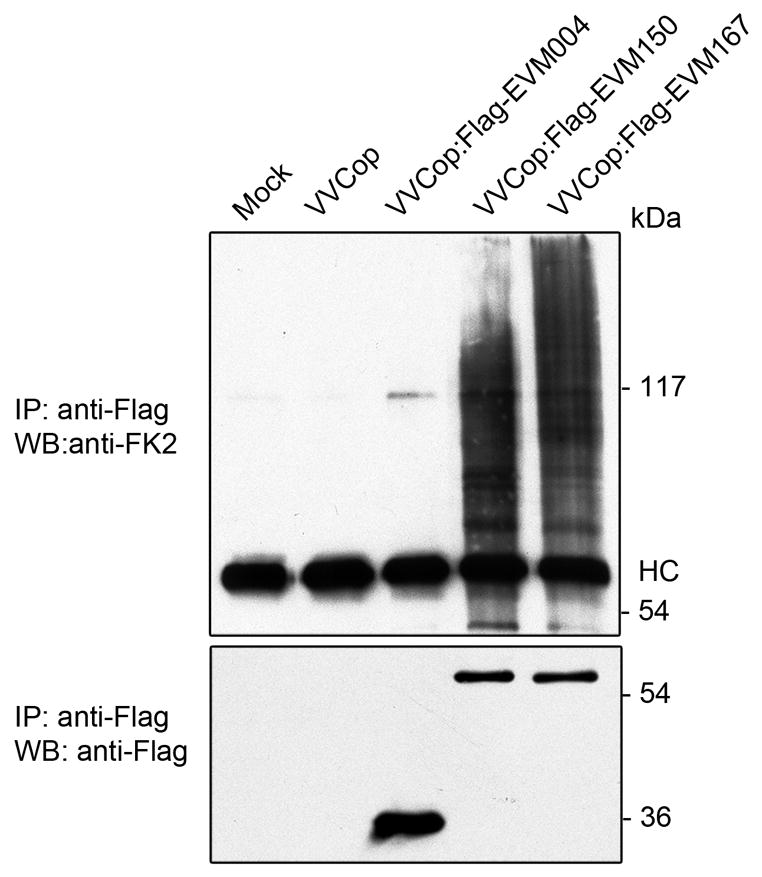
Conjugated ubiquitin co-immunoprecipitates with EVM150 and EVM167. HeLa cells were infected with VV(Cop), VVCop:FlagEVM004, VVCop:FlagEVM150 or VVCop:FlagEVM167 at an MOI 5. Twelve hours post infection the cells were subjected to immunoprecpitation with anti-Flag. Expression of Flag-EVM150 and Flag-EVM167 was detected by immunoprecipitation with anti-Flag followed by western blotting with anti-Flag-HRP. Co-immunoprecipiation of EVM150 or EVM167 with conjugated ubiquitin was detected by immunoprecipitation with anti-Flag followed by western blotting with the anti-ubiquitin antibody (FK2) which detects only conjugated ubiquitin.
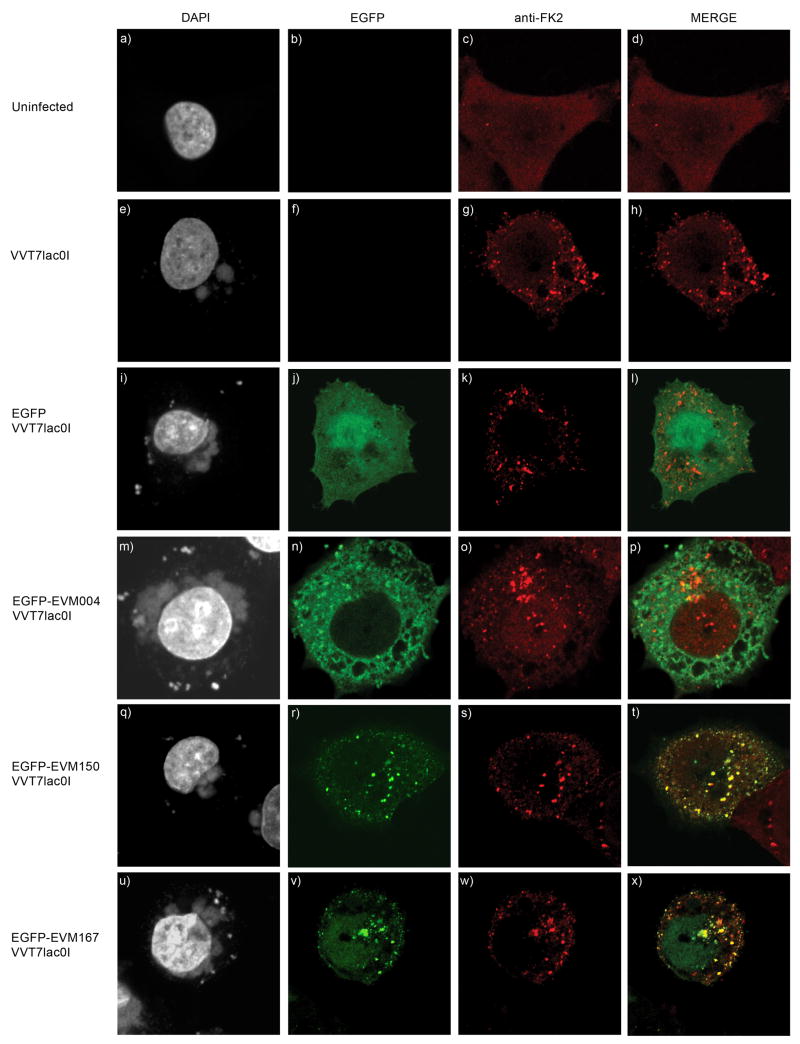
The presence of conjugated ubiquitin was confirmed by confocal microscopy. HeLa cells were infected with VVT7lacOI and transfected with either pEGFP, pEGFP-EVM004, pEGFP-EVM150, or pEGFP-EVM167 and the presence of conjugated ubiquitin was visualized using the FK2 antibody (Fujimuro et al., 1994). Uninfected HeLa cells stained with the FK2 antibody showed a diffuse staining pattern of conjugated ubiquitin (Fig. 10 a–d). Upon infection with VVT7lacOI, which encodes four BTB/kelch proteins, a punctate staining pattern indicative of regions enriched in conjugated ubiquitin was detected (Fig. 10 panel e–h). HeLa cells infected and transfected with EGFP again showed punctate cytoplasmic regions of conjugated ubiquitin, but no co-localization with EGFP (Fig. 10 panel i–l) and a similar lack of co-localization was also seen in cells expressing EVM004 (Fig. 10 panel m–p). Cells infected and transfected with EGFP-tagged EVM150 or EVM167 however, showed regions of obvious co-localization with conjugated ubiquitin (Fig. 9B panel q–x) suggesting that the ectromelia virus BTB/kelch proteins EVM150 and EVM167 support the formation of active cullin-3-based ubiquitin ligases potentiality recruiting unidentified substrates for ubiquitination.
