Abstract
The activity of proteins and their complexes often involves the conversion of chemical energy (stored or supplied) into mechanical work through conformational changes. Mechanical forces are also crucial for the regulation of the structure and function of cells and tissues. Thus, the shape of eukaryotic cells (and by extension, that of the multicellular organisms they form) is the result of cycles of mechanosensing, mechanotransduction, and mechanoresponse. Recently developed single-molecule atomic force microscopy techniques can be used to manipulate single molecules, both in real time and under physiological conditions, and are ideally suited to directly quantify the forces involved in both intra- and intermolecular protein interactions. In combination with molecular biology and computer simulations, these techniques have been applied to characterize the unfolding and refolding reactions in a variety of proteins. Single-molecule mechanical techniques are providing fundamental information on the structure and function of proteins and are becoming an indispensable tool to understand how these molecules fold and work.
Mechanical Force as a New Biochemical Parameter
Modern biochemistry tends to regard the cell as a factory crowded with specialized molecular “nanomachines,” mainly proteins acting as single polypeptides or complexes (1). Bio-nanomachines that use mechanical forces are located throughout the cell (from the cell nucleus to the extracellular matrix) and are involved in processes as diverse as replication, transcription, translation, protein folding, protein and nucleic acid unfolding, protein degradation, nucleic acid and protein translocation, organelle transport, muscle elasticity, cell adhesion, membrane fusion, and cell crawling (2, 3). The molecular mechanisms by which mechanical forces influence the structure and function of molecules, cells, and tissues have been elusive because of the lack of appropriate tools. With the recent advent of single-molecule manipulation techniques such as AFM,3 we can now investigate these new biochemical pathways by directly probing bond dynamics in real time and under physiological conditions. These new techniques allow the use of mechanical force as an additional parameter in a biochemical reaction (Fig. 1), which can dramatically affect its rates in both directions.
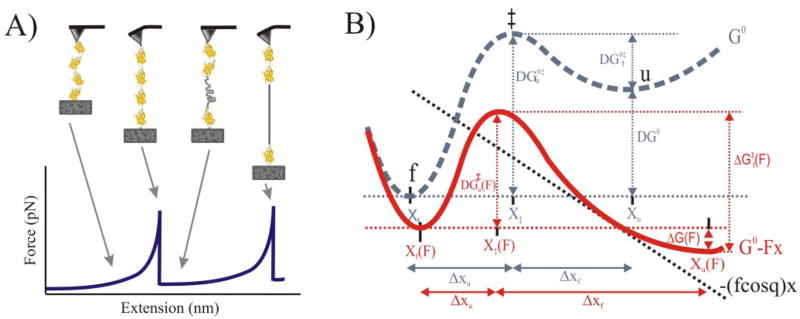
FIGURE 1. Pulling proteins with the AFM and effect of a mechanical force on the unfolding/folding reaction.
A, schematic of the sequence of events during the stretching of a multimeric protein. Stretching the ends of the protein sequentially unfolds the domains, generating a saw-tooth pattern in the force-extension relationship (bottom) that reveals the mechanical properties of the protein (modified from Ref. 11). B, effect of a mechanical force on the free energy diagram of a protein that unfolds following a two-state model (folded (f) and unfolded (u)). The dashed gray curve represents the process in the absence of an applied force. An applied force (black dashed line) tilts the energy diagram of the process, decreasing the barrier to the transition state, ‡ (ΔG‡(F) < ΔG0‡) (red curve). The application of force also lowers the energy of the unfolded state relative to that of the folded state (ΔG(F) < 0). The mechanical reaction coordinate is x (modified from Ref. 3).
Single-molecule Force Spectroscopy of Proteins: Principle and Modes
The AFM was originally developed as a high resolution imaging tool (4) before it began to be used to probe and manipulate atoms and molecules. The so-called “force spectroscopy” or “force-measuring” configuration was designed to record force-extension curves obtained by pulling in a single direction (z axis). Single molecules can be readily analyzed in this way, known as “single-molecule force spectroscopy” (SMFS). In proteins, this technique has been used to characterize the mechanical resistance of both individual polypeptides (intramolecular interactions) and protein-biomolecule bonds (intermolecular interactions) (5).
