Abstract
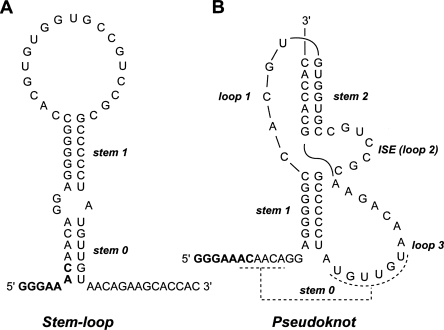
The stimulatory RNA of the Visna-Maedi virus (VMV) −1 ribosomal frameshifting signal has not previously been characterized but can be modeled either as a two-stem helix, reminiscent of the HIV-1 frameshift-stimulatory RNA, or as an RNA pseudoknot. The pseudoknot is unusual in that it would include a 7 nucleotide loop (termed here an interstem element [ISE]) between the two stems. In almost all frameshift-promoting pseudoknots, ISEs are absent or comprise a single adenosine residue. Using a combination of RNA structure probing, site directed mutagenesis, NMR, and phylogenetic sequence comparisons, we show here that the VMV stimulatory RNA is indeed a pseudoknot, conforming closely to the modeled structure, and that the ISE is essential for frameshifting. Pseudoknot function was predictably sensitive to changes in the length of the ISE, yet altering its sequence to alternate pyrimidine/purine bases was also detrimental to frameshifting, perhaps through modulation of local tertiary interactions. How the ISE is placed in the context of an appropriate helical junction conformation is not known, but its presence impacts on other elements of the pseudoknot, for example, the necessity for a longer than expected loop 1. This may be required to accommodate an increased flexibility of the pseudoknot brought about by the ISE. In support of this, 1H NMR analysis at increasing temperatures revealed that stem 2 of the VMV pseudoknot is more labile than stem 1, perhaps as a consequence of its connection to stem 1 solely via flexible single-stranded loops.
Keywords: frameshifting, RNA pseudoknot, translation, retrovirus, Visna-Maedi virus
INTRODUCTION
Programmed −1 ribosomal frameshifting is a translational control mechanism that allows the production of a specific ratio of gene products from two overlapping open reading frames (ORFs), the relative quantities of which depends upon the frameshift efficiency of the particular signal. Frameshifting occurs during the elongation phase of protein synthesis where, in response to elements in the mRNA, the ribosome switches from the zero reading frame to the −1 frame (in a 5′ direction) at a defined position and translation continues in the new frame (Jacks and Varmus 1985; for review, see Farabaugh 1996; Giedroc et al. 2000; Brierley and Pennell 2001; Baranov et al. 2006; Brierley et al. 2007). Several RNA viruses employ frameshifting in the expression of their replicase enzymes. In many retroviruses, for example, frameshifting allows expression of reverse transcriptase (as part of the Gag-Pol polyprotein) from the overlapping gag and pol ORFs (Hatfield and Oroszlan 1990). The mRNA signals that promote frameshifting comprise a slippery sequence, where the frameshift takes place seperated by a short spacer region (typically, 5–9 nucleotides [nt] in length) from a 3′ stimulatory RNA structure. The heptanucleotide slippery sequence usually contains consecutive homopolymeric triplets (XXXYYYZ), with the ribosome-bound tRNAs decoding the P- and A-site codons slipping from the zero frame (XXYYYZ) to the −1 frame (XXXYYY). At some sites, a stem–loop appears to be sufficient as a stimulatory structure, but in the majority an RNA pseudoknot is present. These are H-(hairpin)-type pseudoknots (Pleij et al. 1985; Brierley et al. 2007), although some viral frameshift-promoting pseudoknots have been described as kissing hairpins (Herold and Siddell 1993; Baranov et al. 2005). The mechanism of frameshifting is not fully understood, although a number of models have been proposed to explain how the interaction of the ribosome with the stimulatory RNA leads to a realignment of the tRNAs decoding the slippery sequence into the −1 frame (Plant et al. 2003, and references therein; Namy et al. 2006).
The study of retroviral ribosomal frameshifting signals has provided a wealth of information about the process and has contributed substantially to the development of models. However, many retrovirus signals remain uncharacterized and could potentially offer new insights into the structure and function of frameshift-stimulatory RNAs. Here, we describe an unusual structural feature present within the frameshift signal of the ovine lentivirus Visna-Maedi virus (VMV). The sequence of the gag/pol overlap of VMV harbors a likely slippery sequence GGGAAAC and a region downstream with the potential to fold into a stem–loop or an RNA pseudoknot (Brierley et al. 1989; Le et al. 1991). The proposed pseudoknot is unusual, however, in that coaxial stacking of the two stems would be interrupted by a 7-nt stretch that we refer to here as an interstem element (ISE) (see Fig. 1). Unpaired bases between constituent stems are rare in frameshift-promoting pseudoknots and, if present, usually comprise a single residue (ten Dam et al. 1990). An alternative possibility is that the VMV stimulatory RNA is a stem–loop. In this model (Fig. 1), base-pairing between two sets of complementary regions would form a two-stem helix similar to that found at the HIV-1 frameshift signal (Dulude et al. 2002; Gaudin et al. 2005; Staple and Butcher 2005). The lower helix in this structure could also form in the pseudoknot model (giving a putative stem 0) (Fig. 1). To distinguish between these folding possibilities, we analyzed the VMV signal by site-directed mutagenesis coupled with in vitro and in vivo frameshift assays, RNA secondary structure probing, and 1H NMR methods. The results reveal that that VMV stimulatory RNA is indeed a pseudoknot, conforming closely to the predicted structure and containing the functionally essential ISE. The significance of this secondary structure element to proposed models of frameshifting is discussed in comparison with other functionally important pseudoknot features.
FIGURE 1.
The stimulatory RNA of the VMV frameshifting signal is proposed to form a stem–loop (A) or an RNA pseudoknot (B). Stem 0 of the model in A could also be present in the pseudoknot model (dashed lines). The slippery sequence (GGGAAAC) is emboldened. The interstem element (ISE) between the two stems of the pseudoknot model (B) is indicated.
