Abstract
Five members of the RecQ subfamily of DEx-H-containing DNA helicases have been identified in both human and mouse, and mutations in BLM, WRN and RECQ4 are associated with human diseases of premature aging, cancer, and chromosomal instability. Although a genetic disease has not been linked to RECQ1 mutations, RECQ1 helicase is the most highly expressed of the human RecQ helicases, suggesting an important role in cellular DNA metabolism. Recent advances have elucidated a unique role of RECQ1 to suppress genomic instability. Embryonic fibroblasts from RECQ1-deficient mice displayed aneuploidy, chromosomal instability, and increased load of DNA damage.1 Acute depletion of human RECQ1 renders cells sensitive to DNA damage and results in spontaneous γ-H2AX foci and elevated sister chromatid exchanges, indicating aberrant repair of DNA breaks.2 Consistent with a role in DNA repair, RECQ1 relocalizes to irradiation-induced nuclear foci and associates with chromatin.2 RECQ1 catalytic activities3 and interactions with DNA repair proteins2,4,5 are likely to be important for its molecular functions in genome homeostasis. Collectively, these studies provide the first evidence for an important role of RECQ1 to confer chromosomal stability that is unique from that of other RecQ helicases and suggest its potential involvement in tumorigenesis.
Keywords: RECQ1 (RECQL), helicase, genomic instability, cancer, DNA repair, recQ, replication, homologous recombination, gene silencing
Introduction
Helicases catalytically unwind structured nucleic acid substrates in a nucleoside-triphosphate and directionally specific manner. Collectively, DNA and RNA helicases represent a vital group of cellular enzymes that are essential for a diverse set of processes including replication, DNA repair, recombination, transcription, chromosome segregation, and RNA stability, processing, and translation.6–13 The RecQ subfamily of DEx-H containing 3′→5′ DNA helicases, of which E. coli RecQ is the prototype, contain an approximately 450 amino acid domain with seven conserved helicase motifs. E. coli RecQ helicase is implicated in homologous recombination (HR) and double-strand break (DSB) repair mediated by the RecF pathway and suppression of illegitimate recombination.14 Other RecQ-related proteins include Saccharomyces cerevisiae SGS1 that participates in recombination, chromosome partitioning, and genome maintenance,15,16 and Schizosaccharomyces pombe RQH1 which prevents recombination and suppresses inappropriate recombination.17
Unlike E. coli, S. cerevisiae and S. pombe, which only have one RecQ helicase, higher eukaryotes express multiple RecQ-related helicases.8,12 In humans, the first described was RECQL or RECQL1,18–20 hereafter designated RECQ1, followed by BLM,21,22 WRN,23 RECQL4 (RECQ4)24,25 and RECQL5 (RECQ5).24 Of these five, three human RecQ helicases have been implicated in distinct genetically inherited homozygous recessive diseases. Bloom syndrome (BS), defective in the BLM helicase, is associated with a very high incidence of different types of cancers, both solid tumours and leukaemia, and is also manifested by skin disorders, sunlight sensitivity, proportional dwarfism, immunodeficiency, male sterility, and genomic instability marked by an elevated level of sister chromatid exchange (SCE).21 Werner syndrome (WS), due to mutation in the WRN helicase, is characterized by premature aging with an elevated risk of age-associated diseases such as cancer, atherosclerotic cardiovascular disease, diabetes mellitus (Type II), osteoporosis, and genomic instability manifested by chromosomal deletions and translocations.23 Rothmund-Thomson syndrome (RTS), also known as poikiloderma congenitale, defective in the RECQ4 helicase, displays growth deficiency, photosensitivity with poikilodermatous skin changes, early greying and hair loss, juvenile cataracts, skeletal dysplasias, a predisposition to malignancy, especially osteogenic sarcomas, and chromosomal instability.25 RECQ4 mutations were also detected in Finnish patients with an autosomal recessive disorder RAPADILINO syndrome [radial hypoplasia/aplasia, patellae hypoplasia/aplasia and cleft or highly arched palate, diarrhoea and dislocated joints, little size (height at least 2 standard deviations smaller than the average height) and limb malformation, nose slender and normal intelligence].26 Although many features of the two genetic disorders overlap, poikiloderma, a hallmark of RTS, has been described as being generally absent from RAPADILINO syndrome. A third genetic disorder associated with RECQ4 mutations was found in a subgroup of patients with Baller-Gerold syndrome, a rare autosomal recessive condition with radial aplasia/hypoplasia and craniosynostosis.27
The two other human RecQ helicases, RECQ1 and RECQ5, are not yet genetically linked to a disease. Conceivably, mutations in RECQ1 or RECQ5 might also lead to a hereditary chromosomal instability disorder or predispose individuals to cancer. The RECQ1 (RECQL/RECQL1) gene, originally cloned independently by two groups,18,20 is located on chromosome 12p11–12 and encodes a 649 amino acid protein with a molecular mass of 73 kDa. In addition to the helicase core domain, RECQ1 contains a conserved RecQ C-terminal (RecQ-Ct) motif implicated in DNA binding and protein interactions and an N-terminal highly acidic region.12 Despite the fact that RECQ1 was the first human RecQ helicase protein to be identified and the most abundant, little was known until very recently about its genetic functions in mammalian cells.
Here we summarize our current understanding of the molecular and cellular functions of RECQ1. We propose that RECQ1 has a unique and important role to suppress genomic instability. Importantly, the findings from our work and others strongly suggest that RECQ1, like the other human RecQ helicases, has critical biological functions through its involvement in the DNA damage response and fundamental processes of nucleic acid metabolism.
Biological Importance of RECQ1
In order to assess the biological importance of RECQ1, model systems have been devised using organisms in which a RECQ1 homolog existed since no information is available on the phenotypes of human RECQ1 mutant cells. Characterization of human cells depleted of RECQ1 by RNA interference has revealed a new appreciation of its physiological importance. We will describe the findings and conclusions derived from genetic and/or cellular studies with model systems.
Regulatory Role of Human RECQ1 for Active Cell Proliferation
A common feature of RecQ helicase disorders is growth retardation and cancer predisposition, suggesting a role of RecQ helicases in the regulation of cellular proliferation. Earlier work had demonstrated that RecQ helicases are differentially regulated in a cell cycle and cell proliferation dependent manner. The expression of RECQ1 is upregulated in actively proliferating cells and also upon cellular transformation by EBV or SV40 T antigen.28 Acute depletion of RECQ1 by siRNA knockdown in HeLa cells resulted in reduced proliferative survival and growth retardation.2 Colony forming assays demonstrated a significant reduction in both the size and number of colonies when cells were inhibited for the expression of RECQ12 (Fig. 1A).
Figure 1.
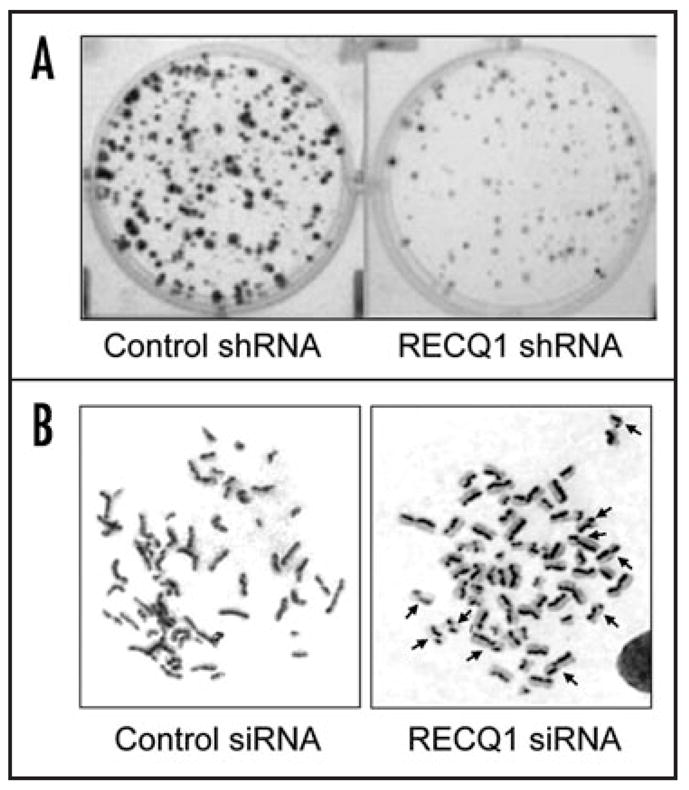
Reduced cell growth and elevated sister chromatid exchange in RECQ1-depleted cells. (A) Proliferation of HeLa cells transfected with control- or RECQ1-shRNA plasmid was determined by colony forming assay. (B) Sister chromatid exchanges were assayed in BrdU labeled, giemsa-stained chromosome spreads from control- or RECQ1-siRNA transfected HeLa cells. See ref. 2.
Interestingly, a cancer-specific role of RECQ1 in cell proliferation was suggested based on the observation that RECQ1 silencing in cancer cells resulted in mitotic catastrophe whereas RECQ1 downregulation in normal human fibroblasts did not affect their proliferation potential.29 However, we did not observe a growth defect in primary embryonic fibroblasts from Recql-null mice,1 suggesting that manifestation of cellular or organismal phenotypes in Recql-null mice may be masked by other genetic factors. Compensatory mechanisms might operate in the case of germline deletion of RECQ1 that are distinct from the proliferation defects in somatic cells when RECQ1 is acutely depleted.
We reported that RECQ1 deficiency in transformed HeLa cells leads to perturbation of normal cell cycle progression and compromised cellular ability to maintain G2/M checkpoint.2 A defective G2 arrest may allow cells to enter mitosis without proper repair of damaged DNA. It is likely that RECQ1 is required during cellular proliferation when actively replicating cells cope with endogenous DNA damage such as replication errors, stalled replication forks, and other recombinogenic intermediates. Mitotic catastrophe observed in RECQ1-silenced cells might result when these cells enter mitosis without the repair of endogenous DNA damage.
