Abstract
Idiopathic hypogonadotropic hypogonadism (IHH) with anosmia (Kallmann syndrome; KS) or with a normal sense of smell (normosmic IHH; nIHH) are heterogeneous genetic disorders associated with deficiency of gonadotropin-releasing hormone (GnRH). While loss-of-function mutations in FGF receptor 1 (FGFR1) cause human GnRH deficiency, to date no specific ligand for FGFR1 has been identified in GnRH neuron ontogeny. Using a candidate gene approach, we identified 6 missense mutations in FGF8 in IHH probands with variable olfactory phenotypes. These patients exhibited varied degrees of GnRH deficiency, including the rare adult-onset form of hypogonadotropic hypogonadism. Four mutations affected all 4 FGF8 splice isoforms (FGF8a, FGF8b, FGF8e, and FGF8f), while 2 mutations affected FGF8e and FGF8f isoforms only. The mutant FGF8b and FGF8f ligands exhibited decreased biological activity in vitro. Furthermore, mice homozygous for a hypomorphic Fgf8 allele lacked GnRH neurons in the hypothalamus, while heterozygous mice showed substantial decreases in the number of GnRH neurons and hypothalamic GnRH peptide concentration. In conclusion, we identified FGF8 as a gene implicated in GnRH deficiency in both humans and mice and demonstrated an exquisite sensitivity of GnRH neuron development to reductions in FGF8 signaling.
Introduction
Vertebrate reproduction depends upon the secretion of gonadotropin-releasing hormone (GnRH), a decapeptide produced by a small number of GnRH neurons in the preoptic area (1). The coordinated pulsatile release of GnRH from this neural network directs the synthesis and secretion of the gonadotropins, which in turn stimulate gonadal steroidogenesis and gametogenesis (2). Although all mammalian species depend upon this unique pathway to initiate reproduction, the genetic program underlying GnRH neuronal ontogeny is poorly understood. GnRH neurons originate in the olfactory placode and undergo a remarkable axophilic migration along the scaffold of olfactory, vomeronasal, and terminal nerves into the forebrain. Ultimately, the GnRH neurons dissociate from their olfactory guiding fibers to reach the preoptic area, where their axons extend into the median eminence (3). These complex developmental events are tightly regulated by specific spatiotemporal expression patterns of growth factors such as FGFs (4), adhesion molecules, and/or diffusible attractants and repellents. Disruption of any of these complex events leads to defects in GnRH secretion and, in severe cases, idiopathic hypogonadotropic hypogonadism (IHH).
IHH is a rare genetic condition characterized by the failure of sexual maturation and infertility (5). This condition can occur in association with anosmia (Kallmann syndrome; KS), or with a normal sense of smell (normosmic IHH; nIHH). The majority of cases are congenital. There is also a small group of patients in whom hypogonadotropic hypogonadism occurs after normal pubertal development; this condition is termed adult-onset hypogonadotropic hypogonadism (AHH; ref. 6). Although IHH was previously thought to be a lifelong condition, a small subset of patients undergoes reversal of hypogonadotropic hypogonadism in adulthood (7). The IHH disease model represents an opportunity to identify genes controlling human reproduction. To date, several loci have been implicated in human GnRH deficiency, including KAL1 (OMIM 308700), GnRH receptor (GNRHR; OMIM 138850), nasal embryonic LHRH factor (NELF; OMIM 608137), FGF receptor 1 (FGFR1; OMIM 136350), G protein–coupled receptor 54 (GPR54; OMIM 604161), prokineticin 2 (PROK2; OMIM 607002), and prokineticin receptor 2 (PROKR2; OMIM 607123). The genetic complexity of this disorder has increased through the recent discovery of an oligogenic inheritance pattern in certain cases (i.e. genetic insults at more than one locus; refs. 8, 9). Furthermore, our experience to date suggests that only 30% of IHH subjects have at least 1 known gene defect, suggesting the existence of unknown loci.
The demonstration that loss-of-function mutations in FGFR1 caused IHH disclosed a key role for the FGF signaling system in GnRH ontogeny (10, 11). Mammalian FGF signaling commences at the cell surface through tightly regulated interactions of 7 principal FGFR isoforms, 18 FGFs, heparan sulfate (HS) proteoglycans, and, in certain instances, other coreceptors such as klotho proteins (12). At least 11 different FGF ligands are capable of activating FGFR1 (13); however, to our knowledge, no specific FGF has been implicated in GnRH ontogeny.
The coalescence of several findings led us to hypothesize that FGF8 is a key ligand for FGFR1c in the etiology of GnRH deficiency. First, a proband was identified with KS and cleft palate who harbors the L342S mutation in the FGFR1c gene (8). This mutation has previously been shown to dramatically reduce the binding affinity of the receptor for FGF8b with minor loss in binding affinity for FGF1 and FGF2 (8). Second, defective nasal cavity development and olfactory bulb dysgenesis in Fgf8 hypomorphic mice was previously described (14). Third, we noted overlapping patterns of FGF8 and FGFR1 expression in the brain in the Genomics Institute of the Novartis Research Foundation (GNF) tissue atlas ( http://symatlas.gnf.org/SymAtlas/). FGF8 was first identified as a mitogen (15), but a number of subsequent studies have indicated that it acts as a neural morphogen regulating expression of downstream genes that control neural patterning (16–19). More recently, FGF8 was shown to be involved in survival of developing neuronal cells (20, 21). In the present study, we demonstrated a critical role of FGF8 in GnRH neuron ontogeny in humans and mice and established FGF8 as what we believe to be a novel locus for IHH.
Results
FGF8 mutational analysis
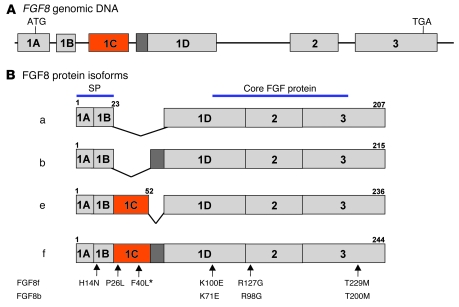
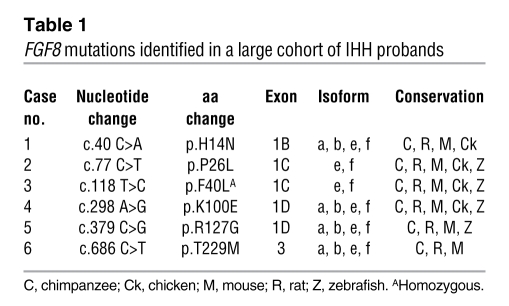
A schematic of the genomic structure of the human FGF8 gene and its 4 alternatively spliced products are shown in Figure 1. An alternative splicing event at the N terminus of the ligand gives rise to 4 isoforms — a, b, e, and f — in humans (22). Of the 461 unrelated IHH probands screened, we identified 6 FGF8 mutations (<2%), specifically 3 familial cases (2 KS and 1 nIHH) and 3 sporadic cases (2 nIHH and 1 AHH; Tables 1 and 2 and Figures 1 and 2). The FGF8 mutations are numbered according to the FGF8f isoform, the largest FGF8 ligand, which is encoded by all exons. The 4 common mutations are also numbered according to FGF8b isoform (Figure 1). These 4 common mutations are a heterozygous p.H14N mutation within the signal peptide region and 3 heterozygous mutations, p.K100E (K71E in FGF8b), p.R127G (R98G in FGF8b), and p.T229M (T200M in FGF8b), that map to the FGF constant region (Figure 1). The heterozygous p.P26L and homozygous p.F40L mutations are located in exon 1C and thus affect both FGF8e and FGF8f isoforms. None of these mutations were detected in 180 ethnically matched controls, and all were highly conserved across species (Table 1) and within the FGF8 subfamily.