RESULTS
Secondary structure probing of the VMV frameshift signal
The 5′ boundary of the VMV frameshift signal is almost certainly the GGGAAAC heptamer located within the gag/pol overlap region (genomic coordinates 1763–1769), the only candidate slippery sequence in the region (Brierley et al. 1989). Scrutiny of potential base-pairing interactions downstream of this stretch suggests the formation of either a stem–loop or an RNA pseudoknot structure (Fig. 1). The stem–loop would comprise a two-stem helix totaling 13 base pairs (bp) in length with a loop of 19 nt. The base-paired regions (stem 0 and stem 1) are separated by a purine-rich interior loop with stem 0 incorporating some of the slippery sequence. A shorter version could also exist in which stem 0 does not form and a spacer of typical length (6 nt) seperates slippery sequence and stimulatory RNA. The proposed pseudoknot would also begin 6 nt downstream of the GGGAAAC motif and would possess two stems of 7 bp, linked by loops of 5 nt (loop 1) and 14 nt (loop 3). An ISE of 7 nt is present between the two stems. (It should be noted that in the early pseudoknot literature, most examples did not possess unpaired residues between the two stems [for review, see Brierley et al. 2007], thus the convention was to name the groove-spanning loops L1 and L2 [now L1 and L3]). In this article we have adopted the L1, L2, L3 nomenclature, but L2 will be referred to as an ISE to highlight the unusual presence of this loop in a frameshift-promoting pseudoknot).
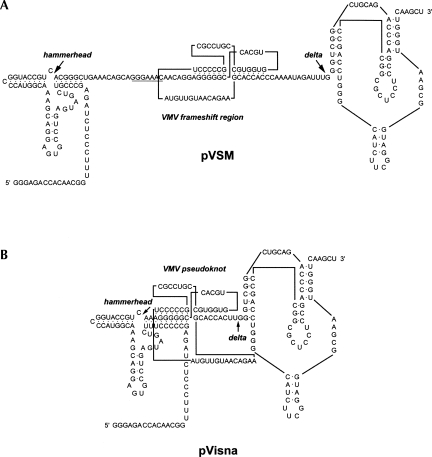
In an effort to distinguish between the stem–loop and pseudoknot models, RNA structure probing was performed. VMV sequences comprising the putative pseudoknot, flanked by 29 and 13 nt at the 5′ and 3′ ends, respectively, were cloned into a ribozyme-based transcription vector such that a cleaved VMV RNA species of 97 nt (Fig. 2) could be purified and would allow resolution of the whole structure on a single gel. This was end labeled and subjected to chemical and enzymatic digestion prior to analysis on denaturing polyacrylamide gels. Four enzymatic probes were used; RNases CL3, T1, and U2, which preferentially cleave single-stranded C, G, and A bases respectively, and RNase CV1 which targets double-stranded and stacked bases. The chemical probes imidazole and lead acetate were also employed, which show specificity for single-stranded regions. In these experiments, the Mg2+ level was kept at 2 mM, which is the approximate concentration of this ion in the rabbit reticulocyte lysate in vitro translation system (RRL) (Jackson and Hunt 1983).
FIGURE 2.
The sequence and proposed secondary structures present in the T7 RNA polymerase-derived transcripts of plasmids pVSM/HindIII (A; for RNA structure mapping) and pVisna/HindIII (B; for NMR analysis). The VMV frameshift region contains the slippery sequence GGGAAAC (underlined) and a downstream stimulatory RNA shown as a pseudoknot. In each transcript, the VMV sequences are flanked by hammerhead and delta ribozymes that cleave the RNA at the sites indicated by the large arrows. In pVisna, the right-hand portion of the hammerhead ribozyme was extended compared to that of pVSM to allow ribozyme folding to compete effectively with the folding of stem 1 of the pseudoknot in the primary transcript (indicated as shared base-pairs in B).
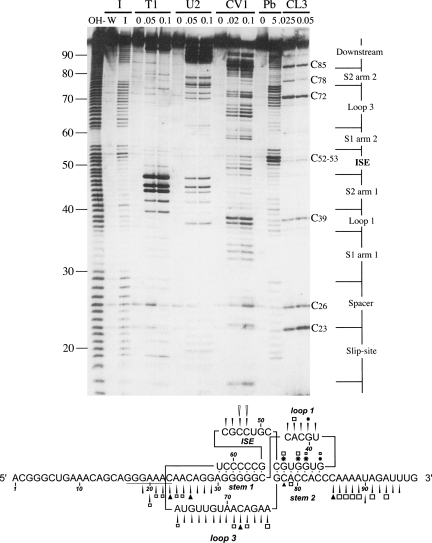
The results of the chemical probing analysis were strongly supportive of the pseudoknot model (Fig. 3). The cleavage pattern of imidazole especially was highly consistent with the formation of a pseudoknot, with cleavage occurring at all regions predicted to be single stranded and no cleavage within the stems. Lead acetate gave a similar cleavage pattern, although there were two differences in comparison to imidazole. First, loop 3 showed differential reactivity, with the first 6 nt (bases 63–68) showing more resistance to lead-induced cleavage than the last 8 (bases 69–76). This raised the possibility that bases 63–68 may be base-paired, although more weakly than stems 1 and 2. An obvious partner for such a pairing are those bases in the slippery sequence/spacer region that would form stem 0 of the two-helix stem–loop model, or the putative stem 0 in the pseudoknot model (Fig. 1). Indeed, these spacer bases also showed somewhat reduced accessibility to lead ions. Secondly, the C bases in the proposed ISE (C52, C53) were extremely sensitive to lead-induced cleavage. Such hyperreactivity may reflect a particular conformation of this loop (see Discussion). As with the chemical probing, the enzymatic probing data were consistent with the proposed pseudoknot, with two exceptions. First, there was no obvious specificity of cleavage of RNase CV1 within the predicted double-stranded regions of either model. Although some cuts were seen in the proposed stem regions, particularly within stem 1 and toward the 3′ end of stem 2 arm 2 (C81–C83), there were also cuts within regions proposed to be single stranded. While most of the latter are likely to be a consequence of cleavage of single-stranded bases in stacked conformation, the CV1 data were not particularly informative. The most noticable feature of the enzymatic probing, however, was the extreme reactivity of the G bases in the first arm of stem 2 to RNase T1 and to a lesser extent, RNase U2, with G44, G45, and G47 being the most reactive bases in the probed frameshift region. It could be argued that these particular cleavages support the stem–loop model, since the G bases involved would lie within the 19-nt loop region of this model (Fig. 1). However, in previous studies, we have observed an unusual reactivity of RNase T1 with this (double-stranded) region of a variety of frameshift-promoting pseudoknots, perhaps indicative of unusual accessibility (Manktelow et al. 2005; I. Brierley, unpubl.). Furthermore, RNase CL3 exhibited little or no reactivity toward C bases within the proposed pseudoknot stem regions (including arm 2 of stem 2) in comparison to those of single-stranded regions. Further experiments (detailed below) also strongly support the pseudoknot model. Significantly, the proposed ISE appears to form as predicted, showing reactivity to single-stranded probing reagents, particularly plumbous ions (see above). However, the extent of CL3 cleavage was less dramatic in the ISE than that seen in other single-stranded regions (spacer, loop 1, loop 3, downstream region), which may reflect some hindrance to enzyme accessibility.