For these SMFS studies, individual protein molecules or supramolecular protein complexes are first immobilized between the substrate (a glass coverslip) and the force sensor (Fig. 1A). Proteins analyzed are typically made of multiple repeats (or pseudo-repeats) of domains: either naturally occurring modular proteins (e.g. titin, fibronectin, tenascin, spectrin, ankyrin) or engineered polyproteins (6). The periodicity of these proteins provides an unequivocal fingerprint to identify single molecules. As in a lilliputian medieval rack, the protein is then unfolded by moving apart the AFM positioner, which is mechanically coupled to the substrate. This imposes a specific reaction coordinate (i.e. the end-to-end distance) on the unfolding process. By retracting the AFM positioner, the protein can also be refolded in the presence or in absence of mechanical force. These experiments are typically done under non-equilibrium conditions, and two basic AFM modes are currently used depending on the variable being controlled: the more common length clamp, which yields a force-extension curve, and the force clamp, which yields an extension-time curve.
SMFS allows the direct measurement of the mechanical stability of the barriers that a protein offers to its stretching as well as their location. It can measure forces of tens of piconewtons and changes in length with nanometer resolution. Also on the basis of conventional transition state theory, we can estimate the kinetic parameters of the process of forced unfolding/re-folding (7, 8). An applied mechanical force tilts the energy diagram of the process, decreasing the barrier to the transition state and increasing the rate of the forward reaction (Fig. 1B).
We can calculate the probability density for unfolding, which predicts the most likely force of unfolding (mechanical stability) in terms of the spontaneous unfolding rate constant as follows (3, 7, 8): . Using an analytical solution of this type, it is possible to calculate the kinetic parameters for the process: (spontaneous rate of unfolding) and Δxu (width of the activation energy barrier: distance on the reaction coordinate over which the force must be applied to reach the transition state). This equation predicts that the mechanical stability of a protein (F) depends on the unfolding distance Δxu), the height of the barrier , which depends on ), thermal energy, and the loading rate used during extension of the protein (r = dF/dt = k·v, where v is the pulling speed). Also, by relaxing the tethered polyprotein before it breaks and waiting appropriate periods of time, we can also perform refolding experiments (in the presence of force or in its virtual absence) and extract the equivalent parameters of the folding process. Finally, SMFS has recently been used to gather detailed structural information on proteins through a method called “mechanical triangulation” (9) and to probe dynamic rearrangements within the active site of an enzyme with unprecedented resolution (10).
Complementary Techniques: Simulations and Protein Engineering
Typical SMFS experiments can measure the forces required for the mechanical unfolding of protein molecules and can resolve the changes in length with single amino acid resolution (6, 11). However, the basic technique does not typically provide detailed structural information. To this end, computer simulations based on molecular dynamics have proven to be very important for the atomic analysis of this process, as the synergy between experiment and simulation has proven very powerful (12).
One problem in SMFS is that force peaks can originate from a variety of sources other than the unfolding of single protein domains such as the detachment of other molecules from any of the two anchoring points, protein-protein interactions, and disentanglement of molecules or from multiple molecules in parallel. This important drawback was overcome by using modular proteins (13, 14) or recombinant polyproteins (6, 15), in which their periodicity has been successfully used to unambiguously follow the unfolding and refolding pathways of single proteins.