Figure 10.
EVM150 and EVM167 co-localize with conjugated ubiquitin. HeLa cells were infected with VVT7lacOI (MOI 5) and transfected with pEGFP, pEGFP-EVM004, pEGFP-EVM150, or pEGFP-EVM167. Expression was induced by treatement with IPTG and twelve hours post-infection cells were fixed and stained with DAPI to visualize nuclei and virus factories and conjugated ubiquitin was detected with an antibody (FK2) that detects conjugated ubiquitin. (A-D) Mock infected HeLa cells display a diffuse staining pattern of conjugated ubiquitin. (E-H) HeLa cells infected with VVT7lacOI demonstrate a punctate localization of conjugated ubiquitin. (I-L) EGFP expression during virus infection does not co-localize with conjugated ubiquitin. (M-P) EGFP-EVM004 does not co-localize with conjugated ubiquitin. (Q-T) EGFP-EVM150 co-localizes with conjugated ubiquitin. (U-X) EGFP-EVM167 co-localizes with conjugated ubiquitin.
Discussion
The efficient degradation of proteins is crucial for normal cellular function, and the major mechanism that regulates protein destruction is the ubiquitin proteasome pathway (Pickart, 2001; Weissman, 2001). One family of proteins responsible for the transfer of ubiquitin to the substrate protein is the cullin-based ubiquitin ligase family (Petroski and Deshaies, 2005). By interacting with a variety of substrate adapters, cullin-based ubiquitin ligases have the capacity to direct the precise ubiquitination of a wide range of proteins (Petroski and Deshaies, 2005). Recently, cellular BTB/kelch proteins have been shown to function as substrate specific adapters for cullin-3 ubiquitin ligases (Furukawa et al., 2003; Geyer et al., 2003; Pintard et al., 2003b; Xu et al., 2003). BTB/kelch proteins were first identified in the genome of poxviruses more than 10 years ago; however, a specific function for these viral proteins has not been established (Koonin et al., 1992; Senkevich et al., 1993). Here we show that two of the BTB/kelch proteins encoded by ectromelia virus, EVM150 and EVM167, interact with cullin-3, suggesting that the viral proteins may function similarly to cellular BTB/kelch proteins as substrate specific adapters for cullin-3 ubiquitin ligases.
Ectromelia virus strain Moscow, the causative agent of mousepox, contains four genes, EVM018, EVM027, EVM150, and EVM167, that are predicted to encode proteins with both BTB and kelch domains (Chen et al., 2003) (Fig. 1 and 2A). Using a series of transient transfection and co-immunoprecipitation experiments we found that EVM150 and EVM167 reproducibly interacted with cullin-3 and that the BTB domain of EVM150 and EVM167 was necessary and sufficient for this interaction (Fig. 3 and 4). Similar to cellular BTB-containing proteins, a conserved domain within the N-terminus of cullin-3 was responsible for interaction with EVM150 and EVM167, while a C-terminal region of cullin-3 responsible for binding the RING-finger protein Roc1 was not required (Furukawa et al., 2003) (Fig. 5). Furthermore, both EVM150 and EVM167 co-localized with ectopically expressed cullin-3 during infection, as assessed by confocal microscopy (Fig. 7), and interacted with endogenous cullin-3 and Roc1 (Fig. 8).
Confocal microscopy demonstrated that both EVM150 and EVM167 displayed a punctate distribution in the cytoplasm of infected cells (Fig. 6). In an attempt to discern the role of cellular structures in the distribution of EVM150 and EVM167 we checked for possible co-localization with lysosomes, early and late endosomes or the Golgi complex and no co-localization was apparent, providing no clue to the unique localization of these proteins (B. Wilton and M. Barry, unpublished data). An examination for the presence of ubiquitinated proteins during virus infection depicted a similar pattern of localization to EVM150 and EVM167 (Fig. 10). Further analysis demonstrated co-localization with EVM150 and EVM167 and conjugated ubiquitin (Fig. 10). BTB domains are known to mediate the formation of protein complexes through homo- and hetero-oligomeric interactions (Ahmad et al., 1998; Albagli et al., 1995; Bardwell and Treisman, 1994; Katsani et al., 1999; Li et al., 1999; Muller et al., 1999; Takenaga et al., 2003), and these large complexes have been observed with a number of cellular BTB/kelch proteins including Kel-8 in C. elegans, the Drosophila KELCH protein, and the promyelocytic leukemia (PML) gene product (Li et al., 1999; Robinson and Cooley, 1997; Schaefer and Rongo, 2006). The unique sub-cellular localization of EVM150 and EVM167 combined with the co-localization to the same regions may therefore be the result of protein complexes formed through BTB-domain oligomerization and the concordant formation of organized, cullin-3 ligase-containing ubiquitinating centers.
Although EVM150 and EVM167 interact with cullin-3, we were unable to detect an interaction between cullin-3 and the other BTB/kelch proteins, EVM018, EVM027, or the BTB-only protein EVM004 (Fig. 3 and 8). The lack of an interaction may be due to the presence of the large N-terminal EGFP tag which interferes with the ability of EVM018, EVM027 and EVM004 to interact with cullin-3. Additionally, EVM018 lacks the N-terminal alpha helix and beta sheet, which could account for the inability of EVM018 to interact with cullin-3 (Fig. 1A). Alternatively, given that BTB domains function in a range of cellular processes other than ubiquitination, it is possible that EVM018, EVM027 and EVM004 have functions independent of cullin-3 interaction and ubiquitination (Ahmad et al., 2003; Bomont et al., 2000; Melnick et al., 2000; Minor et al., 2000; Ziegelbauer et al., 2001).