FIGURE 3.
Structure probing of the VMV frameshift signal. RNA derived by transcription of pVSM/Hind III with T7 RNA polymerase was 5′-end labeled with [γ-33P]-ATP and subjected to limited RNase or chemical cleavage using structure-specific probes. Sites of cleavage were identified by comparison with a ladder of bands created by limited alkaline hydrolysis of the RNA (OH−) and the position of known RNase U2 and T1 cuts, determined empirically. Products were analyzed on a 10% acrylamide/7 M urea gel containing formamide. Data were also collected from 6% and 15% gels (gels not shown). Enzymatic structure probing was with RNases CL3, T1, U2, and CV1. Uniquely cleaved nucleotides were identified by their absence in untreated control lanes (0). The number of units of enzyme added to each reaction is indicated. Chemical structure probing was with imidazole (4 h, I) or lead acetate (Pb2+; millimolar concentration in reaction). The water lane (W) represents RNA that was dissolved in water, incubated for 4 h and processed in parallel to the imidazole-treated sample. The sequence of the probed RNA and the inferred secondary structure is shown below the gel. The reactivies of the imidazole (black arrow), T1 (asterisk), U2 (square), lead (white arrow, only hyperreactive bases shown), and CL3 (black triangle) probes are marked.
Site-directed mutagenesis of the VMV frameshift signal
We sought further support for the pseudoknot model by mutational analysis and functional assays. The VMV frameshift signal, either wild-type or modified, was inserted between the Renilla (R-luc) and firefly luciferase (F-luc) genes of the dual reporter plasmid p2luc (Grentzmann et al. 1998; see Materials and Methods) in such a way that expression of the R-luc:F-luc fusion protein (analogous to Gag:Pol) was dependent upon frameshifting at the inserted VMV signal. In vitro frameshift assays were carried out by translating mRNAs in RRL, and nonframeshifted (37 kDa) and frameshift (99 kDa) products quantified by densitometry. In in vivo assays, frameshift plasmids were transfected into mammalian cells and luciferase activities measured 24 h post-transfection. In order to quantify frameshifting efficiency in vivo, a “100% frameshift” control plasmid was prepared in which the R-luc and F-luc sequences were aligned in frame by insertion of a C base immediately after the slippery sequence. The in vivo experiments confirmed the pattern of mutational effects seen in vitro, although the frameshift efficiencies were generally lower in vivo, as seen by others (e.g., Parkin et al. 1992; Reil et al. 1993). The results presented below refer to the in vitro frameshift values unless specifically stated.
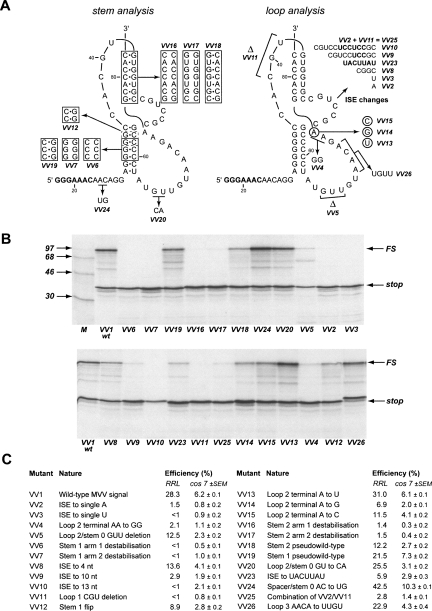
Figure 4 shows the mutations introduced into the frameshift region within p2luc/VV1 (Fig. 4A, VV2–VV26;), in vitro translations of the mutant constructs (Fig. 4B) and a summary of the frameshift efficiencies measured for each construct both in RRL and in transfected tissue culture cells (Fig. 4C, cos 7). Analysis of the proposed stem regions involved the introduction of complementary and compensatory changes within stems 1 and 2. Three mutations were prepared for each stem. Two were destabilizing mutations, introduced into each arm of the relevant stem by changing either the three central base pairs (stem 1) or all base pairs (stem 2) to their complementary Watson–Crick bases. The third was a double mutation leading to a “pseudowild-type” structure, in which both changes were made and should be compensatory. In RRL, the frameshifting efficiency of the wild-type VMV signal was 28.3% (VV1). Destabilization of either stem 1 (VV6, VV7) or stem 2 (VV16, VV17) reduced frameshifting dramatically (to <2%), but in the double mutant, pseudowild-type constructs, significant frameshifting was observed supporting the belief that both stems form and are required for frameshifting. In the double mutant stem 1 construct, frameshifting was restored to close to the wild-type value (VV19, 21.5%). In the equivalent stem 2 construct, about half of the wild-type frameshift efficiency was recovered (VV18, 12.2%). The incomplete restoration may be a consequence of an altered stability of the new stem 2 or an effect of flipping the G77-C48 pair at the stem–stem junction, which could influence the conformation of this functionally important region (see below).
FIGURE 4.