Proteins Studied
“Mechanical proteins” (i.e. proteins with a mechanical function) generate, transmit, or use mechanical forces to carry out their functions and fall into two main subclasses: proteins that generate mechanical forces (biomolecular motors, probed mainly by optical tweezers) and proteins that are subjected to the mechanical forces generated by biomolecular motors or from the environment (e.g. cytoskeletal and cell adhesion proteins, probed mainly by AFM). To analyze the underlying molecular mechanisms involved, “protein nanomechanics” analyzes their mechanics at the single-molecule level by studying the forces, distances, motions, energies, and deformations involved in individual proteins or protein complexes, typically in the submicrometer and subnanonewton ranges. Mechanical forces have also been used to probe the mechanical strength of “non-mechanical proteins” (i.e. those with no mechanical function known). Some of the protein structures analyzed so far include 1) “all-β” structures of the β-sandwich type, including “Ig-like” domains (e.g. from titin, fibronectin, filamin) (6, 14, 16–21), and the β-barrel type (green fluorescent protein) (22, 23); 2) “α + β” proteins (e.g. T4 lysozyme, barnase, ubiquitin, Top7) (24–32); 3) “all-α” structures (e.g. spectrin) (33–35); and 4) several unstructured proteins (e.g. elastin, titin PEVK and N2B) (36–39). In addition to single polypeptides, a few protein complexes have also been studied (27, 40–42).
Molecular Determinants of Mechanical Stability in Proteins
The number of proteins analyzed so far by SMFS is still relatively small (~55 Protein Data Bank structures), and they have been analyzed with a different degree of detail. Although the molecular basis underlying the mechanical resistance of proteins is still unclear, several determinants have been identified through these studies: amino acid sequence, mechanical topology, unloaded unfolding rate constant, and pulling geometry. Some tendencies are already emerging.
(a) Proteins have widely different mechanical stability when pulled in the N → C direction, ranging from below the limit of resolution of the AFM (typically ~10 pN; e.g. calmodulin) to ~330 pN (e.g. titin Ig domains). Interestingly, mechanical proteins that must resist force tend to be more mechanically stable than both non-mechanical and elastomeric proteins. (b) Unstructured and β-spiral proteins (e.g. elastin) are among the less mechanically stable proteins. (c) α-Helical proteins (e.g. calmodulin, T4 lysozyme) have a relatively low mechanical stability, although β-helical bundles (e.g. spectrin (33–35), myosin II tail (40)) and solenoids (e.g. ankyrin B (24)) are more stable. (d) β-Stranded proteins tend to unfold at higher forces than α-helical ones. (e) The mechanical stability of most mechanical proteins tends to be determined by a mechanical clamp usually formed by a patch of highly localized mechanical hydrogen bonds (43). However, in some cases, the hydrophobic core contributes also to mechanical resistance (44). In addition to secondary structure-based elasticity, there is tertiary (e.g. ankyrin B solenoid (24)) and quaternary (e.g. myosin II tail (40), adhesive pili (41)) structure elasticity. (f) The mechanical stability and mechanical unfolding pathways depend also on the pulling geometry, which is affected by both the topology at the breakpoint and the point of application of the force. Hence, β-stranded proteins with a shear mechanical topology at the breakpoint (where the force vector is orthogonal to the hydrogen bonds) are more mechanically stable than zipper β-stranded proteins (where the force vector is parallel to the hydrogen bonds). The points of application of the force to a protein are also relevant, as they can substantially alter its mechanical stability (21, 28), implying that proteins have “Achilles’ heels.” (g) The mechanical stability is a kinetic property that, in general, is not correlated with thermodynamic stability (ΔG) or melting temperature (Tm = ΔG/ΔS) (45). (h) The mechanical stability can be modulated by ligand binding (27, 46, 47) and disulfide bond formation (10, 48–51).
The molecular structure of a protein poses constraints on the location of the transition state in the mechanical unfolding pathway. Tertiary interactions are thought to have shorter distances to their transition states than secondary structures, and they tend to be more brittle (i.e. breaking at high forces and after small deformations) than secondary interactions, which are more compliant (breaking at low forces and after large deformations). Furthermore, tertiary interactions may require more time to equilibrate than secondary ones, and therefore, they often present hysteresis in the pulling-relaxation cycle (3). Most proteins show a high degree of connectivity, and as a result, their unfolding seems to be highly cooperative. Because of the local action of the applied force, their mechanical stability tends to be related to localized molecular structures near the mechanical “breakpoint” rather than to the global structure (3, 43). A massive survey using simplified simulation methods was recently conducted in proteins for which there is atomic structure to identify these mechanical clamps and to classify the available protein structures based on their mechanical stability (52).
