Abstract
The human malaria parasite Plasmodium falciparum utilizes a mechanism of antigenic variation to avoid the antibody response of its human host and thereby generates a long-term, persistent infection. This process predominantly results from systematic changes in expression of the primary erythrocyte surface antigen, a parasite-produced protein called PfEMP1 that is encoded by a repertoire of over 60 var genes in the P. falciparum genome. var genes exhibit extensive sequence diversity, both within a single parasite’s genome as well as between different parasite isolates, and thus provide a large repertoire of antigenic determinants to be alternately displayed over the course of an infection. While significant work has recently been published documenting the extreme level of diversity displayed by var genes found in natural parasite populations, little work has been done regarding the mechanisms that lead to sequence diversification and heterogeneity within var genes. In the course of producing transgenic lines from the original NF54 parasite isolate, we cloned and characterized a parasite line, termed E5, which is closely related to but distinct from 3D7, the parasite used for the P. falciparum genome nucleotide sequencing project. Analysis of the E5 var gene repertoire, as well as that of the surrounding rif and stevor multi-copy gene families, identified examples of frequent recombination events within these gene families including an example of a duplicative transposition indicating recombination events play a significant role in the generation of diversity within the antigen encoding genes of P. falciparum.
Keywords: gene conversion, recombination, antigenic diversity, malaria
1. INTRODUCTION
The most severe form of human malaria is caused by the protozoan parasite Plasmodium falciparum. This parasite is capable of inducing a persistent infection that is typified by recurrent waves of parasitemia, with each wave representing the growth of a subpopulation of parasites displaying distinct antigenic and cytoadherent characteristics(Miller et al., 1994). The waves of parasitemia are the result of a process of antigenic variation, through which the parasite displays different variants of the primary immune antigen displayed on the infected cell surface, a protein called PfEMP1. By generating subpopulations of parasites that vary the form of PfEMP1 that is exposed to the immune system, the parasite is able to avoid complete clearance from the circulation and thereby perpetuate a persistent infection that is difficult for the infected individual to clear. In addition, each variant of PfEMP1 has distinct cytoadhesive characteristics, thus switches in the form of PfEMP1 displayed on the infected cell surface also result in changes in the pathogenicity of the infection, providing an explanation for the often variable nature of malaria severity.
PfEMP1 is encoded by the multi-copy var gene family(Baruch et al., 1995; Smith et al., 1995; Su et al., 1995). The genome of each parasite contains approximately 60 var genes located either within the subtelomeric regions of most chromosomes or within clusters of tandemly arranged genes in the more central chromosomal regions(Gardner et al., 2002).The genes are expressed in a mutually exclusive manner, with the expressed copy determining the form of PfEMP1 displayed on the infected cell surface and as a consequence, the cytoadhesive and antigenic phenotype of the infected cell. There is also evidence of a link between expression of particular var genes and the pathogenicity of the disease, with expression of certain var subtypes associated with more severe disease outcomes(Jensen et al., 2004). In particular, expression of a single, conserved var gene, called var2csa, has been associated with pregnancy associated malaria(Salanti et al., 2003; Salanti et al., 2004), leading to the prospects of using the PfEMP1 encoded by this gene as a vaccine specifically targeting malaria in pregnancy(Smith and Deitsch, 2004). A number of recent studies have begun to shed light on the regulation of var gene transcription, in particular how var gene silencing, mutually exclusive expression and switching are controlled(Frank and Deitsch, 2006; Duraisingh et al., 2005; Freitas-Junior et al., 2005; Voss et al., 2006; Dzikowski et al., 2006; Frank et al., 2006; Frank et al., 2007; Voss et al., 2007; Dzikowski et al., 2007). However, little work has focused on how diversity within the var gene family is generated. The complete sequencing and annotation of the entire genome for the 3D7 isolate(Gardner et al., 2002), as well as additional sequencing projects for several other isolates have shown that the var gene sequences possessed by any given parasite are extremely diverse, although certain subfamilies of var genes can be identified based on the structure of several domains within the encoded PfEMP1(Lavstsen et al., 2003; Kraemer and Smith, 2003; Robinson et al., 2003; Kraemer et al., 2007). In addition, sampling of var gene repertoires from parasites isolated from geographically distant regions has shown that the diversity that exists when comparing the var gene complement between different field isolates is immense, suggesting that var genes possess a mechanism for creating sequence changes at an accelerated rate(Kyes et al., 1997; Fowler et al., 2002; Barry et al., 2007; Kraemer et al., 2007). This is in contrast to housekeeping genes which have been shown to be highly conserved between strains from different geographic regions(Mu et al., 2007; Volkman et al., 2007; Jeffares et al., 2007). Analysis of genome sequences from various laboratory and wild parasite lines have provided evidence that var genes may be subject to specific recombinational processes that result in “shuffling” of different portions of the coding regions, leading to rapid alterations in the encoded proteins(Taylor et al., 2000b; Tami et al., 2003; Ward et al., 1999). Indeed, analysis of the progeny of an experimental genetic cross between two laboratory lines identified an ectopic recombination event that resulted in the duplication of a specific region of one var gene(Freitas-Junior et al., 2000).
Similar to var genes, the rif and stevor genes are found as multiple copies located adjacent to var gene clusters. The genes are also highly diverse and in the case of rif, the proteins they encode have been suggested to be important antigens in the development of clinical immunity(Abdel-Latif et al., 2002; Abdel-Latif et al., 2003). However, while PfEMP1 is known to be placed on the infected cell surface and mediate cytoadherence, the functions of the proteins encoded by rifs and stevors is unknown. Nonetheless, the chromosomal location of these gene families as well as their divergent genetic make up suggest a common mechanism for recombination and the generation of diversity amongst these variant multi-copy gene families.
In the course of characterizing transformed clonal lines derived from the NF54 isolate, we identified a parasite line that is closely related to, but genetically distinct from 3D7, the parasite line that was isolated from NF54 and used in the original P. falciparum genome nucleotide sequencing project. Here we describe an analysis of the var gene repertoire of this parasite line, termed E5, which indicates that it shares more than half of its var genes with 3D7, while the remainders are unique to the new clone. More detailed analysis of DBLα sequences of the unique var genes identified examples of duplicative transpositions of sequence fragments from 3D7 var genes into the newly identified var genes. Such gene conversion events resulted in “shuffling” of the amino acid sequences, leading to the production of hybrid sequences that are presumably unique to the new clone. Similar “shuffling” between family members was also found when analyzing sequences of the rif and stevor gene families. The data gathered suggest that frequent recombination events, including gene conversions, represent a major pathway for the generation of diversity within certain gene families of P. falciparum.