Figure 1. Genomic structure and differential splicing of the human FGF8 gene.
(A) Structure of the FGF8 gene. Boxes denote exons; lines denote introns. (B) Schematic of the 4 FGF8 isoforms identified in humans, which differ with regard to the inclusion of exon 1C and part of exon 1D. Most of the conserved FGF core is encoded by exons 2 and 3. Numbers above exons denote aa numberings for each isoform. The mutations identified to date are indicated by arrows numbered according to the FGF8f and FGF8b protein isoforms. Asterisk denotes the homozygous change.
Table 1 .
FGF8 mutations identified in a large cohort of IHH probands
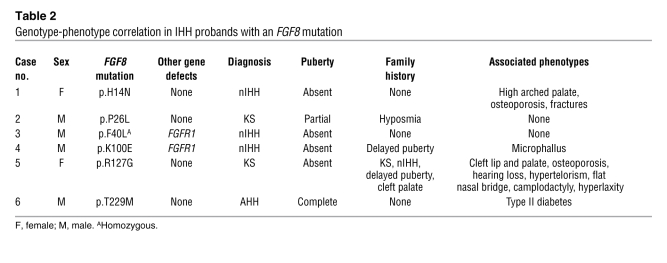
Table 2 .
Genotype-phenotype correlation in IHH probands with an FGF8 mutation
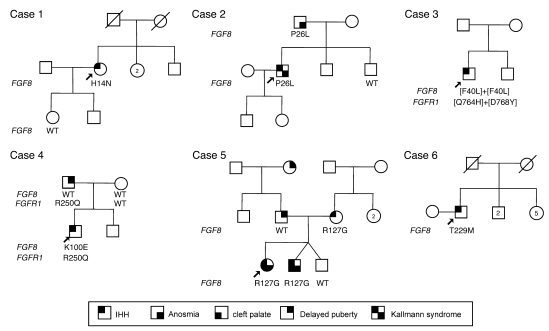
Figure 2. Pedigrees of probands carrying an FGF8 mutation.
Arrows indicate the probands. Circles denote females; squares denote males; struck-through symbols denote deceased subjects; numbers within symbols denote number of individuals. Phenotypes are as described in Results.
Oligogenicity in IHH
Probands harboring an FGF8 mutation were screened for other loci underlying IHH. We identified an additional double-heterozygous FGFR1 mutation (c.2292 G>T + c.2302 G>T, p.[Q764H+D768Y]) in case 3 and an additional heterozygous FGFR1 mutation (c.794 G>A, p.R250Q) in case 4.
Genotype-phenotype correlation
Case 1; p.H14N.
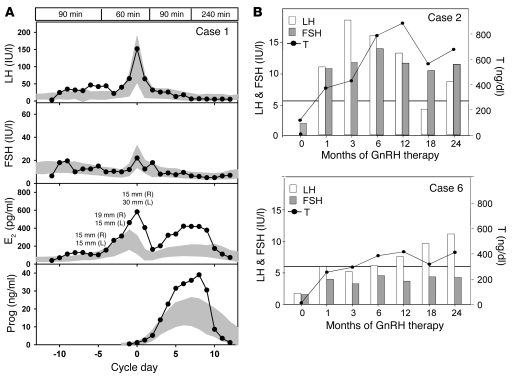
The 32-yr-old nIHH female proband of mixed European descent (Table 2) presented at age 17 yr with primary amenorrhea, absent puberty, eunuchoidal proportions (arm span, 179 cm; height, 170 cm), high arched palate, and hyperlaxity of the digits. Initial radiologic studies revealed a prepubertal uterus and very small ovaries, normal kidneys, normal formal smell testing (sixtieth percentile), and markedly delayed bone age (13 yr 6 mo for a chronologic age of 17 yr). MRI indicated a small pituitary gland with normal olfactory structures. Her neuroendocrine evaluation revealed an apulsatile luteinizing hormone (LH) secretion pattern (mean LH, 2.55 IU/l), undetectable serum estradiol (E2; <20 pg/ml), and low follicle-stimulating hormone (FSH; 5.3 IU/l). Interestingly, while insulin tolerance test induced appropriate hypoglycemia (nadir glucose, 41 mg/dl), she exhibited a suboptimal growth hormone response (0.5 to 2.3 IU/l) with an otherwise normal pituitary reserve. She was diagnosed with nIHH. Subsequently, a wrist fracture after a minor trauma led to a bone density scan revealing very low bone density (lumbar T-score, –3.5). At age 18, she received pulsatile GnRH at a dose of 100 ng/kg at a physiologic frequency, producing 2 dominant follicles and ovulation (Figure 3A). On this therapy, she conceived 2 children. Genetic screening revealed a p.H14N FGF8 mutation. H14 is located within the hydrophobic signal peptide of the protein (Table 1 and Figures 1 and 2) and is highly conserved across multiple mammals and other vertebrates. The proband’s daughter, WT for FGF8, initiated pubertal development at age 11 yr, and her son was prepubertal at age 10 yr (Figure 2).
Figure 3. Response to GnRH therapy in 3 probands harboring an FGF8 mutation.
(A) Pulsatile GnRH was administered at a dose of 100 ng/kg i.v. at a physiologic frequency to the proband described in case 1, demonstrating a normal increase in LH and FSH, an increase in E2 consistent with development of 2 follicles, and an increase in progesterone (Prog) consistent with ovulation. Data are centered to the day of ovulation (0 d); for E2, follicle diameters are indicated; shaded areas represent mean ± 1 SD hormone levels in 109 control women; boxed regions at top denote GnRH pulse frequency. (B) LH, FSH, and T responses to GnRH therapy in 2 male patients (cases 2 and 6). Horizontal lines denote lowest value of the normal range for adult serum T levels. To convert serum T values from ng/dl to nmol/l, divide by 28.84.
Case 2; p.P26L.
The male KS proband of mixed European descent was diagnosed with hypogonadism at age 16 yr (Table 2). Subsequent testosterone (T) therapy induced virilization; at age 28, he presented to MGH for fertility treatment. At that time he was eunuchoidal (weight, 83 kg; height, 178 cm; arm span, 183 cm), had testicular volumes of 9 ml, and had a decreased sense of smell (less than fifth percentile) consistent with partial KS. His 12-h neuroendocrine evaluation revealed apulsatile LH secretion (mean LH, 1.8 IU/l), undetectable FSH (<1.6 IU/l), and a serum T level of 13 ng/dl, with otherwise normal anterior pituitary function. Brain MRI revealed a partial empty sella and bilateral hypoplastic olfactory bulbs and tracts, while renal ultrasound and bone density were normal. On pulsatile GnRH therapy, serum T level reached the normal range within 2 mo (Figure 3B); he developed sperm in his ejaculate (range, 18 × 106 to 80 × 106 sperm/ml) and subsequently conceived 2 children. He was found to carry the heterozygous c.77 C>T; p.P26L FGF8 mutation (Table 1 and Figure 1). Notably, his father carried the same FGF8 mutation and had a history of decreasing olfaction, while the proband’s mother was WT for FGF8 and asymptomatic. The proband has 2 brothers with a history of normal pubertal development. The proband’s 5-yr-old son was diagnosed with Asperger syndrome. His 3 yr-old daughter had seemingly normal development (Figure 2).