The co-localization and interaction of EVM150 and EVM167 with cullin-3 strongly suggested that, similar to cellular BTB/kelch proteins, these viral proteins function as substrate adapters for cullin-3 ubiquitin ligases. Strengthening this idea is the co-immunoprecipitation of Roc1, a necessary component of the cullin-3 ligase complex (Furukawa et al., 2002). In support of this, conjugated ubiquitin co-localized with EVM150 and EVM167 and this observation was confirmed by the co-immunoprecipitation of EVM150 and EVM167 with conjugated ubiquitin (Fig. 9 and 10). Alternatively, the viral BTB/kelch proteins may function by simply sequestering cullin-3, thereby inhibiting the ubiquitination of cellular proteins that are normally regulated by cullin-3. For example, sequestration of cullin-1 by HIV encoded Vpu prevents IκB ubiquitination and degradation thereby inhibiting NFκB signaling (Margottin et al., 1998). If EVM150 and EVM167 function by sequestering cullin-3, the co-localization with conjugated ubiquitin may be due to ubiquitination of components of the cullin-3 ubiquitin ligase or perhaps EVM150 or EVM167, themselves. Indeed, evidence indicates that components of cullin ligases are regulated at the level of ubiquitination (Scaglione et al., 2007; Zhou and Howley, 1998). However, since multiple distinct BTB/kelch proteins are encoded within poxvirus genomes we speculate that the BTB/kelch proteins are not simply sequestering cullin-3 but may be functioning as cullin-3 specific substrate adapters. EVM150 and EVM167 differ within the kelch regions, suggesting the potential for each BTB/kelch protein to recruit specific substrates to cullin-3-based ubiquitin ligases. Evidence clearly indicates an important function for the BTB/kelch proteins during poxvirus infection. For example, deletion of BTB/kelch genes from the genome of cowpox virus and sheeppox virus dramatically altered virus pathogenicity (Balinsky et al., 2007; Kochneva et al., 2005). Additionally, deletion of the BTB/kelch proteins, C2L, F3L and A55R, from vaccinia virus, resulted in altered pathogenesis following intradermal inoculation of mice (Beard et al., 2006; Froggatt et al., 2007; Pires de Miranda et al., 2003).
To date, only a limited number of targets have been characterized for cullin-3 based ubiquitin ligases. These include MEI-1 in C. elegans, which regulates the transition from meiosis to mitosis (Furukawa et al., 2003; Geyer et al., 2003; Pintard et al., 2003a; Pintard et al., 2003b; Xu et al., 2003); ionotropic glutamate receptors in the neurons of C. elegans (Schaefer and Rongo, 2006); RhoBTB2, a candidate tumor suppressor deleted in breast and lung cancer (Wilkins et al., 2004); NRF2, a regulator of oxidative stress (Cullinan et al., 2004; Furukawa and Xiong, 2005; Kobayashi et al., 2004; Zhang et al., 2004a); topoisomerase I; and cyclin E (Singer et al., 1999; Zhang et al., 2004b). Although few substrates for cullin-3-based ubiquitin ligases have been definitively identified, the majority of BTB containing-proteins examined thus far interact with cullin-3 (Furukawa et al., 2003; Geyer et al., 2003; Kobayashi et al., 2004; Xu et al., 2003). Combined with the observation that the human genome is predicted to encode over 200 BTB-containing proteins, this raises the possibility that a wide range of cellular proteins may be specifically targeted to cullin-3 ubiquitin ligases for ubiquitination. Given that EVM150 and EVM167 co-localize with conjugated ubiquitin and interact with cullin-3, we hypothesize that EVM150 and EVM167 may recruit cellular or viral proteins to cullin-3 ubiquitin ligases for degradation. In fact, other viruses have evolved mechanisms to specifically recruit cellular proteins to cullin-based ubiquitin ligases (Barry and Fruh, 2006). The Vif protein of HIV directs the degradation of APOBEC3G through a cullin-5 based ubiquitin ligase (Bishop et al., 2004; Harris and Liddament, 2004; Yu et al., 2003), paramyxovirus V protein targets STAT proteins for degradation via cullin-4a (Horvath, 2004), and adenovirus E4orf6 and E1B55K induce p53 degradation through cullin-5 mediated ubiquitination (Blanchette et al., 2004; Querido et al., 2001). The future identification of any proteins targeted by the poxvirus BTB/kelch proteins will be highly informative, aiding in the potential elucidation of cellular anti-viral pathways.
Manipulation of the ubiquitin degradation pathway is emerging as a common theme during poxvirus infection. We have recently demonstrated that a RING-finger protein, p28, found in some strains of vaccinia virus, myxoma virus, and ectromelia virus, functions as a bona fide ubiquitin ligase (Huang et al., 2004; Nerenberg et al., 2005). To date, p28 substrates have not been identified, but deletion of p28 from ectromelia virus results in attenuated virulence, indicating an important role for this protein in pathogenesis (Senkevich et al., 1994). The avian poxviruses contain an expanded family of genes encoding RING-finger proteins that are predicted to function as ubiquitin ligases (Afonso et al., 2000; Tulman et al., 2004). Additionally, canarypox and entomopoxviruses encode their own ubiquitin molecules (Bawden et al., 2000; Tulman et al., 2004). Myxoma virus, the causative agent of myxomatosis in rabbits, encodes a RING-finger domain protein, M153R, that reduces surface expression of MHC class I and CD4 by targeting both for degradation (Guerin et al., 2002; Mansouri et al., 2003). More recently, the myxoma virus MT5 protein, an ankyrin repeat protein that also contains an F-box-like domain, has been shown to interact with cullin-1 (Johnston et al., 2005). Numerous other proteins encoded by poxviruses contain F-box-like domains, suggesting that these proteins interact with cullin-1 to modulate the ubiquitin pathway (Afonso et al., 2006; Mercer et al., 2005). Our data predicts the existence of yet another important mechanism that poxviruses have evolved to modulate the ubiquitin pathway through cullin-3-based ubiquitin ligases.
The presence of multiple BTB/kelch proteins encoded in almost every genus of the Chordopoxvirinae suggests an evolutionary important function during poxvirus infection. Although the specific biological function of EVM150 and EVM167 remains unknown, the observation that these proteins interact with cullin-3 and co-localize with cullin-3 is the first described interaction for these proteins. Given that poxviruses encode multiple discrete BTB/kelch proteins and that EVM150 and EVM167 co-localize with conjugated ubiquitin, we speculate that cellular or viral proteins are specifically recruited to cullin-3-based ubiquitin ligases and targeted for degradation by the 26S proteasome. These observations lay the groundwork for future studies aimed at identifying and characterizing potential cullin-3 substrates targeted by poxviral BTB/kelch proteins that may play a role in cellular anti-viral pathways.