2. MATERIALS AND METHODS
Isolation of clonal P. falciparum cultures, extraction of genomic DNA and Southern
Clonal populations of cultured NF54 parasites were isolated by limiting dilution in 96 well plates as described (Kirkman et al., 1996). The presence of parasites was determined using blood smears during media changes on days 21, 25 and 30 and positive wells were expanded to 20 ml cultures and used for gDNA isolation as described by Frank et al.(Frank et al., 2006). Transformation of NF54 parasites, isolation and analysis of clonal lines that had integrated the plasmid constructs into either the hrp2 or var locus is described in detailed by Frank et al.(Frank et al., 2006). Southern blots were performed as described previously(Frank et al., 2006; Dzikowski et al., 2006).
Real-time PCR amplification of 3D7 specific var and stevor genes
Real-time quantitative RT-PCR was used to detect the presence of all var genes found in the 3D7 genome using the primer set of Salanti et. al.(Salanti et al., 2003) with the modifications described by Dzikowski et al.(Dzikowski et al., 2006). The primer set was supplemented with an additional primer pair for PF08_0107 (5′ - CCTAAAAAGGACGCAGAAGG - 3′ and 5′ - CCAGCAACACTACCACCAGT - 3′) and alternative primer pairs for PFD1005c (5′ - ACGATTGGTGGGAAACAAAT - 3′ and 5′ - CCCCATTCTTTTATCCATCG - 3′) and PFD1015c (5′ - AAAGGAATTGAGGGGGAAAT - 3′ and 5′ - TAAACCACGAAACGGACTGA - 3′). Mal8P1.220 was not annotated at the time of analysis and thus not included in the primer set used. The stevor primer set used was identical to that published in Lavazec et al. (2007) and the specific rif primers are presented in Table 1 of the Supplementary Data. All reactions included three control genes published by these authors; namely, seryl-tRNA synthetase (PF07_0073), fructose biphsphate aldolase (PF14_0425) and actin (PFL2215w); plus an additional two control sets, arginyl-tRNA synthetase (PFL0900c) and glutaminyl-tRNA synthetase (PF13_0170) as described(Dzikowski et al., 2006; Lavazec et al., 2007). Amplification efficiencies were verified by performing amplifications using different concentrations of genomic DNA as template. Reactions were performed at a final primer concentration of 0.5 μM using Biorad ITAQ SYBR green Supermix® in 20 μl reactions on an ABI Prism® 7900HT. All runs were done in triplicate and yielded virtually identical Ct values.
Table 1. Determination of the presence or absence of 3D7 var and stevor genes within the E5 genome.
Assay of E5 genomic DNA for the presence or absence of specific genes within the 3D7 var gene repertoire. The presence of all the var genes annotated within the 3D7 genome was assessed by quantitative real-time PCR using a panel of gene specific PCR primers and E5 genomic DNA as template. The left hand column shows the chromosomal location of each var gene, the middle column lists genes that could not be detected, and the right hand column shows genes that amplified as single copy genes. As controls, amplifications using 3D7 gDNA as well as amplifications of single copy housekeeping genes were also performed
| Chromosomal location | “Missing” var genes | Intact var genes | “Missing” stevor genes | Intact stevor genes |
|---|---|---|---|---|
| Chr 1, subtelomeric | PFA0005w, PFA0015c, PFA0765c | PFA0090c, PFA0750w, PFA0705c, PFA0105w | ||
| Chr 2, subtelomeric | PFB0010w | PFB1055c | PFB0050c, PFB0025c | PFB1020w, PFB0955w, PFB0065w |
| Chr 3, subtelomeric | PFC0005w, PFC1120c | PFC0025c, PFC1105w | ||
| Chr 4,subtelomeric | PFD0020c, PFD0005w, PFD1235w, PFD1245c | PFD0035c, PFD1220w | PFD0065w, PFD0125c | |
| Chr 4,internal | PFD0995c, PFD1000c, PFD1005c, PFD1015c | PFD0615c, PFD0625c, PFD0630c, PFD0635c | ||
| Chr 5, subtelomeric | PFE0005w | PFE0030c | ||
| Chr 6, subtelomeric | PFF0010w, PFF0020c, PFF1580c, PF1595c | PFF0850c, PFF1550w | ||
| Chr 6, internal | PFF0845c | |||
| Chr 7, subtelomeric | MAL7P1.187 | MAL7P1.212 | PF07_0130 | MAL7P1.218, MAL7P1.310, MAL7P1.223, MAL7P1.227 |
| Chr 7, internal | MAL7P1.50, MAL7P1.55, MAL7P1.56, PF07_0048, PF07_0049, PF07_0050, PF07_0051 | |||
| Chr 8, subtelomeric | PF08_0140, PF08_0141, PF08_0142 | MAL8P1.217 | ||
| Chr 8, internal | PF08_0103, PF08_0106, PF08_0107 | |||
| Chr 9, subtelomeric | PFI0005w, PFI1820w, PFI1830c | PFI0080w, PFI0045c | ||
| Chr 10, subtelomeric | PF10_0001 | PF10_0406 | PF10_0395 | PF10_0009 |
| Chr 11, subtelomeric | PF11_0007, PF11_0008 | PF11_0521 | PF11_0013 | PF11_0516 |
| Chr 12, subtelomeric | PFL0005w, PFL0020w, PFL0030c, PFL2665c | PFL2610w, PFL2620w, PFL2635w | ||
| Chr 12, internal | PFL0935c, PFL1950w, PFL1955w, PFL1960w | |||
| Chr 13, subtelomeric | PF13_0001, PF13_0003 | MAL13P1.356 | MAL13P1.7, MAL13P1.505 | MAL13P1.490 |
| Chr 14, subtelomeric | PF14_0007 | PF14_0767, PF14_0771 |
Amplification and sequencing of var-DBLα, rif and stevor segments