RecQ helicases like RECQ129 and WRN30,31 provide a growth advantage to actively proliferating cancer cells. This appears paradoxical to the proposed tumor suppressor functions of RecQ helicases. A role of WRN in supporting oncogenic proliferation was identified through its functional link with the MYC oncoprotein.32,33 Indeed, expression of RecQ helicases is increased upon cellular transformation and a high level of RECQ1 is present in cancer cells.28,29 It is plausible that cancer cells maintain a high copy number of RECQ1 to resolve and repair the elevated load of DNA intermediates that are generated due to active replication. Silencing RecQ helicases in cancer cells, which are already compromised for their checkpoint functions, may reduce their oncogenic proliferative capacity and, in the case of RECQ1, result in mitotic death. Therefore, the cancer cell specific mitotic death induced by RECQ1 silencing is a potential strategy for anti-cancer therapy.29
Genomic Instability and Spontaneous DNA Damage in RECQ1-Deficient Cells
A critical role of RECQ1 in the repair of endogenous DNA damage during cellular replication was revealed by the cytogenetic analysis of RECQ1-deficient cells. Primary embryonic fibroblasts from Recql-null mice display aneuploidy, spontaneous chromosomal breakage, and frequent translocation events.1 In addition, the RECQ1-deficient mouse and human cells were hypersensitive to ionizing radiation and exhibited an increased load of DNA damage.1,2 Strikingly, a transient knockdown of RECQ1 in HeLa cells resulted in significantly elevated spontaneous SCEs2,29,34 (Fig. 1B). Increased SCEs are a hallmark of BS cells.35,36 These apparently reciprocal exchanges arise primarily due to HR events that occur during repair phases of the cell cycle. The of DNA damage arising in the S or G2 elevated SCE observed in RECQ1-deficient mouse and human cells may represent unsuccessful attempts to “repair” damaged replication forks by HR at DSBs. Moreover, the spontaneously elevated Rad51 foci and γ-H2AX, an early marker of DSB formation, in RECQ1-deficient cells adds further support to a proposed role for RECQ1 in HR repair at sites of chromosomal DNA damage.1,2 Constitutively high numbers of Rad51 foci have also been detected in WS and BS cells.37,38 The presence of Rad51 foci corresponding to nucleoprotein filaments that are necessary for early strand invasion during HR suggests incomplete resolution of recombination intermediates as a consequence of the absence of RECQ1. These findings suggest that RECQ1 repairs DSBs or prevent DSBs from occurring in the first place.2 Furthermore, there is a unique requirement for RECQ1 to suppress SCEs and maintain genomic stability in mammalian cells.
Genetic Analysis of RECQ1 Using the Chicken DT40 System
Genetic studies using the chicken B lymphocyte DT40 cell line demonstrated that recql1 cells did not display a growth deficiency, sensitivity to DNA damage, or elevated SCE; however, recql1 blm cells grew more slowly compared to blm cells due to an increased population of dead cells, indicating that RECQ1 is involved in cell viability under the BLM-impaired condition.39 In addition, the double mutant recql1 blm cells exhibited elevated mitomycin C-induced SCE compared with the blm cells, suggesting that RECQ1 and BLM may substitute for each other when DNA damage blocks replication fork progression. However, RECQ1 does not perfectly substitute for BLM because blm cells still display elevated SCE, indicating that the requirement for BLM may be more stringent.
Since RECQ1, RECQ5, and BLM were all reported to interact with topoisomerase IIIα (TOP3α),40–43 Seki and colleagues investigated genetic interactions between these three RecQ helicases in DT40 cells.44 Triple mutants of recql1 recql5 blm were viable, and grew at a similar rate as the double mutants. Methylmethanesulfonate or UV sensitivity and frequency of damage-induced mitotic chiasma in the triple mutant cells was similar to those of blm cells, suggesting that RECQ1 and RECQ5 have only a minor role, if any, compared to BLM in the repair of or tolerance to DNA damage. SCE frequency in recql1 recql5 blm cells is approximately the same as recql5 blm cells, suggesting that RECQ1 has little to no role in suppressing SCE in chicken B lymphocyte DT40 cells.44 Based on the results showing that acute depletion of RECQ1 in human cells results in growth retardation, sensitivity to DNA damaging agents, accumulation of DNA damage, and chromosomal instability, we propose that RECQ1 has more specialized biological functions in mammalian systems.
Structural Properties, Catalytic Activities and Protein Interactions of RECQ1
In light of its known structural properties, catalytic activities, DNA substrate preference (Table 1), and protein interactions (Table 2), we suggest that RECQ1 is well equipped to facilitate biological pathways important for chromosomal stability maintenance.
Table 1.
DNA Substrate Specificity of RECQ1 Helicase
Table 2.
RECQ1 protein interactions
| RECQ1 Partner | Physical Interaction | Functional Interaction | Reference |
|---|---|---|---|
| RPA | Yes, via RPA70 | RPA stimulates RECQ1 helicase | 4,46 |
| EXO-1 | Yes | RECQ1 stimulates EXO-1 nuclease | 5 |
| MSH2/6 | Yes | MSH2/6 stimulates RECQ1 helicase | 5 |
| MLH1/PMS2 | Yes | ND | 5 |
| Top3α | Yes | NR | 41 |
| Imporfin-α(Qip1, Rch1) | Yes | NR | 110 |
| Rad51 | Yes | ND | 2 |
ND, None detected; NR, None reported, See text for details.
DNA Substrate Specificity of RECQ1 Helicase
The first biochemical studies of RECQ1 were performed using an endogenous source of the RECQ1 protein purified from nuclear extracts of human cells.45 These studies revealed that RECQ1 ATPase activity is strongly stimulated by the presence of a DNA cofactor, and that longer single-stranded DNA (ssDNA) molecules serve as better effectors for RECQ1 ATP hydrolysis, suggesting that RECQ1 translocates processively along ssDNA. RECQ1, like other RecQ helicases characterized to date,12 unwinds duplex DNA substrates with a 3′ to 5′ directionality as determined by a classic linearized M13 partial duplex substrate45 or an oligonucleotide-based substrate with a 3′ ssDNA overhang46 (Table 1). Increasing the length of the 3′ ssDNA tail to lengths of 25 or 75 nucleotide (nt) compared to a 10 nt 3′ tailed substrate significantly improved the ability of RECQ1 to unwind the adjacent duplex DNA,46 suggesting that RECQ1 requires a minimal ssDNA tail length for optimal loading or assembly state formation. RECQ1 failed to unwind a short RNA-RNA duplex substrate under conditions that it efficiently unwound 3′ tailed duplex DNA or bubble DNA substrates with 3′ and 5′ duplex regions flanking an internal ssDNA segment.46
To characterize the effect of duplex length on RECQ1 helicase activity, a series of M13 partial duplex DNA substrates of increasing lengths were tested.4 The apparent unwinding rates for the 25 and 50 base pair (bp) partial duplexes were almost identical. The concentration of RECQ1 required for unwinding the 25 and 50 bp partial duplex substrates was proportional to the length of the duplex to be unwound. However, RECQ1 helicase activity was dramatically reduced with longer DNA duplexes. In fact, only a small fraction of the 110 bp duplex could be unwound by RECQ1 and no unwinding was detected for the 216 bp duplex, even at the highest RECQ1 concentration tested. Therefore, in the absence of any auxiliary factor, RECQ1 is able to efficiently unwind only short DNA substrates in vitro.
An analysis of DNA substrate specificity demonstrated that recombinant human RECQ1 purified from insect cells, which has similar activity compared to that of the native RECQ1,46 preferentially unwound forked duplex substrates compared to those that have only a 3′ tail3 (Table 1). RECQ1 was also shown to unwind (in order or preference) 3′-flap, 5′-flap, and synthetic replication fork structures.3 Thus, RECQ1 unwinds conventional duplex DNA substrates representing key replication and repair intermediates that lack single-strand character in the 3′, 5′, or both arms adjacent to the duplex. RECQ1, like WRN,47 unwinds a replication fork structure with heterologous fully duplex leading and lagging strand arms in the direction of the fork.3 In contrast, Drosophila RECQ5β unwinds the lagging strand of a synthetic replication fork,48 suggesting some apparent differences among the RecQ helicases. The recent demonstration that WRN49,50 and BLM49,51 catalyze fork regression of a model DNA replication fork with homologous arms raises the experimental question if RECQ1 might also catalyze fork regression. The ability of RECQ1 to stably bind multi-stranded branched DNA molecules that represent DNA repair and replication intermediates3 suggests that its intrinsic DNA binding activity might be utilized for a novel function.