Mechanical Dissection of a Model System: Titin I27 Module
The model system most commonly used to study mechanical unfolding/refolding in proteins is the I27 module from titin. Titin is a gigantic multimodular protein responsible for the passive elasticity of muscle (53). The I27 module is an 89-amino acid long Ig-like β-sandwich fold (Fig. 2). Molecular dynamics simulations of its stretching identified two patches of backbone hydrogen bonds as barriers with different mechanical resistance: a low force barrier involving two hydrogen bonds between β-strands A and B and a high force barrier involving six hydrogen bonds between β-strands A′ and G (43). Hydrogen bonds in both barriers are perpendicular to the direction of the force vector (a “shear” mechanical topology: bonds are arranged “in parallel”), whereas the remaining hydrogen bonds in the structure are parallel to the force vector (a “zipper” mechanical topology: bonds are arranged in series) and, like the hydrophobic core, seem to offer low resistance to extension (Fig. 2).
FIGURE 2. Model system in protein nanomechanics: titin I27 domain.
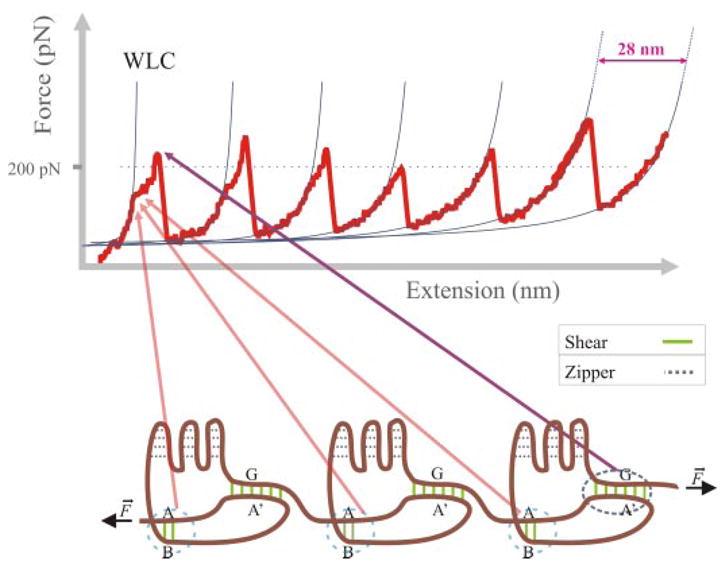
Upper, force-extension curve obtained after stretching an I27 polyprotein. The thin lines correspond to fits to the worm-like chain (WLC) model. Lower, mechanical architecture of the I27 module. The schematic representation of an I27 polyprotein shows patches of backbone hydrogen bonds in both zipper (dashed gray lines) and shear (AB, A′G; solid green lines) mechanical topologies.
These predictions were consistent with experimental data obtained from SMFS experiments: an intermediate found at low force (~100 pN, at 0.6 nm/ms) was associated with the rupture of the AB patch, whereas a high force peak (~200 pN, at 0.6 nm/ms) was found to depend exclusively on the A′G patch and the associated side chain-packing interactions between strands A′ and G (17). The hypothetical role of the AB and A′G patches was tested by loop insertion (11) and proline mutagenesis (54); these mutants also provided the first mechanical phenotypes.
