Abstract
The evolutionary interaction between influenza A virus and the human immune system, manifest as ‘antigenic drift’ of the viral haemagglutinin, is one of the best described patterns in molecular evolution. However, little is known about the genome-scale evolutionary dynamics of this pathogen. Similarly, how genomic processes relate to global influenza epidemiology, in which the A/H3N2 and A/H1N1 subtypes co-circulate, is poorly understood. Here through an analysis of 1,302 complete viral genomes sampled from temperate populations in both hemispheres, we show that the genomic evolution of influenza A virus is characterized by a complex interplay between frequent reassortment and periodic selective sweeps. The A/H3N2 and A/H1N1 subtypes exhibit different evolutionary dynamics, with diverse lineages circulating in A/H1N1, indicative of weaker antigenic drift. These results suggest a sink-source model of viral ecology in which new lineages are seeded from a persistent influenza reservoir, which we hypothesize to be located in the tropics, to sink populations in temperate regions.
Influenza is one of the most important respiratory infections of humans, responsible for 250,000 to 500,000 deaths annually1. Of the three types of influenza virus, type A is the most virulent and is associated with seasonal (winter) epidemics in temperate regions, more persistent transmission in the tropics2, and occasional large-scale global pandemics characterized by increased morbidity and mortality.
Since the global pandemic of 1918 caused by a subtype H1N1 influenza A virus, five genome segments have maintained an unbroken evolutionary history within humans—those encoding the nucleocapsid protein (NP), the matrix proteins (M1 and M2) and the nonstructural proteins (NS1 and NS2), and two encoding polymerase proteins (PB2 and PA)3,4. In contrast, new haemagglutinin (HA) and neuraminidase (NA) surface glycoproteins, as well as the PB1 polymerase, have been acquired by human influenza A virus through reassortment with avian influenza viruses. These acquisitions coincided with global pandemics; the HA subtype H2 and the NA subtype N2 appeared in 1957, the HA subtype H3 emerged in 1968, and a new PB1 segment was acquired in both 1957 and 1968. Although A/H1N1 viruses re-emerged in 1977 (ref. 5) and continue to circulate, seasonal epidemics of influenza A virus since 1968 have been dominated by A/H3N2 viruses6, and characterized by punctuated antigenic evolution7.
Despite the recent availability of complete genome sequence data8, many aspects of the evolutionary and epidemiological dynamics of influenza A virus remain opaque. In particular, there has been no rigorous measurement of viral diversity across time, across space and among subtypes. Additionally, most studies of evolutionary dynamics have focused on single segments, without exploring the interactions among them. Although comparative studies have revealed that reassortment occurs frequently within A/H3N2 (refs 9, 10), within and between the A/H2N2 and A/H1N1 subtypes11-13, and among avian influenza viruses14, the full extent of reassortment, and how it relates to antigenic evolution, has not been determined. At the epidemiological scale, although it is apparent that A/H3N2 and A/H1N1 experience oscillating seasonal dynamics, the forces that determine this periodicity, and how they vary spatially, are unknown.
To help to resolve these issues, we examined in detail the evolutionary dynamics of influenza A virus at the genomic and epidemiological scales. Using a data set of 1,302 A/H3N2 and A/H1N1 isolates sampled over a 12-yr period and that is illustrative of large populations in temperate regions from the Northern (New York state, USA) and Southern (New Zealand) Hemispheres, we quantify the genetic diversity of influenza A virus among subtypes, genome segments and geographic locations.
Dynamics of influenza genetic diversity
The changing patterns of genetic diversity in viral isolates from New York state and New Zealand clearly reveal the seasonal dynamics of influenza A in individual temperate populations (Fig. 1). In the case of the better-sampled A/H3N2 subtype, the pattern exposed by our coalescent-based analysis is of an annual series of peaks in genetic diversity interspersed by strong genetic bottlenecks at the end of most influenza seasons. As expected, genetic diversity of the New York state epidemics peak in the Northern Hemisphere winter, whereas those of New Zealand are offset by approximately 6 months, corresponding to the Southern Hemisphere winter. (A similar pattern was observed when 123 genome sequences from Australia were added, suggesting that these patterns are common to other temperate populations, see Supplementary Information.) Furthermore, the genetic diversity of A/H3N2 is usually lower in New Zealand than in New York state, probably reflecting the smaller host population in New Zealand (most isolates were sampled from Canterbury, South Island). Differences in the size of the host population may also explain why A/H3N2 in New Zealand is sometimes less diverse than A/H1N1 in New York state, even though A/H3N2 is usually epidemiologically dominant. The absolute amount of genetic diversity, even at seasonal peaks, is small compared to other rapidly evolving viruses that infect far fewer people15,16, suggesting that strong natural selection, in addition to periodic bottlenecks, reduces the level of diversity that co-circulates at any time. Simulations demonstrate that our reconstructions of genetic diversity are robust to the sampling protocol (Supplementary Information).