Identification of new var gene sequences from the E5 genome was performed according to the procedure described by Taylor et al.(Taylor et al., 2000a). Briefly, degenerate primers specific to DBLα, called αAF (5′-GCACG(A/C)AGTTTTGC-3′) and αBR (5′-GCCCATTC(G/C)TCGAACCA-3′), were used to amplify var sequences from genomic DNA extracted from E5. The amplified products were then cloned into pGEM-T (Promega) or pCR-TOPO (Invitrogen) using the manufacturer’s protocols and individual clones sequenced by the BioResource Center of Cornell University. Additional sequences were generated using a degenerate primer designed for the upstream sequence of B type var genes- (5′-ATATAGATAGAGAGAAACGGAAGAA(G/C)ATATTT-3′) paired with αBR. The derived sequences were compared to the known 3D7 genome sequence using BLAST program on PlasmoDB (www.plasmodb.org), and unique sequences were translated to confirm that the encoded amino acid sequences contained motifs typical of PfEMP1 DBLα. A similar strategy was employed to identify new rif and stevor sequences, except the following primer sets were used: universal stevor primers (5′-CAAAAGGAAGAGATAAGTAT-3′ and 5′-GTTTCTTGCATTCATGTTTCC-3′); rif degenerate primer (5′-TTAAGCGAATG(C/T)GA(A/C)(A/C/T)T-3′ and 5′-(A/C)TTCATTTT(C/T)TT(C/T)TTTCG(A/G/T)C(G/T)ATAACG-3′). Genotyping with MSP1, MSP2 and GLURP was performed using published primers(Snounou et al., 1999; Ranford-Cartwright et al., 1997). All PCR reactions were performed on a BIORAD MyCycler® PCR machine using QuiagenTaq Polymerase® under the following conditions: final primer concentration 0.5μM, 94°C for 5 minutes followed by 35 cycles of: melting temperature 94°C, annealing temperature 55°C, extension 60 °C and a final extension step of 3 minutes. PCR fragments were visualized on a 1% agarose gel with sybr green 1 nucleic acid gel stain® (Molecular Probes) as described by the manufacturer. The var2csa upstream region (UpsE) was amplified using primers 5′-GGTACCTGAACGCTTAAAGAAACAAGG-3′ and 5′-CTGCAGCATTTTGTCCAACCATTTACA-3′.
3. RESULTS
3.1 Identification of a unique parasite clone distinct from 3D7
The parasite line NF54 was isolated from a malarial patient living near the Schipol Airport in Amsterdam, The Netherlands (Ponnudurai et al., 1981), and from this source the clonal line 3D7 was isolated by limiting dilution (Walliker et al., 1987). Analysis of these two cultured parasite lines indicated that they were isogenic, in particular with regard to their var gene complements (Salanti et al., 2003). However here we provide evidence that the NF54 line is in fact polyclonal, a conclusion that stems from observations of profound genetic differences among clonal transgenic parasite lines that were created from transformations of an uncloned culture of NF54 parasites. In the process of generating these transgenic lines, numerous clones were isolated that had integrated the transfecting plasmid into the genome at either the hrp2 locus in the subtelomeric region of chromosome 8 or within the intron of var gene PFB1055c in the subtelomeric region of chromosome 2 (Frank et al., 2006). As part of the analysis to characterize these clones, genomic DNA was isolated and a complete set of var gene primer pairs (derived by Salanti et al.(Salanti et al., 2003)) was used to amplify by quantitative realtime PCR (Q-PCR) gene specific DNA fragments corresponding to each var gene in the 3D7 genome. Whereas this primer set amplified all genes as single copy equivalents from the DNA of clonal lines that had integrated the construct at the hrp2 locus, as well as from DNA isolated from the original uncloned NF54 culture, 23 of the primer pairs failed to amplify DNA fragments from multiple clonal lines that had integrated the construct at the var locus (Table 1). This indicated that 23 var loci were either altered or missing from the genome of the latter apparently isogenic clones, hereafter denoted “E5”, which otherwise had var gene complements identical to 3D7. To determine if a large portion of the var gene repertoire had simply been deleted in E5, a Southern blot of genomic DNA from 3D7 and E5 was hybridized to a probe corresponding to var gene exon 2. This probe recognizes a conserved region shared by all var genes and thus displays a complex banding pattern on Southern blots that reflects the number of genes in the parasite genome(Su et al., 1995). Changes were observed in the position of several bands, consistent with these two parasites lines possessing slightly different genotypes, whereas the total number of bands and thereby the completeness of the var gene repertoire is similar in both lines (Figure 1). Thus the E5 line likely possesses a complete but partially unique repertoire of var genes and the lack of amplification using 23 of the 3D7 var specific primers was not due to chromosomal deletion. It is therefore likely that these two parasites lines, E5 and 3D7, represent two distinct genotypes that exist within the original NF54 isolate.
Figure 1.
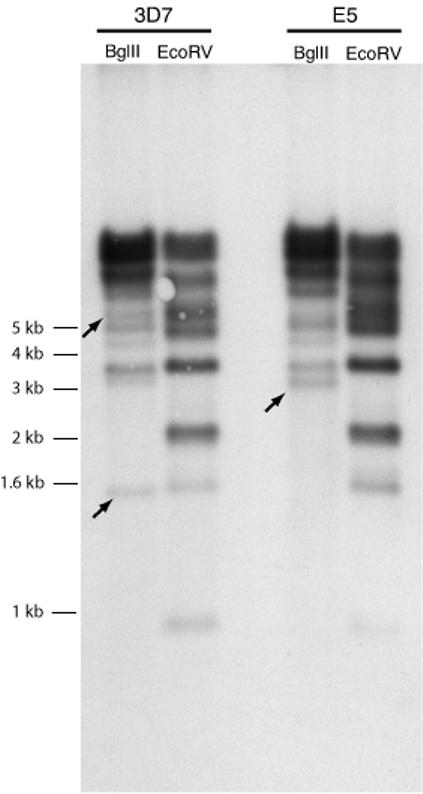
Southern blot showing that E5 possesses an essentially complete repertoire of var genes. Genomic DNA from E5 and 3D7 was digested with different restriction enzymes, blotted and hybridized with a conserved var exon 2 probe to detect DNA fragments harboring var genes. The source of the genomic DNA and the restriction enzyme used in each digestion are shown at the top of each lane. The complex banding pattern indicates that the number of var genes is similar between E5 and 3D7. Unique bands are marked with arrows.