Case 3; p.[F40L]+[F40L].
The 19-yr-old male nIHH proband was evaluated at age 15.5 yr for delayed puberty (Table 2). He was unvirilized and overweight (BMI, 32), with prepubertal testes (1–2 ml) and pubic hair at Tanner stage II. Subsequent reevaluation revealed normal brain MRI, hypogonadal serum T level, and undetectable serum gonadotropins, with minimal increase after GnRH stimulation (LH, 2.2 IU/l; FSH, 4.5 IU/L). Initiation of T therapy induced virilization, and after 1.5 yr of T therapy his testicular volume increased to 6–8 ml. The proband carried a homozygous FGF8 mutation, p.[F40L]+[F40L] (Table 1 and Figure 1). In addition, he also carried an additional double-heterozygous FGFR1 mutation, p.[Q764H/D768Y]. This subject was lost to follow-up.
Case 4; p.K100E.
The nIHH male proband of mixed European descent was born with microphallus (Table 2). At 5.5 mo of age, his serum T level and gonadotropins were undetectable (<6 ng/dl and <1.6 IU/l; respectively), consistent with IHH. T therapy for 6 mo induced phallus growth. The proband was 10 yr of age at the termination of this study and had normal growth, development, and olfaction (smell testing, twenty-ninth percentile). He carried a de novo heterozygous FGF8 mutation, p.K100E (Table 1 and Figure 1), because his parents were both WT for that gene (Figure 2). Interestingly, both the proband and his father — who exhibited a normal sense of smell (forty-third percentile), bilateral hearing loss, and a history of delayed puberty — carried an additional heterozygous FGFR1 mutation, p.R250Q. It is notable that several second cousins on the father’s side had a history of delayed puberty. The mother had menarche at age 13 and had a normal sense of smell (thirty-ninth percentile). The proband’s deceased maternal grandfather had 3 kidneys. The proband’s prepubertal brother had no evident clinical anomalies at age 8 yr (Figure 2).
Case 5; p.R127G.
The KS female proband of mixed European descent was born with cleft lip and palate and presented at age 14 with primary amenorrhea, no breast development, short stature (Z-score for height, –3.5), hypertelorism, flattened bridge of the nose, hyperlaxity of the digits, mild scoliosis, and camplodactyly (Table 2). Further evaluation revealed anosmia (smell testing, less than fifth percentile), color blindness, and bilateral hearing loss. Serum E2 and gonadotropins were undetectable (<20 pg/ml and <1.6 IU/l, respectively), and imaging studies revealed apparent normal olfactory bulbs and nerves, normal renal ultrasound, and very low bone density (lumbar Z-score, –2.6). At age 19, she underwent a 3-mo washout from contraceptive therapy and remained amenorrheic, with undetectable serum gonadotropins and low serum E2 (23 pg/ml), consistent with a severe form of KS. She carried a heterozygous FGF8 mutation, p.R127G, located in the core region of FGF (Table 1 and Figure 1). The proband’s mother, who harbored the FGF8 mutation, had nIHH (Figure 2). On gonadotropin therapy, she conceived twice. The dizygotic twins had markedly different phenotypes: one harbored the same FGF8 mutation and had severe KS with microphallus, undescended testes, absent puberty, and cleft lip/palate. The other twin underwent a normal puberty, but with short stature, and did not carry the FGF8 mutation. The proband’s father, WT for FGF8, had a normal sense of smell and a history of delayed puberty. Interestingly, the proband’s paternal grandmother also had a history of delayed puberty (Figure 2).
Case 6; p.T229M.
The AHH male proband of mixed European descent presented at age 40 for infertility and decreased libido (Table 2). He denied exposure to toxins as well as history of depression or stressors and reported normal puberty, with documented normal semen analyses up to 2 yr prior to diagnosis. At presentation, he was well virilized, with a BMI of 28.4 and a testicular volume of 11 ml bilaterally. His 12-h neuroendocrine evaluation revealed undetectable serum gonadotropins (<1.6 IU/l) and frankly hypogonadal T levels (40 mg/dl). Radiologic studies revealed normal renal ultrasound, brain MRI, and bone density. He was diagnosed with AHH and initiated pulsatile GnRH therapy, achieving normal serum gonadotropins and T levels within 1 mo (Figure 3) and developing normal sperm counts (range, 8 × 106 to 92 × 106 sperm/ml) while on therapy. Subsequently, he was diagnosed with Graves disease, type II diabetes, and hypertension. He carried a heterozygous FGF8 mutation, p.T229M, located in the C-terminal tail of FGF8 (Table 1 and Figure 1). He had no family history of reproductive or olfactory defects. His maternal aunt had type I diabetes (Figure 2).
Structural and biochemical analysis of FGF8 mutations demonstrates that they reduce the function of FGF8
Structural analysis using FGF8b isoform.
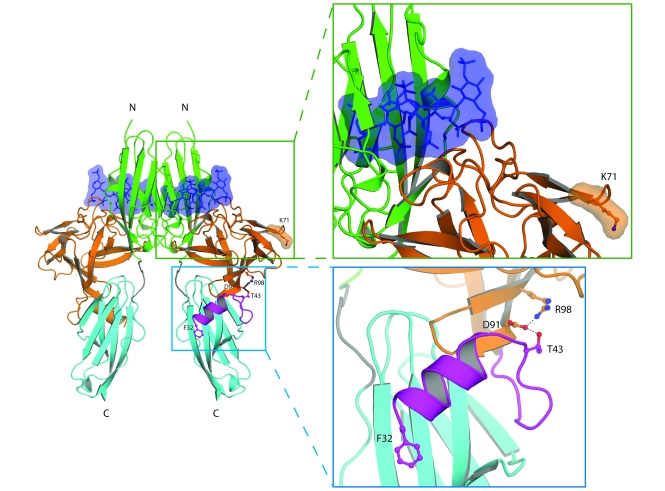
The crystal structure of the FGF8b/FGFR2c complex has previously been solved (23), and this served as a template to predict the functional impact of 2 mutations shared by all 4 isoforms: K71E and R98G in FGF8b (Figure 4). To study the potential impact of these 2 mutations on heparin/HS binding affinity of FGF8b, we constructed a 2:2:2 FGF8b/FGFR2c/heparin model by superimposing FGF8b from the FGF8b/FGFR2c complex onto each of the 2 FGF2 molecules in the 2:2:2 FGF2/FGFR1c/heparin structure (Protein Data Bank identifier 1FQ9; ref. 24). In the model, K71 is located at the periphery of the heparin binding site of FGF8, and the K71E mutation can negatively impact interactions of FGF8 with the reducing end of longer heparin/HS chains. R98 is located in the β5 strand and makes 3 intramolecular hydrogen bonds. One of these bonds is with the side chain of D91 (in the β4 strand), and the others are with the backbone oxygen of residues 122 and 123 in the loop between the β7 and β8 strands. D91, in turn, engages in hydrogen bonding with T43 at the N terminus of FGF8b, which indicates that R98 indirectly contributes to the conformational stability of the N terminus. The R98G mutation reduces binding affinity to FGFR by introducing flexibility into the N terminus of the ligand, a region that plays a critical role in binding of FGF8 to FGFRs (23). Alternatively, loss of the intramolecular hydrogen bonds destabilizes FGF8b protein as a whole. The T200M mutation maps to the very C-terminal end of FGF8b, which is not present in the crystallized FGF8b protein. The functional consequence of the mutation is not clear, because deletion of the last C-terminal residues was previously found to have no impact on receptor binding in surface plasmon resonance experiments (23).