Materials and Methods
Cells and viruses
HEK293T, CV-1, HuTK−143B and HeLa cells were obtained from American Type Culture Collection, and grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U of penicillin/ml, 50 μg of streptomycin/ml and 200 μM glutamine (Invitrogen Corporation). Baby green monkey kidney (BGMK) cells were routinely grown in DMEM supplemented with 10% newborn calf serum, 50 U of penicillin/ml, 50 μg of streptomycin/ml and 200 μM glutamine (Invitrogen Corporation). Recombinant vaccinia virus strain Western Reserve expressing T7 polymerase (VVT7lacOI) was a gift from Dr. B. Moss, National Institute of Allergy and Infectious Diseases (Alexander et al., 1992; Fuerst et al., 1986). VV(Cop) expressing lacZ was a gift from Dr. G. McFadden, University of Florida. Recombinant ectromelia virus strain Moscow (EVM) expressing β-galactosidase was a gift from Dr. M. Buller, St. Louis University. All viruses were propagated in BGMK cells and purified as previously described (Stuart et al., 1991). HeLa or HEK293T cells were infected at a multiplicity of infection (MOI) of 5 or 10 plaque forming units per cell in 500 μl DMEM at 37°C. One hour after infection, cells were supplemented with DMEM containing 10% FBS.
Antibodies
Mouse anti-Flag-M2 and anti-Flag-M2-horseradish peroxidase (HRP) conjugate were purchased from Sigma-Aldrich Company. Rabbit anti-green fluorescence protein (GFP) and goat anti-GFP antibodies were gifts from Dr. L. Berthiaume, University of Alberta. Mouse anti-GFP (clone B34) was purchased from Covance Incorporated. Mouse anti-Myc antibody (clone 9E10) was a gift from Dr. T. Hobman, University of Alberta. Rabbit anti-cullin-3 and rabbit anti-Roc1 were generated as previously described (Furukawa et al., 2003; Ohta et al., 1999b). Mouse anti-FK2 was purchased from BIOMOL International. Donkey anti-mouse- and donkey anti-rabbit-horseradish peroxidase-conjugated antibodies were purchased from Jackson Laboratories. Alexa Fluor 546 goat anti-mouse, Alexa Fluor 546 goat anti-rabbit and Alexa Fluor 488 goat anti-mouse were purchased from Invitrogen Corporation.
Alignments and secondary structure prediction
Protein alignments for the ectromelia BTB/kelch proteins and the cellular BTB/kelch protein Keap1 were performed via Clustral W (1.82) (http://www.ebi.ac.uk/clustalW/#) (Chenna et al., 2003). To predict secondary structure, each protein was separately submitted to 3D-PSSM version 2.6.0 and PYRE version 0.2 (http://www.sbg.bio.ic.ac.uk/~3dpssm/index2.html) (Kelley et al., 2000). Keap 1 was used as a control and verified by comparison to previous published data (Stogios et al., 2005). Predicted secondary structures were then assigned within the amino acid sequences.
RT-PCR
RNA transcripts for EVM004, EVM018, EVM027, EVM150 and EVM167 were analyzed by reverse transcription polymerase chain reaction (RT-PCR). CV-1 cells (1 × 106) were infected with ectromelia virus strain Moscow at an MOI 5 in the absence or presence of 80 μg/ml of cytosine β-D-arabinofuranoside hydrochloride (AraC) (Sigma-Aldrich Company). At 4, 8, 12 and 24 hours post infection cells were collected and RNA was extracted with TRIZOL (Invitrogen Corporation) according to the manufacturers’ directions. To exclude contaminating viral DNA, RNA was treated with 1Unit/μl DNase (Invitrogen Corporation). Reverse transcription was subsequently performed using 1μg of RNA and 200 units of SuperScriptII reverse transcriptase (Invitrogen Corporation) and Oligo dT (Invitrogen Corporation). The RT reaction was used as template for PCR using primers specific for the BTB domains of EVM004, EVM018, EVM027, EVM150 and EVM167. EVM004 forward primer 5′-(EcoRI)GAATTC TCATGAGTGATTACTATTTT-3′, reverse primer 5′-(BamHI)GGATCCTTAATAATACCTA GAAAATAT-3′; EVM018 forward primer 5′-(EcoRI)GAATTCTCATGGAAAGCGTGATA TTT-3′, reverse 5′-(BamHI)GGATCCTTACATCCTTATACAATTTGTGG-3′; EVM027 forward primer 5′-(BglII)AGATCTCATGCCGATATTTGTCAAT-3′, reverse 5′-(EcoRI)GAA TTCTTACTCGACACAATATTCCTTTCT-3′; EVM150 forward primer 5′-(EcoRI)GAATT C TCATGAATAACAGCAGTGAA-3′; reverse 5′-(BamHI)GGATCCTTAATCGATACAGTTTC TAGA-3′; EVM167 forward primer 5′-(EcoRI)GAATTCTCATGGATATTGATGATATT-3′; reverse 5′ (BamHI)GGATCCTTAATATATACAGGTATCATG-3′. To show the inhibition of late gene expression in the presence of AraC, primers specific to EVM058 (I5L) were used 5′-(EcoRI) GAATTCATGGTGGATGCTATAACC-3′, reverse 5′-(BamHI)GGATCCACTTTTCA TTAATAGGGA-3′. As a positive control, β-actin was amplified using primers 5′-GCACCACACCTTCTACAATGAG-3′ and 5′-AAATAGCACAGCCTGGATAGCAAC-3′.