Comparison of the “missing” var genes in the E5 genotype with the chromosomal location within the known 3D7 genome nucleotide sequence determined that all of the missing var genes were grouped into specific chromosomal clusters (Figure 2). In all cases, if a particular var gene failed to be amplified using gene specific primers, all adjacent var genes within the same chromosomal location also failed to be amplified, indicating that all of the genes within these chromosomal regions were either missing or contained significantly different sequences. Similarly, if a particular var gene was detected in the genome, all the adjacent var genes were also detected, indicating that the two parasite genotypes shared some chromosomal regions while differing at others.
Figure 2.
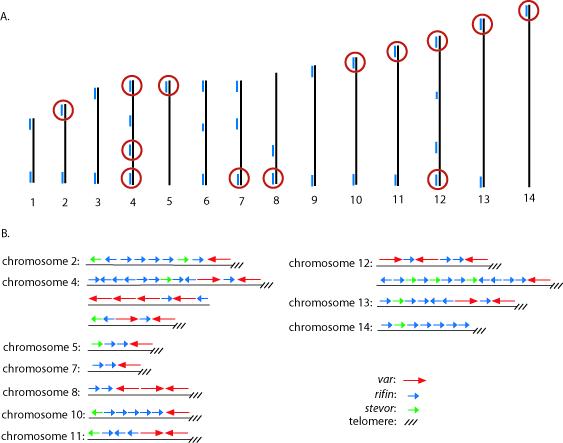
A. Schematic of the 3D7 genome structure showing the chromosomal locations of var genes that are missing or intact in the E5 genome. The 14 chromosomes of P. falciparum are represented by numbered black lines, while the blue lines represent clusters of var, rif, and stevor genes. Red circles denote clusters of genes that could not be detected by PCR using gene specific PCR primers designed to amplify sequences specific to 3D7. B. Expanded view showing the arrangement of var, rif and stevor genes from the chromosomal clusters circled in red in A.
To determine if the pattern of gene alteration or deletion extended to the neighboring gene families that are typically localized adjacent to the var genes, a newly derived primer set to the stevor family(Lavazec et al., 2007) as well as 6 different primers pairs for specific rif genes were used to amplify PCR products from E5 genomic DNA. As expected, in most cases genes located adjacent to var genes that were shared with 3D7 were efficiently amplified, whereas those adjacent to “predicted E5 specific” var genes did not produce a diagnostic PCR product. Primers designed to housekeeping genes around the “unique” var gene clusters on chromosome 4 amplified the same product in both parasite lines (data not shown). Other genes that have been described as variable yet relatively more conserved than the var, rif or stevor families include, var2csa, the type 3 var genes, as well as MSP1, MSP2 and GLURP, which are often used for genotyping(Snounou et al., 1999; Ranford-Cartwright et al., 1997). MSP1 and MSP2 PCR products were the same size in both 3D7 and E5, whereas the GLURP products were different (Supplemental Figure S1). var2csa (PFL0030c) did not amplify with the original primer set of Salanti et al, however additional primers did identify the 5′ end of the coding region and the unique, conserved upstream region (called UpsE, supplemental data), indicating that this gene is present in the E5 genotype. The observed pattern of blocks of common versus dissimilar inheritance is what would be expected from Mendelian inheritance, suggesting that 3D7 and E5 are two sibling parasites present in the NF54 isolate.
3.2 var, rif, and stevor gene sequences from the E5 genotype
Based on the real-time PCR data, E5 and 3D7 were predicted to share 36 identical var genes and differ in 23. To identify var genes that are unique to E5, a sampling of var gene sequences from both the 3D7 and E5 genotypes was performed using PCR amplification with degenerate DBLα primers that are designed to amplify without gene-specific bias the first DBL region of most var genes (Taylor et al., 2000a). The resulting PCR products were cloned and sequenced, and the sequences compared to the previously annotated 3D7 genome sequence (see Supplemental data). As expected, all of the sequences derived from the subcloned parasite lines that displayed the 3D7-like amplification pattern by real-time PCR were identical to sequences found in the genome database, confirming that these parasites are identical to 3D7. In contrast, while approximately half of the sequences derived from the E5 parasite lines were identical to sequences found in the 3D7 genome, many sequences were unique and did not match any sequences previously deposited in the non-redundant database. Translations to the predicted amino acid sequences confirmed that they indeed encode PfEMP1 DBL regions and therefore represented likely E5 var genes. The sequences amplified from E5 that were identical to genes previously identified in 3D7 all corresponded to var genes that were successfully detected by the real-time PCR assay, confirming that these genes are present in the E5 genome and consistent with the schematic map shown in Figure 2.
The sequencing analysis performed for var genes was extended to the rif and stevor gene families. For each of these gene families, a sampling of the gene sequences for both the E5 and 3D7 genotypes was generated by PCR amplification using conserved, degenerate primers that specifically amplify hyper-variable regions found in the stevor(Lavazec et al., 2006) and rif gene families. DNA fragments amplified from E5 and 3D7 genomic DNA were then cloned and sequenced, and the sequences compared to the annotated 3D7 genome sequence. Similar to the results obtained from the var gene analysis, all of the sequences derived from the 3D7-type parasite clones exactly matched sequences found in the 3D7 nucleotide sequence database, once again confirming that these parasites are isogenic with 3D7. For the E5 clone, as expected approximately half of the sequences were exact matches with sequences in the 3D7 database, while the remaining sequences were unique (see Supplemental data). All of the matching sequences came from genes that were found in regions of the chromosomes that were predicted to be identical to 3D7 based on the schematic diagram in Figure 2, again consistent with the model of E5 and 3D7 sharing specific chromosomal regions.