Nucleotide-Induced Conformational Change in RECQ1 Provides a Molecular Switch from a Strand Annealing to a DNA Unwinding Mode
In addition to unwinding duplex DNA, a growing number of RecQ helicases, including all five human RecQ helicases,3,52–55 have been reported to catalyze strand annealing of complementary ssDNA molecules. Strand annealing by RECQ1 is quite efficient and occurs in the absence of ATP.3 RECQ1-promoted strand annealing was dependent on RECQ1 concentration; however, RECQ1 concentrations >20 nM inhibited strand annealing. RECQ1 has comparable ability to unwind the forked duplex substrate (in the presence of ATP) or to catalyze annealing of complementary single strands (in the absence of ATP) at low RECQ1 concentrations. ATP or the poorly hydrolyzable analog ATPγS strongly prevented RECQ1 strand annealing whereas ADP had no effect.3 Analyses of strand annealing utilizing ATP-binding/hydrolysis mutants revealed that nucleotide binding inhibits strand annealing catalyzed by RECQ1. Furthermore, partial proteolysis studies demonstrated that ATP binding induces a conformational change in RECQ1 protein, which serves as a molecular switch from a strand-annealing to a DNA-unwinding mode.3
The biological importance of the dual enzymatic activities catalyzed by RECQ1 is currently not known. Conceivably, strand annealing and/or DNA unwinding catalyzed by RECQ1 may be important for replication fork regression (formation of a ‘chicken foot’ structure at blocked DNA replication forks), directional migration of a D-loop or Holliday Junction (HJ) structure, or a pathway of HR repair such as synthesis-dependent strand annealing (SDSA). In SDSA, the D-loop is dissolved after some DNA synthesis, and the disengaged invading strand reanneals with the second end of the DSB, always forming a non-crossover. Crossover avoidance is one of the key features of SDSA as crossovers have the potential to generate genomic rearrangements. A role for RECQ1 in SDSA is relevant since mitotic recombination in somatic cells is rarely associated with crossovers and thought to proceed through SDSA.56 This proposed role of RECQ1 in SDSA, if true, would explain the elevated SCEs in RECQ1-deficient cells. The spontaneous γ-H2AX foci and elevated SCE in RECQ1-deficient cells,1,2 indicating aberrant repair of DNA breaks, are consistent with a role of RECQ1-associated activities in a pathway of HR repair.
Structural Aspects of RECQ1
There has been considerable interest in the assembly state for various RecQ helicases since quaternary structure is important for the mechanism of DNA unwinding.12 To provide further structural insight to the regulation of DNA unwinding and strand annealing, Vindigni and co-workers examined the importance of RECQ1 assembly states for catalytic activity.57 Two quaternary forms of RECQ1 exist: (1) higher-order oligomers consistent with pentamers or hexamers; (2) smaller oligomers consistent with monomers or dimers. Importantly, size exclusion chromatography and transmission electron microscopy (EM) demonstrated that the equilibrium between the two quaternary states is modulated by ssDNA and ATP binding, in which ATP or ATPγS favors the smaller oligomeric form. The RECQ1 EM reconstructions of the higher order oligomeric form revealed a cage-like structure of approximately 120 Å × 130 Å. The body of the helicase assembly is arranged in two ring-like densities interconnected by diagonally extending densities to a larger middle ring structure, giving the appearance of a spiral. Together, the three rings form a hollow channel with an inner diameter of 20 Å.
Under conditions in which the oligomeric (pentamer or hexamer) form is stabilized, RECQ1 efficiently catalyzes strand annealing whereas competition experiments with ATPase-deficient RECQ1 mutants indicated that RECQ1 monomers or tightly bound dimers mediate DNA unwinding.57 Thus, a functional explanation for the role of the different assembly states of RECQ1 (and perhaps other RecQ helicases) was provided, consistent with the theme that ATP binding to RECQ1 triggers a molecular switch from strand annealing to DNA unwinding mode.
Recently, the crystal structure of an E. coli expressed truncated human RECQ1 protein (amino acids 49–616) in complex with Mg2+-ADP was solved at 2.0 Å (Pike A, Burgess Brown N, Shrestha B, King O, Niesen FH, Muzzolini L, Vindigni A and Gileadi O, unpublished observations; Protein Database Code 2V1X) (Fig. 2A). protein appears as a dimer in The catalytically active RECQ149–616 the crystal, a result that was confirmed by equilibrium and sedimentation velocity analytical centrifugation and dynamic light scattering. Amino acids 9–48 are required for RECQ1 to assemble as a higher order oligomeric form that may correspond to a hexamer. The crystal structure of RECQ149–616 contains all the sub-domains of E. coli RecQ:58 the two RecA-like domains, the Zn2+-binding RecQ-Ct domain, and a C-terminal winged helix (WH) domain. However, the WH domain is re-oriented relative to that in the bacterial protein (Fig. 2B). In particular, the surface suggested to be involved in DNA binding in the E. coli enzyme58 is not accessible in RECQ149–616.
Figure 2.
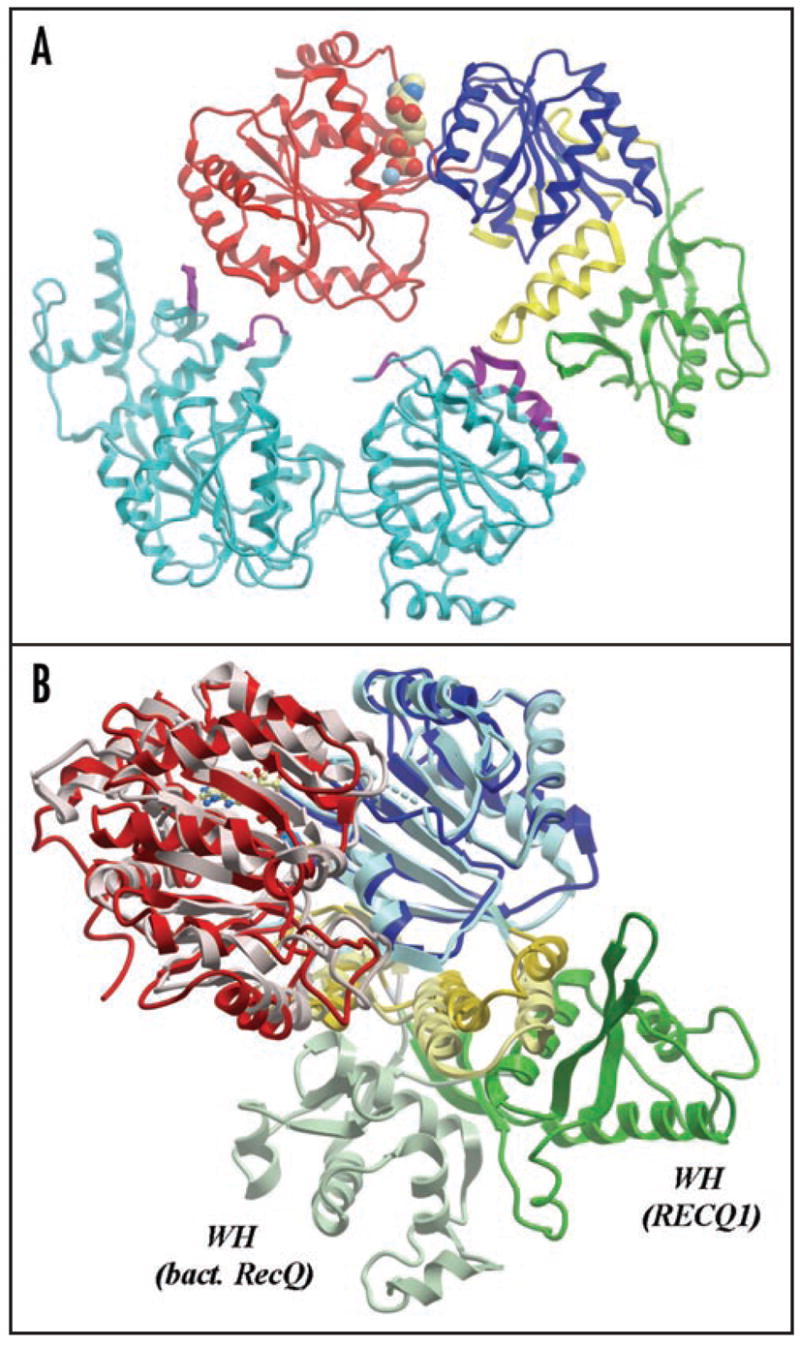
RECQ1 crystal structure. (A) RECQ1 dimer. Two molecules of RECQ1 in the crystal structure (Protein Database Code 2V1X). A Mg2+-ADP molecule (space-filling model) is bound in the cleft between the two RecA-like domains (colored red and blue). The Zn2+-binding and winged-helix (WH) domains are marked in yellow and green, respectively. The polypeptide chain of the second monomer is shown in light blue, with the regions involved in dimer interaction marked in purple. (B) RECQ1-RecQ overlay. The structure of RECQ1 (colored as in the previous image) aligned with that of E. coli RecQ (pale colors), maximizing the overlap of the RecA-like domains. While the RecA domains and Zn2+-binding domains are well aligned, the WH assumes very different orientations in the two protein structures.