Figure 4. Structural modeling of K71E and R98G FGF8 mutations predicts that they are loss-of-function mutations.
The locations of the mutated FGF8 residues are mapped onto the ribbon diagram of 2:2:2 FGF8b/FGFR2c/heparin complex. Orange and purple denote the core and N terminus, respectively, of FGF8b. Green, cyan, and gray denote D2, D3, and D2-D3 linker, respectively, of the extracellular FGFR2c ligand-binding region. The 2 heparin oligosaccharides are rendered as blue sticks and surface representations. Also shown are detailed views of the regions where the mutated FGF8 residues are located. Selected residues are shown as ball-and-stick representations, as is F32 of FGF8b N terminus to underscore the importance of the FGF8b N terminus in FGF8b-FGFR binding. Blue balls, nitrogen atoms; red balls, oxygen atoms. Hydrogen bonds are shown as dashed lines. The N and C termini of polypeptide chains are indicated.
Expression of FGF8 mutants.
Of the 4 human FGF8 isoforms, only FGF8a and FGF8b have previously been studied in brain patterning (23). FGF8b is a stronger morphogen than FGF8a due to higher affinity for its cognate FGFRs (including FGFR1c) generating a stronger signal. These findings have been traced to the additional contacts between F32 at the alternatively spliced N terminus of FGF8b and D3 of FGFR (23), while FGF8a lacks F32. Notably, F32 is present in FGF8f, but not in FGF8e, consistent with the stronger mitogenic activity of FGF8f in vitro (25).
We therefore studied mutations in the common shared region using the FGF8b protein and the 2 mutations in exon 1C using the FGF8f protein (Figure 1). WT or mutant proteins were expressed in E. coli as inclusion bodies. The expression of WT FGF8b protein was greater than that of WT FGF8f, while mutant proteins had expression levels similar to those of their respective WT proteins. Expressed proteins were refolded and purified using heparin affinity. Attempts to further purify by ion exchange and size exclusion chromatographies reduced the protein yields, due to the limited solubility of FGF8 proteins. In particular, we observed that FGF8f was substantially less soluble than FGF8b (data not shown). Therefore, we decided to use the semipurified protein from the heparin column for the cell-based assay. Of note, the purity of FGF8b proteins following heparin affinity purification was markedly greater than that of FGF8f proteins.
Impact on FGFR1c activation.
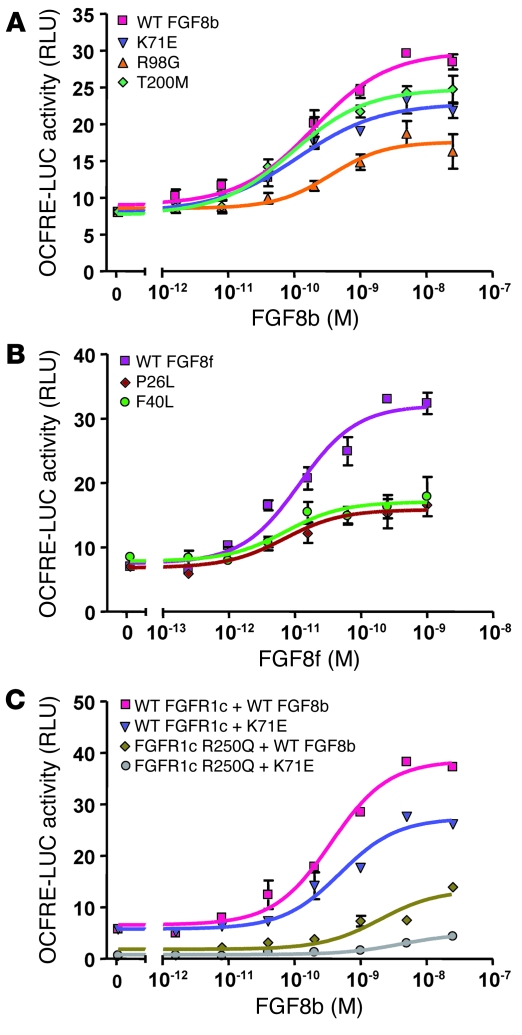
The functionality of the FGF8 mutants was evaluated in vitro using a transcriptional reporter assay. Rat L6 myoblast cells were transiently transfected with WT FGFR1c and induced with increasing concentrations (0–25 nM) of WT or mutant FGF8b or FGF8f. Receptor activation was quantified using a luciferase reporter gene driven by an osteocalcin FGF response element (OCFRE; ref. 8). Both WT FGF8b and WT FGF8f induced a dose-dependent response with a maximal 4-fold induction (Figure 5, A and B). While all 3 FGF8b mutants exhibited significantly decreased activity relative to WT (42%–76% of WT, P < 0.0001; Figure 5A), the R98G mutant was the most severely effected (42% of WT), consistent with the prediction from the structural analysis. The 2 FGF8f mutants were also observed to be loss-of-function mutations, displaying activity levels 34%–39% that of WT (P < 0.0001; Figure 5B).
Figure 5. FGF8b mutants are loss-of-function mutations, consistent with structural prediction.
L6 myoblasts were transiently transfected with WT and mutant FGFR1 cDNA along with the OCFRE luciferase reporter, and then stimulated with increasing doses of WT or mutant FGF8. Luciferase activity was plotted as mean ± SEM of 3 triplicates, and a dose-response curve was fitted as described in Methods. The results of a representative experiment is shown. (A) Reporter activity of FGF8b WT and mutants. (B) Reporter activity of FGF8f WT and mutants. (C) Reporter activity of FGF8b WT and K71E mutant assayed on cell transfected with either WT or R250Q mutant FGFR1c.
Using the FGF reporter assay, we further evaluated the effects on FGF signaling from FGF8 and FGFR1c mutants combined. In these experiments, the reporter activity was assayed using a combination of WT or mutant FGF8b and FGFR1c. While the combination of K71E FGF8b and WT FGFR1 evoked 70% of the WT/WT signaling activity, the paired ligand/receptor mutants showed severely compromised activity (Figure 5C).