3.3 Identification of potential gene conversion events within var and rif gene sequences
The unique var gene fragments amplified from E5 were not identical to any var genes found in the 3D7 complete genome nucleotide sequence. Gene specific primers made to these unique sequences amplified the correct DNA fragments when E5 genomic DNA was used as template in a PCR reaction, but failed to amplify any sequences when applied to 3D7 genomic DNA, confirming that these genes are indeed unique to the E5 genotype and are not found in 3D7 (Figure 3). Similarly, gene specific primers for var genes predicted to be missing in E5 amplified products from 3D7 gDNA but not from E5. All primer pairs specific to either E5 or 3D7 var genes successfully amplified products using genomic DNA from a cryo-preserved early stock of NF54, indicating that both genotypes were present in the original isolated population of parasites. Quantitative real-time PCR amplification however showed that not all E5 sequences amplified with equal efficiency from NF54 gDNA, suggesting the possibility of additional genotypes that share some of the E5 specific sequences (Table S2).
Figure 3.
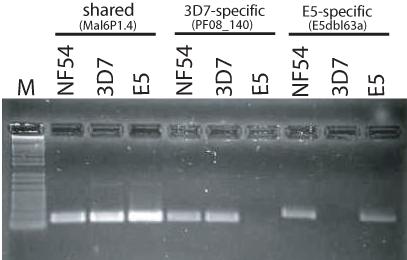
Detection of var genes that are specific to 3D7, E5, or are shared by both clones. PCR reactions designed to specifically amplify from genomic DNA a var gene shared in both clones (PFF1580c, also called MAL6P1.4); a var gene only found in 3D7 (PF08_140); and a var gene only found in E5 (E5dbl63a, see Supplemental data). It was possible to amplify products for all three genes using genomic DNA extracted from an early isolate of NF54, indicating that this isolate contains parasites with both genotypes.
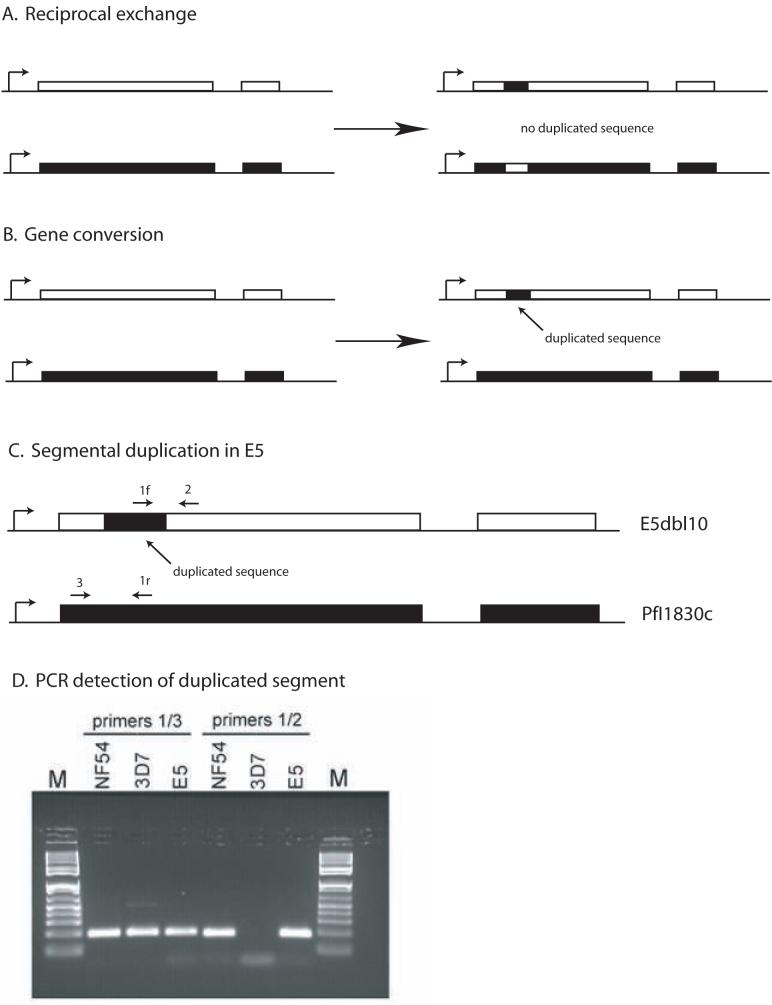
Closer examination of the var gene sequences unique to E5 verified that while none of the DBLα fragments amplified from E5 were found in their entirety in the 3D7 genome, it was possible to identify several relatively long stretches of sequence that were 100% identical with 3D7 var gene sequences, but inserted within the context of sequences unique to E5 (Figure 4). For example, a 105 bp fragment of complete sequence identity to the var gene PFI1830c was found inserted into a unique var gene in E5. The length and perfect identity of these stretches of sequence strongly indicate that they represent recent transposition from one position in the genome into another, either via reciprocal recombination between genes or through duplicative transposition (also referred to as gene conversion) (Figure 5). In the case of the fragment from PFI1830c, it is possible to distinguish whether the shared sequence is the likely result of reciprocal exchange or gene conversion because both the original PFI1830c gene and the unique var gene containing the identical 105 bp fragment are found in the E5 genome. A reciprocal recombination event would have resulted in an exchange of sequences between PFI1830c and the var gene unique to E5, thus altering both sequences and resulting in the 105 bp fragment remaining as a single copy in the genome. In contrast, a gene conversion event would leave PFI1830c intact and result in a duplication of the 105 bp fragment in the genome of E5. PCR amplification using primers specific to the 105 bp fragment verified that this sequence is in fact duplicated in E5. While this does not conclusively rule out other recombination mechanisms (for instance a double cross over in a previous generation) it provides strong support for a gene conversion mechanism in the production of new var genes (Figure 5D).
Figure 4.

Evidence for recombination events in var and rif genes. The sequences of var genes (A-C) and a rif gene (D) from the E5 clone are shown. The red letters show stretches that are 100% identical to sequences found within the 3D7 genome recombined with sequences unique to the E5 genotype (black letters). In A, 105 bp of PFI1830c is inserted within the E5 specific sequence E5dbl10. The partial amino acid sequence of the translated PFEMP1s are shown in B. In C, a stretch of sequence identical to PF10_0406 is shown inserted into the E5 specific sequence E5DBL35. In D, an E5 specific rif gene (E5rif3) is shown that contains 341 bp of sequence identical to the rif gene PFB0055c from 3D7 (red letters).