Figure 1. Population dynamics of genetic diversity in influenza A virus.
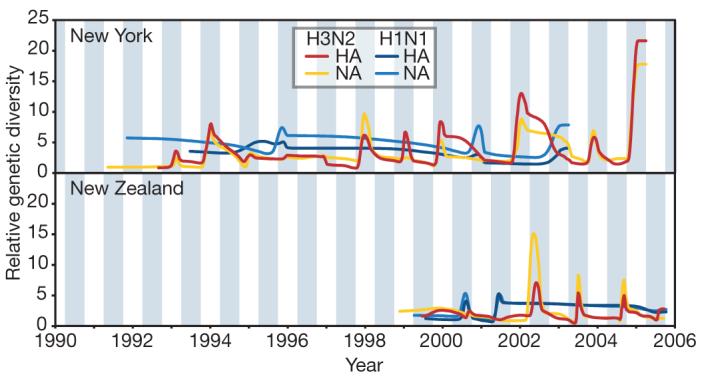
Bayesian skyline plots of the HA and NA segments for the A/H3N2 and A/H1N1 subtypes in New York state (top) and New Zealand (bottom). The horizontal shaded blocks represent the winter seasons. The y-axes represent a measure of relative genetic diversity (see Methods for details). The shorter timescale of New Zealand skyline plot is due to the shorter sampling period.
More notable is that, in both populations, A/H1N1 seasons exhibiting highly defined peaks in diversity typically coincide with weakly defined peaks in A/H3N2 diversity (that is, the measures of the epidemic ‘peakedness’ for A/H3N2 and A/H1N1 are negatively correlated; Wilcoxon signed-rank test, W = 348, n = 32, P < 0.002; Supplementary Figs 1 and 2). This implies an evolutionary interaction between subtypes; for example, that A/H1N1 epidemics are suppressed by herd immunity when A/H3N2 is dominant, or that A/H3N2 out-competes A/H1N1, perhaps owing to greater replicative fitness. Consistent with our observation, A/H1N1 only dominates in seasons following unusually mild H3N2 epidemics17, and infection with one subtype can protect against reinfection with the other in sequential epidemics18. Furthermore, since 1977, A/H1N1 epidemics exhibit lower mortality rates than A/H3N2 epidemics (refs 19 and 20) and are less spatially synchronized21-23. In both hemispheres, A/H1N1 seems less prone to the seasonal genetic bottlenecks that characterize A/H3N2, suggesting that genetically diverse A/H1N1 lineages are better able to coexist. This might indicate that antigenic selection acts with less potency on A/H1N1, manifest as lower rates of amino acid fixation in HA1 (ref. 17), so that the selective turnover of lineages occurs more slowly17,24.
The persistence of viral diversity between epidemic peaks for A/H3N2 and A/H1N1 has two explanations: that chains of infection are surviving in each population and across inter-epidemic intervals, or that genetic diversity is imported into temperate populations each year. Phylogenetic evidence strongly weighs against the former, because there are few direct phylogenetic links between influenza A viruses sampled in successive seasons from specific locations, as expected if in situ evolution was commonplace25,26. Furthermore, a high risk of stochastic extinction makes the repeated survival of small chains of infection between successive epidemics unlikely. Additionally, the dates of segment ancestry in the New York state 1999-2000 season reappeared precisely in the 2002-2003 season, after a major bottleneck in the A/H1N1-dominant 2000-2001 season, strongly suggesting that diversity was maintained in a reservoir population.
Extrapolating the evolutionary dynamics of influenza in New York state and New Zealand to other comparable populations in the Northern and Southern Hemispheres leads us to propose a ‘source-sink’ model for the global ecology of influenza A virus (Fig. 2). Continual—but largely unidirectional—gene flow from a common source population (or a linked network of source populations24,27) provides the viruses that ignite each epidemic in populations of the Northern and Southern Hemispheres. Although southern China has been proposed as the epicentre of influenza A virus28, it is possible that tropical regions generally represent ideal source populations because of extended viral transmission2. A necessary consequence of this model is that selection-driven antigenic drift will be much more efficient in the source population, the long-term effective size of which is maintained by a high background infection rate and by the absence of the severe population bottlenecks associated with temperate localities2. Hence, the observation of antigenic change in temperate populations is a secondary effect of selection within, and gene flow from, the source population. This explains the paradox of there being frequent positive selection on HA across seasons at a global scale29-31 but little evidence for antigenic drift at the scale of individual seasonal epidemics in those temperate populations studied so far26,32. To test this hypothesis, it will be essential to obtain more influenza virus samples from tropical regions, such as South-East Asia.
Figure 2. A ‘source-sink’ model for the evolution of influenza A virus.