RPA Interacts with RECQ1 Helicase and Improves its Unwinding Efficiency
A key player in virtually all aspects of cellular DNA metabolism is the ssDNA binding protein Replication Protein A (RPA), a heterotrimeric protein composed of three subunits : (1) RPA70, (2) RPA30; (3) RPA14.59 Each RPA subunit contains at least one OB-fold (oligosaccharide/oligonucleotide binding fold) domain that is found in many ssDNA binding proteins and facilitates ssDNA binding. RPA interacts with RECQ1 and the physical interaction is mediated by the RPA70 subunit, as demonstrated by Far Western analysis.4 The direct DNA-independent interaction between RECQ1 and RPA was substantiated by ELISA, yielding an apparent dissociation constant (Kd) of 6.2 nM for the RECQ1–RPA complex,4 similar to that previously determined for the interaction of RPA with other DNA helicases.60–63 Co-immunoprecipitation experiments using human nuclear extracts confirmed that RECQ1 and RPA interact in vivo.4 Importantly, RPA dramatically stimulates the ability of RECQ1 to unwind duplex DNA substrates.4,46 RPA stimulation of RECQ1 helicase activity is biochemically important since RECQ1 is not a very processive helicase and even under multiple turnover conditions, RECQ1 fails to unwind DNA duplexes exceeding 100 bp.4 However, in the presence of RPA, RECQ1 unwinds much longer duplex DNA substrates such as a 500 bp partial duplex.4 Stimulation of RECQ1 helicase activity by RPA is specific since the heterologous E. coli single-stranded DNA binding protein (SSB) failed to stimulate RECQ1 helicase activity at concentrations up to 10-fold higher than that used for RPA.4
RPA, but not other SSBs, was also reported to stimulate the DNA unwinding reactions catalyzed by other human RecQ helicases,60,61,64 suggesting a similar mechanism for the functional interaction observed between RECQ1 and RPA. Using information from mapping analyses, evidence was presented that the physical interaction between RPA and WRN or BLM helicases is required for RPA stimulation of helicase-catalyzed DNA unwinding.62 It will be informative to determine if RECQ1 interacts with RPA via an acidic region outside the helicase domain, and if this region is important for the functional interaction with RPA, as reported for WRN helicase.62 The molecular details underlying the mechanism for stimulation of DNA helicases by SSBs has been a topic of interest.12,65 For example, it is still unknown if a given SSB helps to load a helicase on to ssDNA during an ongoing unwinding reaction, or if the helicase loads the SSB on to the unwound strands in a coordinated event. In a recent study, it was reported that the SSB stabilizes the well characterized Hepatitis C virus helicase at the unwinding junction and prevents its dissociation, suggesting a potential mechanism for improving helicase processivity.66 It may be relevant that SSBs such as RPA strongly inhibit the ability of RECQ1 to efficiently catalyze annealing between complementary ssDNA molecules.3 This effect of RPA distinguishes the strand-annealing reaction catalyzed by RECQ1 helicase from that of Rad52-mediated strand annealing which functions co-operatively with RPA.67 It is conceivable that RPA may bind to and stabilize the smaller oligomeric form of RECQ1 that is catalytically active as a helicase,57 and this interaction is also a mechanistic component of the stimulatory effect of RPA on RECQ1 helicase activity.
Understanding how the interactions of RPA with RECQ1 and other RecQ helicases are regulated in a biological setting is an even greater challenge. The observations that both RECQ12 and RPA59 are post-translationally modified in response to DNA damage raises the possibility that a protein modification such as phosphorylation will affect the cellular role of RECQ1 by modulating its interaction with RPA or other protein partners.
RECQ1 Catalytically Unwinds Homologous Recombination Intermediates
A growing body of work provides compelling evidence that the genomic instability in a variety of organisms carrying a mutation in a gene of the RecQ helicase family is a consequence of inappropriate processing of DNA replication or recombination intermediates. RecQ helicases are proposed to play central roles in HR and repair of blocked or damaged replication forks.8,12,13,68 Upon replication fork stalling, HR pathways may be elicited to deal with the structural anomaly. A stalled replication fork can be converted to a four-way junction resembling a HJ by branch migration and re-annealing of nascent DNA strands.69,70 A potential role of RECQ1 or a related RecQ helicase in the rescue of a stalled fork is to catalyze reverse branch migration past the lesion to reset the replication fork, and the lesion can be subsequently corrected by DNA repair.71 Localization of RecQ helicases (WRN, BLM, and Sgs1) at stalled DNA replication forks72–74 is consistent with their role in metabolizing DNA replication or recombination intermediates at blocked replication forks through a non-recombinogenic pathway or Rad51-mediated HR. RECQ1,3 like a number of other RecQ helicases (e.g., WRN, BLM, and Sgs1),12 unwinds a synthetic HJ structure (Table 1) to generate a splayed arm product in a reaction dependent on ATP hydrolysis. Consistent with the possibility that RECQ1 catalyzes disruption of the HJ by branch fork migration, preincubation of the HJ substrate with increasing concentrations of RuvA protein, an E. coli protein implicated in HR that has a very high affinity for the HJ,75 inhibited RECQ1-catalyzed branch migration in a RuvA concentration-dependent manner.3 Thus, RECQ1 catalyzes branch migration of the HJ by initiating unwinding at the junction crossover.
RECQ1 unwinds another distinct intermediate of HR, the three-stranded D-loop structure3 (Table 1). The D-loop forms during an early step in HR when a recombinanse protein binds ssDNA and mediates invasion of the ssDNA molecule into a homologous duplex.76,77 This strand invasion event results in the displacement of one strand of the recipient duplex with the concomitant formation of Watson-Crick base pairing between the complementary strand and the incoming single strand.78,79 RECQ1 unwinds D-loops with either a protruding single-stranded 3′- or 5′-tail by releasing the invading third strand from D-loop structures in an ATP- and protein concentration-dependent manner; however, preferential unwinding of the D-loop with a protruding single-stranded 3′-tail was observed over the entire range of RECQ1 protein concentrations.3 Most recently, RECQ1 was shown to promote unidirectional branch migration of D-loops in a 3′ to 5′ polarity, and as a result, specifically disrupt joint molecules that may represent dead-end intermediates of HR (Bugreev DV, Brosh RM Jr., and Mazin AV, unpublished data). Indeed, the Recql null mouse embryonic fibroblasts display elevated Rad51 foci that might represent intermediates of HR.1 The catalytic action of RECQ1 on D-loop substrates may be important as an anti-recombinase function, as proposed for other RecQ helicases.80–83 Alternatively, efficient unwinding of D-loops by RECQ1 may play a role in telomere maintenance, as proposed for WRN helicase,84 or in DNA repair by a SDSA pathway that requires multiple rounds of strand invasion/resynthesis for the complementary strand to anneal to its partner strand.
Understanding the relationship between the molecular functions of RECQ1 helicase and its role in the DNA damage response and maintenance of chromosomal stability will require innovative approaches and model systems. As discussed in two very recent reviews (reviewed in refs. 85 and 86), three major themes have emerged over the last decade in terms of DNA processing pathways that insure genomic stability: (1) the processes of DNA replication, recombination, and repair cooperate with each other to maintain genome homeostasis and cell viability; (2) the DNA damage response is mediated by a sophisticated cast of players that orchestrate programs to repair DNA and resolve problems at the replication fork; (3) post-translational protein modifications regulate these genome maintenance pathways. Regulation of RECQ1 function by post-translational modifications has not been explored in detail yet, and will be an important area of study. Our cellular studies demonstrate that RECQ1 is indeed phosphorylated upon cellular exposure to DNA damaging agents.2 Protein phosphorylation may regulate RECQ1 catalytic activities and/or its interactions with DNA repair factors which will affect the physiological role of RECQ1 in the DNA damage response. It will be of paramount importance to determine the biochemical and physiological significance of RECQ1 post-translational modifications such as protein phosphorylation.
A number of models have been proposed to describe the mechanisms whereby DNA helicases limit recombination through their catalytic activities. For example, genetic and biochemical results provide evidence that the yeast Srs2 helicase disrupts Rad51 presynaptic filaments to restrict recombination.87,88 BLM81 and RECQ589 also disrupt Rad51 filaments, suggesting a mechanism whereby they suppress recombination. In addition to unwinding DNA recombination intermediates, RECQ1 may utilize its motor ATPase function to displace proteins from HR DNA repair intermediates. However, neither RECQ181 nor WRN,89 even at relatively high protein concentrations, were capable of disrupting Rad51 protein filaments or inhibiting the Rad51-mediated D-loop formation. An alternative strategy for RECQ1 to actively participate in HR repair may be to confer directionality of branch migration of recombination intermediates via the coordinate action of RECQ1 helicase and strand annealing activities.
RECQ1 Interacts with Mismatch Repair Factors that Regulate Genetic Recombination
To identify protein partners of human RECQ1 that exist in vivo, co-immunoprecipitation experiments were performed using an antibody raised against a 20-amino acid peptide of a unique sequence in the C-terminal region of RECQ1. The anti-RECQ1 antibody co-immunoprecipitated RECQ1 with three human DNA repair proteins (EXO-1, MSH2/6 and MLH1).5 Through its direct protein interaction, RECQ1 stimulated the incision activity of EXO-1,5 a 5′ to 3′ exonuclease and structure-specific endonuclease that has been implicated in mismatch correction, mitotic and meiotic recombination, and DSB repair.90–94 Yeast genetic studies identified a role of EXO-1 in spontaneous mitotic and meiotic recombination between direct repeats.90 The 5′ to 3′ exonuclease activity of EXO-1 is proposed to generate 3′ ssDNA tails that can be used by homologous pairing proteins for recombination.
EXO-1 also affects the metabolism of telomeres in yeast,95 and EXO-1-dependent ssDNA at telomeres is required for cell cycle arrest in budding yeast ku70 deletion mutants.96 A deletion of EXO-1 in mammalian cells impairs DNA damage signaling.97 Genetic and biochemical studies have demonstrated the importance of certain RecQ helicases in telomere metabolism.12 It is conceivable that RECQ1 stimulates EXO-1 cleavage of DNA structures in sub-telomeric regions to stabilize the telomeric end or facilitate its replication. EXO-1 nucleolytic activity may serve to create long telomeric ssDNA tails that promote recombination-driven lengthening of telomeres when telomerase or telomere binding proteins are absent,95,98 or activate a DNA damage checkpoint.96 Proposed roles of EXO-1 in a mismatch repair-independent mutation avoidance pathway99,100 or repair/restart of stalled replication forks101 may also involve RecQ helicases such as RECQ1 or WRN.102
In addition to EXO-1, RECQ1 was observed to be associated with proteins (MSH2/6, MLH1-PMS2) implicated in mismatch repair.5 MSH2/MSH6 (MutSα) is a protein heterodimeric complex that binds with high affinity to base-base mismatches103 and G-quadruplexes.104 MutLα, a protein complex of MLH1 and PMS2, is also required for mismatch repair103 and was recently reported to have an endonucleolytic function in humans105 and yeast.106 From a functional standpoint, MutSα was found to stimulate RECQ1 helicase activity.5 The interaction of RECQ1 with mismatch repair factors that regulate genetic recombination is provocative because mouse1 and human2 cells deficient of RECQ1 display genomic instability and an increased sensitivity to genotoxic agents. Studies of the sole RecQ homolog in Saccharomyces cerevisiae have suggested that Sgs1 functions to prevent HR15,107 or act as a sensor for DNA damage.108 A model was proposed in which Sgs1 interacts with the MutSα complex to unwind DNA recombination intermediates that contain mismatches.109 It is conceivable that RECQ1 collaborates with mismatch repair proteins in a specialized pathway of HR to suppress erroneous strand invasion events at DSBs, or facilitates DNA processing events at repetitive DNA elements or specialized chromosomal structures such as telomeres. Further studies will be needed to fully understand the molecular mechanisms by which RECQ1 and the mismatch repair factors work together to maintain genomic stability.
