Abstract
The cardiac proteasome is increasingly recognized as a complex, heterogeneous and dynamic organelle contributing to the modulation of cardiac function in health and diseases. The emerging picture of the proteasome system reveals a highly regulated and organized molecular machine integrated into multiple biological processes of the cell. Full appreciation of its cardiovascular relevance requires an understanding of its proteolytic function as well as its underlying regulatory mechanisms; of which assembly, stoichiometry, post-translational modification and the role of the associating partners are increasingly poignant.
Keywords: protein degradation, complex assembly, proteolytic activity
Introduction
The mammalian protein degradation machinery is dominated by the proteasome, as it endoproteolytically cleaves over 70% of intracellular proteins (Rock et al. 1994). The core of this multimeric protease is a duplex of two sets of fourteen subunits, housing duplicate sites of trypsin-like, caspase-like and chymotrypsin-like peptidase activities. Termed the 20S proteasome, its gated pores maintain the complex in a latently active state, permitting only limited proteolysis, possibly through the exposed hydrophobic residues of oxidized and denatured proteins (Widmer et al. 2007). Additional protein complexes such as the 19S and P131 bind to the 20S proteasomes to respectively activate or inhibit the complex (Zais et al. 2002), while the binding of PA200, PA28 and PR39 changes the proteolytic cleavage patterns (Kloetzel 2004, Cascio and Goldberg 2005, Gaczynska and Osmulski 2005). As work turns towards the proteasomes’ complexity and regulation, it is increasingly apparent that they are engaged in a fundamental and specialized role in the cardiovascular system. Of particular interest are the perturbations in proteasome activities associated with cardiovascular disease phenotypes. Despite several controversial reports, there is an emerging consensus that injured myocardium (e.g., myocardial ischemia reperfusion injury, left ventricular dysfunction) is concomitant with an attenuated proteolytic function in the heart (Bulteau et al. 2001, Gurusamy et al. 2007, Voortman and Giaccone 2006, Luss et al. 2002, Pye et al. 2003, Stansfield et al. 2007).
Functional Proteasomes Require a Highly Regulated Assembly
The assembly process begins with single subunits. Expression of the proteasome subunits is a coordinated and controlled process orchestrated through a number of signaling mechanisms and cellular sensors. Under physiological conditions, the Rpn4 subunit of the 26S proteasomes binds to the proteasome-associated control element (PACE) through a zinc finger motif, activating expression of α, β and 19S subunits (Mannhaupt and Feldmann 2007). With a half-life of 2 min, Rpn4 is degraded by the proteasomes and enables the subunit to dynamically stabilize proteasome levels within the cell (Ju et al. 2001). Proteasome transcription is also governed by the proteolysis-inducing factor (PIF), a sulphated glycoprotein excreted by tumors that regulates muscle mass (Russell et al. 2007). In murine myotubes, PIF induces proteasome expression through 14(S)-hydroxyeicosatetraenoic acid and the subsequent PKCα mediated activation of nuclear factor-κB DNA-binding activity (Wyke et al. 2004). By modulating proteasome expression, the cell produces a pool of free subunits from which proteasome assembly can rapidly respond to dynamic regulatory input.
Mature proteasomes require a highly regulated assembly process that must compliment the tissue type and its multitude of functions (Figure 1). Some associating proteins serve as the molecular scaffolds and chaperones that regulate the assembly of the 20S and 26S proteasomes, providing another mechanism for controlling cellular proteasome levels (Schmidt et al. 2005). The proteasome assembly chaperones 1 and 2 (PAC1 and PAC2) dimerize, and then bind α5 and α7 to initiate the formation of the α ring (Hirano et al. 2006). Subsequent arrangement of the α ring and the initial β subunits is aided by a dimer of proteasome assembly chaperones 3 and 4 (PAC3 and PAC4) as they bind α2, β3 and β4 (Le Tallec et al. 2007). At this point, incorporation of either constitutive or inducible β subunits may take place, thus altering the proteolytic characteristics of the proteasomes (Fuchs et al. 2007). A protein that associates with proteasomes in yeast (Ump1p) and humans (hUmp1p, proteassemblin, POMP) is believed to assist with the formation of the remaining β subunits to form the half proteasome (Jayarapu and Griffin 2004). Maturation of the complex through the transitional 13S and 16S intermediates is assisted by hUmp1p and stabilized by the heat shock protein HSC73 (Schmidtke et al. 1997). Proteolytic processing of the constitutive or inducible β propeptides follows the junction of two half-proteasomes forming the proteolytically active 20S proteasome (Jayarapu and Griffin 2004).