Muscle Elasticity Explained at the Single-molecule Level
A remarkable feat achieved through SMFS has been the reconstruction of the passive elasticity of intact myofibrils by simply scaling up from the mechanical properties of single titin molecules (supplemental Fig. S1) (55). The mechanics of single titin proteins was reconstructed in turn from the mechanical properties of representative elements of its elastic region (i.e. N2, PEVK, and the proximal and distal Ig regions), showing that titin behaves very differently from a simple Hookean spring. Instead, in response to axial tension, titin behaves as a multistage spring that adjusts both its length and apparent stiffness by virtue of its particular modular design. At low forces, the entropic springs (i.e. N2, PEVK, and Ig straightening) dominate, whereas at a high force (i.e. under non-physiological conditions), the enthalpic springs (partial and total unfolding of a few Ig domains) would act as “shock absorbers” to prevent damage of the sarcomere.
Mechanical proteins tend to be modular, with their modules frequently having distinct mechanical stabilities. The shock absorber effect found in titin Ig domains is also present in other modular proteins such as the cell adhesion protein tenascin, in which it was proposed to extend the range and life time of cell-cell interactions (14).
How Well Do SMFS Experiments Mimic in Vivo Protein Mechanics?
In the case of extension machines like the sarcomere, SMFS seems to adequately mimic the natural linear pulling geometry because proteins are pulled apart from both ends of the polypeptide chain. This may also be the case for other cytoskeletal machineries, the adhesion machinery, some mechanosensitive ion channels, and some chaperonins, which may also pull apart their protein substrates in a similar way prior to their refolding (56). Protein translocases (from the mitochondrion, chloroplast, and endoplasmic reticulum) and compartmental proteases such as the proteasome are also thought to work mechanically. Nevertheless, the evidence here favors a different geometry involving a single attachment point from which the pulling would be done by threading the protein toward the entrance of a narrow channel. This model is based mainly on the fact that the susceptibility of substrate proteins to unfolding by these nanomachines (in vivo) correlates more closely with the mechanical stability obtained by mechanical unfolding (using SMFS) than with thermodynamic or kinetic stability (measured in vitro by bulk chemical or heat denaturation). In the case of compartmental proteases, the AAA+ ATPase motor involved in the pulling process seems to unfold the structure adjacent to the degradation tag by trapping local unfolding fluctuations. Global unfolding then occurs immediately, driven by the cooperativity of the protein unfolding process (57–60). Similarly, protein import by the mitochondrial translocase depends on the N-terminal targeting sequence and the local structure of the adjacent protein, more akin to the vectorial nature of AFM pulling experiments than to solution or heat denaturation experiments (61); indeed, mechanical hypomorphic mutations also accelerate mitochondrial import (62).
Mechanical Folding/Unfolding: New Insights into Protein Folding
One of the attractive aspects of our new capacity to unfold/refold proteins by force is that it can give new insights into the protein folding problem. Force-clamp SMFS has been used to directly examine the mechanical folding pathways of I27, ubiquitin, and projectin molecules (16, 63–65). In these experiments, the protein was first unfolded and extended at a high force and then relaxed at lower forces so that refolding could be monitored by measuring the changes in the end-to-end length of the protein. Under these conditions, the folding collapse was marked by large fluctuations in the end-to-end length of the protein, which have been interpreted as the folding of the chain through many continuous steps. By controlling the end-to-end length of a single protein with subnanometer resolution, these studies are providing a unique perspective on how to analyze protein folding trajectories.
Additional technical improvements have been required to observe the mechanical refolding of other proteins. Thus, refolding of filamin was observed using mechanical double-jump experiments (20, 40), and optical tweezers (which use lower spring constants and hence correspondingly lower loading rates) have been used to study the mechanical unfolding/folding of RNase H, identifying a folding intermediate and defining the energy landscape of the process (30).
Concluding Remarks and Perspectives
In just one decade after the feat of pulling the first protein by SMFS, a new discipline has emerged that has provided a wealth of information on the molecular elasticity of proteins, a fundamental property in many biological processes. This new methodology is unveiling the mechanical properties of many proteins and is giving new insights into the protein folding problem.