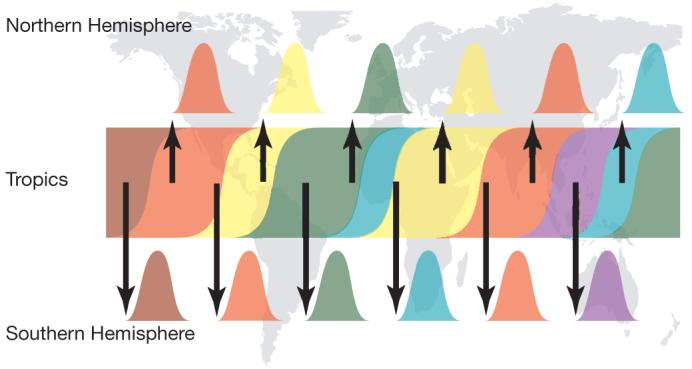
Viral genetic and antigenic diversity (shown by different colours) is continuously generated in a reservoir, or ‘source’ population, perhaps represented by the tropics, before being exported to ‘sink’ populations in the Northern and Southern Hemispheres as shown by the arrows. The continuous transmission of influenza A virus in the source population, and hence its larger effective population size, allows natural selection for antigenic diversity to proceed more efficiently than in the sink populations that are afflicted by major seasonal bottlenecks.
Genomic diversity and reassortment
To determine the genome-wide evolutionary dynamics of human influenza A, we inferred the population genetic history of each segment of A/H3N2 viruses sampled from New York state (the largest data set). This analysis demonstrated that, at any given time, individual segments can differ substantially in their relative genetic diversity and hence in phylogenetic history (Supplementary Fig. 3).
To determine the causes of these differences in genetic diversity, we estimated the time to the most recent common ancestor (TMRCA) of each segment for each influenza season (Fig. 3a; the highest posterior density intervals for each estimate are shown in Supplementary Fig. 4). Most TMRCAs fall well before the start of the season from which they were sampled, such that multiple lineages persist across multiple epidemic troughs, consistent with our source-sink model. TMRCAs also vary among segments and among years, reflecting the interacting processes of genomic reassortment, natural selection and gene flow. For example, the TMRCAs of the NP segment are usually greater than those of the HA segment, indicating that genetic diversity persists for longer in the former. These changing patterns of diversity provide a unique insight into genome-wide evolutionary processes. In particular, the 2001-2002 season is characterized by reduced TMRCAs across the entire A/H3N2 genome (following a dominant A/H1N1 season), such that all segments have approximately the same TMRCA, strongly suggesting a genome-wide selective sweep. In the following 2002-2003 season, genetic diversity reappears at the genomic scale, so that all segments have TMRCAs dating no later than 1999-2000. Subsequently, in 2003-2004, the HA segment alone has a reduced TMRCA, indicating that it underwent a selective sweep, with reassortment maintaining the diversity of other segments. Another genome-wide selective sweep is reflected in the tight TMRCAs of 2004-2005.
Figure 3. Population genetic history of human influenza A virus.
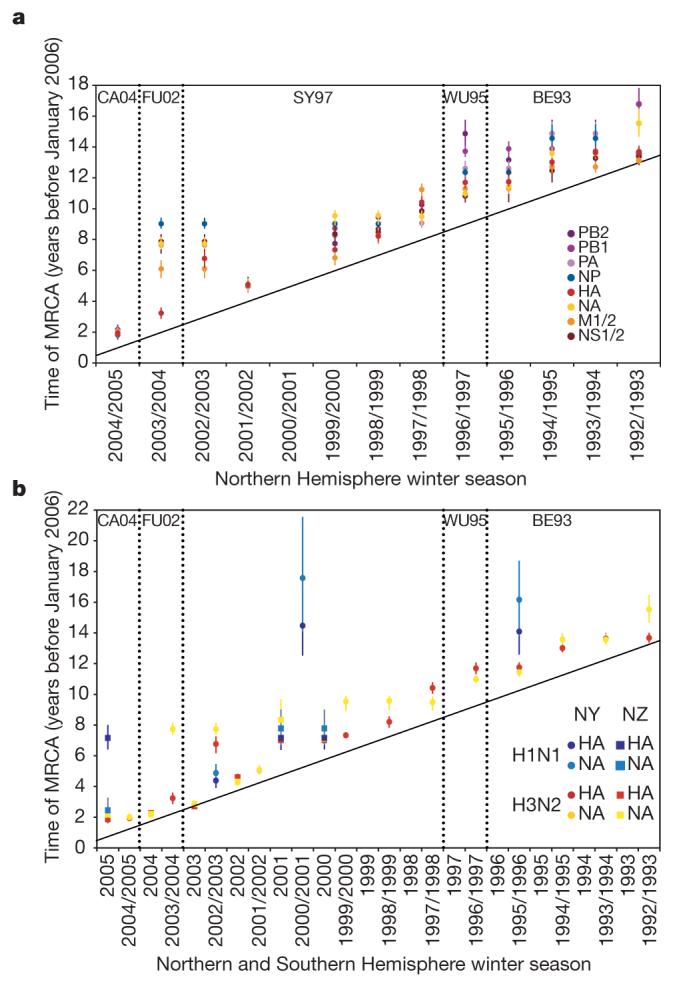
a, b, Time to the most recent common ancestor (TMRCA) of each genomic segment for A/H3N2 isolates circulating each season in New York state (a) and HA and NA genomic segments for the A/H3N2 and A/H1N1 isolates circulating in New York state and New Zealand (b). The values shown represent the mean and 95% highest posterior density intervals for TMRCAs estimated across the trees sampled using bayesian MCMC analyses. Explicit posterior probabilities of whether any season has a TMRCA that is older than that of each preceding season are given in Supplementary Table 1. The diagonal line goes through 1st January of each season (1st July for New Zealand), approximating the seasonal mid-point. The timescale of major changes in antigenic (HA) type in the United States is also depicted. In the Northern Hemisphere, individual influenza seasons straddle two years, whereas in the Southern Hemisphere they are contained within one calendar year.