Plasmid constructs
Flag- and Myc-tagged cullin-3 constructs and Myc-cullin-3 N41 and Myc-cullin-3 Roc, which contain deletions of amino acids 34 to 74 and 597 to 615, respectively have been previously described (Furukawa et al., 2003). Flag-cullin-3(200–768) was generated by PCR using the forward primer 5′-GTCTATGAAGAAGATTTTGAGGCT-3′ and the reverse primer 5′-TTATGCTACATATGTGTATA-3′ and subcloned into pGEMT (Promega Corporation). EVM004, EVM018, EVM027, EVM150 and EVM167 were amplified by PCR from ectromelia virus DNA using PWO polymerase (Roche Diagnostics Corporation) and gene specific primers. The amplified products were subsequently cloned into pGemT (Promega Corporation). To generate pGEMT-EVM004, forward primer 5′ (EcoRI)GAATCCTCATG AGTGATTACTATTTT3′ and reverse primer 5′ (BamHI)GGATCCTTAATAATACC TAGAAAATAT3′ were used. To generate pGEMT-EVM018, forward primer 5′-(EcoRI)GAATTCTCATGGAAAGCGTGATATTT-3′ and reverse primer 5′-(BamHI)GGATCCCTATTGTAGGAATTTTTT-3′ were used. To generate pGemT-EVM027, forward primer 5′-(BglII)AGATCTCATGCCGATATTTGTCAAT-3′ and reverse primer 5′-(EcoRI)GAATTCTTACCATCTTATCCCATT-3′ were used. To generate pGemT-EVM150, forward primer 5′-(EcoRI)GAATTCTCATGAATAACAGCAGTGAA-3′and reverse primer 5′-(BamHI)GGATCCTCAACTACCTATAAAACT-3′ were used. To generate pGemT-EVM167, forward primer 5′-(EcoRI)GAATTCTCATGGATATTGATGATATT-3′ and reverse primer 5′ ′-(BamHI)GGATCCTTAGTAGATGGGTAGTGT-3′ were used. To generate N-terminal EGFP-tagged version of EMV004, EVM018, EV0M27, EVM150 and EVM167 each gene was subcloned into pEGFP-C3 (Becton, Dickinson and Company) to generate pEGFP-EVM004, pEGFP-EVM018, pEGFP-EVM027, pEGFP-EVM150 and pEGFP-EVM167. To generate EGFP-tagged BTB-only constructs for EVM150 and EVM167 the same forward primers described above were used, and reverse primers were generated based upon BTB conserved domain alignments generated by NCBI reverse position specific (rsp) BLAST (Marchler-Bauer and Bryant, 2004). Amplification of the BTB-only domains used the following reverse primers; EVM150BTB reverse 5′-(BamHI)GGATCCTTAATCGATACAGTTTCTAGA-3′; EVM167BTB reverse 5′-(BamHI)GGATCCTTAATATATACAGGTATCATG-3′. Following amplification, the genes were cloned into pGemT and subsequently subcloned into pEGFP-C3 to generate pEGFP-EVM150BTB and pEGFP-EVM167BTB. To generate EGFP-tagged kelch-only constructs for EVM150 and EVM167, the gene specific reverse primers described above were used in combination with forward primers which were created based upon kelch domain alignments using NCBI rspBLAST (Marchler-Bauer and Bryant, 2004). To amplify the kelch regions of EVM150 and EVM167 the following forward primers were used; EVM150kelch 5′-(EcoRI)GAATTCATGATATCTTCACGTGGATA-3′, and EVM167kelch 5′-(EcoRI)GAATTC ATGGAATACAATACCATTTAC-3′. The DNA was amplified, cloned into pGem-T and subsequently subcloned into pEGFP-C3 to generate pEGFP-EVM150kelch and pEGFP-EVM167kelch. pSC66-Flag-EVM004, pSC66-Flag-EVM150 and pSC66-Flag-EVM167, which places genes under the control of a poxvirus promoter (Davison and Moss, 1990; Mackett et al., 1982), were generated by PCR amplification using primers that incorporated the Flag sequence. To express proteins during VVT7lacOI infection EGFP-tagged constructs were amplified by PCR using an EGFP forward primer 5′ (KpnI)GGTACCATGGTGAGCAAGGGCGAG-3′ and the gene specific reverse primers for EVM004, EVM150 and EVM167. The amplified DNA was cloned into pGemT (Promega Corporation) placing the genes under control of the T7 promoter to generate pGEMT-EGFP-EVM004, pGEMT-EGFP-EVM150 and pGEMT-EGFP-EVM167. pEGFP-FPV039BH1 encoding a truncated version of the fowlpox virus Bcl-2 homologue was generated as previously described (Banadyga et al., 2007). Construction of pSC66-EGFP was previously described (Wasilenko et al., 2003).
Generation of recombinant viruses
To generate recombinant viruses, CV-1 cells were infected with VV strain Copenhagen (VVCop) and transfected with pSC66-Flag-EVM004, pSC66-Flag-EVM150 or pSC66-Flag-EVM167. Recombinant viruses were selected in HuTK−143B cells in the presence of 25 μg/ml bromodeoxyuridine and plaque purified using 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal). The presence of Flag-tagged EVM004, EVM150 or EVM167 was analyzed by western blot.
Immunoprecipitations
To determine interaction between the ectromelia virus BTB/kelch proteins and cullin-3, HEK293T cells (1×106) were co-transfected with either 2 μg of DNA of the appropriate construct and 3μg pcDNA3-Flag-cullin-3, pcDNA-Myc-cullin-3, pcDNA-Myc-cullin-3ΔRoc1, or Myc-cullin-3 N41 using Lipofectamine 2000 (Invitrogen Corporation). Co-transfected cells were lysed in NP-40 lysis buffer containing 150 mM NaCl, 1% NP-40, 50 mM Tris (pH 8.0) and EDTA-free protease inhibitors (Roche Diagnostics Corporation). EGFP-tagged proteins were immunoprecipitated with goat anti-GFP followed by the addition of protein G sepharose (GE Healthcare) and co-immunoprecipitation was analyzed by western blotting. Reciprocal immunoprecipitations were performed using mouse anti-Flag M2 and analyzed by western blot. To determine if Flag-EVM150, or Flag-EVM167 interacted with endogenous cullin-3 and Roc1 during virus infection, 1×106 HEK293T cells were infected with VV(Cop), VV-Flag-EVM004, VV-Flag-EVM150 or VV-Flag-EVM167 at an MOI of 5. Twelve hours post infection cells were lysed in NP-40 lysis buffer and supernatants were subjected to immunoprecipitation with rabbit anti-cullin-3 or anti-Flag. To detect ubiquitinated proteins that co-immunoprecipitate with either Flag-EVM150 or Flag-EVM167, immunoprecipitates were blotted with mouse anti FK-2 (Biomol International) which detects conjugated ubiquitin but not free ubiquitin (Fujimuro et al., 1994).
Infection/Transfection Assays
To express proteins during poxvirus infection, cells were infected with a recombinant vaccinia virus expressing T7 polymerase, VVT7lacOI, under the control of the lac repressor (Alexander et al., 1992; Fuerst et al., 1986). Following infection, cells were transfected with plasmids containing genes of interest under the control of a T7 promoter and T7 polymerase expression was induced by treating cells with 10mM IPTG. Alternatively, cells were infected with either vT7lacOI or VV(Cop) and transfected with pSC66 plasmids.