Figure 5.
Schematic showing the results of reciprocal exchange or gene conversion. The genes shown display the two exon structure typical of var genes, with the promoter denoted by a bent arrow. A. Reciprocal exchange results in changes in both genes, but does not lead to either duplication or deletion of any sequences. B. Gene conversions, also referred to as duplicative transpositions, result in the duplication of a stretch of sequence, which then replaces a similar sequence in a related gene, leading to a corresponding deletion. C. Schematic of the duplication of a segment of the var gene PfI1830c into a different var gene within the genome of E5. The positions of gene specific primers that can be used to detect the duplication event are shown and numbered 1-3. Primer 1f and primer 1r are reverse complements and thus function as primers in opposite orientations. D. Ethidium bromide stained agarose gel showing PCR detection of the duplicated segment of PfI1830c. Primer pair 1/3 detects the original PfI1830c gene in NF54, 3D7 and E5 while primer pair 1/2 detects the duplicated segment in NF54 and E5, but not in 3D7.
Examination of rif and stevor sequences also identified similar stretches as evidence for recombination within these sequences, including 341 bp of sequence from PFB0055c adjoined to a unique sequence (Figure 4D). In this case, only one end of the recombination event was recovered in our sampling of DNA sequences, suggesting that it could also represent the site of a single recombination cross over. Examination of genes surrounding PFB0055c in 3D7 determined that in E5, genes on the centromeric side of this gene are present, while those toward the telomere are all undetectable, suggesting that this sequence represents the site of a single cross over within this particular rif gene.
4. DISCUSSION
The cloning and identification of E5, a sibling parasite line to 3D7, provides valuable opportunities for comparative analysis of recombination events, particularly within the var, rif and stevor gene families. E5 might also be useful in studies investigating phenotypic differences, and it might be possible to isolate additional closely related parasites from the original, uncloned NF54 population, greatly increasing the power of such analyses. Interestingly, while E5 was a rather minor portion of the NF54 parasite population grown in the lab, it was nonetheless isolated relatively frequently in the process of isolating clonal, stably transformed lines of transgenic parasites (Frank et al., 2006). This implies that parasites with the E5 genotype must have possessed a growth advantage when selecting for plasmid integration in this particular study, although the nature of this advantage remains unknown. In addition, integration events in the E5 genetic background all happened at a var locus, while integration into the genome of 3D7 occurred within the hrp2 gene, suggesting that these two closely related parasite lines might have different recombination propensities.
The generation of gene conversion events within var genes described here is consistent with a previous report of a similar gene conversion event that included var gene sequences (Freitas-Junior et al., 2000). The length and perfect identity recombinant segments imply that they are the result of relatively recent events, perhaps occurring at some point within the last transmission cycle of the parasites. Whether such events occur primarily in meiosis during sexual development or in mitosis during asexual replication is not yet clear, although the event described by Junior et al. as well as similar events described by Taylor et al. occurred during an experimental cross, implicating meiosis in those cases(Taylor et al., 2000b; Freitas-Junior et al., 2000).
Mu et al. identified major recombination hotspots in subtelomeric and central parts of P. falciparum chromosomes in parasite isolates from different geographic regions, chromosomal areas that also harbor the var, rif and stevor gene families(Mu et al., 2005). Whether these recombination hotspots are a result of high cross over rates at these positions or rather a reflection of immune-mediated diversifying selection remains to be established (Volkman et al., 2002). However, there are reports that P. falciparum infections can last as long as 480 days, a length of time that would seem to require an antigenic repertoire larger than the ∼60 var genes thought to be found in most parasite genomes. The potential ability for parasites to generate new var genes over the course of an individual infection might therefore contribute to long-term persistent infections, and would have implications for understanding the dynamics of host-parasite interactions. Future studies using progeny from the different genetic crosses and from patient isolates of long-term chronic infections are necessary to determine the relative contribution of meiosis vs mitosis to the genetic diversity observed within the multi-copy variant antigen families of P. falciparum. Additionally, more comprehensive analysis using pulse-field gel electrophoresis to examine chromosomal alterations would be useful.
Gene conversions as a mechanism for generating diversity within gene families encoding the antigenic determinants of pathogenic organisms has been demonstrated for a number of other infectious agents including the protozoan parasites Trypanosoma brucei (Borst et al., 1997), and Babesia bovis (al Khedery and Allred, 2006), as well as bacteria of the genera Neisseria (Criss et al., 2005) and Treponema (Centurion-Lara et al., 2004). The molecular mechanisms underlying gene conversions are thought to involve the homologous repair pathway used by organisms to repair double strand breaks in chromosomal DNA. This pathway relies on sequence similarity in choosing a template for the conversion event, and thus can result in shuffling of similar sequences between different gene copies. Preferential choice of template for gene conversions between var genes may explain why the three main var gene types have diverged from one another into separate subgroups (Kraemer and Smith, 2003; Lavstsen et al., 2003). Also the propensity of certain regions of the chromosomes to selectively pair with one another, for instance in telomere clusters or “bouquets”, might also influence template choice for gene conversions and result in the generation of subgroups based on chromosomal location(Figueiredo et al., 2002; Figueiredo et al., 2002; O’Donnell et al., 2002; Marty et al., 2006). Such a mechanism has been previously proposed for increasing the probability of recombination within subtelomeric regions. If gene conversion events play a significant role in generating diversity within the var gene family, one question that remains is why conserved var gene like var2csa and the so called “type 3” vars are not subject to this process and remain relatively conserved between isolates. Future work into the frequency of gene conversions and the parameters that determine template choice will likely shed light on how these genes remain conserved throughout wild parasite isolates.