An analysis of the HA and NA from A/H1N1 in New York state and New Zealand reveals a similar pattern of temporal fluctuations in diversity (Fig. 3b). However, the TMRCAs are often much greater than those observed for A/H3N2 in the same year, providing further evidence that greater variation persists among epidemic seasons in A/H1N1 than in A/H3N2. This may be due to a smaller effective population size or weaker immune selection in A/H1N1, such that natural selection is less effective at fixing advantageous variants.
The occurrence of reassortment and natural selection also coincides with changes in HA antigenicity (Fig. 3a). The earliest antigenic transition in our data is between Beijing/1993 (BE93)-like and Wuhan/1995 (WU95)-like viruses7. This is associated with a sharp increase in the TMRCA of PB2 in the 1996-1997 season, owing to the reappearance of a lineage present until 1994-1995, coupled with reassortment. Similarly, the WU95 to Sydney/1997-like virus (SY97) transition coincides with a marked reduction in the genetic diversity of all segments other than M1/2, in which reassortment has unlinked this segment from the rest of the genome, whereas a new HA lineage is acquired at the time of the SY97 to Fujian/2002-like virus (FU02) antigenic change33. Finally, the genome-wide selective sweep of 2004-2005 coincides with the FU02 to California/2004-like virus (CA04) antigenic transition.
Evolutionary interactions among segments
We used multivariate statistics to summarize the differences in phylogenetic history among segments for A/H3N2 viruses from New York state (Fig. 4 and Supplementary Fig. 12). Each cloud of points of equal colour corresponds to a specific segment of the influenza virus genome, with the spread of points within a cloud representing the statistical uncertainty in the phylogenetic history of that segment. Clearly, the differences in history among segments are greater than the estimation error for each segment. The most prominent result is the divergent position of HA, particularly in relation to NA, even though both encode surface glycoproteins. These differences in evolutionary history are compatible with frequent reassortment, suggesting that the HA segment is continually placed in new genomic backgrounds, perhaps greatly affecting virus fitness.
Figure 4. Differences in phylogenetic history among segments of H3N2 influenza A virus.
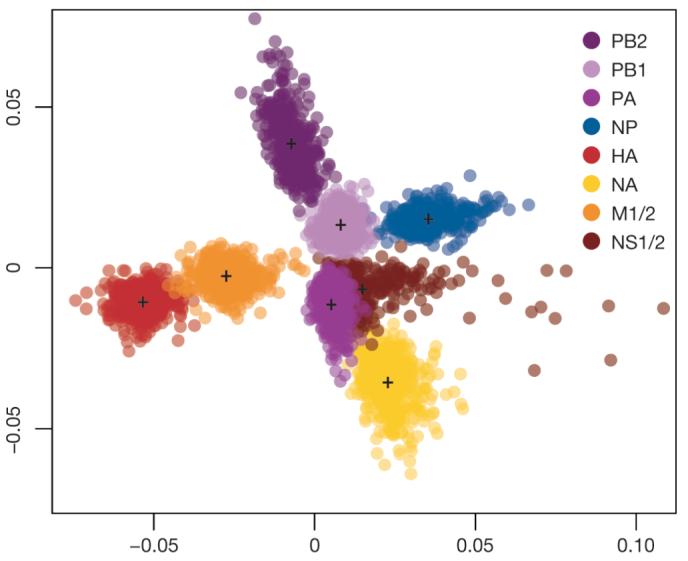
A multi-dimensional scaling plot of distances between samples of 500 trees for each segment of A/H3N2 viruses sampled from New York state. The tree-to-tree distances are calculated as one minus the correlation coefficient of the vector of TMRCAs for each season. Each cloud of points of equal colour represents the statistical uncertainty in the phylogenetic history of an individual genomic segment (with the centroids of each segment distribution indicated). Considering each comparison in a pairwise manner between segments reveals a similar pattern (Supplementary Fig. 12).