Confocal Microscopy
For fixed cell confocal microscopy, coverslips were seeded with 2.5 × 105 HeLa cells. Cells were infected with either VV(Cop), VVCop:Flag-EVM004, VVCop:Flag-EVM150, or VVCop:Flag-EVM167 at an MOI of 5 for 12 hours, fixed with 4% paraformalydehyde and permeabilized with 1% NP-40. Coverslips were incubated with anti-Flag antibody (1:200) for 1 hour followed by staining with Alexa 488 goat anti-mouse. Coverslips were washed with PBS containing 1%FBS and mounted with 4 mg/ml of N-propyl gallate (Sigma Aldrich) in 50% glycerol containing 250 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen Corporation) to visualize nuclei and cytoplasmic viral factories. To determine co-localization with Flag-cullin-3, HeLa cells were infected with vT7lacOI for 12 hrs at an MOI of 5 and co-transfected with either 2 μg of pEGFP, pEGFP-EVM150, pEGFP-EVM167 and 3 μg of pcDNA-Flag-cullin-3 or pGemT-Flag-cullin-3(200–768). Cells were fixed with 4% paraformaldehyde and coverslips were stained with anti-Flag antibody (1:200) for 1 hour followed by staining with Alexa 546 goat anti-mouse. Flag-cullin-3 was detected at 543nm and EGFP fluorescence was detected at 488nm. To detect the presence of ubiquitinated proteins by confocal microscopy, HeLa cells were stained with anti-FK2 (1:500 dilution) (Biomol International) in PBS containing 1%FBS and Alexa 546 goat-anti-mouse to visualize FK2 localization (Fujimuro et al., 1994). Confocal experiments were performed using a Zeiss Axiovert laser scanning confocal microscope.
Western blotting
Proteins were subjected to SDS-PAGE analysis and transferred to nitrocellulose membranes (Fischer Scientific) using a semi-dry transfer apparatus (Tyler Research Corporation) for 2 hours at 420 mA. Membranes were blocked overnight in 5% skim milk containing 0.1% Tween-20, 50 mM Tris (pH7.6) and 150 mM NaCl. EGFP was detected using either rabbit anti-GFP at a dilution of 1:20,000 or mouse anti-GFP at a dilution of 1:5,000. Flag-tagged proteins were detected by HRP-conjugated anti-Flag antibody at a dilution of 1:2,000. Anti-Myc (clone 9E10) was used at a dilution of 1:5,000 to detect Myc-tagged proteins. Anti-cullin-3 and anti-Roc1 were used at 1:2000. Ubiquitinated proteins were detected using anti-ubiquitin (FK2) (1:2,000) which detects conjugated ubiquitin and not free ubiquitin (Fujimuro et al., 1994). Membranes were treated with either donkey anti-mouse- or donkey anti-rabbit-HRP-conjugated antibody at 1:10,000. Proteins were visualized with a chemiluminescence detection system (GE Healthcare).
Acknowledgments
We thank Logan Banadyga and John Taylor for critical comments on the manuscript and Klaus Fruh for insightful discussions. Work in the authors’ laboratories is supported by grants to M. B. from the Canadian Institutes of Health Research (CIHR), the Howard Hughes Medical Institute (HHMI), and an NIH grant GM067113 to Y.X. S.C. is the recipient of a Canada Graduate Scholarship. M.B. is a CIHR New Investigator, an Alberta Heritage Foundation for Medical Research Senior Scholar and a HHMI International Research Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10(1):17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- Afonso CL, Tulman ER, Delhon G, Lu Z, Viljoen GJ, Wallace DB, Kutish GF, Rock DL. Genome of crocodilepox virus. J Virol. 2006;80(10):4978–91. doi: 10.1128/JVI.80.10.4978-4991.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso CL, Tulman ER, Lu Z, Zsak L, Kutish GF, Rock DL. The genome of fowlpox virus. J Virol. 2000;74(8):3815–31. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad KF, Engel CK, Prive GG. Crystal structure of the BTB domain from PLZF. Proc Natl Acad Sci U S A. 1998;95(21):12123–8. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad KF, Melnick A, Lax S, Bouchard D, Liu J, Kiang CL, Mayer S, Takahashi S, Licht JD, Prive GG. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12(6):1551–64. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6(9):1193–8. [PubMed] [Google Scholar]
- Alexander WA, Moss B, Fuerst TR. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66(5):2934–42. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balinsky CA, Delhon G, Afonso CL, Risatti GR, Borca MV, French RA, Tulman ER, Geary SJ, Rock DL. Sheeppox virus kelch-like gene SPPV-019 affects virus virulence. J Virol. 2007 doi: 10.1128/JVI.01093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banadyga L, Gerig J, Stewart T, Barry M. Fowlpox Virus Encodes a Bcl-2 Homologue That Protects Cells from Apoptotic Death through Interaction with the Pro-Apoptotic Protein Bak. J Virol. 2007 doi: 10.1128/JVI.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks L, Pim D, Thomas M. Viruses and the 26S proteasome: hacking into destruction. Trends Biochem Sci. 2003;28(8):452–9. doi: 10.1016/S0968-0004(03)00141-5. [DOI] [PubMed] [Google Scholar]
- Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8(14):1664–77. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- Barry M, Fruh K. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci STKE 2006. 2006;(335):pe21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28(1):263–6. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawden AL, Glassberg KJ, Diggans J, Shaw R, Farmerie W, Moyer RW. Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses. Virology. 2000;274(1):120–39. doi: 10.1006/viro.2000.0449. [DOI] [PubMed] [Google Scholar]
- Beard PM, Froggatt GC, Smith GL. Vaccinia virus kelch protein A55 is a 64 kDa intracellular factor that affects virus-induced cytopathic effect and the outcome of infection in a murine intradermal model. J Gen Virol. 2006;87(Pt 6):1521–9. doi: 10.1099/vir.0.81854-0. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004;305(5684):645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- Blanchette P, Cheng CY, Yan Q, Ketner G, Ornelles DA, Dobner T, Conaway RC, Conaway JW, Branton PE. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol Cell Biol. 2004;24(21):9619–29. doi: 10.1128/MCB.24.21.9619-9629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomont P, Cavalier L, Blondeau F, Ben Hamida C, Belal S, Tazir M, Demir E, Topaloglu H, Korinthenberg R, Tuysuz B, Landrieu P, Hentati F, Koenig M. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet. 2000;26(3):370–4. doi: 10.1038/81701. [DOI] [PubMed] [Google Scholar]