FGF8 is required for GnRH neuron development
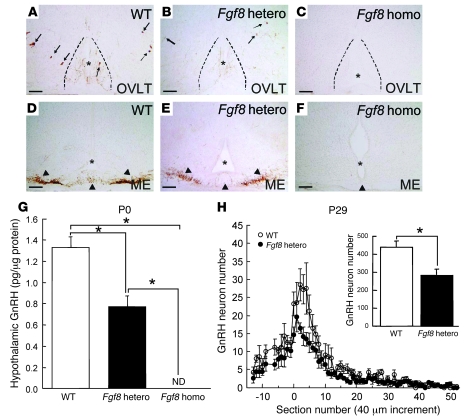
To further establish the role of FGF8 in GnRH ontogeny, we studied Fgf8 hypomorphic mice. The homozygous Fgf8 hypomorphs were previously reported to have a 55% reduction in the functional FGF8 protein levels and to die within 1 d of birth (14). Immunohistochemistry (IHC) studies showed that at P0, GnRH neuron perikarya were completely absent in regions ranging from the preoptic area to the hypothalamus (Figure 6C). GnRH fibers were also absent in the median eminence (Figure 6F) and other brain regions. The WT littermates, in contrast, possessed a normal complement of GnRH neurons and fibers (Figure 6, A and D), and the heterozygous Fgf8 hypomorphs exhibited a reduced number of GnRH neurons (Figure 6, B and E). RIA data confirmed the absence of GnRH peptide in the hypothalamic extracts of the P0 homozygous mutants (Figure 6G). Interestingly, GnRH peptide concentrations were reduced by about 40% in the heterozygous hypomorph mutants (Figure 6G), demonstrating a sensitive and dose-dependent effect of FGF8. The loss of the GnRH system in mutant heterozygotes was irreversible and persisted until P29 (Figure 6H). Although reduced in number (Figure 6H, inset), the distribution of these GnRH neurons in the brains of heterozygous mutants was largely similar to that of the WT littermates (Figure 6H).
Figure 6. Study of GnRH neurons in Fgf8 hypomorphic mice.
(A–F) GnRH ICC illustrating the complete absence of GnRH neuron fibers and cell bodies in P0 homozygous Fgf8 hypomorphic mice (C and F) in the preoptic area and median eminence (ME) compared with WT controls (A and D) and heterozygous mice (B and E). Arrows denote GnRH neuron cell bodies; arrowheads denote GnRH fibers. Asterisks and dotted lines delineate the medial aspects of the brain. OVLT, organum vasculosum of the lamina terminalis. Scale bars: 100 μm. (G and H) The level of immunoreactive GnRH markedly decreased in Fgf8 heterozygotes at all postnatal ages measured. (H) Distribution of GnRH neurons in the preoptic-hypothalamic area of P29 WT controls and Fgf8 heterozygotes. Numbers in the x axis indicate increments of 40-μm sections. Negative and positive numbers indicate sections rostral and caudal to the organum vasculosum of the lamina terminalis (denoted 0), respectively. The distribution of GnRH neurons was largely normal in the transgenic mice, except that there were fewer GnRH neurons. Fgf8 heterozygotes had markedly fewer GnRH neurons in the preoptic-hypothalamic area compared with age-matched controls. ND, not detected. Data are mean ± SEM. *P < 0.05.
Discussion
Loss-of-function mutations in FGF8 underlie both KS and nIHH. Therefore, we believe FGF8 to be a novel locus for human GnRH deficiency. These mutations manifest in variable onsets and severities of GnRH deficiency, ranging from failure to initiate pubertal development to reproductive failure after completion of sexual maturation (i.e., AHH). Further support for a critical role of FGF8 in GnRH neuronal development was provided by our studies of Fgf8 hypomorphic mice, which retained approximately 50% of Fgf8 expression, survived up to birth, and lacked any GnRH neurons and fibers in the preoptic area and median eminence. Furthermore, the heterozygous Fgf8 hypomorphic mice survived through adulthood, but exhibited markedly decreased numbers of forebrain GnRH neurons, indicating an exquisite sensitivity of the decreased FGF8 signaling for GnRH neuron development.
We documented loss-of-function FGF8 mutations in subjects with KS who presented with variable degrees of GnRH deficiency. Our observations in these clinical cases indicate that the role of FGF8 in the ontogeny of GnRH neurons is intimately tied to olfactory epithelium and/or bulb development. We demonstrated that Fgf8 hypomorphic mice, which exhibit no olfactory bulbs (14), lacked GnRH neurons in the hypothalamus. Furthermore, in mice lacking Fgf8 expression in the FoxG1 domain, the olfactory placode forms, but fails to differentiate into an olfactory epithelium because of massive apoptosis of neuronal stem cells during the early stages of olfactory pit development (26). Therefore, decreased FGF8 signaling could disrupt olfactory bulb morphogenesis and lead to defects in GnRH neuronal migration. In addition, because both GnRH and olfactory neurons arise from the olfactory placode (27), a reduction in FGF8 signaling could also alter GnRH fate cell specification. Further studies on Fgf8 hypomorphic or deficient mice will shed light on the mechanisms by which decreased FGF8 signaling leads to KS.
On the other hand, some probands with IHH harboring loss-of-function FGF8 mutations exhibited apparently normal olfactory function and displayed GnRH deficiency before and after completion of puberty. These data suggest that FGF8 signaling must play an additional role in GnRH ontogeny beyond patterning the olfactory epithelium and/or bulbs. Interestingly, both FGFRs and FGFs are expressed in the olfactory placode, along the GnRH neuron migratory path, and within the adult hypothalamus (ref. 28 and the GNF tissue atlas). Furthermore, FGF signaling has also been implicated in GnRH neurite outgrowth and appears to play a role in GnRH axon guidance to the median eminence (29), which suggests that FGF8 could also act as a chemoattractant. Thus, decreased FGF8 signaling could lead to an isolated defect in GnRH neuron migration and/or axon targeting without affecting the olfactory system. Finally, our discovery of a naturally occurring FGF8 mutation in a subject with the rare AHH established a genetic component in the pathogenesis of AHH (6). Interestingly, FGF8 signaling is involved in the survival of neuronal cells (20, 21). Taken together, these data and our present results suggest that decreased FGF8 signaling, either alone or in combination with other genetic and/or environmental insults, modulates long-term GnRH neuron survival.
Beyond the reproductive and olfactory phenotypes, several other clinical characteristics — including cleft palate, synkinesia, scoliosis, and low bone mass — were noted in probands and family members carrying FGF8 mutations. These are also consistent with the wide expression pattern of FGF8 during embryogenesis and adulthood (14, 30, 31). Both KS siblings in the family of case 5 carrying the p.R127G mutation have cleft lip/palate. Reminiscent of pedigrees harboring FGFR1 KS mutations (10, 11, 32), the family demonstrated incomplete penetrance of the FGF8 mutation for cleft lip/palate. Of note, cleft palate has been documented in the Fgf8 hypomorphic mice (14), and a report of a putative FGF8 mutation in the pathogenesis of human cleft palate has recently been published (33). Additional bone phenotypes were seen in 2 IHH probands harboring a FGF8 mutation, with low bone mass and a history of fractures from minor trauma. While the association of osteopenia and GnRH deficiency has been previously attributed to hypogonadal sex steroid levels (34), the severity of the bone phenotype in these probands was particularly striking and suggests a possible additional effect of decreased FGF8 signaling on bone development. Interestingly, FGF8 is critical for normal limb formation (35). Furthermore, FGF8 stimulates differentiation of osteoblasts and bone formation in vitro (36).
We documented variable phenotypic expressivity within family members carrying the same FGF8 mutation. Furthermore, both heterozygous and homozygous FGF8 mutations underlaid severe GnRH deficiency. Recently, we have demonstrated that IHH may be caused by or modulated by more than one gene defect (8). Consistent with this model of oligogenicity, we identified an additional FGFR1 mutation in 2 IHH probands. In case 4, mutations in both the receptor and ligand (FGFR1 and FGF8) clearly synergized to cause severe GnRH deficiency in the IHH proband (Figure 5C). As decreases in both FGFR1 (37) and FGF8 signaling caused a reduction in the number of GnRH neurons in the hypothalamus, it is conceivable that combined defects in this pathway result in a further decrease in GnRH neuron number and/or fitness and ultimately severe human GnRH deficiency. Further examples of synergistic mutations within a pathway have previously been described in diseases such as Hirschsprung disease (RET and GDNF; ref. 38), or retinitis pigmentosa (retinal outer segment membrane protein 1 and retinal degeneration slow gene; ref. 39).