The NF54 isolate is frequently used in laboratory studies because of its robust ability to produce gametocytes and mediate transmission to mosquitoes. This study should serve as caution that NF54 is not clonal and harbors at least one sibling parasite, E5, in addition to the predominant population that is isogenic with 3D7. Moreover, it is known from other studies that the chromosome ends of cultured parasites are fragile and deletions of up to 100 kb or more are common(Ribacke et al., 2007; Lavazec et al., 2007; Pologe and Ravetch, 1988). Because parasite transformation methodologies routinely incorporate a cloning step, it must be born in mind for NF54 and all laboratory adapted parasite lines that the observed phenotypes of such resulting clones may not be due to the genetic manipulations intended by the investigator, but by the inadvertent isolation of a rare genotypically variant parasite that was present in the source culture. For this reason it is prudent to assess phenotypes of multiple, independently isolated clones representing separate genetic events.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Julian Gabor for his assistance in the genotyping experiments. This work was supported National Institutes of Health Grants AI 52390 and AI 54580 to KWD and TJT, respectively, and by a grant from the Ellison Medical Foundation to KWD. The Department of Microbiology and Immunology at Weill Medical College of Cornell University acknowledges the support of the William Randolph Hearst Foundation. KWD and TJT are Stavros S. Niarchos Scholars. LAK is supported by NIH training grant AI 07613 to the Division of International Medicine and Infectious Diseases.
Reference List
- Abdel-Latif MS, Dietz K, Issifou S, Kremsner PG, Klinkert MQ. Antibodies to Plasmodium falciparum rifin proteins are associated with rapid parasite clearance and asymptomatic infections. Infect. Immun. 2003;71:6229–6233. doi: 10.1128/IAI.71.10.6229-6233.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Latif MS, Khattab A, Lindenthal C, Kremsner PG, Klinkert MQ. Recognition of variant Rifin antigens by human antibodies induced during natural Plasmodium falciparum infections. Infect. Immun. 2002;70:7013–7021. doi: 10.1128/IAI.70.12.7013-7021.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al Khedery B, Allred DR. Antigenic variation in Babesia bovis occurs through segmental gene conversion of the ves multigene family, within a bidirectional locus of active transcription. Mol. Microbiol. 2006;59:402–414. doi: 10.1111/j.1365-2958.2005.04993.x. [DOI] [PubMed] [Google Scholar]
- Barry AE, Leliwa-Sytek A, Tavul L, Imrie H, Migot-Nabias F, Brown SM, McVean GA, Day KP. Population Genomics of the Immune Evasion (var) Genes of Plasmodium falciparum. PLoS. Pathog. 2007;3:e34. doi: 10.1371/journal.ppat.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Borst P, Rudenko G, Blundell PA, Van Leeuwen F, Cross MA, McCulloch R, Gerrits H, Chaves IM. Mechanisms of antigenic variation in African trypanosomes. Behring Inst. Mitt. 1997:1–15. [PubMed] [Google Scholar]
- Centurion-Lara A, LaFond RE, Hevner K, Godornes C, Molini BJ, Van Voorhis WC, Lukehart SA. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol. Microbiol. 2004;52:1579–1596. doi: 10.1111/j.1365-2958.2004.04086.x. [DOI] [PubMed] [Google Scholar]
- Criss AK, Kline KA, Seifert HS. The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae. Mol. Microbiol. 2005;58:510–519. doi: 10.1111/j.1365-2958.2005.04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium faiciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- Dzikowski R, Frank M, Deitsch K. Mutually Exclusive Expression of Virulence Genes by Malaria Parasites Is Regulated Independently of Antigen Production. PLoS. Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, Patel S, Wellems TE, Deitsch KW. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo LM, Freitas-Junior LH, Bottius E, Olivo-Marin JC, Scherf A. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. Embo Journal. 2002;21:815–824. doi: 10.1093/emboj/21.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler EV, Peters JM, Gatton ML, Chen N, Cheng Q. Genetic diversity of the DBLalpha region in Plasmodium falciparum var genes among Asia-Pacific isolates. Mol. Biochem. Parasitol. 2002;120:117–126. doi: 10.1016/s0166-6851(01)00443-1. [DOI] [PubMed] [Google Scholar]
- Frank M, Deitsch K. Activation, silencing and mutually exclusive expression within the var gene family of Plasmodium falciparum. Int. J. Parasitol. 2006;36:975–985. doi: 10.1016/j.ijpara.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Frank M, Dzikowski R, Amulic B, Deitsch K. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol. Microbiol. 2007;64:1486–1498. doi: 10.1111/j.1365-2958.2007.05736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Dzikowski R, Constantini D, Amulic B, Burdougo E, Deitsch K. Strict pairing of var promoters and introns is required for var gene silencing in the malaria parasite plasmodium falciparum. J. Biol. Chem. 2006;281:9942–9952. doi: 10.1074/jbc.M513067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, Kyes S, Krishna S, Newbold C, Dermitzakis ET, Berriman M. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat. Genet. 2007;39:120–125. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ATR, Magistrado P, Sharp S, Joergensen L, Lavstsen T, Chiucciuini A, Salanti A, Vestergaard LS, Lusingu JP, Hermsen R, Sauerwein R, Christensen J, Nielsen MA, Hviid L, Sutherland C, Staalsoe T, Theander TG. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. Journal of Experimental Medicine. 2004;199:1179–1190. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman LA, Su XZ, Wellems TE. Plasmodium falciparum: isolation of large numbers of parasite clones from infected blood samples. Exp. Parasitol. 1996;83:147–149. doi: 10.1006/expr.1996.0058. [DOI] [PubMed] [Google Scholar]
- Kraemer SM, Kyes SA, Aggarwal G, Springer AL, Nelson SO, Christodoulou Z, Smith LM, Wang W, Levin E, Newbold CI, Myler PJ, Smith JD. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC. Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer SM, Smith JD. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Molecular Microbiology. 2003;50:1527–1538. doi: 10.1046/j.1365-2958.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- Kyes S, Taylor H, Craig AG, Marsh K, Newbold CI. Genomic representation of var gene sequences in Plasmodium falciparum field isolates from different geographic regions. Molecular and Biochemical Parasitology. 1997;87:235–238. doi: 10.1016/s0166-6851(97)00071-6. [DOI] [PubMed] [Google Scholar]