Figure 1. Flowchart of proteasome assembly.
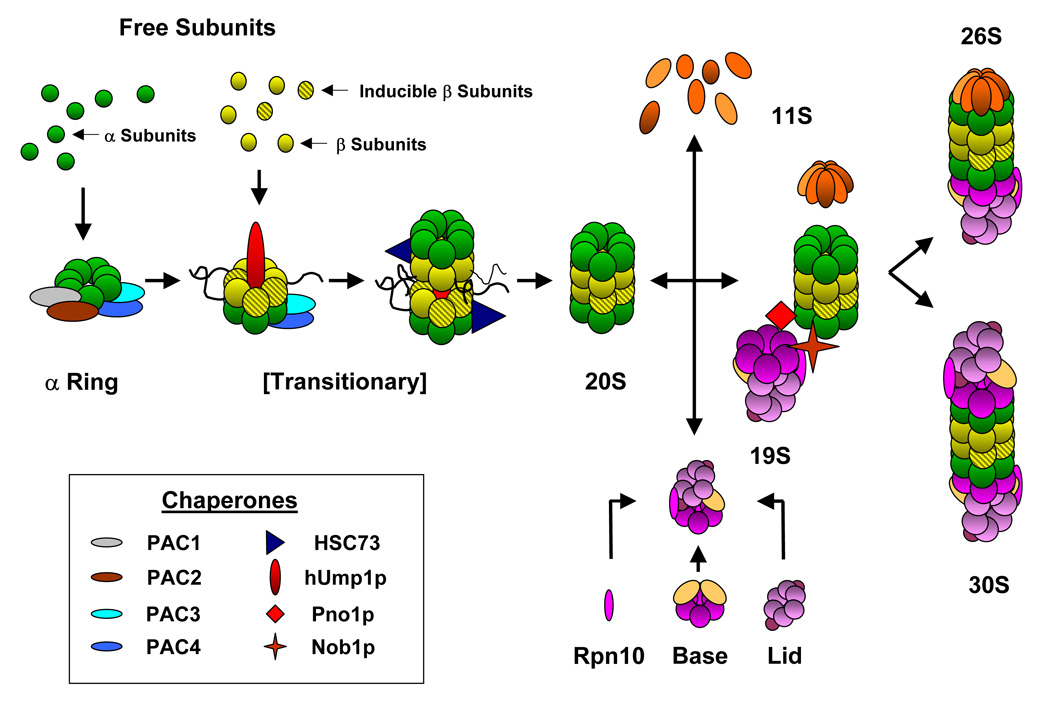
Proteasome assembly is mediated by a growing list of molecular chaperones. The currently accepted view of proteasome assembly begins with the recruitment of free α subunits into an α ring by the pac1/pac2 heterodimer (Hirano et al. 2006); subsequently, completion of the α ring and addition of the constitutive or inducible β subunits to form a transitionary half-proteasome is aided by the pac3/pac4 dimer (Le Tallec et al. 2007) In addition, Hsc73 and hUmp1p aid in the junction of two half-proteasome complexes and the proteolytic processing of the propeptides on some of β subunits yield a latently proteolytically active 20S (Jayarapu and Griffin 2004, Schmidtke et al. 1997). Finally, the regulatory complexes, namely the 11S, 19S, or PA200, may be mated to either end of the 20S proteasome to form functional complexes (26S or 30S). For example, the 19S complex is formed from a base of ATPase subunits, Rpn10 and a lid of non-ATPase subunits. Combination of the 19S with the 20S proteasome is facilitated by the Pno10 and Nob1p chaperones and can interface once to form the 26S or twice to form the 30S proteasomes (Tone and Toh-E 2002). The 11S heptameric complex is composed of α, β, and γ subunits and can also interface with the 20S proteasome, forming hybrid complexes of alternative function (Kloetzel 2004). Inset: chaperones that have been putatively identified to aid in proteasome assembly.