We identified 5 loss-of-function FGF8 mutations in the mature FGF8 peptide. Of these, 4 mapped to the shared region of the FGF8 isoforms, thus implicating all 4 FGF8 isoforms, while 2 mutations implicated either the FGF8e or the FGF8f isoforms in GnRH development. Further studies are needed to elucidate the physiologic significance of the multiple splice variants in FGF8 and their specific role in GnRH ontogeny.
Several lines of evidence revealed that FGF8 signaling through FGFR1c plays a key role in GnRH neuronal development. First, loss-of-function mutations in both FGFR1 and FGF8 cause KS and nIHH (10, 11). Second, both FGF8 and FGFR1 are essential for normal telencephalic development, and both mice with targeted ablation of Fgfr1expression in the telencephalon and Fgf8 hypomorphic mice lack olfactory bulbs (14, 40). Third, both mice expressing a dominant-negative form of Fgfr1 in their GnRH neurons (37) and Fgf8 hypomorphic mice exhibit a decrease in the number of, or the absence of, GnRH neurons in the hypothalamus. Despite these similarities in phenotype, there are subtle differences that may be of critical importance in understanding the complexity of the FGFR/FGF system for GnRH ontogeny. Mice deficient in Fgf8 in the Foxg1 domain lack nasal cavities and olfactory epithelium (26), whereas these structures develop in mice deficient in Fgfr1 expression in the Foxg1 domain (40), although their GnRH neuron status in unknown. Therefore, FGF8 may act through other FGFRs during olfactory placode development. FGFR1–FGFR4 are expressed in the olfactory epithelium (28). In addition, surface plasmon resonance and mitogenic studies have previously shown that FGF8b also binds and signals through FGFR2c, FGFR3c, and FGFR4 (23, 25).
Taken together, the results of our clinical human studies and mouse genetic studies demonstrated a critical role for FGF8 signaling in GnRH neuronal development and a great sensitivity of GnRH ontogeny to reduced FGF8 signaling. Our human genetic studies also pointed to a critical role for FGF8f or FGF8e protein isoform in GnRH ontogeny. It is hoped that this report will provide the foundation for further examination of the complex biology of FGF signaling in GnRH neuron development.
Methods
Subjects.
The Human Research Committee of Massachusetts General Hospital approved this study, and all subjects provided written informed consent before participation. Included in the study were 237 KS, 193 normosmic IHH, and 21 AHH probands from unrelated pedigrees. KS and nIHH subjects had failed to initiate or complete puberty by 18 yr of age. Two subjects were diagnosed earlier in life because of microphallus and failed activation of the hypothalamo-pituitary-gonadal axis during the first 6 mo of age (41). AHH subjects had normal pubertal development, with GnRH deficiency diagnosed later in adulthood (6). Additionally, all subjects had the following: (a) serum T 100 ng/dl for men or E2 20 pg/ml for women in association with low or normal levels of serum gonadotropins; (b) otherwise normal pituitary function; (c) normal serum ferritin concentrations; and (d) normal MRI of the hypothalamic-pituitary region. The control population of mixed European descent consisted of 180 healthy adult volunteers as assessed by history and clinical examination (n = 360 chromosomes).
Mutation analysis of FGF8.
Sequencing of the coding regions of the FGF8 gene (GenBank accession no. NM 033163) was performed as previously described (42). Amplified products were sequenced in both directions using the AmpliTaq Dye Terminator Cycle Sequencing kit and an ABI Prism 377 DNA sequencer (Perkin-Elmer). All sequence variations were found on both strands and confirmed in a separate PCR reaction. All genes and proteins are described using standard nomenclature (43). Nonsense changes resulting in a truncated protein, frameshift, insertion, or deletion were categorized as definite mutations. Nucleotide changes that were absent from the SNP and expressed sequence tag NCBI databases, that were absent in 360 ethnically matched control chromosomes, that were evolutionarily conserved across species, and that resulted in aa substitutions were also identified as disease-causing mutations.
Genetic screening of other IHH loci.
Subjects found to harbor a FGF8 mutation were also screened for other loci underlying IHH: KAL1 (GenBank accession no. M97252; ref. 44), GNRHR (NM_000406; ref. 45), NELF (NM_015537; ref. 8), FGFR1 (BC018128; ref. 46), GPR54 (AY253981; ref. 47), PROK2 (NM_021935; ref. 48), and PROKR2 (NM_144773; ref. 9).
Clinical and biochemical studies.
A detailed individual and family history of pubertal development and associated reproductive and nonreproductive phenotypes (i.e., anosmia, dental agenesis, cleft lip/palate, hearing loss, and synkinesia) was obtained. Probands underwent a complete physical examination, including Tanner staging and testicular volume measurement using a Prader orchidometer. Classification of absent, partial, or complete pubertal development was made based on clinical history, testicular size, or both (49). Olfactory acuity was assessed by clinical history and confirmed with quantitative smell testing when possible (50). When possible, subjects underwent a detailed neuroendocrine evaluation at the MGH General Clinical Research Center. This involved an overnight 12-h frequent blood-sampling study for LH (every 10 min for 12 h) after withdrawal of hormonal therapy. The pattern of LH secretion was analyzed using a validated modification of the Santen and Bardin method (51). A pool of the 12-h sampling was also assayed for FSH, T, E2, and inhibin B. In men able to produce an ejaculate, seminal fluid analysis was performed. Radiologic phenotyping included visualizing the olfactory system with MRI (32), renal ultrasound, and bone densitometry via dual energy X-ray absorptiometry. Additionally, as part of a separate research protocol, some subjects received pulsatile GnRH therapy as previously described (52, 53). Hormonal and gonadal responses to therapy were assessed in serial clinical visits. Gonadotropin results are expressed in IU/l as equivalents of the second international reference preparation of human menopausal gonadotropins (49).
Predictions of the functional impact of FGF8 mutations using the FGF8b/FGFR2c crystal structure.
The K71E and R98G mutations were mapped onto the FGF8b/FGFR2c crystal structure to predict their effect on FGF8 function (23). In order to study the potential impact of these mutations on the heparin/HS binding activity of FGF8b, we constructed a 2:2:2 FGF8b/FGFR2c/heparin model by superimposing FGF8b from the FGF8b/FGFR2c complex onto each of the 2 FGF2 molecules in the 2:2:2 FGF2/FGFR1c/heparin structure (protein data bank identifier 1FQ9; ref. 24).
Expression and purification of WT and mutant FGF8b and FGF8f ligands.
The DNA fragment encoding mature FGF8b (residues 23–215) and FGF8f (residues 23–244) were amplified by PCR and subcloned into the pET-28a bacterial expression vector. The introduction of K71E, R98G, and T200M mutations into the WT FGF8b construct, and the P26L and F40L mutations into the WT FGF8f construct, was carried out using the QuikChange site-directed mutagenesis kit (Stratagene). The WT and mutant proteins were expressed in E. coli as inclusion bodies, refolded in vitro, and purified using heparin affinity (23). The concentration of FGF8b proteins was determined by 280 nm spectroscopy and confirmed by Coomassie Blue staining. Because of very low yields in FGF8f proteins and the presence of impurities, FGF8f concentrations were estimated using Coomassie Blue staining.