Physical Interactions of RECQ1 with Top3α, Rad51 and Importin-α
The first identified RECQ1 protein interaction was with the human importin-α homologs Qip1 and Rch1.110 Mapping studies revealed that Qip1 binds to the nuclear localization sequence motif of RECQ1, suggesting that the importin-α homolog may function in the transport of RECQ1 to the nucleus. RECQ1 was recently found to interact with the Rad51 recombinase.2 Although an effect of Rad51 on RECQ1 helicase activity was not detected, it seems likely that the physical interaction may be important in Rad51-mediated invasion into homologous duplex DNA to generate joint molecules (D-loops) in an early step of HR. The sensitivity of RECQ1-deficient cells1,2 to ionizing radiation which induces strand breaks may reflect a defect in HR repair in which processed DNA ends are not properly acted upon by the concerted action of RECQ1 and Rad51. This idea is further supported by the elevated γ-H2AX foci in RECQ1-deficient cells.1,2
The interaction of BLM helicase with Top3α is proposed to be important in the suppression of SCE.111 The elevated SCE in RECQ1-deficient human2 and mouse1 cells suggests that RECQ1 functions to prevent cross-overs. Although RECQ1 was demonstrated to be associated with Top3α by co-immunoprecipitation from human lymphoblastoid cell extracts,41 no functional interaction has been reported. Moreover, RECQ1 failed to substitute in vitro for BLM to act with Top3α to catalyze double HJ dissolution reaction,112 a proposed mechanism for the exclusive resolution of double HJs into non-crossover recombinant products.111 One possibility is that in mammalian cells RECQ1 operates by a mechanism different from that of BLM; however, it remains to be determined if the role of RECQ1 in SCE suppression is Top3α-dependent or Top3α-independent. Much work remains to fully understand the mechanistic functions of RECQ1 with its protein partners.
Does RECQ1 Play a Role in Gene Expression?
The reduced cellular proliferation, metabolic activity, and growth in human cells depleted of RECQ1 indicate that RECQ1 is required for normal cellular physiology.2 Non-coding DNA sequences within the human genome, formerly referred to as “junk DNA”, satisfy vital functions in chromatin assembly and regulation of gene expression.113 Mounting evidence from model organisms indicate that small noncoding RNAs transcribed from transgenic sequences participate in gene silencing programs that affect cell growth and differentiation. From a more global perspective, evidence demonstrates that gene silencing of repetitive or complex sequences is important in a variety of biological processes that affect development, genome defense against transposons and viruses, and mechanisms of inheritance (reviewed in refs. 113–117). In addition to a DNA repair function, it is possible that RECQ1 may play a role in regulating gene expression. We will discuss evidence supporting this hypothesis and place this in the broader context of RECQ1 as an important RecQ helicase to maintain genomic stability.
RECQ1 Resides in a piRNA Complex Implicated in Transcriptional Gene Silencing
Recently, Lau and colleagues identified a Piwi protein and RECQ1 as co-existing members of a piRNA protein complex that is important for gene silencing.118 Essentially, rat testis extract was fractionated by ion exchange, and a novel ribonucleoprotein complex containing RNA of 25–31 nt was identified and the associated proteins further purified. Mass spectrometry analysis revealed the presence of rat homologs to a protein of the Piwi class of Argonaute proteins implicated in gene silencing and also the RECQ1 helicase. The piRNA protein complex contained ATPase and DNA helicase activities as well as RNA cleavage activity that would be predicted to be catalyzed respectively by RECQ1 and a conserved Argonaute protein responsible for RNA-guided cleavage of target RNAs.
piRNAs are a unique class of small (24–30 nt) noncoding RNAs that are generated by a mechanism distinct from the classic Dicer endonuclease processing of a single long double-stranded RNA molecule. The size range, single-stranded nature, and strand specificity of piRNAs and their clustered origin suggest a process of piRNA biogenesis that is similar to the process of DNA replication.119 The mechanistic similarities between piRNA synthesis and DNA replication suggest that RECQ1 and associated DNA replication proteins are involved in the biogenesis of small dsRNA molecules that serve to silence the expression of genes at the post-transcriptional level. Evidence in support of this notion was recently garnered by the identification of another DNA replication protein that interacts with a RECQ1 homolog and an RNA polymerase that are implicated in gene silencing in Neurospora (see below).
A Role for a RECQ1 Homolog in Gene Silencing
The closest RecQ homologs of the model fungus Neurospora crassa QDE-3 helicase are RECQ1 and BLM.120,121 Phylogenetic analysis revealed that the other three mammalian RecQ helicases (WRN, RECQ4, RECQ5) are less similar to QDE-3 and are separated from the QDE-3/RECQ1/BLM cluster, reducing the probability they are QDE-3 orthologs.121 QDE-3 has been implicated in gene silencing, known as quelling, in Neurospora.120 Three classes of quelling defective mutants (qde-1, qde-2, qde-3) have been isolated.122 In order for gene silencing to occur in Neurospora, transgenic loci must be transcribed. The quelling deficiency of qde-1 mutants defective in RNA-dependent RNA polymerase prompted Cogoni and coworkers to investigate protein interactions of QDE-1. Using an immunoaffinity purification approach, it was found that QDE-1 interacts with the RPA70 subunit of the RPA heterotrimer.123 The essential requirement of RPA in replication led the researchers to explore the question if gene silencing is linked to DNA replication. It was observed that QDE-1 was specifically recruited into the repetitive transgene loci and that the accumulation of short interfering RNAs (siRNAs) implicated in gene silencing required ongoing DNA synthesis.123
These findings suggest a direct coupling of DNA replication of repeated sequences to targeted silencing mediated by a post-transcriptional gene silencing protein complex. As suggested by the authors of the study,123 the well documented interaction of RPA with RecQ helicases may be relevant to the observation that QDE-3, a protein with greatest sequence similarity to RECQ1, is essential for gene silencing and operates upstream of dsRNA production in the process. It was proposed that RPA plays a role in differentiating transgenic sequences from endogenous genes and through its interaction with QDE-1 targets transgenic sequences for silencing by the in situ production of dsRNA. The requirement for transgenic sequences to be inserted in tandem in order to trigger silencing raises the possibility that during replication the tandem repeats form slippage intermediates that resemble cruciform structures which stall the replication fork.
The abundant evidence that RecQ helicases act at stalled or blocked replication forks to maintain genomic stability by preventing erroneous recombination events would be consistent with a role of a RecQ helicase to help a cell cope with potential replication slippage events. RecQ helicases recognize aberrant DNA structures (HJ, three- and four-stranded DNA structural intermediates associated with the replication fork).12 Furthermore, the direct physical and functional interactions of RECQ1 and related RecQ helicases (WRN, BLM, RECQ5) with RPA point toward a model in which the helicase and SSB collaborate during replication of slippage intermediates to prevent illegitimate recombination events.12 Conceptually, RPA and a RecQ helicase (QDE-3 in Neurospora) also interact with QDE-1 at the repeat sequences to facilitate the biogenesis of dsRNA molecules that operate in gene silencing (Fig. 3). In this model, a number of details remain to be fleshed out including the origin of the RNA used as a template by QDE-1 to produce dsRNA. Nonetheless, the collective evidence that RecQ helicases, including RECQ1, play an instrumental role in preserving genomic stability through their catalytic activities and protein interactions suggest that their unique position at the crossroads of DNA replication, repair, recombination, and transcription is consistent with a proposed role of these specialized motor ATP-dependent DNA unwinding enzymes in gene silencing.
Figure 3.
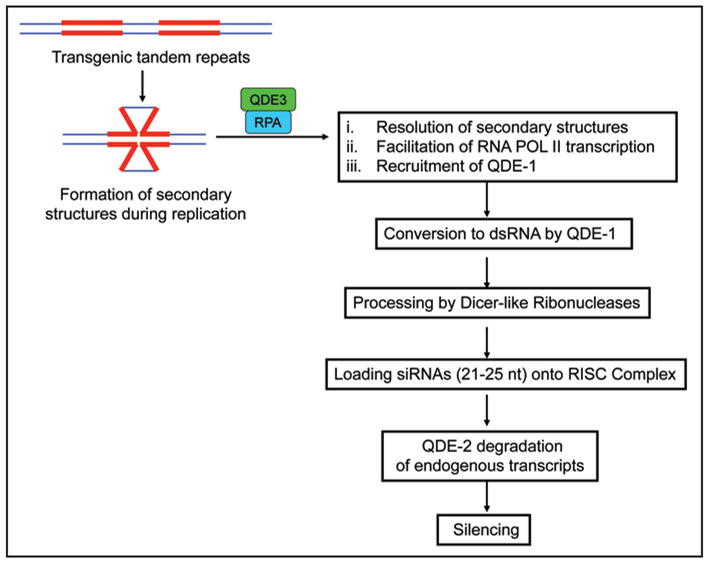
Quelling in Neurospora with an emphasis on potential roles of QDE-3 (RECQ1) homolog with RPA in the pathway. QDE-3: RPA complex may serve in several functions: (1) to resolve secondary structure that forms during replication of tandem repeats; (2) to facilitate transcription of transgenic locus; (3) to recruit QDE-1 to nascent transgenic RNA.