Still just a few protein folds have been analyzed to date, and many improvements are still necessary such as more specific immobilization methods for soluble proteins to improve the efficiency and sample control of these experiments. These immobilization methods should ideally be compatible with the quasi-simultaneous imaging of proteins in the same sample (66). We also need single-molecule reporters to conduct more reliable studies on intermolecular interactions between proteins. Moreover, there is a need for a sensor that could report forces inside the living cell, although important advances in this way have been reported recently (67–69).
Single-molecule mechanical techniques are providing fundamental information on the structure and function of proteins and are becoming an indispensable tool to understand how proteins fold and work in real time. With the newfound capacity to manipulate and look at the “secret life” of single molecules, we should be prepared for many surprises from protein mechanochemistry. We are entering a new and exciting age of biology, which, in combination with the knowledge generated in this proteomic era, is likely to move us closer to understanding the inner workings of proteins.
Supplementary Material
Footnotes
This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009. This work was supported by Grant R01DK073394 from the National Institutes of Health, the John Sealy Memorial Endowment Fund for Biomedical Research, and Grant 116a2r from the Polycystic Kidney Foundation (to A. F. O.) and by Grant BIO2007-67116 from the Spanish Ministry of Science and Education, Grant S-0505/MAT/0283 from the Consejería de Educación of the Madrid Community, and Grant 200620F00 from the Spanish Research Council (to M. C.-V.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and an additional reference.
The abbreviations used are: AFM, atomic force microscopy/microscope; SMFS, single-molecule force spectroscopy; pN, piconewtons
References
- 1.Alberts B. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Associates, Inc; Sunderland, MA: 2001. [Google Scholar]
- 3.Bustamante C, Chemla YR, Forde NR, Izhaky D. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 4.Binnig G, Quate CF, Gerber C. Phys Rev Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 5.Bustamante C, Macosko JC, Wuite GJ. Nat Rev Mol Cell Biol. 2000;1:130–136. doi: 10.1038/35040072. [DOI] [PubMed] [Google Scholar]
- 6.Carrión-Vázquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernández JM. Proc Natl Acad Sci U S A. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell GI. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 8.Evans E, Ritchie K. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietz H, Rief M. Proc Natl Acad Sci U S A. 2006;103:1244–1247. doi: 10.1073/pnas.0509217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiita AP, Perez-Jimenez R, Walther KA, Grater F, Berne BJ, Holmgren A, Sanchez-Ruiz JM, Fernández JM. Nature. 2007;450:124–127. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrión-Vázquez M, Marszalek PE, Oberhauser AF, Fernández JM. Proc Natl Acad Sci U S A. 1999;96:11288–11292. doi: 10.1073/pnas.96.20.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao M, Sotomayor M, Villa E, Lee EH, Schulten K. Phys Chem Chem Phys. 2006;8:3692–3706. doi: 10.1039/b606019f. [DOI] [PubMed] [Google Scholar]
- 13.Rief M, Gautel M, Oesterhelt F, Fernández JM, Gaub HE. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 14.Oberhauser AF, Marszalek PE, Erickson HP, Fernández JM. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 15.Steward A, Toca-Herrera JL, Clarke J. Protein Sci. 2002;11:2179–2183. doi: 10.1110/ps.0212702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bullard B, Garcia T, Benes V, Leake MC, Linke WA, Oberhauser AF. Proc Natl Acad Sci U S A. 2006;103:4451–4456. doi: 10.1073/pnas.0509016103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrión-Vázquez M, Oberhauser AF, Díez H, Hervás R, Oroz J, Fernández J, Martínez-Martín D. In: Advanced Techniques in Biophysics. Arrondo JLR, Alonso A, editors. Springer-Verlag; Heidelberg, Germany: 2006. pp. 163–245. [Google Scholar]