In vitro reporter gene assay.
These assays were carried out as previously described using L6 myoblasts, a cell line largely devoid of endogenous FGFs and FGFRs (8). Cells were transiently transfected at 15% confluence with the full-length human WT FGFR1c expression vector together with the OCFRE luciferase reporter (obtained from D.M. Ornitz, Washington University in St. Louis, St. Louis, Missouri, USA), using Fugene6 reagent (Roche). Following serum starvation, cells were treated with 0–25 nM WT or mutant FGF8 isoform for 16 h in serum-free medium. Luciferase activity was assayed using a Promega Luciferase Assay System. Transfection experiments were performed in triplicate and repeated at least 3 times. A 4-parameter logistic dose-response curve was fitted to each raw data set, and the maximal activity of each mutant was assessed compared with WT activity using Prism software (version 4; GraphPad).
FGF8 hypomorphic mice.
Transgenic mice that were hypomorphic for the Fgf8 allele (14) were maintained as heterozygotes. These hypomorphs have a neomycin resistance gene inserted upstream of the Fgf8 coding sequence. Because the neo cassette contains cryptic splice sites, the level of functional Fgf8 transcript is diminished. All mice were housed under a 12-h light, 12-h dark photoperiod and fed water and rodent chow ad libitum. All animal procedures were approved by the Animal Care and Use Committee of the University of Colorado at Boulder.
GnRH IHC and RIA.
GnRH IHC and RIA were conducted as previously described (37) on animals of mixed sex. Briefly, for IHC, heads of P0 pups were removed, skinned, and immersion-fixed in 4% paraformaldehyde for 6 h. P29 animals were fixed by transcardial perfusion with 4% paraformaldehyde followed by postfixation of the brain in the same fixative for 2 h. Tissues were cryoprotected in 30% sucrose and cut on a cryostat. Thaw-mount coronal sections (20 μm; from P0) or floating sections (40 μm; from P29) were collected and processed for IHC using a rabbit anti-GnRH antiserum (LR5; gift of R. Benoit, Montreal General Hospital, Montreal, Quebec, Canada). GnRH neurons were scored in serial brain sections ranging from the anterior preoptic area to the beginning of the median eminence. Every section was scored by an observer blinded to the identity of the slide. GnRH RIA of hypothalamic extracts followed the protocol described previously (54) using an antiserum (R1245) provided by T.M. Nett (Colorado State University Fort Collins, Fort Collins, Colorado, USA).
Statistics.
For in vivo reporter gene assays, the maximal activity of mutants compared with WT (top of the dose-response curve) was compared using the F test option of the nonlinear curve fitting module of Prism software (version 4; GraphPad). A P value less than 0.05 was considered significant.
Acknowledgments
This work was supported by NIH grants R01 HD015788-21, U54HD028138-16, R01 DE13686-08 (to M. Mohammadi), R01 HD42708, and M01-RR-01066; by the Helsingin Sanomat Foundation; by the Paulo Foundation; and by the Foundation for Pediatric Research of Finland.
Footnotes
Nonstandard abbreviations used: AHH, adult-onset hypogonadotropic hypogonadism; E2, estradiol; FGFR, FGF receptor; FSH, follicle-stimulating hormone; GnRH, gonadotropin-releasing hormone; HS, heparan sulfate; IHC, immunohistochemistry; IHH, idiopathic hypogonadotropic hypogonadism; KS, Kallman syndrome; LH, luteinizing hormone; nIHH, normosmic IHH; OCFRE, osteocalcin FGF response element; T, testosterone.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:2822–2831 (2008). doi:10.1172/JCI34538
Wilson C.J. Chung and Andrew Beenken contributed equally to this work.
References
- 1. Herbison, A.E. 2006. Physiology of the GnRH neuronal network. InKnobil and Neill’s physiology of reproduction. 3rd edition. J.D. Neill, editor. Academic Press. San Diego, California, USA. 1415–1482. [Google Scholar]
- 2.Knobil E., Neill J.D., Johansson E.D. Influence of hypophysectomy, sham hypophysectomy and other surgical procedures on luteal function in the rhesus monkey. Endocrinology. 1968;82:410–415. doi: 10.1210/endo-82-2-410. [DOI] [PubMed] [Google Scholar]
- 3.Bless E.P., et al. Live view of gonadotropin-releasing hormone containing neuron migration. Endocrinology. 2005;146:463–468. doi: 10.1210/en.2004-0838. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Martinez D., Hu Y., Bouloux P.M. Ontogeny of GnRH and olfactory neuronal systems in man: novel insights from the investigation of inherited forms of Kallmann’s syndrome. Front. Neuroendocrinol. 2004;25:108–130. doi: 10.1016/j.yfrne.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Seminara S.B., Hayes F.J., Crowley W.F., Jr. Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann’s syndrome): pathophysiological and genetic considerations. Endocr. Rev. . 1998;19:521–539. doi: 10.1210/er.19.5.521. [DOI] [PubMed] [Google Scholar]
- 6.Nachtigall L.B., Boepple P.A., Pralong F.P., Crowley W.F., Jr. Adult-onset idiopathic hypogonadotropic hypogonadism-a treatable form of male infertility. N. Engl. J. Med. 1997;336:410–415. doi: 10.1056/NEJM199702063360604. [DOI] [PubMed] [Google Scholar]
- 7.Raivio T., et al. Reversal of idiopathic hypogonadotropic hypogonadism. N. Engl. J. Med. 2007;357:863–873. doi: 10.1056/NEJMoa066494. [DOI] [PubMed] [Google Scholar]
- 8.Pitteloud N., et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J. Clin. Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodé C., et al. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dode C., et al. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat. Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 11.Pitteloud N., et al. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6281–6286. doi: 10.1073/pnas.0600962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohammadi M., Olsen S.K., Ibrahimi O.A. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X., et al. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyers E.N., Lewandoski M., Martin G.R. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka A., et al. Cloning and characterization of an androgen-induced growth factor essential for the androgen-dependent growth of mouse mammary carcinoma cells. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8928–8932. doi: 10.1073/pnas.89.19.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuchi-Shimogori T., Grove E.A. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- 17.Garel S., Huffman K.J., Rubenstein J.L. Molecular regionalization of the neocortex is disrupted in Fgf8 hypomorphic mutants. Development. 2003;130:1903–1914. doi: 10.1242/dev.00416. [DOI] [PubMed] [Google Scholar]
- 18.Irving C., Mason I. Signalling by FGF8 from the isthmus patterns anterior hindbrain and establishes the anterior limit of Hox gene expression. Development. 2000;127:177–186. doi: 10.1242/dev.127.1.177. [DOI] [PubMed] [Google Scholar]
- 19.Trainor P.A., Ariza-McNaughton L., Krumlauf R. Role of the isthmus and FGFs in resolving the paradox of neural crest plasticity and prepatterning. Science. 2002;295:1288–1291. doi: 10.1126/science.1064540. [DOI] [PubMed] [Google Scholar]