Involvement of Neurospora RECQ1 Homolog in DNA Repair and Genomic Stability
In support of conserved roles of Neurospora QDE-3 helicase and mammalian homologs (mouse and human RECQ1) in the DNA damage response, qde-3 mutants were found to be hypersensitive to a variety of DNA mutagens.120,124,125 The dramatic sensitivity of qde-3 mutants to the S-phase-specific type I topoisomerase inhibitor camptothecin120 suggests a role of QDE-3 during replication. qde-3 mutants were subsequently demonstrated to be sensitive to alkylating agents (methylmethanesulfonate, N-methyl-N′-nitro-N-nitrosoguanidine) and other agents (e.g., 4-nitroquinoline-1-oxide, hydroxyurea) that induce replicational stress,124,125 further pointing toward a role of the RECQ1 homolog to repair DNA lesions during replication or help the cell to cope with aberrant replication structures that arise during fork stalling. Epistatic genetic analysis revealed that the qde-3 gene is assigned to the uvs-6 recombination repair pathway and the uvs-2 postreplication repair pathway.124
In addition to QDE-3, Neurospora has a second RecQ homolog known as RECQ-2.120,124,125 Neurospora mutants with genetic deficiencies in both qde3 and recq2 were found to display a severe growth defect that was dependent on the presence of a wild-type rad51 gene, suggesting that HR is responsible for the growth defect of the qde3 recq2 double mutant.126 A mutator phenotype characterized mostly by deletions in a qde3 recq2 double mutant is suppressed by a mutation in the mus-52 gene, a homolog of the human KU80 gene.126 This may mean that nonhomologous end-joining machinery that is actively repairing DSBs in the qde3 recq2 mutant is responsible for the mutator phenotype. Collectively, the genetic evidence indicates that the Neurospora RECQ1 homolog has a broader role in the DNA damage response and chromosomal stability in addition to its gene silencing function.
Dual Functions of RECQ1 in Genome Stability and Gene Silencing?
As discussed earlier, genomic instability and DNA damage sensitivity is also detected in mouse or human cells deficient in RECQ1, suggesting pleiotropic and important functions of mammalian RECQ1 homologs in cellular nucleic acid metabolism. However, a role for RECQ1 (as well as WRN and BLM) in RNAi-mediated silencing was not observed using an assay to measure short term RNAi-mediated silencing in mice.121 It is possible that mouse RECQ1 plays a role in long term RNAi-mediated silencing, or that the RecQ-dependent pathway operates differently in humans and mice. The lack of sequence conservation between many mouse and human piRNAs117 suggest that differences in piRNA metabolism between the two species may operate. Similarly, some distinctions between the cellular phenotypes of RECQ1-deficient mouse and human cells exist, such as the reduced cellular proliferation observed in RECQ1-depleted human cells2 not readily detected in the recql mouse embryonic fibroblasts.1
The human RECQ1 gene is localized to chromosome 12p11–12,18,19 a region of instability in testicular germ-cell tumors.127 RECQ1 expression in mouse is highest in the testis.1 The purification of a RECQ1 ribonucleoprotein complex from rat testis with testis-specific small non-coding piRNAs118 is likely to be relevant since piRNAs and piRNA complexes have also been detected in testis of other mammals such as mice and humans.128 Mouse orthologs of Piwi are required for spermatogenesis, and it is suggested that germ-line-specific expression of piRNAs plays a role.128 This raises the possibility that the piRNA complex in which RECQ1 resides regulates male meiosis by modulating genome organization or stability. Understanding the precise mechanism whereby RECQ1 and its associated piRNA complex regulates the germline is an important challenge. High priorities will be to define the inter-connected roles of RECQ1 homologs, such as QDE-3, in gene silencing and chromosomal stability maintenance and to determine if mammalian RECQ1 functions by a conserved mechanism. It was proposed that the piRNA pathway might have a conserved function in silencing retrotransposons and preventing DNA damage in the germline.116 Clearly, the potentially dual yet complementary functions of piRNA associated proteins like RECQ1 in the DNA damage response and piRNA metabolism are tantalizing and provoke a novel interest in the biological functions of RecQ helicases in the context of gene expression and genomic stability.
A number of studies have suggested that piRNAs have a primary function in maintaining germline DNA stability and that piRNA mutations are a secondary consequence of a defective DNA damage response (reviewed in ref. 115). Therefore it was postulated that piRNAs (and by inference piRNA associated proteins) directly promote DNA repair, induce proper chromosome assembly to avoid DNA damage, or suppress the expression of euchromatic genes that lead to DSBs. It seems reasonable that RECQ1 may serve as a member of the piRNA complex to facilitate the proper DNA damage response and maintain germline stability.
Prospective Importance of RECQ1 in Human Health and Cancer Therapy
From a human health perspective, it will be important to decipher the elusive role(s) of RECQ1 in genome homeostasis and determine precisely the involvement of RECQ1 in the DNA damage response and processes of chromosomal nucleic acid metabolism. Although a genetic disorder has not yet been linked to a mutation in RECQ1, the significant reduction in cellular proliferation due to RECQ1 depletion raises the possibility that RECQ1 is essential for viability in humans. In somatic cells, recombination defects leading to chromosomal instability due to a RECQ1 deficiency may predispose individuals to cancer (Fig. 4). Conversely, the presence of RECQ1 confers error-free DNA repair, proper replication restart, and a stable genome. Upregulated RECQ1 expression may provide growth advantage in transformed or actively proliferating cells. Cytogenetic data suggest that RECQ1 may be associated with cancer predisposition. Loss of heterozygosity of 12p12, the chromosomal location of the Recq1 gene,18–20 is a frequent event in a wide range of hematological malignancies and solid tumors, suggesting the presence of a tumor suppressor locus. Allelic losses on chromosome 12p12–13 are associated with childhood acute lymphoblastic leukemia and several solid neoplasms.129 Chromosome 12p12 deletion has been reported in a rare chronic myeloid leukemia-like syndrome case in a Li-Fraumeni syndrome family.130 RECQ1 is highly expressed in the lungs,24 and deletions at chromosome 12p12 have been reported in bronchial epithelia of patients with primary non-small-cell lung cancer,131 suggesting that RECQ1 sequence alterations may influence the risk of lung cancer. Microsatellite instability in the polyguanine repeat (G)9 in the Recq1 gene is frequently observed in mismatch repair-deficient human nonpolyposis colorectal cancer.132
Figure 4.
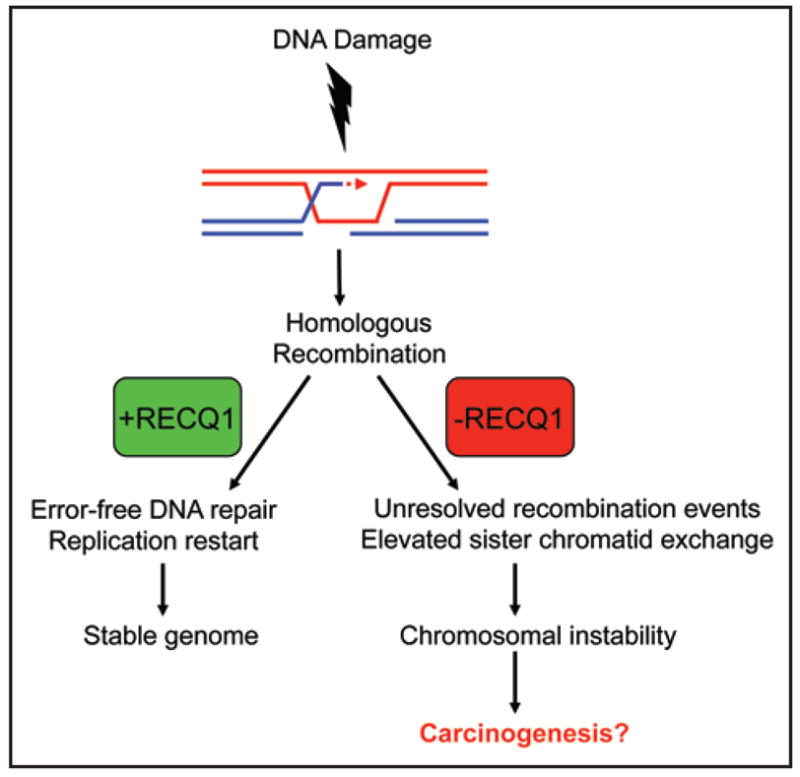
RECQ1 preserves genomic integrity through its proposed role in homologous recombinational repair. See text for details.
Recent analyses of Recq1 single-nucleotide polymorphisms (SNPs) have identified an association of RECQ1 with a reduced survival of pancreatic cancer patients.133,134 Only one RECQ1 A159C SNP allele is required to significantly decrease overall pancreatic cancer survival, suggesting a predictive and prognostic role for RECQ1 SNPs. RECQ1 SNPs displayed significant genetic interaction with SNPs in the HR repair genes ATM, RAD54L, XRCC2 and XRCC3. A role for RECQ1 in HR is supported by the findings that SNPs in RECQ1 negatively affect the response to the anticancer drug gemcitabine-induced radiosensitization that selectively requires functional HR.133,134 Thus, variant alleles of RECQ1 (and other helicases) may impact the efficacy of chemotherapy treatments. DNA repair helicases have been proposed to be potential targets for cancer therapies mediated by DNA damaging agents or radiation.135–137 In the future, the importance of polymorphic variation in RECQ1 and other human RecQ helicases for human diseases and cancer therapies will continue to be an important area of collaboration for clinicians and biomedical researchers.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging. We thank Dr. Opher Gileadi and colleagues (University of Oxford, UK) for kindly providing crystal structure figure of RECQ1 dimer (Protein Database Code 2V1X) and the figure showing an overlay of RECQ1 and E. coli RecQ.