- Boutell C, Sadis S, Everett RD. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol. 2002;76(2):841–50. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5(9):739–51. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chen N, Danila MI, Feng Z, Buller RM, Wang C, Han X, Lefkowitz EJ, Upton C. The genomic sequence of ectromelia virus, the causative agent of mousepox. Virology. 2003;317(1):165–86. doi: 10.1016/s0042-6822(03)00520-8. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L, Ganem D. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J Clin Invest. 2001;107(12):1599–606. doi: 10.1172/JCI12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24(19):8477–86. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Moss B. New vaccinia virus recombination plasmids incorporating a synthetic late promoter for high level expression of foreign proteins. Nucleic Acids Res. 1990;18(14):4285–6. doi: 10.1093/nar/18.14.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers A, Osborne J, Slack S, Roper RL, Upton C. Poxvirus Orthologous Clusters (POCs) Bioinformatics. 2002;18(11):1544–5. doi: 10.1093/bioinformatics/18.11.1544. [DOI] [PubMed] [Google Scholar]
- Esteban DJ, Buller RM. Ectromelia virus: the causative agent of mousepox. J Gen Virol. 2005;86(Pt 10):2645–59. doi: 10.1099/vir.0.81090-0. [DOI] [PubMed] [Google Scholar]
- Everett RD. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays. 2000;22(8):761–70. doi: 10.1002/1521-1878(200008)22:8<761::AID-BIES10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124(4):767–82. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Froggatt GC, Smith GL, Beard PM. Vaccinia virus gene F3L encodes an intracellular protein that affects the innate immune response. J Gen Virol. 2007;88(Pt 7):1917–21. doi: 10.1099/vir.0.82815-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986;83(21):8122–6. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994;349(2):173–80. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5(11):1001–7. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- Furukawa M, Ohta T, Xiong Y. Activation of UBC5 ubiquitin-conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J Biol Chem. 2002;277(18):15758–65. doi: 10.1074/jbc.M108565200. [DOI] [PubMed] [Google Scholar]
- Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25(1):162–71. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12(3):783–90. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- Guerin JL, Gelfi J, Boullier S, Delverdier M, Bellanger FA, Bertagnoli S, Drexler I, Sutter G, Messud-Petit F. Myxoma virus leukemia-associated protein is responsible for major histocompatibility complex class I and Fas-CD95 down-regulation and defines scrapins, a new group of surface cellular receptor abductor proteins. J Virol. 2002;76(6):2912–23. doi: 10.1128/JVI.76.6.2912-2923.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4(11):868–77. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Nakayama KI. U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun. 2003;302(4):635–45. doi: 10.1016/s0006-291x(03)00245-6. [DOI] [PubMed] [Google Scholar]
- Horvath CM. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur J Biochem. 2004;271(23–24):4621–8. doi: 10.1111/j.1432-1033.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- Huang J, Huang Q, Zhou X, Shen MM, Yen A, Yu SX, Dong G, Qu K, Huang P, Anderson EM, Daniel-Issakani S, Buller RM, Payan DG, Lu HH. The poxvirus p28 virulence factor is an E3 ubiquitin ligase. J Biol Chem. 2004;279(52):54110–6. doi: 10.1074/jbc.M410583200. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A. 1995;92(7):2563–7. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishido S, Wang C, Lee BS, Cohen GB, Jung JU. Downregulation of major histocompatibility complex class I molecules by Kaposi’s sarcoma-associated herpesvirus K3 and K5 proteins. J Virol. 2000;74(11):5300–9. doi: 10.1128/jvi.74.11.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102(5):549–52. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Johnston JB, Wang G, Barrett JW, Nazarian SH, Colwill K, Moran M, McFadden G. Myxoma virus M-T5 protects infected cells from the stress of cell cycle arrest through its interaction with host cell cullin-1. J Virol. 2005;79(16):10750–63. doi: 10.1128/JVI.79.16.10750-10763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsani KR, Hajibagheri MA, Verrijzer CP. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. Embo J. 1999;18(3):698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, MacCallum RM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299(2):499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85(6):829–39. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130–9. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva G, Kolosova I, Maksyutova T, Ryabchikova E, Shchelkunov S. Effects of deletions of kelch-like genes on cowpox virus biological properties. Arch Virol. 2005;150(9):1857–70. doi: 10.1007/s00705-005-0530-0. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Senkevich TG, Chernos VI. A family of DNA virus genes that consists of fused portions of unrelated cellular genes. Trends Biochem Sci. 1992;17(6):213–4. doi: 10.1016/0968-0004(92)90379-n. [DOI] [PubMed] [Google Scholar]
- Li X, Peng H, Schultz DC, Lopez-Guisa JM, Rauscher FJ, 3rd, Marmorstein R. Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 1999;59(20):5275–82. [PubMed] [Google Scholar]
- Lorenzo ME, Jung JU, Ploegh HL. Kaposi’s sarcoma-associated herpesvirus K3 utilizes the ubiquitin- proteasome system in routing class major histocompatibility complexes to late endocytic compartments. J Virol. 2002;76(11):5522–31. doi: 10.1128/JVI.76.11.5522-5531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackett M, Smith GL, Moss B. Vaccinia virus: a selectable eukaryotic cloning and expression vector. Proc Natl Acad Sci U S A. 1982;79(23):7415–9. doi: 10.1073/pnas.79.23.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri M, Bartee E, Gouveia K, Hovey Nerenberg BT, Barrett J, Thomas L, Thomas G, McFadden G, Fruh K. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J Virol. 2003;77(2):1427–40. doi: 10.1128/JVI.77.2.1427-1440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32(Web Server issue):W327–31. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1(4):565–74. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- Mathias N, Johnson SL, Winey M, Adams AE, Goetsch L, Pringle JR, Byers B, Goebl MG. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-toS-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16(12):6634–43. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick A, Ahmad KF, Arai S, Polinger A, Ball H, Borden KL, Carlile GW, Prive GG, Licht JD. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol Cell Biol. 2000;20(17):6550–67. doi: 10.1128/mcb.20.17.6550-6567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AA, Fleming SB, Ueda N. F-Box-Like Domains are Present in Most Poxvirus Ankyrin Repeat Proteins. Virus Genes. 2005;31(2):127–33. doi: 10.1007/s11262-005-1784-z. [DOI] [PubMed] [Google Scholar]