Binding of the 19S activator protein complex to either end of the 20S forms the 26S and 30S proteasomes, a process that is facilitated by the Pno1p and Nob1p chaperones (Tone and Toh-E 2002). Metabolic control of the 19S is believed to dynamically regulate the 26S proteasomes. Phosphorylation of the ATPase subunits, ser120 of RPT6 in particular, by PKA correlates with increased chymotryptic and tryptic activity, and is reversible by treatment with serine/threonine phosphatase PP1γ. Alternatively, O-GlcNAcylation of Rpt2 can act as a master switch, shutting off proteolytic activity upstream of proteasome phosphorylation (Zhang et al. 2007). Targeting of poly-ubiquitinated proteins through additional protein complexes further integrates the proteasomes’ activities with the varying cellular machinery of the heart and different tissues (Dong 2004 et al., Verma et al. 2004). Although the proteasome assembly process is beginning to be unraveled, the impact it has on proteasome activity or its regulatory relevance remains to be determined.
Stoichiometry and Quantification of the Proteasome Complexes
Elucidating the relative amounts of all the components of the ubiquitin proteasome system is necessary for mapping its regulation. Establishing the ratios of any of these proteins as they pertain to the proteasomes has yet to be fully accomplished, but the attempts to date provide for an excellent groundwork. Much of the information regarding the 20S stoichiometry and quantification has come from the more traditional biochemical and gel based approaches. Two – dimensional gels of immuoprecipitated 20S proteasomes from Hela cells, utilized incorporated 3H-leucine to identify all 14 constitutive subunits and approximate, via the counts per minute, that all subunits appear in equal stoichiometry (Hendil et al. 1993). The subunit ratios of the 26S proteasomes have been documented to change in the gastrocnemius muscle of mice as they age. Expression levels from Northern blots suggests that the relative amount of the α2, α3, β6 and Rpt2 subunit transcripts increases over the course of 34 months, with α2 increasing over 900% (Bardag-Gorce et al. 1999). It was also noted by Western blot that the absolute amount of α1 increased by 40% at 29 months before returning back to normal levels by 34 months.
Utilizing a combination of LC-ESI/MS, LC-MALDI/MS, two-dimensional electrophoresis and the Isotope Labeled Affinity Tags (ICAT), levels of rat liver 20S proteasome subunits were compared to those in erythrocyte and U937 cells (Schmidt et al. 2005). The ratios of 20S proteasome subunits by ESI/MS in the rat liver gave values with little significant difference, and no values for β2, β5 or the inducible subunits. The MALDI/MS values gave similar ratios with no values for α2, α5, β2 β5, β7 or the inducible subunits, suggesting that most of the subunits are present in equal amounts. Burlet-Schiltz’s group has also used a quantitative mass spectrometry approach in conjunction with two-dimensional electrophoresis to investigate 20S proteasome heterogeneity between human erythrocytes and U937 cells (Froment et al. 2005). Their use of cleavable ICAT (cICAT) labeled peptides of purified erythrocyte 20S proteasomes produced values ~1% off the expected 0.2 13C/12C and 20% off the 1.0 13C/12C; however, the values were consistent between the different ratios, suggesting a 1:1 stoichiometry of the subunits (β7, β1i, β2i, β5i were never detected). Comparison of 20S proteasomes between erythrocyte and U937 cells, α1, α3, α4, α5, α7 and β6 all showed the expected 1:1 ratio, while α2, β1 and β4 in U937 cells were only 15–20% as abundant. Although the subunit ratios are now well established for the proteasome, the lack of quantitative data between tissues and organisms will, we hope, lessen with more accurate and reliable methods of quantification.