Analysis of the VMV frameshift signal by site-directed mutagenesis. (A) Mutations were introduced into the VMV frameshift region to modify the proposed pseudoknot. Deleted bases are indicated by Δ. (B) Wild-type p2luc/VV1 or mutant derivatives were digested with HpaI and transcribed with T7 RNA polymerase, and transcripts were translated in RRL at a concentration of ∼50 μg/mL. Products were labeled with 35S-methionine, separated on a 12.5% SDS/polyacrylamide gel and detected by autoradiography. The nonframeshifted (stop) and frameshifted (FS) species are marked with arrows. M represents 14C protein markers (Amersham Pharmacia Biotech). (C) Summary of the mutations made and the resulting frameshift efficiencies in RRL and cos 7 cells.
To investigate the potential pairing between the spacer and loop 3 regions (Fig. 1, stem 0), two bases in each region were changed to their complementary Watson–Crick partners; A25/C26 to U25/G26 in the spacer (VV24) and G65/U66 to C65/A66 in loop 3 (VV20). Each mutation would be predicted to destabilize stem 0, leading to a reduced efficiency of frameshifting, but this was not seen, arguing against the formation of stem 0. The loop 3 substitutions had very little effect on frameshift efficiency (VV20, 25.5%) but the spacer mutation led to a noticeable enhancement (VV24, 42.5%). The explanation for the latter observation is unclear, but related findings have been reported (Kim et al. 2001; Bertrand et al. 2002; Bekaert et al. 2003) and the phenomenon could be relevant to models of frameshifting (Plant et al. 2003).
We next targeted the ISE, decreasing or increasing its length in triplet steps to give a range of ISEs of 1–13 nt. As shown in Figure 4, frameshifting was optimal at the natural length of 7 nt (Fig. 4C, VV1, 28.3%) but reduced to varying extents in the mutants. For the shortest ISE (1 nt), frameshifting was greatly diminished (to <2%) irrespective of whether the base was an A (VV2) or a U (VV3). An ISE of 4 nt retained about half of the activity of the wild type (Fig. 4C, VV8; 13.6%), but the longer ISEs exhibited substantially reduced frameshifting (Fig. 4C, VV9, 10 nt, 2.9%, VV10, 13 nt, <1%). Changing the sequence of the ISE to the alternate pyrimidine/purine bases while retaining its length (i.e., 5′-CGUCCGC-3′ to 5′-UACUUAU-3′) also reduced the capacity of the pseudoknot to induce frameshifting (Fig. 4C, VV23, 5.9%). We do not know whether the inhibitory effects seen upon changing the length or sequence composition of the ISE reflect conformational constraints on global pseudoknot folding or disruption of local tertiary interactions. However, a mutation that reduced the length of loop 1 from 5 to 2 nt reduced frameshifting significantly (VV11, <1%), despite the fact that the remaining 2 nt have been shown to be sufficient to span a 7 bp stem 2 in model building studies (Pleij et al. 1985) and in pseudoknots with a stem 2 of similar length (see Michiels et al. 2001 and references therein). The presence of the ISE may therefore necessitate a loop 1 length of >2 nt. Based on this, we prepared a construct in which the ISE was reduced to a single A residue in the context of the shorter loop 1. While an increase in frameshifting was seen with this mutant (Fig. 4C, VV25, 2.8%) in comparison to the individual changes (Fig. 4C, VV2, ISE is a single A, 1.5%, VV11, loop 1 of 2 nt, <1%), the stimulatory RNA still had a considerable deficit of function compared to the wild-type structure.
Loop 3 of the VMV pseudoknot could potentially form minor groove interactions with stem 1, as seen, for example, in the pseudoknot of beet western yellows virus (BWYV; loop 3 is 7 nt) (Su et al. 1999), a variant of the simian retrovirus 1 (SRV-1) gag/pro stimulatory RNA (pk103; loop 3 is 9 nt) (Michiels et al. 2001) and a synthetic pseudoknot (pKA-A; loop 3 is 8 nt) (Liphardt et al. 1999) derived from the coronavirus infectious bronchitis virus (IBV) signal. In BWYV and pk103, triplexes are formed primarily through adenines in the loop, including a conserved stretch 5′-AACAA-3′. A similar sequence is present in loop 3 of the VMV pseudoknot (5′-AACA-3′), but when this was changed to 5′-UUGU-3′, only a small reduction in frameshifting was seen (Fig. 4C, VV26, 22.9%). Thus the minor groove triplex interactions of the kind seen with BWYV and pk103 do not appear to be necessary for frameshifting in VMV. This is likely to be a consequence of the longer loop 3 in VMV; maintenance of the 5′-AACAA-3′ sequence in loop 3 of the the wild-tpe SRV-1 gag/pro pseudoknot (where loop 3 is longer at 12 nt) does not appear to be necessary for efficient frameshifting (ten Dam et al. 1995). The loop–helix interaction documented in pKA-A is mediated at least in part by the ultimate base of loop 3 (Liphardt et al. 1999). In contrast to what was seen with this pseudoknot, where only an A was functional at this location, variants of the VMV pseudoknot retained significant activity, whether the A was replaced with G (VV14, 76.9%), C (VV15, 1311.5%), or U (VV13, 31%). Again, this difference is probably due to the longer length of the VMV loop 3. When loop 3 of the pKA-A pseudoknot was increased to 14 nt (identical in length to that of VMV), the identity of the nucleotide at the end of loop 3 became less important, with all four bases promoting frameshifting to a similar level (Liphardt et al. 1999). At present, however, we cannot rule out that the VMV pseudoknot engages in some form of loop–helix interaction since replacement of the ultimate and penultimate A residues at the end of loop 3 by G residues was significantly inhibitory to frameshifting (Fig. 4C, VV4, 2.1%). While this mutation could be impacting upon another aspect of pseudoknot architecture or function, for example, the conformation of the junction between the two stems, it could also be modulating interactions with stem 1. It may be significant that a flip mutation that reversed two G-C base pairs in stem 1, although having no effect on predicted helix stability (Turner et al. 1988), inhibited frameshifting conspicuously (Fig. 4C, VV12, 8.9%), perhaps by affecting contacts with the 3′ end of loop 3. To ascertain whether a reduction in loop 3 length could be tolerated, 3 nt were deleted close to the 5′ end of the loop, retaining the 5′-AACA-3′ motif and the A residues at the 3′ end of the loop. This mutation showed a deficit in frameshifting (Fig. 4C, VV5, 12.5%), suggesting a minimal loop length is required. However, in some frameshift-promoting pseudoknots it has been revealed that certain bases at the junction of stem 1 and loop 3 can affect frameshifting despite no obvious involvement in secondary or tertiary interactions (for example, see Kim et al. 2000; Pallan et al. 2005). How this is mediated is uncertain, but it may be that the VV5 deletion impinges on this function.