- Minor DL, Lin YF, Mobley BC, Avelar A, Jan YN, Jan LY, Berger JM. The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel. Cell. 2000;102(5):657–70. doi: 10.1016/s0092-8674(00)00088-x. [DOI] [PubMed] [Google Scholar]
- Muller SA, Sasaki T, Bork P, Wolpensinger B, Schulthess T, Timpl R, Engel A, Engel J. Domain organization of Mac-2 binding protein and its oligomerization to linear and ring-like structures. J Mol Biol. 1999;291(4):801–13. doi: 10.1006/jmbi.1999.2996. [DOI] [PubMed] [Google Scholar]
- Nerenberg BT, Taylor J, Bartee E, Gouveia K, Barry M, Fruh K. The poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. J Virol. 2005;79(1):597–601. doi: 10.1128/JVI.79.1.597-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999a;3(4):535–41. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Xiong Y. Association with cullin partners protects ROC proteins from proteasome-dependent degradation. Oncogene. 1999b;18(48):6758–66. doi: 10.1038/sj.onc.1203115. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pintard L, Kurz T, Glaser S, Willis JH, Peter M, Bowerman B. Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol. 2003a;13(11):911–21. doi: 10.1016/s0960-9822(03)00336-1. [DOI] [PubMed] [Google Scholar]
- Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, Peter M. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitinligase. Nature. 2003b;425(6955):311–6. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- Pires de Miranda M, Reading PC, Tscharke DC, Murphy BJ, Smith GL. The vaccinia virus kelch-like protein C2L affects calcium-independent adhesion to the extracellular matrix and inflammation in a murine intradermal model. J Gen Virol. 2003;84(Pt 9):2459–71. doi: 10.1099/vir.0.19292-0. [DOI] [PubMed] [Google Scholar]
- Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW, Branton PE. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15(23):3104–17. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Cooley L. Drosophila kelch is an oligomeric ring canal actin organizer. J Cell Biol. 1997;138(4):799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione KM, Bansal PK, Deffenbaugh AE, Kiss A, Moore JM, Korolev S, Cocklin R, Goebl M, Kitagawa K, Skowyra D. SCF E3-Mediated Autoubiquitination Negatively Regulates Activity of Cdc34 E2 but Plays a Nonessential Role in the Catalytic Cycle In Vitro and In Vivo. Mol Cell Biol. 2007;27(16):5860–70. doi: 10.1128/MCB.01555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer H, Rongo C. KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol Biol Cell. 2006;17(3):1250–60. doi: 10.1091/mbc.E05-08-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, Koonin EV, Buller RM. A poxvirus protein with a RING zinc finger motif is of crucial importance for virulence. Virology. 1994;198(1):118–28. doi: 10.1006/viro.1994.1014. [DOI] [PubMed] [Google Scholar]
- Senkevich TG, Muravnik GL, Pozdnyakov SG, Chizhikov VE, Ryazankina OI, Shchelkunov SN, Koonin EV, Chernos VI. Nucleotide sequence of XhoI O fragment of ectromelia virus DNA reveals significant differences from vaccinia virus. Virus Res. 1993;30(1):73–88. doi: 10.1016/0168-1702(93)90017-h. [DOI] [PubMed] [Google Scholar]
- Shackelford J, Pagano JS. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 2005;41:139–56. doi: 10.1042/EB0410139. [DOI] [PubMed] [Google Scholar]
- Shchelkunov S, Totmenin A, Kolosova I. Species-specific differences in organization of orthopoxvirus kelch-like proteins. Virus Genes. 2002;24(2):157–62. doi: 10.1023/a:1014524717271. [DOI] [PubMed] [Google Scholar]
- Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13(18):2375–87. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson PG, Efstathiou S, Doherty PC, Lehner PJ. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc Natl Acad Sci U S A. 2000;97(15):8455–60. doi: 10.1073/pnas.150240097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6(10):R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Graham K, Schreiber M, Macaulay C, McFadden G. The target DNA sequence for resolution of poxvirus replicative intermediates is an active late promoter. J Virol. 1991;65(1):61–70. doi: 10.1128/jvi.65.1.61-70.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaga M, Hatano M, Takamori M, Yamashita Y, Okada S, Kuroda Y, Tokuhisa T. Bcl6-dependent transcriptional repression by BAZF. Biochem Biophys Res Commun. 2003;303(2):600–8. doi: 10.1016/s0006-291x(03)00396-6. [DOI] [PubMed] [Google Scholar]
- Tortorella D, Gewurz BE, Furman MH, Schust DJ, Ploegh HL. Viral subversion of the immune system. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- Totmenin AV, Kolosova IV, Shchelkunov SN. [Orthopoxvirus genes for Kelch-like proteins. I. Analysis of species specific differences by gene structure and organization] Mol Biol (Mosk) 2002;36(4):610–6. [PubMed] [Google Scholar]
- Tulman ER, Afonso CL, Lu Z, Zsak L, Kutish GF, Rock DL. The genome of canarypox virus. J Virol. 2004;78(1):353–66. doi: 10.1128/JVI.78.1.353-366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulane CM, Horvath CM. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology. 2002;304(2):160–6. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- Wasilenko ST, Stewart TL, Meyers AF, Barry M. Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc Natl Acad Sci U S A. 2003;100(24):14345–50. doi: 10.1073/pnas.2235583100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2(3):169–78. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Wilkins A, Ping Q, Carpenter CL. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev. 2004;18(8):856–61. doi: 10.1101/gad.1177904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425(6955):316–21. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302(5647):1056–60. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004a;24(24):10941–53. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HF, Tomida A, Koshimizu R, Ogiso Y, Lei S, Tsuruo T. Cullin 3 promotes proteasomal degradation of the topoisomerase I-DNA covalent complex. Cancer Res. 2004b;64(3):1114–21. doi: 10.1158/0008-5472.can-03-2858. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zhou P, Howley PM. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell. 1998;2(5):571–80. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- Ziegelbauer J, Shan B, Yager D, Larabell C, Hoffmann B, Tjian R. Transcription factor MIZ-1 is regulated via microtubule association. Mol Cell. 2001;8(2):339–49. doi: 10.1016/s1097-2765(01)00313-6. [DOI] [PubMed] [Google Scholar]