Extending beyond the proteasome, a considerable amount of work has focused on quantifying some of the ancillary proteins that interact and potentially regulate the proteasomes. Lan Huang’s group quantified the 26S proteasomes and its associating partners in arginine auxotrophic yeast strains using stable isotope labeling with amino acids (SILAC) (Guerrero et al. 2006). Heavy and light 26S proteasomes were purified with the use of tandem affinity columns, mixed, digested in solution and subjected to LC-MS/MS analysis. The relative abundance ratios of the peptides (L/H) were calculated by either monoisotopic peak intensity or area. Of the proteasome subunits, the 19S Rpt6 showed a ratio of 5.1 and the 20S α1 was 3.2, a difference suggesting a difference in growth between wild type and auxotrophic strains. A second effort from their line of work focused on quantifying the interacting proteins of the proteasomes. By differentiating between proteins that were pulled down with the 26S proteasomes and those that interacted with the complex after purification, their group identified 35 stable interacting proteins and 16 dynamic interacting proteins. Establishing the abundance and stoichiometry of the associating partners will enable the sorting of intersecting and regulatory pathways of the proteasomes in any tissue.
Current Understanding of the Mammalian Cardiac Proteasomes
A model of the cardiac proteasomes is emerging with the complexity and organization expected from such an integral molecular machine (Figure 2). A novel approach separating 20S proteasomes using in-solution isolectric focusing in a laminar flow indicates that the heart maintains a wide variety of proteasomes, differing in molecular composition and proteolytic activity from those in the liver. A majority of these cardiac proteasomes have an isolectric point of 5.26, compared to a pI of 5.05 for most of the proteasomes, and are largely reflective of their phosphorylation complement. The 20S proteasome subpopulations within each tissue also demonstrated unique proteolytic activity profiles and inducible subunit compositions (Drews et al. 2007). Proteasomes exhibiting distinctly varying hydrophobic and basic protease activities with different targets during oxidative stress injury of the rat heart may also suggest specific proteasome functions associated with individual subpopulations (Gurusamy et al. 2007). Evidence suggesting that proteasomes exist as a diverse and distinct range of forms within the cardiomyocyte is supported by functional proteomic studies of the 26S proteasomes. All inducible subunits were profiled in purified 26S cardiac proteasomes as well as alternate splicing of Rpn10, N-terminal acetylation of Rpn1, Rpn 5, Rpn 6, Rpt 3, Rpt 6, α2, α5, α7, β3 and β4, myristolation of Rpt 2 and phosphorylation of α7 (Gomes et al. 2006). Another element of proteasome heterogeneity in the heart and intimately related to the post-translational state of the proteasome is the involvement of associating partners. Of the many proteins that have been found to stably interact with the proteasome, PKA and PP2A have been found to respectively increase and decrease the three proteolytic activities of the cardiac 20S in a substrate-specific fashion (Zong et al. 2006). As the capacity of the cardiac proteasomes emerges, studying the varying array of proteasomes within the cell and between tissue types will be critical to understanding protein quality control in mammalian systems. Maintaining a wide variety of proteasome subpopulations affords a cell the simultaneous versatility and regulation that contributes to all tissue types.
Figure 2. The model of cardiac proteasomes.