- 18.Oberhauser AF, Badilla-Fernandez C, Carrión-Vázquez M, Fernández JM. J Mol Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 19.Qian F, Wei W, Germino G, Oberhauser A. J Biol Chem. 2005;280:40723–40730. doi: 10.1074/jbc.M509650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwaiger I, Schleicher M, Noegel AA, Rief M. EMBO Rep. 2005;6:46–51. doi: 10.1038/sj.embor.7400317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brockwell DJ, Paci E, Zinober RC, Beddard GS, Olmsted PD, Smith DA, Perham RN, Radford SE. Nat Struct Biol. 2003;10:731–737. doi: 10.1038/nsb968. [DOI] [PubMed] [Google Scholar]
- 22.Dietz H, Rief M. Proc Natl Acad Sci U S A. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Jimenez R, Garcia-Manyes S, Ainavarapu SR, Fernández JM. J Biol Chem. 2006;281:40010–40014. doi: 10.1074/jbc.M609890200. [DOI] [PubMed] [Google Scholar]
- 24.Lee G, Abdi K, Jiang Y, Michaely P, Bennett V, Marszalek PE. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 25.Yang G, Cecconi C, Baase WA, Vetter IR, Breyer WA, Haack JA, Matthews BW, Dahlquist FW, Bustamante C. Proc Natl Acad Sci U S A. 2000;97:139–144. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Best RB, Li B, Steward A, Daggett V, Clarke J. Biophys J. 2001;81:2344–2356. doi: 10.1016/S0006-3495(01)75881-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ainavarapu SR, Li L, Badilla CL, Fernández JM. Biophys J. 2005;89:3337–3344. doi: 10.1529/biophysj.105.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrión-Vázquez M, Li H, Lu H, Marszalek PE, Oberhauser AF, Fernández JM. Nat Struct Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- 29.Schlierf M, Li H, Fernández JM. Proc Natl Acad Sci U S A. 2004;101:7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cecconi C, Shank EA, Bustamante C, Marqusee S. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 31.Cao Y, Li H. Nat Mater. 2007;6:109–114. doi: 10.1038/nmat1825. [DOI] [PubMed] [Google Scholar]
- 32.Sharma D, Perisic O, Peng Q, Cao Y, Lam C, Lu H, Li H. Proc Natl Acad Sci U S A. 2007;104:9278–9283. doi: 10.1073/pnas.0700351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rief M, Pascual J, Saraste M, Gaub HE. J Mol Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 34.Law R, Carl P, Harper S, Dalhaimer P, Speicher DW, Discher DE. Biophys J. 2003;84:533–544. doi: 10.1016/S0006-3495(03)74872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batey S, Randles LG, Steward A, Clarke J. J Mol Biol. 2005;349:1045–1059. doi: 10.1016/j.jmb.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 36.Urry DW, Hugel T, Seitz M, Gaub HE, Sheiba L, Dea J, Xu J, Parker T. Philos Trans R Soc Lond B Biol Sci. 2002;357:169–184. doi: 10.1098/rstb.2001.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Oberhauser AF, Redick SD, Carrión-Vázquez M, Erickson HP, Fernández JM. Proc Natl Acad Sci U S A. 2001;98:10682–10686. doi: 10.1073/pnas.191189098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar A, Caamano S, Fernández JM. J Biol Chem. 2005;280:6261–6264. doi: 10.1074/jbc.C400573200. [DOI] [PubMed] [Google Scholar]
- 39.Leake MC, Grutzner A, Kruger M, Linke WA. J Struct Biol. 2006;155:263–272. doi: 10.1016/j.jsb.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 40.Schwaiger I, Sattler C, Hostetter DR, Rief M. Nat Mater. 2002;1:232–235. doi: 10.1038/nmat776. [DOI] [PubMed] [Google Scholar]