In contrast, there is clear similarity in the phylogenetic histories of the HA and M1/2 segments, compatible with the observations that HA interacts structurally with M1 in viral assembly and that the M2 ion channel balances the pH for optimal HA fusion34-38. There is also a similarity in phylogenetic history among the NS and NP segments, and to a lesser extent among the PB2, PB1 and PA polymerase complex segments, suggesting that they are subject to varying degrees of physical linkage. Elucidating the physiological basis for these linkage patterns will undoubtedly provide information about the emergence of pandemic strains of influenza virus.
Further insights into segment interactions are provided by an analysis of evolutionary rates in A/H3N2 isolates from New York state (Supplementary Fig. 5). We identified a major rate difference between those segments expressed on the surface of the virion (high rates) and those that only have internal functions (low rates); the exception to this was the NS segment, for which only the NS2 protein is a minor virion component. As expected given its role in immune evasion, the HA protein exhibits the highest overall rate of evolutionary change (mean of 5.72 × 10-3 nucleotide substitutions per site per year (subs per site per year), 95% highest posterior density interval 5.17, 6.28 × 10-3 subs per site per year), as well as the highest rate of nonsynonymous substitution, indicated by the relative rates at the first and second versus the third codon positions. More surprisingly, the M1/2, NS1/2 and NA segments evolve at rates overlapping those of HA (mean of 5.20 × 10-3 subs per site per year (95% highest posterior density interval 4.36, 6.07 × 10-3 subs per site per year), 5.39 × 10-3 (4.48, 6.35 × 10-3) and 5.41 × 10-3 (4.81, 5.99 × 10-3), respectively), although with lower rates of nonsynonymous change. In contrast, the three segments that constitute the viral polymerase, as well as the RNA-binding NP, exhibit markedly lower rates at all sites and at the first and second codon positions only, revealing stronger selective constraints against amino acid variation.
Because the NA and HA segments have different phylogenetic histories, their similarities in overall evolutionary rate are unlikely to be wholly due to genetic linkage. Hence, the NA is also likely to be subject to relatively strong positive selection, either for antigenic variation or for functional compatibility with HA39. Immunemediated selection may also explain the increased rates in M1/2 and NS1/2. The extracellular domain of the M2 ion channel is a key target in the development of a ‘universal’ influenza vaccine40, whereas the M1 protein is the major component of the viral capsid and a potential target of cellular immune responses41. Likewise, NS1 encodes a pleiotropic, nonstructural protein that downregulates double-stranded-RNA-induced antiviral responses42, or that perhaps co-evolves with the PB1 segment as observed in the last two pandemics30,43.
Conclusions
Our analyses suggest that the evolutionary dynamics of influenza A virus are shaped by a complex interplay between rapid mutation, frequent reassortment, widespread gene flow, natural selection (occasionally generating genome-wide selective sweeps), functional interactions among segments, and global epidemiological dynamics. Although the increased rates of evolution in HA support models in which this protein is subject to strong selection for immune escape, the high rates of evolutionary change in other virion-associated proteins (that have different phylogenetic histories to HA) indicate that they are likewise subject to strong positive selection. It is possible that a significant component of this adaptation involves optimizing the functional compatibility of segments; for example, a new HA variant may not increase fitness unless it is linked to functionally compatible segments. Such a phenomenon may explain why the lineage leading to the FU02 antigenic type did not dominate the viral population until a few years after its initial appearance and co-incident with an HA reassortment event33.
Equally notable was the high degree of genomic reassortment and its association with periodic selective sweeps. It is therefore crucial to consider the process of antigenic drift within the context of frequent reassortment and genome-wide epistatic interactions. Reassortment places antigenically novel HA variants into different genomic back-grounds, a fraction of which may restore, or even increase, the viral replicative fitness that may have been lost as a result of the change in HA. Although it is unlikely that the reassortment rate per se varies among segments, it is probable that most reassortments involving certain segment combinations are deleterious—such as those involving HA and M1/2, which seem to be relatively tightly linked—and are removed by purifying selection, giving rise to the similarities and differences in phylogenetic history discovered here.
In addition to genome-wide interactions, it is essential to consider the complex spatial epidemiological dynamics of influenza if we are to fully understand antigenic evolution. We observed consistent dynamical patterns in two populations illustrative of temperate regions in the Northern and Southern Hemispheres, together with the persistence of viral lineages across multiple epidemics. To resolve these apparently contradictory observations, we propose the existence of a continuous reservoir or source population, within which the strong selection for antigenic change takes place. Such complexity necessarily means that the long-term success of any individual lineage of influenza virus is dependent not only on its antigenic properties but also on its replicative capacity, its transmissibility and the environmental factors that perhaps underlie the seasonality of influenza in temperate regions. In this context, it is important to determine the precise reasons why antigenic evolution proceeds at a reduced pace in A/H1N1, such that multiple lineages co-circulate globally, even though this subtype is occasionally the major cause of seasonal influenza.