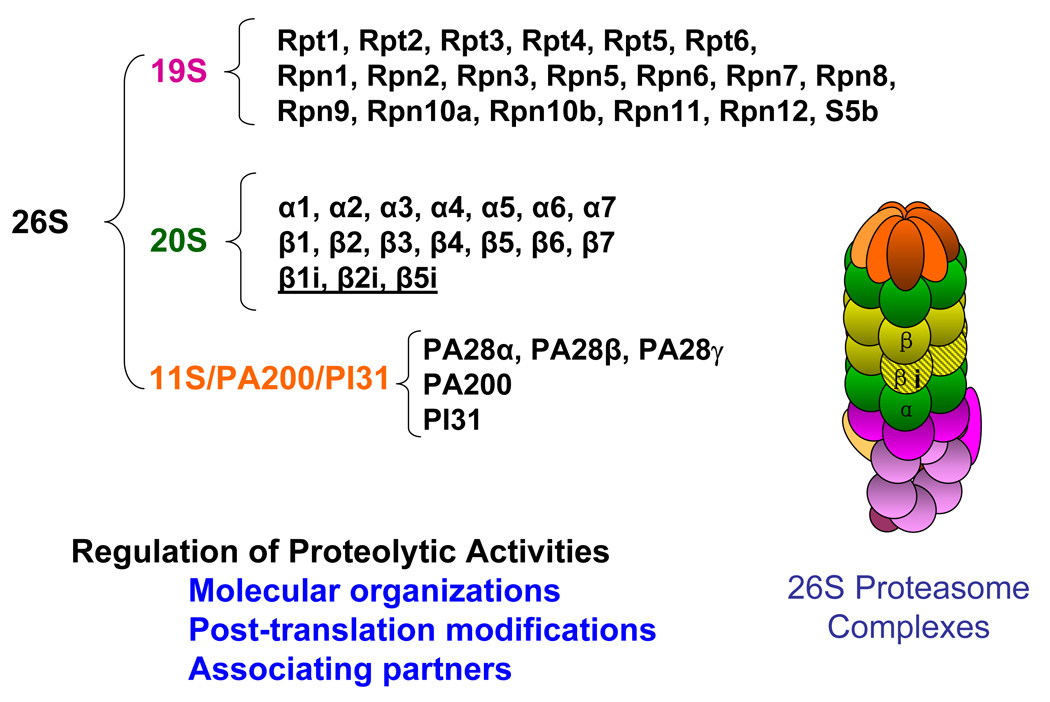
Cardiac proteasome complexes were purified from the murine heart by multidimensional chromatography and analyzed by mass spectrometry to yield a profile of components. The mammalian heart displays a heterogeneous mix of proteasomes, as all14 constitutive subunits (α1, α2, α3, α4, α5, α6, α7, β1, β2, β3, β4, β5, β6, β7) and all three of the inducible subunits (β1i, β2i, β5i) were found in the 20S proteasomes. Of the 19S proteasome complex, a total of 19 subunits have been identified. The six ATPase subunits include Rpt1, Rpt2, Rpt 3, Rpt4, Rpt5 and Rpt 6. The remaining 19S subunits are all non-ATPases and include Rpn1, Rpn2, Rpn3, Rpn4, Rpn5, Rpn6, Rpn7, Rpn8, Rpn9, Rpn10a and its isoform Rpn10b, Rpn11, Rpn12 and S5b. In addition to the 19S activator, the three subunits of the 11S activator (PA28α, PA28β, PA28γ), PI31 inhibitor and the PA200 regulatory complexes were also found associated with the purified proteasomes. These proteomic analyses of proteasome complexes helped us understand the mechanistic insights of this protein degradation machinery. Our investigations demonstrate that cardiac proteolytic function is regulated via at least three mechanisms, i.e., molecular organization, post-translational modification and the associating partners of the cardiac proteasomes (Gomes et al. 2006).