NMR analysis of the VMV stimulatory RNA
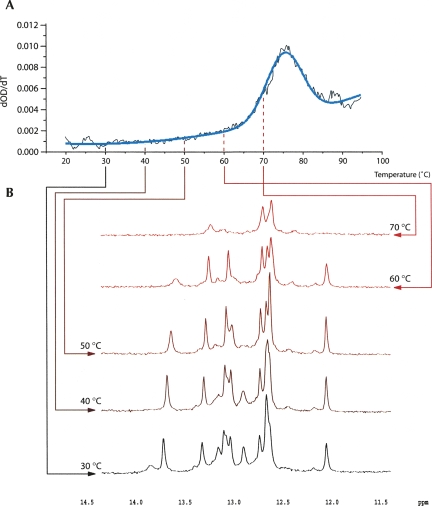
Naturally, an atomic model of the VMV pseudoknot would be of considerable value and would aid interpretation of the structure probing and mutagenesis data. To this end, we attempted to crystallize the molecule. Milligram quantities of a homogeneous RNA containing the VMV pseudoknot flanked by two unpaired residues at either end was purified from pVisna-derived transcripts using a dual ribozyme method (see Materials and Methods) (Fig. 2). However, despite extensive screening, no crystals were obtained under any of the conditions tested over a range of temperatures. However, sufficient material was available to allow a preliminary analysis of the structure by UV melting and 1D and 2D NMR. Initial UV melting experiments indicated a single melting transition beginning at ∼60°C with a midpoint at 76.8°C (Fig. 5). This could represent, in principle, melting of either or both helices of the two-stem helix or the pseudoknot, depending on how the stems unfolded (in a single or two melting transitions). The situation was clarified by NMR analysis. 1D proton NMR experiments were performed in 10% D2O at temperatures between 30°C and 70°C and the data are shown in Figure 5. NMR spectra recorded at 30°C yielded at least 13 peaks, although precise quantification is difficult due to extensive overlap between the peaks, where some of the larger peaks contain overlapping signals from 2 or 3 bp. This information alone did not discriminate between the two-stem helix and pseudoknot models (with 13 and 14 bp in each structure, respectively). However, only two of the peaks had a chemical shift indicative of an A-U base pair (see below), instead of the five predicted by the two-stem helix model, suggesting that the predominant species is the pseudoknot. With increasing temperature, the base-pair count was slowly reduced until 60°C, at which point only seven pairs appear to remain, with a single A-U peak, consistent with the maintenance of pseudoknot stem 1. The spectrum at 70°C has only three peaks, as would be expected from its position within the main melting transition. As discussed below, peaks can be assigned to bases in the G-rich stem throughout the temperature range and therefore are indicative of melting of the other stem, almost certainly stem 2 of the pseudoknot.
FIGURE 5.
Probing the VMV pseudoknot structure by 1D NMR. (A). First derivative UV melting curve of the VMV pseudoknot displaying a single melting transition with a midpoint at 76.8°C. Smoothed raw data are shown in black and fitted data in blue. Arrows link to the corresponding spectra. (B) 1D NMR spectra of the imino proton resonances of the VMV pseudoknot at 10°C intervals from 30°C to 70°C. For a full description, see main text.
The idea that stem 1 is present and unwinds last comes from a tentative peak assignment for nonexchangeable protons in the 60°C spectrum. One peak is present with a chemical shift >13.5 ppm. This peak is broad, and imino protons in Watson–Crick base pairs at the end of helical regions are known to exchange more rapidly with solvent (Varani et al. 1989). A strong NOE cross-peak correlates this resonance to the one of an adenine H2 in a 2D 1H-1H NOESY spectrum of the RNA recorded at 30°C (data not shown), indicating that a stable A-U base pair is forming. Imino-amino correlations observed for all other resonances in the same spectrum identify them as G imino protons within G-C base pairs (data not shown). The three close peaks between 12.5 and 12.7 ppm (Fig. 5) could derive from the three central G-C base pairs in stem 1, given that these base pairs are in a similar structural context. The single sharp resonance at 12.1 ppm, showing similar imino-to-amino correlations, is likely to derive from the closing C-G pair of stem 1. An upfield shift in the imino of the base pair closing a stem has been noted previously in the frameshift-promoting pseudoknot of mouse mammary tumor virus gag/pro (MMTV) and has been attributed to the stacking of adjacent bases, with the first base of loop 3 stacking onto the bottom of stem 1 (Shen and Tinoco 1995). In the VMV case, stacking may be present with bases in the ISE or loop 1, given the possibility of an open conformation of the stem–stem junction afforded by the presence of the ISE. This peak also shows some broadening relative to the others with increasing temperature, possibly indicating a position at the end of a helix, and may become subject to faster solvent proton exchange as interactions across the stem–stem junction are melted. Finally, the two remaining strong resonances at 13.0 and 13.2 ppm are expected to derive from the two remaining G-C pairs in stem 1. In the NOESY spectrum (data not shown), sequential imino correlations link the A-U pair and the three subsequent G-C base pairs, which remain in the 60°C spectrum, consistent with the proposed secondary structure of stem 1.
A final comment concerns the stability of the two stems. At temperatures between 30°C and 60°C, the base-pair count is reduced with increasing temperature, indicating gradual melting or breathing of stem 2. This may explain the lack of a defined stem 2 melting transition by UV and also the sensitivity of stem 2 bases to cleavage by RNase T1 described above. Together, these data are most consistent with the pseudoknot model with a breathing stem 2.
DISCUSSION
Inspection of the sequence of the Visna-Maedi virus ribosomal frameshifting signal suggested that the RNA may fold into either a structure resembling the two-stem helix employed at the HIV-1 gag/pol frameshift signal (Dulude et al. 2002) or an RNA pseudoknot with a unusual 7-nt insertion (loop 2 or ISE) between the two stems. By using a combination of RNA structure probing, site-directed mutagenesis, and NMR, we show that the stimulatory RNA is a pseudoknot and that the ISE is a critical functional element.