METHODS SUMMARY
Complete genome sequences of A/H3N2 and A/H1N1 influenza viruses from New York state, USA, and New Zealand, were collated from the NCBI Influenza Database as part of the Influenza Genome Sequencing Project8,44. Sequence alignments representing each genomic segment, viral subtype and sampling location were assembled. Evolutionary dynamics were estimated for each data set using an established bayesian Markov chain Monte Carlo (MCMC) approach45,46 that incorporates the exact day of viral sampling. Each MCMC analysis resulted in marginal posterior estimates of: the rate of nucleotide substitution (Supplementary Fig. 5); the relative evolutionary rate at the first and second versus the third codon positions (Supplementary Fig. 5); and the dynamics of population genetic diversity through time (see Fig. 1 and Supplementary Fig. 4). Additionally, an estimate of the posterior distribution of genealogies that relate the sequence data was inferred concurrently for each alignment. From these distributions, we obtained the marginal posterior estimate of the TMRCA of each genomic segment in each influenza season (Fig. 3a, b). Differences between the node-height distributions of each posterior set of genealogies were visualized using multidimensional scaling (Fig. 4). The node heights and topological structure of each genealogical distribution was summarized using the maximum clade support approach (Supplementary Fig. 3).
Supplementary Material
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, Fogarty International Center, the National Institute of Allergy and Infectious Diseases and the National Institute of General Medical Sciences. A.R. and O.G.P. are supported by The Royal Society of London. A.R. works as a part of the Interdisciplinary Centre for Human and Avian Influenza Research (ICHAIR).
APPENDIX
METHODS
To examine the interaction between the H1N1 and H3N2 subtypes of human influenza A virus and the seasonal fluctuations in the Northern and Southern Hemispheres, we compiled the HA and NA genes (coding regions only) for A/H1N1 in New York state (81 isolates sampled between 1995 and 2003) and in New Zealand (127 isolates sampled between 2000 and 2005), as well as for A/H3N2 in New York state (687 isolates sampled between 1993 and 2005) and in New Zealand (407 isolates between 2000 and 2005). Influenza is strongly seasonal in both localities, with few reported cases outside of their respective winters (data available at the World Health Organization ‘FluNet’ surveillance system; http://gamapserver.who.int/GlobalAtlas/home.asp). To examine genome-wide evolutionary processes in more detail, we conducted an equivalent analysis on each segment of the 687 isolates of A/H3N2 sampled from New York state. For each segment, the protein-coding regions were extracted and aligned. For segments with more than one gene (M1/2 and NS1/2), both genes were concatenated, with overlapping codons included only once (see Supplementary Information). In all cases, alignment was unambiguous, resulting in the following data sets: PB2 (2,277 nucleotides), PB1 (2,271 nucleotides), PA (2,148 nucleotides), HA (1,698 nucleotides), NP (1,494 nucleotides), NA (1,407 nucleotides), M1/2 (979 nucleotides) and NS1/2 (835 nucleotides). A full list of viral isolates, along with their GenBank accession numbers, is available as Supplementary Information.
We used a flexible demographic model to examine the seasonal changes in genetic diversity over the sampling time span47. This approach, based on the coalescent48, allows the overall genetic diversity of a population to be estimated from a small sample of genetic sequences. An estimate of the change in diversity over time can be obtained for a given genealogy47. Individual estimates obtained from many different genealogies are combined, with genealogies and parameter values sampled according to their relative probability, given the sequence data45,46. In the absence of natural selection, the genetic diversity measure obtained reflects the change in effective number of infections over time (Neτ, where τ is the average generation time). Because strong natural selection has previously been demonstrated for influenza A virus, we interpret these plots as measures of relative genetic diversity. The MCMC approach implemented in the BEAST package has the advantage that it is not conditioned on a single, and potentially unrepresentative, estimate of the underlying genealogy. It provides marginal posterior estimates of all the parameters of the substitution process including the overall rate of molecular evolution and the times of all the nodes in the genealogy—a timescale that is informed by the known dates (days) of sampling of the sequences and the amount of genetic change between them45,49. Indeed, the use of sequences that have been sampled over many seasons allows the reconstruction of fluctuating dynamics limited by the date of the earliest sample (see Supplementary Information).
For these analyses, we used a nucleotide-substitution model that accommodates the different rates of and constraints on evolution at the different codon positions50. This allows us to estimate the overall rate of molecular evolution for each gene and the relative rate of first and second codon position to the third—a measure of the constraint against amino acid change that correlates strongly with the ratio of nonsynonymous to synonymous nucleotide substitutions50.