The significance of the proteasomes in the cardiovascular system and their relevance to disease is just now beginning to be revealed. Myocardial ischemia reperfusion injury and the corresponding oxidative modification of the proteasome have both been shown to alter the proteolytic activity (Post et al. 2006, Bulteau et al. 2001). The relationship between oxidized or ubiquitinated proteins and the proteasome directly correlate with postischemic recovery (Powell et al. 2005), while some cardioprotective proteins are stimulated during proteasome inhibition (Stangl et al. 2002, Townsend et al. 2004). Proteasome inhibitors during I/R injury have also been found to decrease myocardial infarct size, reduce leukocyte accumulation and almost entirely eliminate coronary contractile dysfunction (Pye et al. 2003, Campbell et al. 1999). Collectively, multiple lines of evidence documented a relationship of altered proteasome function in diseased myocardium. However, it remains controversial whether reduced proteasome function is beneficial or detrimental (Luss et al. 2002, Voortman and Giaccone 2006, Stansfield et al. 2007). In our view, such inconsistencies arise from the deficit in our current knowledge of the proteasome complexes, specifically with respect to their molecular structure and underlying regulatory mechanisms. This paucity of information makes elucidating its complete role in the cardiovascular system imperative for advancements in cardiac biology and medicine.
Concluding Remarks
In summary, alterations in proteasome assembly or activity may be achieved by manipulating the ratios or availability of assembled subunits. The emerging realization of the proteasomes’ significance to cardiovascular health underscores the importance of a careful examination of this organelle. A preliminary model of cardiac proteasome complexes suggests multiple regulatory facets, each of which remains to be fully elucidated. Every molecule involved in assembling the proteasome, or altering its activities and functions, provides the complex with additional levels of regulation and increases its functional capacity within the cell. With the addition of post-translational modifications and cellular localization, the proteasome achieves a functional diversity crucial to cellular activities. Gaining insight into the precise stoichiometry and quantities of the complex in each tissue will aid in the elucidation of its regulation and response to pathology. Advances in mass spectrometry and the corresponding methods in protein quantification will provide great insight into the regulation of the proteasome and its binding partners.
Acknowledgments
Source of Funding This work is supported by the NIH grant HL-80111 (P.P.) a Laubisch Endowment at he University of California, Los Angeles and AHA 0715004Y (G.Y.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bardag-Gorce F, Farout L, Veyrat-Durebex C, et al. Changes in 20S proteasome activity during ageing of the LOU rat. Mol Biol Rep. 1999;1–2:89–93. doi: 10.1023/a:1006968208077. [DOI] [PubMed] [Google Scholar]
- Bulteau AL, Lundberg KC, Humphries KM, et al. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. 2001;276:30057–30063. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- Campbell B, Adams J, Shin YK, et al. Cardioprotective effects of a novel proteasome inhibitor following ischemia and reperfusion in the isolated perfused rat heart. J Mol Cell Cardiol. 1999;31:467–476. doi: 10.1006/jmcc.1998.0880. [DOI] [PubMed] [Google Scholar]
- Cascio P, Goldberg AL. Preparation of hybrid (19S-20S-PA28) proteasome complexes and analysis of peptides during protein degradation. Methods Enzymol. 2005;398:336–352. doi: 10.1016/S0076-6879(05)98028-2. [DOI] [PubMed] [Google Scholar]