Structure of the VMV frameshift-promoting pseudoknot
The RNA structure probing and in vitro translation studies strongly support the pseudoknot model of Figure 1. The overall fold was especially obvious from the single-stranded chemical probing experiments, which clearly delineated the single- and double-stranded regions. While stem 2 was contrarily sensitive to the single-strand G-specific RNase T1, such unusual reactivity has been seen with other pseudoknots, and it is clear from the other probing reagents and from NMR that stem 2 does form. Indeed, mutations that abrogated base-pairing in either pseudoknot stem were inhibitory to frameshifting, yet frameshifting was restored in pseudowild-type double mutants with restored base-pairing. Although the stem 1 double mutant regained close to full activity, the stem 2 double mutant regained only about 50% of wild-type activity. This is likely to be a consequence of the flipping of the stem 2 base pair at the stem–stem junction, as such junction changes have the capacity to influence frameshifting (for examples, see Kim et al. 1999; Cornish et al. 2006). Structure–function analysis of a number of frameshift-promoting pseudoknots has revealed a complexity of interactions between loops and stems and at the junction between component stems and has identified some exposed regions not involved in tertiary contacts that can affect frameshifting, including bases at the junction of stem 1 and loop 3 (Kim et al. 1999, 2000; Liphardt et al. 1999; Su et al. 1999; Michiels et al. 2001; Nixon et al. 2002a,b; Wang et al. 2002; Cornish et al. 2005; Pallan et al. 2005). Our studies indicate that the MVV pseudoknot does not engage in the kind of extended loop 3–stem 1 helix minor groove interaction typified by the short luteovirus pseudoknots (e.g., BWYV), since these occur primarily through adenines, and removal of a block of these (VV26) does not greatly impinge on function.
Further support for the pseudoknot model comes from phylogenetic sequence analysis. An alignment of the frameshift region of published small ruminant lentiviruses, including 71 Swiss caprine and ovine lentiviruses (Shah et al. 2004), reveals excellent conservation of pseudoknot features (Supplemental Fig. 1). As expected for a (dual) coding region, the overall length of the pseudoknot is conserved. Almost all of the variation between isolates is present in the spacer region, the first two bases of loop 1 and within loop 3. From this, we observe that stem 0 is not conserved between isolates, as one would predict from our mutational analysis. The sequence of loop 3, with the exception of the gag termination codon (UAA; bases 69–71) and following residue (C72), shows considerable variation. Importantly, the penultimate A residues are not conserved, arguing against a role for these bases in loop–helix interactions. The inhibitory effect of changing these residues to Gs (VV4) could thus be related to effects at the stem–stem junction. From the alignment, it can be seen that the stems are highly conserved. Within the 7-bp stem 1, the central 5 bp are preserved, although in several cases the central GC pair is replaced by GU. The first and seventh base pairs are not as well conserved, however, and stem 1 in some isolates is predicted to be only 5 bp long. This suggests that the pseudoknot can function with a shorter stem and can tolerate single nucleotide increases to the length of the spacer, loop 1, ISE, and loop 3. In many isolates, the top base pair of stem 1, CG, is replaced by the phylogenetically conserved AU pair. The sequences that make up stem 2 are identical in all isolates bar one, where the central GC pair is flipped to CG, indicating conservation of this stem. The ISE is fully conserved in all isolates.
Role of the ISE
Frameshift-promoting pseudoknots with a single unpaired adenosine between the stems have been described previously (typified by MMTV gag/pro [Shen and Tinoco 1995], and a few possess a long, highly structured ISE, for example, the stimulatory RNA of Rous sarcoma virus [RSV; ISE is 52 nt] [Marczinke et al. 1998] and the closely related pseudoknot of gill-associated virus [GAV; ISE is 64 nt] [Cowley et al. 2000], but most lack an ISE [van Batenburg et al. 2001; Brierley et al. 2007]). Indeed, among pseudoknots in all RNAs described to date, >85% do not possess a loop 2 (van Batenburg et al. 2001; Kim et al. 2004). However, an examination of the sequences present at viral frameshift sites reveals two other potential examples of frameshift-promoting pseudoknots with short ISEs, a 3-nt ISE in the pseudoknot of the 1a/1b frameshift signal of the arterivirus lactate dehydrogenase-elevating virus, strain C (Godeny et al. 1993) and a 2-nt ISE at the pro/pol overlap of human T-lymphotrophic virus type II (R. Girnary and I. Brierley, unpubl.).
The presence of the 7-nt ISE in the VMV pseudoknot is unexpected from the perspective of function. Our previous studies with the IBV frameshift signal revealed that the artificial introduction of an ISE (of 3 nt) between the component stems of the pseudoknot was inhibitory to function (Brierley et al. 1991). Similarly, increasing the length of the ISE of the MMTV gag/pro pseudoknot from 1 to 2 nt also severely compromises activity (Chen et al. 1995). The ISE of the VMV pseudoknot, in contrast, is essential for frameshifting and must therefore be accommodated into the global fold without compromising pseudoknot activity. While reducing the length of this element to 4 nt was tolerated to a degree (VV8), further reduction or an increase in length led to a dramatic inhibition of frameshifting. The strong reactivity of the ISE to lead probing and the apparent sequence requirement (VV23) suggests a specific structural role. The ISE might be expected to increase the flexibility of the pseudoknot; as discussed earlier, loop 1 is unusually long for a pseudoknot of this size, a feature that may be required to accommodate the increased flexibility. The ISE would also be expected to affect junction conformation and the thermodynamics of unfolding of the pseudoknot. Without detailed structural information, however, we do not know whether the ISE is engaged in any local interactions at the pseudoknot junction, so its precise role in pseudoknot conformation and function is unknown. Furthermore, we do not know whether the ISE engages in any interactions in trans with components of the translation apparatus.