- Lavazec C, Sanyal S, Templeton TJ. Hypervariability within the Rifin, Stevor and Pfmc-2TM superfamilies in Plasmodium falciparum. Nucleic Acids Res. 2006;34:6696–6707. doi: 10.1093/nar/gkl942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavazec C, Sanyal S, Templeton TJ. Expression switching in the stevor and Pfmc-2TM superfamilies in Plasmodium falciparum. Mol. Microbiol. 2007;64:1621–1634. doi: 10.1111/j.1365-2958.2007.05767.x. [DOI] [PubMed] [Google Scholar]
- Lavstsen T, Salanti A, Jensen ATR, Arnot DE, Theander TG. Subgrouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malaria Journal. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty AJ, Thompson JK, Duffy MF, Voss TS, Cowman AF, Crabb BS. Evidence that Plasmodium falciparum chromosome end clusters are cross-linked by protein and are the sites of both virulence gene silencing and activation. Mol. Microbiol. 2006;62:72–83. doi: 10.1111/j.1365-2958.2006.05364.x. [DOI] [PubMed] [Google Scholar]
- Miller LH, Good MF, Milon G. Malaria Pathogenesis. Science. 1994;264:1878–1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- Mu J, Awadalla P, Duan J, McGee KM, Joy DA, McVean GA, Su XZ. Recombination hotspots and population structure in Plasmodium falciparum. PLoS. Biol. 2005;3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Awadalla P, Duan J, McGee KM, Keebler J, Seydel K, McVean GA, Su XZ. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat. Genet. 2007;39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- O’Donnell RA, Freitas LH, Preiser PR, Williamson DH, Duraisingh M, McElwain TF, Scherf A, Cowman AF, Crabb BS. A genetic screen for improved plasmid segregation reveals a role for Rep20 in the interaction of Plasmodium falciparum chromosomes. Embo Journal. 2002;21:1231–1239. doi: 10.1093/emboj/21.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pologe LG, Ravetch JV. Large deletions result from breakage and healing of P. falciparum chromosomes. Cell. 1988;55:869–874. doi: 10.1016/0092-8674(88)90142-0. [DOI] [PubMed] [Google Scholar]
- Ponnudurai T, Leeuwenberg AD, Meuwissen JH. Chloroquine sensitivity of isolates of Plasmodium falciparum adapted to in vitro culture. Trop. Geogr. Med. 1981;33:50–54. [PubMed] [Google Scholar]
- Ranford-Cartwright LC, Taylor J, Umasunthar T, Taylor LH, Babiker HA, Lell B, Schmidt-Ott JR, Lehman LG, Walliker D, Kremsner PG. Molecular analysis of recrudescent parasites in a Plasmodium falciparum drug efficacy trial in Gabon. Trans. R. Soc. Trop. Med. Hyg. 1997;91:719–724. doi: 10.1016/s0035-9203(97)90539-3. [DOI] [PubMed] [Google Scholar]
- Ribacke U, Mok BW, Wirta V, Normark J, Lundeberg J, Kironde F, Egwang TG, Nilsson P, Wahlgren M. Genome wide gene amplifications and deletions in Plasmodium falciparum. Mol. Biochem. Parasitol. 2007;155:33–44. doi: 10.1016/j.molbiopara.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Robinson BA, Welch TL, Smith JD. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Molecular Microbiology. 2003;47:1265–1278. doi: 10.1046/j.1365-2958.2003.03378.x. [DOI] [PubMed] [Google Scholar]
- Salanti A, Dahlback M, Turner L, Nielsen MA, Barfod L, Magistrado P, Jensen AT, Lavstsen T, Ofori MF, Marsh K, Hviid L, Theander TG. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J. Exp. Med. 2004;200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanti A, Staalsoe T, Lavstsen T, Jensen ATR, Sowa MPK, Arnot DE, Hviid L, Theander TG. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Molecular Microbiology. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Deitsch KW. Pregnancy-associated malaria and the prospects for syndrome-specific antimalaria vaccines. J. Exp. Med. 2004;200:1093–1097. doi: 10.1084/jem.20041974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans. R. Soc. Trop. Med. Hyg. 1999;93:369–374. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- Su X, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JV, Peterson DS, Ravetch JV, Wellems TE. A large and diverse gene family (var) encodes 200-350 kD proteins implicated in the antigenic variation and cytoadherence of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Tami A, Ord R, Targett GA, Sutherland CJ. Sympatric Plasmodium falciparum isolates from Venezuela have structured var gene repertoires. Malar. J. 2003;2:7. doi: 10.1186/1475-2875-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HM, Kyes SA, Harris D, Kriek N, Newbold CI. A study of var gene transcription in vitro using universal var gene primers. Mol. Biochem. Parasitol. 2000a;105:13–23. doi: 10.1016/s0166-6851(99)00159-0. [DOI] [PubMed] [Google Scholar]
- Taylor HM, Kyes SA, Newbold CI. Var gene diversity in Plasmodium falciparum is generated by frequent recombination events. Mol. Biochem. Parasitol. 2000b;110:391–397. doi: 10.1016/s0166-6851(00)00286-3. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Hartl DL, Wirth DF, Nielsen KM, Choi M, Batalov S, Zhou Y, Plouffe D, Le Roch KG, Abagyan R, Winzeler EA. Excess polymorphisms in genes for membrane proteins in Plasmodium falciparum. Science. 2002;298:216–218. doi: 10.1126/science.1075642. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA, Jr., Daily JP, Sarr O, Ndiaye D, Ndir O, Mboup S, Duraisingh MT, Lukens A, Derr A, Stange-Thomann N, Waggoner S, Onofrio R, Ziaugra L, Mauceli E, Gnerre S, Jaffe DB, Zainoun J, Wiegand RC, Birren BW, Hartl DL, Galagan JE, Lander ES, Wirth DF. A genome-wide map of diversity in Plasmodium falciparum. Nat. Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, Reeder JC, Crabb BS, Cowman AF. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- Voss TS, Tonkin CJ, Marty AJ, Thompson JK, Healer J, Crabb BS, Cowman AF. Alterations in local chromatin environment are involved in silencing and activation of subtelomeric var genes in Plasmodium falciparum. Mol. Microbiol. 2007;66:139–150. doi: 10.1111/j.1365-2958.2007.05899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- Ward CP, Clottey GT, Dorris M, Ji DD, Arnot DE. Analysis of Plasmodium falciparum PfEMP-1/var genes suggests that recombination rearranges constrained sequences. Mol. Biochem. Parasitol. 1999;102:167–177. doi: 10.1016/s0166-6851(99)00106-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.