We investigated the effect of reassortment among segments by comparing the correlation of phylogenetic trees sampled independently for each segment. For each tree, we identified the most recent common ancestor of the samples from each season and plotted a marginal posterior probability distribution for the time of these nodes (Fig. 3). Furthermore, we estimated one minus the correlation coefficient of these TMRCAs both within and between a sample of 500 trees from the posterior distribution obtained from the MCMC analysis of each genomic segment. These relationships were plotted using the multi-dimensional scaling dimension reduction technique (Fig. 4) and pair-wise in Supplementary Fig. 12.
For each segment alignment, we used two independent runs of BEAST of 100,000,000 steps sampling trees and parameters every 10,000 steps. The two runs were compared to confirm that both had converged and were sampling the same distributions, and were then combined (having removed 10% as ‘burn-in’). All the results here were summarized from the remaining 18,000 sampled trees or parameter values.
- 47.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 48.Kingman J. The coalescent. Stochastic Process. Appl. 1982;13:235–248. [Google Scholar]
- 49.Rambaut A. Estimating the rate of molecular evolution: Incorporating noncontemporaneous sequences into maximum likelihood phylogenies. Bioinformatics. 2000;16:395–399. doi: 10.1093/bioinformatics/16.4.395. [DOI] [PubMed] [Google Scholar]
- 50.Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.WHO Fact sheet Number 211. Influenza. 2003 http://www.who.int/mediacentre/factsheets/fs211/)
- 2.Viboud C, Alonso WJ, Simonsen L. Influenza in tropical regions. PLoS Med. 2006;3:e89. doi: 10.1371/journal.pmed.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid AH, Taubenberger JK, Fanning TG. Evidence of an absence: the genetic origins of the 1918 pandemic influenza virus. Nature Rev. Microbiol. 2004;2:909–914. doi: 10.1038/nrmicro1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taubenberger JK, et al. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 5.Scholtissek C, von Hoyningen V, Rott R. Genetic relatedness between the new 1977 epidemic strains (H1N1) of influenza and human influenza strains isolated between 1947 and 1957 (H1N1) Virology. 1978;89:613–617. doi: 10.1016/0042-6822(78)90203-9. [DOI] [PubMed] [Google Scholar]
- 6.Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg. Infect. Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 8.Ghedin E, et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- 9.Lindstrom SE, et al. Phylogenetic analysis of the entire genome of influenza A (H3N2) viruses from Japan: evidence for genetic reassortment of the six internal genes. J. Virol. 1998;72:8021–8031. doi: 10.1128/jvi.72.10.8021-8031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schweiger B, Bruns L, Meixenberger K. Reassortment between human A(H3N2) viruses is an important evolutionary mechanism. Vaccine. 2006;24:6683–6690. doi: 10.1016/j.vaccine.2006.05.105. [DOI] [PubMed] [Google Scholar]
- 11.Gregory V, et al. Emergence of influenza A H1N2 reassortant viruses in the human population during 2001. Virology. 2002;300:1–7. doi: 10.1006/viro.2002.1513. [DOI] [PubMed] [Google Scholar]
- 12.Lindstrom SE, Cox NJ, Klimov A. Genetic analysis of human H2N2 and early H3N2 influenza viruses, 1957-1972: evidence for genetic divergence and multiple reassortment events. Virology. 2004;328:101–119. doi: 10.1016/j.virol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Young JF, Palese P. Evolution of human influenza A viruses in nature: recombination contributes to genetic variation of H1N1 strains. Proc. Natl Acad. Sci. USA. 1979;76:6547–6551. doi: 10.1073/pnas.76.12.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macken CA, Webby RJ, Bruno WJ. Genotype turnover by reassortment of replication complex genes from avian influenza A virus. J. Gen. Virol. 2006;87:2803–2815. doi: 10.1099/vir.0.81454-0. [DOI] [PubMed] [Google Scholar]
- 15.Carrington C, Foster J, Pybus O, Bennett S, Holmes E. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J. Virol. 2005;79:14680–14687. doi: 10.1128/JVI.79.23.14680-14687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemey P, Rambaut A, Pybus O. HIV evolutionary dynamics within and among hosts. AIDS Rev. 2006;8:125–140. [PubMed] [Google Scholar]
- 17.Wolf YI, Viboud C, Holmes EC, Koonin EV, Lipman DJ. Long intervals of stasis punctuated by bursts of positive selection in the seasonal evolution of influenza A virus. Biol. Direct. 2006;1:34. doi: 10.1186/1745-6150-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonoguchi T, Naito H, Hara M, Takeuchi Y, Fukumi H. Cross-subtype protection in humans during sequential, overlapping, and/or concurrent epidemics caused by H3N2 and H1N1 influenza viruses. J. Infect. Dis. 1985;151:81–88. doi: 10.1093/infdis/151.1.81. [DOI] [PubMed] [Google Scholar]