Recent cryo-EM images of ribosomes stalled at a coronavirus frameshift-promoting pseudoknot (Namy et al. 2006) have provided support for the long-held belief that the role of the pseudoknot is to resist the ribosomal helicase and affect the movement of mRNA through the ribosome during the elongation cycle (Jacks et al. 1988; Takyar et al. 2005). How such resistance is accomplished is uncertain. A simple “roadblock” action of the pseudoknot has been shown to be oversimplistic (Brierley et al. 1991; Chen et al. 1995), and even pseudoknots containing subtle mutations with remarkably similar overall conformation can display widely differing frameshift efficiencies (see Cornish et al. 2006, and references within). Recent work has revealed that pseudoknots possess greater mechanical stability in comparison to hairpins and a kinetic insensitivity to force (Green et al. 2008), supporting the hypothesis that the extent of −1 frameshifting is related to the difficulty of mechanically unfolding the pseudoknot (Namy et al. 2006; Hansen et al. 2007). Nevertheless, we are not yet at the stage where the activity of a pseudoknot can be foreseen from the sequence and predicted secondary structure. The demonstration in this article of a functional retroviral pseudoknot possessing an ISE adds another layer of complexity to such forecasting.
MATERIALS AND METHODS
Site-directed mutagenesis
Site-specific mutagenesis was carried out using a commercial PCR-based methodology (QuikChange, Stratagene) or by the method of Kunkel (1985) as described previously (Brierley et al. 1989). Mutants were identified by dideoxy sequencing of single-stranded templates (Sanger et al. 1977). Sequencing through G + C-rich regions was facilitated by replacing dGTP with deaza-GTP in the sequencing mixes.
Construction of plasmids
The frameshift reporter plasmid p2luc/VV1 was constructed by inserting three pairs of complementary synthetic oligonucleotides encoding the VMV frameshift region (strain accession number NC001452) between the Renilla and firefly luciferase genes of the dual luciferase reporter plasmid p2luc (Grentzmann et al. 1998) using the SalI and BamHI restriction sites. A “100% frameshift” in-frame control plasmid for in vivo assays, p2luc/VV21, was prepared by inserting a cytosine base immediately downstream of the slippery sequence to align the luciferase frames (GGGAAACCAAC). To perform mutagenesis of the VMV frameshift region, a 1891-bp HindIII/EcoRI fragment from p2luc/VV1 was subcloned into plasmid pING14.2 (Liu et al. 1991) to generate pING14.2/VV1. Following mutagenesis, the HindIII/EcoRI fragments were sequenced in their entirety and reintroduced into p2luc for subsequent frameshift assays. The structure mapping plasmid pVSM was constructed by inserting six pairs of complementary synthetic oligonucleotides into the XbaI and PstI sites of plasmid pUC119∂V (Arnvig et al. 2004), a derivative of pUC18T7Pst∂V (gift of Dr. Sandra Searles, MRC Laboratory of Molecular Biology, Cambridge, UK) (Fig. 2A; Jovine et al. 2000). The cloned VMV sequences comprised the putative pseudoknot flanked by 29 and 13 nt at the 5′ and 3′ ends, respectively. Plasmid pVisna, used to produce unit length transcripts for crystallography trials and NMR, was constructed by inserting six pairs of complementary synthetic oligonucleotides into the XbaI and PstI sites of pUC119∂V (Fig. 2B). The cloned VMV sequences comprised the putative pseudoknot flanked by AA and UU nucleotides at the 5′ and 3′ ends, respectively.
In vitro transcription
Plasmids for in vitro transcription were purified using a commercial kit (Qiagen). Capped mRNAs for in vitro translation were prepared from Hpa I-cut p2luc/VV variants using bacteriophage T7 polymerase and the protocol of Girnary et al. (2007). Large-scale transcriptions of dual ribozyme constructs were performed to generate homogeneous material for structure mapping (pVSM/HindIII), crystallography trials, and NMR (pVisna/HindIII) as described (Ke and Doudna 2004). Transcription reactions (10 mL) were carried out at 37°C for 3 h and typically contained 500 μg Hind III-linearized plasmid DNA, 5 mM of each rNTP, 1 mg of T7 RNA polymerase, and 10 U inorganic pyrophosphatase in a buffer containing 30 mM Tris (pH 8), 25 mM MgCl2, 10 mM DTT, 0.01% Triton X-100, and 2 mM spermidine. After 3 h at 37°C the RNA was heated at 65°C for 10 min, slow cooled to 55°C, and held at this temperature for a further 30 min to promote ribozyme cleavage prior to gel purification of the relevant species. Following purification, the RNA pellet was dissolved in 2 mL 10 mM Tris (pH 8), 5 mM MgCl2, and 0.1 M NaCl and dialyzed (10 kDa cutoff Slide-a-lyser, Pierce) overnight against three changes of 800 mL of the same buffer at 4°C to remove acrylamide contaminants. The RNA was centrifugally concentrated to 5–6 mg/mL (Vivaspin 10 kDa cutoff, Sartorius) and stored at −80°C.
RNA structure probing
Gel-purified pVSM transcripts (10 μg) were 5′-end labeled with [γ-33P]ATP using T4 polynucleotide kinase, repurified from 10% acylamide-urea gels, and dissolved in water. All reactions contained 20,000–100,000 cpm 5′-end-labeled RNA transcript, and structure probing was carried out according to Manktelow et al. (2005). The temperature and salt conditions in the structure probing reactions were ice (enzymes), 25°C (lead acetate), or 37°C (imidazole) in buffers containing 2 mM MgCl2 (all reagents) and either 50 mM Na cacodylate (pH 7) (RNase T1; CL3), 100 mM KCl (RNases CV1, U2), 50 mM KOAc (lead), or 40 mM NaCl (imidazole). RNAs were prepared for analysis on 6%, 10%, or 15% polyacrylamide-7 M urea sequencing-type gels (with or without 20% [v/v] formamide) by dissolution in water and mixing with an equal volume of formamide gel loading buffer (95% [v/v] formamide, 10 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol), prior to heating at 80°C for 3 min.