- Drews O, Wildgruber R, Zong C, et al. Mammalian proteasome subpopulations with distinct molecular compositions and proteolytic activities. Mol Cell Proteomics. 2007;6:2021–2031. doi: 10.1074/mcp.M700187-MCP200. [DOI] [PubMed] [Google Scholar]
- Dong X, Liu J, Zheng H, Glasford JW, et al. In situ dynamically monitoring the proteolytic function of the ubiquitin-proteasome system in cultured cardiac myocytes. Am J Physiol Heart Circ Physiol. 2004;287:H1417–H1425. doi: 10.1152/ajpheart.01233.2003. [DOI] [PubMed] [Google Scholar]
- Froment C, Uttenweiler-Joseph S, Bousquet-Dubouch MP, et al. A quantitative proteomic approach using two-dimensional gel electrophoresis and isotope-coded affinity tag labeling for studying human 20S proteasome heterogeneity. Proteomics. 2005;5(9):2351–2363. doi: 10.1002/pmic.200401281. [DOI] [PubMed] [Google Scholar]
- Fuchs D, Berges C, Opelz G, et al. Increased expression and altered subunit composition of proteasomes induced by continuous proteasome inhibition establish apoptosis resistance and hyperproliferation of Burkitt lymphoma cells. J Cell Biochem. 2007 doi: 10.1002/jcb.21405. (in press) [DOI] [PubMed] [Google Scholar]
- Gaczynska M, Osmulski PA. Characterization of noncompetitive regulators of proteasome activity. Methods Enzymol. 2005;398:425–438. doi: 10.1016/S0076-6879(05)98035-X. [DOI] [PubMed] [Google Scholar]
- Gomes AV, Zong C, Edmondson RD, et al. Mapping the murine cardiac 26S proteasome complexes. Circ Res. 2006;99:362–371. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- Guerrero C, Tagwerker C, Kaiser P, Huang L. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol Cell Proteomics. 2006;2:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- Gurusamy N, Shyamal G, Malik G, Das DK. Oxidative injury induces selective rather than global inhibition of proteasome activity. J Mol Cell Cardiol. 2007 doi: 10.1016/j.yjmcc.2007.10.005. (in press) [DOI] [PubMed] [Google Scholar]
- Hendil KB, Welinder KG, Pedersen D, et al. Subunit stoichiometry of human proteasomes. Enzyme. 1993;47:232–240. doi: 10.1159/000468682. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Hayashi H, Iemura S, et al. Cooperation of multiple chaperones required for the assembly of the mammalian 20S proteasomes. Mol Cell. 2006;24:977–984. doi: 10.1016/j.molcel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Jayarapu K, Griffin TA. Protein-protein interactions among human 20S proteasome subunits and proteassemblin. Biochem Biophys Res Commun. 2005;314:523–528. doi: 10.1016/j.bbrc.2003.12.119. [DOI] [PubMed] [Google Scholar]
- Ju D, Xu H, Wang X, Xie Y. Ubiquitin-mediated degradation of Rpn4 is controlled by a phosphorylation-dependant ubiquitination signal. Biochem Biophys Acta. 2007;1773:1672–1680. doi: 10.1016/j.bbamcr.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Kloetzel PM. The proteasome and MHC class I antigen processing. Biochim Biophys Acta. 2004;1695:225–233. doi: 10.1016/j.bbamcr.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault M, Courbeyrette R, et al. 20S Proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Luss H, Schmitz W, Neumann J. A proteasome inhibitor confers cardioprotection. Cardiovasc Res. 2002;54:140–145. doi: 10.1016/s0008-6363(02)00232-8. [DOI] [PubMed] [Google Scholar]
- Mannhaupt G, Feldmann H. Genomic evolution of the proteasome system among hemiascomycetous yeasts. 2007;65:529–540. doi: 10.1007/s00239-007-9031-y. [DOI] [PubMed] [Google Scholar]
- Post MJ, Sato K, Murakami M, et al. Adenoviral PR39 improves blood flow and myocardial function in a pig model of chronic myocardial ischemia by enhancing collateral formation. Am J Physiol Regul Integr Comp Physiol. 2006;290:R494–R500. doi: 10.1152/ajpregu.00460.2005. [DOI] [PubMed] [Google Scholar]
- Powell SR, Wang P, Katzeff H, et al. Oxidized and ubiquitinated proteins may predict recovery of postischemic cardiac function: essential role of the proteasome. Antioxid Redox Signal. 2005;5–6:538–546. doi: 10.1089/ars.2005.7.538. [DOI] [PubMed] [Google Scholar]