References
- 1.Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM, Jr, Blackshear PJ. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol. 2007;27:1784–94. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma S, Brosh RM., Jr Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS ONE. 2007;2:1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280:28084–272. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 4.Cui S, Arosio D, Doherty KM, Brosh RM, Jr, Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–70. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty KM, Sharma S, Uzdilla LA, Wilson TM, Cui S, Vindigni A, Brosh RM., Jr RECQ1 helicase interacts with human mismatch repair factors that regulate genetic recombination. J Biol Chem. 2005;280:28085–94. doi: 10.1074/jbc.M500265200. [DOI] [PubMed] [Google Scholar]
- 6.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature ageing. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boule JB, Zakian VA. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 2006;34:4147–53. doi: 10.1093/nar/gkl561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–44. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobb JA, Bjergbaek L. RecQ helicases: lessons from model organisms. Nucleic Acids Res. 2006;34:4106–14. doi: 10.1093/nar/gkl557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 11.Jankowsky E, Fairman ME. RNA helicases—one fold for many functions. Curr Opin Struct Biol. 2007;17:316–24. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–37. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 14.Umezu K, Nakayama K, Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci USA. 1990;87:5363–7. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001;27:113–6. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 16.Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–45. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–92. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puranam KL, Blackshear PJ. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–45. [PubMed] [Google Scholar]
- 19.Puranam KL, Kennington E, Sait SN, Shows TB, Rochelle JM, Seldin MF, Blackshear PJ. Chromosomal localization of the gene encoding the human DNA helicase RECQL and its mouse homologue. Genomics. 1995;26:595–8. doi: 10.1016/0888-7543(95)80181-k. [DOI] [PubMed] [Google Scholar]
- 20.Seki M, Miyazawa H, Tada S, Yanagisawa J, Yamaoka T, Hoshino S, Ozawa K, Eki T, Nogami M, Okumura K. Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli Rec Q helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res. 1994;22:4566–73. doi: 10.1093/nar/22.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–66. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 22.Seki T, Wang WS, Okumura N, Seki M, Katada T, Enomoto T. cDNA cloning of mouse BLM gene, the homologue to human Bloom’s syndrome gene, which is highly expressed in the testis at the mRNA level. Biochim Biophys Acta. 1998;1398:377–81. doi: 10.1016/s0167-4781(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 23.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–62. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 24.Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–52. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 25.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–4. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 26.Siitonen HA, Kopra O, Kaariainen H, Haravuori H, Winter RM, Saamanen AM, Peltonen L, Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12:2837–44. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 27.Van Maldergem L, Siitonen HA, Jalkh N, Chouery E, De Roy M, Delague V, Muenke M, Jabs EW, Cai J, Wang LL, Plon SE, Fourneau C, Kestila M, Gillerot Y, Megarbane A, Verloes A. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J Med Genet. 2006;43:148–52. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, Shiratori M, Matsumoto T, Anno K, Sato T, Mitsui Y, Seki M, Enomoto T, Goto M, Ellis NA, Ide T, Furuichi Y, Sugimoto M. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–72. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- 29.Futami K, Kumagai E, Makino H, Goto H, Takagi M, Shimamoto A, Furuichi Y. Induction of mitotic cell death in cancer cells by small interference RNA suppressing the expression of RecQL1 helicase. Cancer Sci. 2008;99:71–80. doi: 10.1111/j.1349-7006.2007.00647.x. [DOI] [PubMed] [Google Scholar]
- 30.Dhillon KK, Sidorova J, Saintigny Y, Poot M, Gollahon K, Rabinovitch PS, Monnat RJ., Jr Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6:53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- 31.Opresko PL, Calvo JP, Von KC. Role for the Werner syndrome protein in the promotion of tumor cell growth. Mech Ageing Dev. 2007;128:423–36. doi: 10.1016/j.mad.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Grandori C, Wu KJ, Fernandez P, Ngouenet C, Grim J, Clurman BE, Moser MJ, Oshima J, Russell DW, Swisshelm K, Frank S, Amati B, Dalla-Favera R, Monnat RJ., Jr Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev. 2003;17:1569–74. doi: 10.1101/gad.1100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grandori C, Robinson KL, Galloway DA, Swisshelm K. Functional link between Myc and the Werner gene in tumorigenesis. Cell Cycle. 2004;3:22–5. [PubMed] [Google Scholar]
- 34.LeRoy G, Carroll R, Kyin S, Seki M, Cole MD. Identification of RecQL1 as a Holliday junction processing enzyme in human cell lines. Nucleic Acids Res. 2005;33:6251–7. doi: 10.1093/nar/gki929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc Natl Acad Sci USA. 1974;71:4508–12. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray JH, German J. Bloom’s syndrome and EM9 cells in BrdU-containing medium exhibit similarly elevated frequencies of sister chromatid exchange but dissimilar amounts of cellular proliferation and chromosome disruption. Chromosoma. 1984;90:383–8. doi: 10.1007/BF00294165. [DOI] [PubMed] [Google Scholar]
- 37.Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner’s syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol Biol Cell. 2001;12:2121–421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276:19375–81. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Seki M, Narita Y, Nakagawa T, Yoshimura A, Otsuki M, Kawabe Y, Tada S, Yagi H, Ishii Y, Enomoto T. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol Cell Biol. 2003;23:3527–35. doi: 10.1128/MCB.23.10.3527-3535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu P, Beresten SF, van Brabant AJ, Ye TZ, Pandolfi PP, Johnson FB, Guarente L, Ellis NA. Evidence for BLM and Topoisomerase IIIalpha interaction in genomic stability. Hum Mol Genet. 2001;10:1287–98. doi: 10.1093/hmg/10.12.1287. [DOI] [PubMed] [Google Scholar]
- 41.Johnson FB, Lombard DB, Neff NF, Mastrangelo MA, Dewolf W, Ellis NA, Marciniak RA, Yin Y, Jaenisch R, Guarente L. Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer Res. 2000;60:1162 –7. [PubMed] [Google Scholar]
- 42.Shimamoto A, Nishikawa K, Kitao S, Furuichi Y. Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res. 2000;28:1647–55. doi: 10.1093/nar/28.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom’s syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275:9636–44. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 44.Otsuki M, Seki M, Inoue E, Abe T, Narita Y, Yoshimura A, Tada S, Ishii Y, Enomoto T. Analyses of functional interaction between RECQL1, RECQL5, and BLM which physically interact with DNA topoisomerase IIIalpha. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbadis.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Seki M, Yanagisawa J, Kohda T, Sonoyama T, Ui M, Enomoto T. Purification of two DNA-dependent adenosinetriphosphatases having DNA helicase activity from HeLa cells and comparison of the properties of the two enzymes. J Biochem. 1994;115:523–31. doi: 10.1093/oxfordjournals.jbchem.a124369. [DOI] [PubMed] [Google Scholar]
- 46.Cui S, Klima R, Ochem A, Arosio D, Falaschi A, Vindigni A. Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human Replication protein A. J Biol Chem. 2003;278:1424–32. doi: 10.1074/jbc.M209407200. [DOI] [PubMed] [Google Scholar]
- 47.Brosh RM, Jr, Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner Syndrome helicase. J Biol Chem. 2002;277:23236–45. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- 48.Ozsoy AZ, Ragonese HM, Matson SW. Analysis of helicase activity and substrate specificity of Drosophila RECQ5. Nucleic Acids Res. 2003;31:1554–64. doi: 10.1093/nar/gkg243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–46. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 50.Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res. 2007;35:5729–47. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–46. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 52.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–41. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–91. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem. 2005;280:23397–407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 55.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–80. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 56.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, Niccolini B, Rappas M, Freemont PS, Vindigni A. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 2007;5:20. doi: 10.1371/journal.pbio.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E. coli RecQ helicase catalytic core EMBO J. 2003;22:4910–21. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–24. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 60.Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–8. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 61.Brosh RM, Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human Replication protein A. J Biol Chem. 1999;274:18341–50. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 62.Doherty KM, Sommers JA, Gray MD, Lee JW, von Kobbe C, Thoma NH, Kureekattil RP, Kenny MK, Brosh RM., Jr Physical and functional mapping of the RPA interaction domain of the Werner and Bloom syndrome helicases. J Biol Chem. 2005;280:29494–505. doi: 10.1074/jbc.M500653200. [DOI] [PubMed] [Google Scholar]
- 63.Gupta R, Sharma S, Sommers JA, Kenny MK, Cantor SB, Brosh RM., Jr FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007;110:2390–8. doi: 10.1182/blood-2006-11-057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen JC, Gray MD, Oshima J, Loeb LA. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998;26:2879–85. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–37. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rajagopal V, Patel SS. Single strand binding proteins increase the processivity of DNA unwinding by the Hepatitis C Virus helicase. J Mol Biol. 2007;376:69–79. doi: 10.1016/j.jmb.2007.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci USA. 1998;95:6049–54. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khakhar RR, Cobb JA, Bjergbaek L, Hickson ID, Gasser SM. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 2003;13:493–501. doi: 10.1016/s0962-8924(03)00171-5. [DOI] [PubMed] [Google Scholar]