- 20.Chi C.L., Martinez S., Wurst W., Martin G.R. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- 21.Storm E.E., Rubenstein J.L., Martin G.R. Dosage of Fgf8 determines whether cell survival is positively or negatively regulated in the developing forebrain. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1757–1762. doi: 10.1073/pnas.0337736100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gemel J., Gorry M., Ehrlich G.D., MacArthur C.A. Structure and sequence of human FGF8. Genomics. 1996;35:253–257. doi: 10.1006/geno.1996.0349. [DOI] [PubMed] [Google Scholar]
- 23.Olsen S.K., et al. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 2006;20:185–198. doi: 10.1101/gad.1365406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlessinger J., et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell. 2000;6:743–750. doi: 10.1016/S1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 25.Blunt A.G., et al. Overlapping expression and redundant activation of mesenchymal fibroblast growth factor (FGF) receptors by alternatively spliced FGF-8 ligands. J. Biol. Chem. 1997;272:3733–3738. doi: 10.1074/jbc.272.6.3733. [DOI] [PubMed] [Google Scholar]
- 26.Kawauchi S., et al. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- 27.Wray S., Nieburgs A., Elkabes S. Spatiotemporal cell expression of luteinizing hormone-releasing hormone in the prenatal mouse: evidence for an embryonic origin in the olfactory placode. Brain Res. Dev. Brain Res. 1989;46:309–318. doi: 10.1016/0165-3806(89)90295-2. [DOI] [PubMed] [Google Scholar]
- 28.Gill J.C., Moenter S.M., Tsai P.S. Developmental regulation of gonadotropin-releasing hormone neurons by Fgf signaling. Endocrinology. 2004;145:3830–3839. doi: 10.1210/en.2004-0214. [DOI] [PubMed] [Google Scholar]
- 29.Gibson M.J., Ingraham L., Dobrjansky A. Soluble factors guide gonadotropin-releasing hormone axonal targeting to the median eminence. Endocrinology. 2000;141:3065–3071. doi: 10.1210/en.141.9.3065. [DOI] [PubMed] [Google Scholar]
- 30.Crossley P.H., Martin G.R. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 31.Sun X., Meyers E.N., Lewandoski M., Martin G.R. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitteloud N., et al. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol. Cell. Endocrinol. 2006;254–255:60–69. doi: 10.1016/j.mce.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 33.Riley B.M., et al. Impaired FGF signaling contributes to cleft lip and palate. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finkelstein J.S., et al. Osteoporosis in men with idiopathic hypogonadotropic hypogonadism. Ann. Intern. Med. 1987;104:777–782. doi: 10.7326/0003-4819-106-3-. [DOI] [PubMed] [Google Scholar]
- 35.Mariani F.V., Ahn C.P., Martin G.R. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature. 2008;453:401–405. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valta M.P., et al. Regulation of osteoblast differentiation: a novel function for fibroblast growth factor 8. Endocrinology. 2006;147:2171–2182. doi: 10.1210/en.2005-1502. [DOI] [PubMed] [Google Scholar]
- 37.Tsai P.S., et al. Targeted expression of a dominant-negative fibroblast growth factor (FGF) receptor in gonadotropin-releasing hormone (GnRH) neurons reduces FGF responsiveness and the size of GnRH neuronal population. Mol. Endocrinol. 2005;19:225–236. doi: 10.1210/me.2004-0330. [DOI] [PubMed] [Google Scholar]
- 38.Parisi M.A., Kapur R.P. Genetics of Hirschsprung disease. Curr. Opin. Pediatr. 2000;12:610–617. doi: 10.1097/00008480-200012000-00017. [DOI] [PubMed] [Google Scholar]
- 39.Kajiwara K., Berson E.L., Dryja T.P. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994;264:1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- 40.Hebert J.M., Lin M., Partanen J., Rossant J., McConnell S.K. FGF signaling through FGFR1 is required for olfactory bulb morphogenesis. Development. 2003;130:1101–1111. doi: 10.1242/dev.00334. [DOI] [PubMed] [Google Scholar]
- 41.Grumbach M.M. A window of opportunity: the diagnosis of gonadotropin deficiency in the male infant. J. Clin. Endocrinol. Metab. 2005;90:3122–3127. doi: 10.1210/jc.2004-2465. [DOI] [PubMed] [Google Scholar]
- 42.Yoshiura K., et al. Genomic structure, sequence, and mapping of human FGF8 with no evidence for its role in craniosynostosis/limb defect syndromes. Am. J. Med. Genet. 1997;72:354–362. doi: 10.1002/(SICI)1096-8628(19971031)72:3<354::AID-AJMG21>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 43.Antonarakis S.E. Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum. Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 44.Oliveira L.M., et al. The importance of autosomal genes in Kallmann syndrome: genotype-phenotype correlations and neuroendocrine characteristics. J. Clin. Endocrinol. Metab. 2001;86:1532–1538. doi: 10.1210/jc.86.4.1532. [DOI] [PubMed] [Google Scholar]
- 45.de Roux N., et al. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N. Engl. J. Med. 1997;337:1597–1602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 46.Pitteloud N., et al. Reversible kallmann syndrome, delayed puberty, and isolated anosmia occurring in a single family with a mutation in the fibroblast growth factor receptor 1 gene. J. Clin. Endocrinol. Metab. 2005;90:1317–1322. doi: 10.1210/jc.2004-1361. [DOI] [PubMed] [Google Scholar]
- 47.Seminara S.B., et al. The GPR54 gene as a regulator of puberty. N. Engl. J. Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 48.Pitteloud N., et al. From the Cover: Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17447–17452. doi: 10.1073/pnas.0707173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pitteloud N., et al. The role of prior pubertal development, biochemical markers of testicular maturation, and genetics in elucidating the phenotypic heterogeneity of idiopathic hypogonadotropic hypogonadism. J. Clin. Endocrinol. Metab. 2002;87:152–160. doi: 10.1210/jc.87.1.152. [DOI] [PubMed] [Google Scholar]
- 50.Doty R.L., Applebaum S., Zusho H., Settle R.G. Sex differences in odor identification ability: a cross-cultural analysis. Neuropsychologia. 1985;23:667–672. doi: 10.1016/0028-3932(85)90067-3. [DOI] [PubMed] [Google Scholar]
- 51.Hayes F.J., McNicholl D.J., Schoenfeld D., Marsh E.E., Hall J.E. Free alpha-subunit is superior to luteinizing hormone as a marker of gonadotropin-releasing hormone despite desensitization at fast pulse frequencies. J. Clin. Endocrinol. Metab. 1999;84:1028–1036. doi: 10.1210/jc.84.3.1028. [DOI] [PubMed] [Google Scholar]
- 52.Hoffman A.R., Crowley W.F., Jr. Induction of puberty in men by long-term pulsatile administration of low-dose gonadotropin-releasing hormone. N. Engl. J. Med. 1982;307:1237–1241. doi: 10.1056/NEJM198211113072003. [DOI] [PubMed] [Google Scholar]
- 53.Santoro N., Filicori M., Crowley W.F., Jr. Hypogonadotropic disorders in men and women: diagnosis and therapy with pulsatile gonadotropin-releasing hormone. Endocr. Rev. 1986;7:11–23. doi: 10.1210/edrv-7-1-11. [DOI] [PubMed] [Google Scholar]
- 54.Pak T.R., Lynch G.R., Tsai P.S. Testosterone and estrogen act via different pathways to inhibit puberty in the male Siberian hamster (Phodopus sungorus). Endocrinology. 2001;142:3309–3316. doi: 10.1210/endo.142.8.8321. [DOI] [PubMed] [Google Scholar]