- 41.Miller E, Garcia T, Hultgren S, Oberhauser AF. Biophys J. 2006;91:3848–3856. doi: 10.1529/biophysj.106.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarkar A, Caamano S, Fernández JM. Biophys J. 2007;92:L36–L38. doi: 10.1529/biophysj.106.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu H, Isralewitz B, Krammer A, Vogel V, Schulten K. Biophys J. 1998;75:662–671. doi: 10.1016/S0006-3495(98)77556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng SP, Rounsevell RW, Steward A, Geierhaas CD, Williams PM, Paci E, Clarke J. J Mol Biol. 2005;350:776–789. doi: 10.1016/j.jmb.2005.04.070. [DOI] [PubMed] [Google Scholar]
- 45.Carrión-Vázquez M, Oberhauser AF, Fisher TE, Marszalek PE, Li H, Fernández JM. Prog Biophys Mol Biol. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 46.Junker JP, Hell K, Schlierf M, Neupert W, Rief M. Biophys J. 2005;89:L46–L48. doi: 10.1529/biophysj.105.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y, Balamurali MM, Sharma D, Li H. Proc Natl Acad Sci U S A. 2007;104:15677–15681. doi: 10.1073/pnas.0705367104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ainavarapu SR, Brujic J, Huang HH, Wiita AP, Lu H, Li L, Walther KA, Carrión-Vázquez M, Li H, Fernández JM. Biophys J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiita AP, Ainavarapu SR, Huang HH, Fernández JM. Proc Natl Acad Sci U S A. 2006;103:7222–7227. doi: 10.1073/pnas.0511035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhasin N, Carl P, Harper S, Feng G, Lu H, Speicher DW, Discher DE. J Biol Chem. 2004;279:45865–45874. doi: 10.1074/jbc.M404103200. [DOI] [PubMed] [Google Scholar]
- 51.Carl P, Kwok CH, Manderson G, Speicher DW, Discher DE. Proc Natl Acad Sci U S A. 2001;98:1565–1570. doi: 10.1073/pnas.031409698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sulkowska JI, Cieplak M. Biophys J. 2008;94:6–13. doi: 10.1529/biophysj.107.105973. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Tskhovrebova L, Trinick J. Nat Rev Mol Cell Biol. 2003;4:679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 54.Li H, Carrión-Vázquez M, Oberhauser AF, Marszalek PE, Fernández JM. Nat Struct Biol. 2000;7:1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Linke WA, Oberhauser AF, Carrión-Vázquez M, Kerkvliet JG, Lu H, Marszalek PE, Fernández JM. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 56.Shtilerman M, Lorimer GH, Englander SW. Science. 1999;284:822–825. doi: 10.1126/science.284.5415.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matouschek A. Curr Opin Struct Biol. 2003;13:98–109. doi: 10.1016/s0959-440x(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 58.Matouschek A, Bustamante C. Nat Struct Biol. 2003;10:674–676. doi: 10.1038/nsb0903-674. [DOI] [PubMed] [Google Scholar]
- 59.Prakash S, Matouschek A. Trends Biochem Sci. 2004;29:593–600. doi: 10.1016/j.tibs.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 60.Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilcox AJ, Choy J, Bustamante C, Matouschek A. Proc Natl Acad Sci U S A. 2005;102:15435–15440. doi: 10.1073/pnas.0507324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato T, Esaki M, Fernández JM, Endo T. Proc Natl Acad Sci U S A. 2005;102:17999–18004. doi: 10.1073/pnas.0504495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernández JM, Li H. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 64.Garcia-Manyes S, Brujic J, Badilla CL, Fernández JM. Biophys J. 2007;93:2436–2446. doi: 10.1529/biophysj.107.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walther KA, Grater F, Dougan L, Badilla CL, Berne BJ, Fernández JM. Proc Natl Acad Sci U S A. 2007;104:7916–7921. doi: 10.1073/pnas.0702179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valbuena A, Oroz J, Vera AM, Gimeno A, Gómez-Herrero J, Carrión-Vázquez M. Rev Sci Instrum. 2007;78:113707. doi: 10.1063/1.2794732. [DOI] [PubMed] [Google Scholar]
- 67.Sarkar A, Robertson RB, Fernández JM. Proc Natl Acad Sci U S A. 2004;101:12882–12886. doi: 10.1073/pnas.0403534101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.