- 69.McGlynn P, Lloyd RG, Marians KJ. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc Natl Acad Sci USA. 2001;98:8235–40. doi: 10.1073/pnas.121007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR. Positive torsional strain causes the formation of a four-way junction at replication forks. J Biol Chem. 2001;276:2790–6. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 71.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM., Jr WRN helicase and FEN-1 form a complex upon replication arrest and together process branch-migrating DNA structures associated with the replication fork. Mol Biol Cell. 2004;15:734–50. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–36. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID, West SC. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Reports. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, Harris CC. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22:1210–22. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parsons CA, Tsaneva I, Lloyd RG, West SC. Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc Natl Acad Sci USA. 1992;89:5452–6. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 77.Sung P, Krejci L, Van KS, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–32. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 78.Haber JE. DNA recombination: the replication connection. Trends Biochem Sci. 1999;24:271–5. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 79.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–65. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 80.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34:2269–79. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–94. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Orren DK, Theodore S, Machwe A. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry. 2002;41:13483–8. doi: 10.1021/bi0266986. [DOI] [PubMed] [Google Scholar]
- 83.van Brabant AJ, Ye T, Sanz M, German JL, III, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–25. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- 84.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–74. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 85.Hanawalt PC. Paradigms for the three rs: DNA replication, recombination, and repair. Mol Cell. 2007;28:702–7. doi: 10.1016/j.molcel.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 86.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 87.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–9. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 88.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–12. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 89.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–84. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–73. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lewis LK, Karthikeyan G, Westmoreland JW, Resnick MA. Differential suppression of DNA repair deficiencies of Yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase) Genetics. 2002;160:49–62. doi: 10.1093/genetics/160.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–33. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qiu J, Qian Y, Chen V, Guan MX, Shen B. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem. 1999;274:17893–900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- 94.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–33. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bertuch AA, Lundblad V. EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics. 2004;166:1651–9. doi: 10.1534/genetics.166.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002;16:1919–33. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K, Schlegelberger B, Schirmacher P, Kunkel TA, Greenberg RA, Edelmann W, Rudolph KL. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130:863–77. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maringele L, Lydall D. EXO1 plays a role in generating type I and type II survivors in budding yeast. Genetics. 2004;166:1641–9. doi: 10.1534/genetics.166.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mansour AA, Tornier C, Lehmann E, Darmon M, Fleck O. Control of GT repeat stability in Schizosaccharomyces pombe by mismatch repair factors. Genetics. 2001;158:77–85. doi: 10.1093/genetics/158.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.von Borstel RC, Savage EA, Wang Q, Hennig UG, Ritzel RG, Lee GS, Hamilton MD, Chrenek MA, Tomaszewski RW, Higgins JA, Tenove CJ, Liviero L, Hastings PJ, Korch CT, Steinberg CM. Topical reversion at the HIS1 locus of Saccharomyces cerevisiae. A tale of three mutants Genetics. 1998;148:1647–54. doi: 10.1093/genetics/148.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–9. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 102.Sharma S, Sommers JA, Driscoll HC, Uzdilla L, Wilson TM, Brosh RM., Jr The exonucleolytic and endonucleolytic cleavage activities of human Exonuclease 1 are stimulated by an interaction with the carboxyl-terminal region of the Werner syndrome protein. J Biol Chem. 2003;278:23487–96. doi: 10.1074/jbc.M212798200. [DOI] [PubMed] [Google Scholar]
- 103.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–9. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Larson ED, Duquette ML, Cummings WJ, Streiff RJ, Maizels N. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr Biol. 2005;15:470–4. doi: 10.1016/j.cub.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 105.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 106.Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O’Donnell M, Kunkel TA, Modrich P. Saccharomyces cerevisiae MutL{alpha} is a mismatch repair endonuclease. J Biol Chem. 2007;282:37181–90. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc Natl Acad Sci USA. 2004;101:9315–20. doi: 10.1073/pnas.0305749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cobb J, Bjergbaek L, Gasser S. RecQ helicases: at the heart of genetic stability. FEBS Lett. 2002;529:43529–43. doi: 10.1016/s0014-5793(02)03269-6. [DOI] [PubMed] [Google Scholar]
- 109.Goldfarb T, Alani E. Distinct roles for the Saccharomyces cerevisiae mismatch repair proteins in heteroduplex rejection, mismatch repair, and non-homologous tail removal. Genetics. 2005;169:563–74. doi: 10.1534/genetics.104.035204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Seki T, Tada S, Katada T, Enomoto T. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem Biophys Res Commun. 1997;234:48–53. doi: 10.1006/bbrc.1997.6535. [DOI] [PubMed] [Google Scholar]
- 111.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–4. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 112.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–87. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hartig JV, Tomari Y, Forstemann K. piRNAs—the ancient hunters of genome invaders. Genes Dev. 2007;21:1707–13. doi: 10.1101/gad.1567007. [DOI] [PubMed] [Google Scholar]
- 114.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 115.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 116.O’Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–23. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 118.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–7. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 119.Bateman JR, Wu CT. DNA replication and models for the origin of piRNAs. Bioessays. 2007;29:382–5. doi: 10.1002/bies.20557. [DOI] [PubMed] [Google Scholar]
- 120.Cogoni C, Macino G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999;286:2342–4. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- 121.Stein P, Svoboda P, Stumpo DJ, Blackshear PJ, Lombard DB, Johnson B, Schultz RM. Analysis of the role of RecQ helicases in RNAi in mammals. Biochem Biophys Res Commun. 2002;291:1119–22. doi: 10.1006/bbrc.2002.6578. [DOI] [PubMed] [Google Scholar]
- 122.Cogoni C, Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci USA. 1997;94:10233–8. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nolan T, Cecere G, Mancone C, Alonzi T, Tripodi M, Catalanotto C, Cogoni C. The RNA-dependent RNA polymerase essential for post-transcriptional gene silencing in Neurospora crassa interacts with Replication protein A. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kato A, Akamatsu Y, Sakuraba Y, Inoue H. The Neurospora crassa mus-19 gene is identical to the qde-3 gene, which encodes a RecQ homologue and is involved in recombination repair and postreplication repair. Curr Genet. 2004;45:37–44. doi: 10.1007/s00294-003-0459-3. [DOI] [PubMed] [Google Scholar]
- 125.Pickford A, Braccini L, Macino G, Cogoni C. The QDE-3 homologue RecQ-2 co-operates with QDE-3 in DNA repair in Neurospora crassa. Curr Genet. 2003;42:220–7. doi: 10.1007/s00294-002-0351-6. [DOI] [PubMed] [Google Scholar]
- 126.Kato A, Inoue H. Growth defect and mutator phenotypes of RecQ-deficient Neurospora crassa mutants separately result from homologous recombination and nonhomologous end joining during repair of DNA double-strand breaks. Genetics. 2006;172:113–25. doi: 10.1534/genetics.105.041756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Suijkerbuijk RF, Sinke RJ, Meloni AM, Parrington JM, van Echten J, de Jong B, Oosterhuis JW, Sandberg AA, Geurts vK. Overrepresentation of chromosome 12p sequences and karyotypic evolution in i(12p)-negative testicular germ-cell tumors revealed by fluorescence in situ hybridization. Cancer Genet Cytogenet. 1993;70:85–93. doi: 10.1016/0165-4608(93)90173-j. [DOI] [PubMed] [Google Scholar]
- 128.Carthew RW. Molecular biology. A new RNA dimension to genome control Science. 2006;313:305–6. doi: 10.1126/science.1131186. [DOI] [PubMed] [Google Scholar]
- 129.Montpetit A, Larose J, Boily G, Langlois S, Trudel N, Sinnett D. Mutational and expression analysis of the chromosome 12p candidate tumor suppressor genes in pre-B acute lymphoblastic leukemia. Leukemia. 2004;18:1499–504. doi: 10.1038/sj.leu.2403441. [DOI] [PubMed] [Google Scholar]
- 130.Guran S, Beyan C, Nevruz O, Yakicier C, Tunca Y. A chronic myeloid leukemia-like syndrome case with del (12) (p12) in a Li-Fraumeni syndrome family. Clin Lab Haematol. 2005;27:135–8. doi: 10.1111/j.1365-2257.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 131.Grepmeier U, Dietmaier W, Merk J, Wild PJ, Obermann EC, Pfeifer M, Hofstaedter F, Hartmann A, Woenckhaus M. Deletions at chromosome 2q and 12p are early and frequent molecular alterations in bronchial epithelium and NSCLC of long-term smokers. Int J Oncol. 2005;27:481–8. [PubMed] [Google Scholar]
- 132.Park J, Betel D, Gryfe R, Michalickova K, Di Nicola N, Gallinger S, Hogue CW, Redston M. Mutation profiling of mismatch repair-deficient colorectal cncers using an in silico genome scan to identify coding microsatellites. Cancer Res. 2002;62:1284–8. [PubMed] [Google Scholar]
- 133.Li D, Frazier M, Evans DB, Hess KR, Crane CH, Jiao L, Abbruzzese JL. Single nucleotide polymorphisms of RecQ1, RAD54L and ATM genes are associated with reduced survival of pancreatic cancer. J Clin Oncol. 2006;24:1720–8. doi: 10.1200/JCO.2005.04.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li D, Liu H, Jiao L, Chang DZ, Beinart G, Wolff RA, Evans DB, Hassan MM, Abbruzzese JL. Significant effect of homologous recombination DNA repair gene polymorphisms on pancreatic cancer survival. Cancer Res. 2006;66:3323–30. doi: 10.1158/0008-5472.CAN-05-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gupta R, Brosh RM., Jr DNA helicases implicated in chromosomal instability disorders as targets for the discovery of novel anti-cancer agents. Frontiers in Drug Design and Discovery (Ed By Bentham Science Publishers) 2007;3:293–316. [Google Scholar]
- 136.Gupta R, Brosh RM., Jr DNA repair helicases as targets for anti-cancer therapy. Curr Med Chem. 2007;14:503–17. doi: 10.2174/092986707780059706. [DOI] [PubMed] [Google Scholar]
- 137.Sharma S, Doherty KM, Brosh RM., Jr DNA helicases as targets for anti-cancer drugs. Curr Med Chem Anti -Canc Agents. 2005;5:183–99. doi: 10.2174/1568011053765985. [DOI] [PubMed] [Google Scholar]