- Pye J, Ardeshirpour F, McCain A, et al. Proteasome inhibition ablates activation of BF-kappa B in myocardial reperfusion and reduces reperfusion injury. AM J Physiol Heart Circ Physiol. 2003;284(4):H919–H926. doi: 10.1152/ajpheart.00851.2002. [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Russell ST, Eley H, Tisdale MJ. Role of reactive oxygen species in protein degradation in murine myotubes induced by proteolysis-inducing factor and angiotensin II. Cell Signal. 2007;19:1797–1806. doi: 10.1016/j.cellsig.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Schmidt F, Dahlmann B, Janek K, et al. Comprehensive quantitative proteome analysis of 20S proteasome subtypes from rat liver by isotope coded affinity tag and 2-D gel-based approached. Proteomics. 2006;16:4622–4632. doi: 10.1002/pmic.200500920. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Hanna J, Elsasser S, Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- Schmidtke G, Schmidt M, Kloetzel PM. Maturation of mammalian 20 S proteasome: purification and characterization of 13S and 16S proteasome precursor complexes. J Mol Biol. 1997;268:95–106. doi: 10.1006/jmbi.1997.0947. [DOI] [PubMed] [Google Scholar]
- Stansfield WE, Tang R, Moss NC, et al. Proteasome inhibition promotes regression of left ventricular hypertrophy. Am J Physiol Heart Circ Physiol. 2007 doi: 10.1152/ajpheart.00196.2007. (in press) [DOI] [PubMed] [Google Scholar]
- Stangl K, Gunther C, Frank T, et al. Inhibition of the ubiquitin-proteasome pathway induces differential heat-shock protein response in cardiomyocytes and renders early cardiac protection. Biochem Biophys Res Com. 2002;291:542–549. doi: 10.1006/bbrc.2002.6476. [DOI] [PubMed] [Google Scholar]
- Tone Y, Toh-E A. Nob1p is required for biogenesis of the 26S proteasome and degraded upon its maturation in Sacharomyces cerevisiae. Genes Dev. 2002;16:3142–3157. doi: 10.1101/gad.1025602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend PA, Cutress RI, Carroll CJ, et al. BAG-1 proteins protect cardiac myocytes from simulated ischemia/reperfusion-induced apoptosis via an alternate mechanism of cell survival independent of the proteasome. J Biol Chem. 2004;279:20723–20728. doi: 10.1074/jbc.M400399200. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Voortman J, Giaccone G. Severe reversible cardiac failure after bortezomib treatment combined with chemotherapy in a non-small cell lung cancer patient: a case report. BMC Cancer. 2006;6:129. doi: 10.1186/1471-2407-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol Cell Proteomics. 2007 doi: 10.1074/mcp.M700261-MCP200. (in press) [DOI] [PubMed] [Google Scholar]
- Widmer R, Kaiser B, Engels M, et al. Hyperammonemia causes protein oxidation and enhanced proteasomal activity in response to mitochondria-mediated oxidative stress in rat primary astrocytes. Arch Biochem Biophys. 2007;464:1–11. doi: 10.1016/j.abb.2007.03.027. [DOI] [PubMed] [Google Scholar]
- Wyke SM, Russel ST, Tisdale MJ. Induction of proteasome expression in skeletal muscle is attenuated by inhibitors of NF-κB activation. Br J Cancer. 2004;91:1742–1750. doi: 10.1038/sj.bjc.6602165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss D, Standera S, Kloetzel P, Sijts A. PI31 is a modulator of proteasome formation and antigen processing. Proc Nat Acad Sci. 2002;99:14344–14349. doi: 10.1073/pnas.212257299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Hu Y, Huang P, et al. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J Biol Chem. 2007;282:22460–22471. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- Zong C, Gomes AV, Drews O, et al. Regulation of murine cardiac 20S proteasomes: role of associating partners. Circ Res. 2006;99:372–380. doi: 10.1161/01.RES.0000237389.40000.02. [DOI] [PubMed] [Google Scholar]