- 19.Kaji M, Watanabe A, Aizawa H. Differences in clinical features between influenza A H1N1, A H3N2, and B in adult patients. Respirology. 2003;8:231–233. doi: 10.1046/j.1440-1843.2003.00457.x. [DOI] [PubMed] [Google Scholar]
- 20.Wright PF, Thompson J, Karzon DT. Differing virulence of H1N1 and H3N2 influenza strains. Am. J. Epidemiol. 1980;112:814–819. doi: 10.1093/oxfordjournals.aje.a113053. [DOI] [PubMed] [Google Scholar]
- 21.Hay AJ, Gregory V, Douglas AR, Lin YP. The evolution of human influenza viruses. Phil. Trans. R. Soc. Lond. B. 2001;356:1861–1870. doi: 10.1098/rstb.2001.0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finkelman B, et al. Global patterns in seasonal activity of influenza A/H3N2, A/H1N1, and B from 1997 to 2005: viral coexistence and latitudinal gradients. PLoS ONE. 2007;2:e1296. doi: 10.1371/journal.pone.0001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viboud C, et al. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science. 2006;312:447–451. doi: 10.1126/science.1125237. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- 25.Nelson MI, Simonsen L, Viboud C, Miller MA, Holmes EC. Phylogenetic analysis reveals the global circulation pattern of seasonal influenza A viruses. PLoS Pathog. 2007;3:e131. doi: 10.1371/journal.ppat.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson MI, et al. Stochastic processes are key determinants of short-term evolution in influenza A virus. PLoS Pathog. 2006;2:e125. doi: 10.1371/journal.ppat.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso WJ, et al. Seasonality of influenza in Brazil: a traveling wave from the Amazon to the subtropics. Am. J. Epidemiol. 2007;165:1434–1442. doi: 10.1093/aje/kwm012. [DOI] [PubMed] [Google Scholar]
- 28.Shortridge KF. Is China an influenza epicentre? Chin. Med. J. (Engl.) 1997;110:637–641. [PubMed] [Google Scholar]
- 29.Bush RM, Fitch WM, Bender CA, Cox NJ. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 1999;16:1457–1465. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- 30.Fitch WM, Leiter JM, Li XQ, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc. Natl Acad. Sci. USA. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shih AC, Hsiao TC, Ho MS, Li WH. Simultaneous amino acid substitutions at antigenic sites drive influenza A hemagglutinin evolution. Proc. Natl Acad. Sci. USA. 2007;104:6283–6288. doi: 10.1073/pnas.0701396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavenu A, et al. Detailed analysis of the genetic evolution of influenza virus during the course of an epidemic. Epidemiol. Infect. 2006;134:514–520. doi: 10.1017/S0950268805005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes EC, et al. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3:e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ali A, Avalos RT, Ponimaskin E, Nayak DP. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 2000;74:8709–8719. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barman S, Ali A, Hui EK, Adhikary L, Nayak DP. Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses. Virus Res. 2001;77:61–69. doi: 10.1016/s0168-1702(01)00266-0. [DOI] [PubMed] [Google Scholar]
- 36.Enami M, Enami K. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J. Virol. 1996;70:6653–6657. doi: 10.1128/jvi.70.10.6653-6657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohuchi M, et al. Rescue of vector-expressed fowl plague virus hemagglutinin in biologically active form by acidotropic agents and coexpressed M2 protein. J. Virol. 1994;68:920–926. doi: 10.1128/jvi.68.2.920-926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi K, Lamb RA. Influenza virus M2 protein ion channel activity stabilizes the native form of fowl plague virus hemagglutinin during intracellular transport. J. Virol. 1994;68:911–919. doi: 10.1128/jvi.68.2.911-919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitnaul LJ, et al. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 2000;74:6015–6020. doi: 10.1128/jvi.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerhard W, Mozdzanowska K, Zharikova D. Prospects for universal influenza virus vaccine. Emerg. Infect. Dis. 2006;12:569–574. doi: 10.3201/eid1204.051020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berkhoff EG, et al. Functional constraints of influenza A virus epitopes limit escape from cytotoxic T lymphocytes. J. Virol. 2005;79:11239–11246. doi: 10.1128/JVI.79.17.11239-11246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krug RM, Yuan W, Noah DL, Latham AG. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309:181–189. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 43.Ludwig S, Schultz U, Mandler J, Fitch WM, Scholtissek C. Phylogenetic relationship of the nonstructural (NS) genes of influenza A viruses. Virology. 1991;183:566–577. doi: 10.1016/0042-6822(91)90985-k. [DOI] [PubMed] [Google Scholar]
- 44.Influenza Genome Sequencing Project. 2007 http://www.niaid.nih.gov/dmid/genomes/mscs/influenza.htm.
- 45.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond AJ, Rambaut A. BEAST: bayesian evolutionary analysis sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