Ribosomal frameshift assays
Messenger RNAs were translated in the rabbit reticulocyte in vitro translation system (RRL) at 30°C as described (Brierley et al. 1987). Translation products were analyzed on SDS-12.5% (w/v) polyacrylamide gels according to standard procedures (Hames 1981). The relative abundance of nonframeshifted and frameshifted products on the gels was determined by direct measurement of [35S]methionine incorporation using a Packard Instant Imager 2024 and adjusted to take into account the differential methionine content of the products. In vivo frameshift assays employed Cos-7 cells, which were maintained in Dulbecco's modification of Eagle's medium supplemented with 10% (v/v) fetal calf serum. The p2luc/VV series of plasmids were transfected using a commercial liposome method (FuGene 6; Roche). Cells were seeded in 60-mm dishes and grown for 18–24 h until 80% confluency was reached. Transfection mixtures (containing plasmid DNA, serum-free medium [Optimem; Gibco-BRL] and FuGene) were set up as recommended by the manufacturer and added directly (dropwise) to the tissue culture cell growth medium. The cells were harvested 24 h post-transfection and reporter gene expression determined using a dual luciferase assay system kit (Promega). Each data point represents the mean value from three separate transfections.
Crystallization trials
The VMV pseudoknot transcript prepared from pVisna was used at a concentration of 4–6 mg/mL. Crystallization experiments employed vapor diffusion and batch methods using commercial sparse matrix and grid screens (Nextal, Qiagen; Hampton Research). Hanging drops were set up in 24-well EasyXtal CrystalSupport trays (Nextal). Batch experiments were dispensed using an Oryx 6 crystallization robot (Douglas Instruments) and imaged using a Rock Imager (Formulatrix).
UV spectroscopy
UV melting experiments were performed on a Jasco V550 UV/Vis spectrometer equipped with an ETC-505T temperature controller. Samples were heated in 10 mM Tris (pH 8), 100 mM NaCl, and 5 mM MgCl2 from 20°C to 95°C in stoppered cuvettes at 1°C per minute and the absorbance at 260 nm read every 0.3–0.4 sec. Data were fitted using ProFit (Quansoft).
NMR spectroscopy
NMR spectra were recorded at 10°C–70°C using Varian UnityPlus and INOVA spectrometers operating at 500, 600, and 800 MHz 1H frequencies and on a Bruker Advance spectrometer operating at 600 MHz 1H frequency equipped with a cryoprobe. The experiments were recorded with 0.3 mM samples of the VMV pseudoknot in a 90% H2O/10% D2O mixture in 10 mM Tris-HCl (pH 8), 100 mM NaCl, and 5 mM MgCl2. Water suppression was achieved by WATERGATE pulse sequences (Piotto et al. 1992). Spectra were referenced internally to the residual water signal with appropriate correction for temperature. 2D NOESY spectra were recorded at 30°C with mixing times of 125 and 300 msec.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We would like to thank Dr. Andres Ramos, Dr. Sasi Conte, and Dr. Steve Martin for helpful discussions. This work was supported by grants from the Biotechnology and Biological Sciences Research Council, UK (I.B.) and the Medical Research Council, UK (S.J.S.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1042108.
REFERENCES
- Arnvig K.B., Pennell S., Gopal B., Colston M.J. A high-affinity interaction between NusA and the rrn nut site in Mycobacterium tuberculosis . Proc. Natl. Acad. Sci. 2004;101:8325–8330. doi: 10.1073/pnas.0401287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P.V., Fayet O., Hendrix R.W., Atkins J.F. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Bekaert M., Bidou L., Denise A., Duchateau-Nguyen G., Forest J.P., Froidevaux C., Hatin I., Rousset J.P., Termier M. Towards a computational model for −1 eukaryotic frameshifting sites. Bioinformatics. 2003;19:327–335. doi: 10.1093/bioinformatics/btf868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand C., Prere M.F., Gesteland R.F., Atkins J.F., Fayet O. Influence of the stacking potential of the base 3′ of tandem shift codons on −1 ribosomal frameshifting used for gene expression. RNA. 2002;8:16–28. doi: 10.1017/s1355838202012086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic −1 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 2001;LXV:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- Brierley I., Boursnell M.E., Binns M.M., Bilimoria B., Blok V.C., Brown T.D., Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterisation of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N.J., Jenner A.J., Inglis S.C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Pennell S., Gilbert R.J.C. Viral RNA pseudoknots: Versatile motifs in gene expression and replication. Nat. Rev. Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Chamorro M., Lee S.I., Shen L.X., Hines J.V., Tinoco I., Jr, Varmus H.E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: Nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish P.V., Hennig M., Giedroc D.P. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. 2005;102:12694–12699. doi: 10.1073/pnas.0506166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish P.V., Stammler S.N., Giedroc D.P. The global structures of a wild-type and poorly functional plant luteoviral mRNA pseudoknot are essentially identical. RNA. 2006;12:1959–1969. doi: 10.1261/rna.199006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus of Penaeus monodon prawns: An invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- Dulude D., Baril M., Brakier-Gingras L. Characterization of the frameshift stimulatory signal controlling a programmed −1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 2002;30:5094–5102. doi: 10.1093/nar/gkf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. Programmed translational frameshifting. Microb. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin C., Mazauric M.H., Traikia M., Guittet E., Yoshizawa S., Fourmy D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J. Mol. Biol. 2005;349:1024–1035. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- Giedroc D.P., Theimer C.A., Nixon P.L. Structure, stability, and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girnary R., King L., Robinson L., Elston R., Brierley I. Structure–function analysis of the ribosomal frameshifting signal of two human immunodeficiency virus type 1 isolates with increased resistance to viral protease inhibitors. J. Gen. Virol. 2007;88:226–235. doi: 10.1099/vir.0.82064-0. [DOI] [PubMed] [Google Scholar]
- Godeny E.K., Chen L., Kumar S.N., Methven S.L., Koonin E.V., Brinton M.A. Complete genomic sequence and phylogenetic analysis of the lactate dehydrogenase-elevating virus (LDV) Virology. 1993;194:585–596. doi: 10.1006/viro.1993.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L., Kim C.H., Bustamante C., Tinoco I., Jr Characterization of the mechanical unfolding of RNA pseudoknots. J. Mol. Biol. 2008;375:511–528. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- Hames B.D. An introduction to polyacrylamide gel electrophoresis. In: Hames B.D., Rickwood D., editors. Gel electrophoresis of proteins—A practical approach. IRL Press; Oxford, UK: 1981. pp. 1–91. [Google Scholar]
- Hansen T.M., Reihani S.N., Oddershede L.B., Sørensen M.A. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl. Acad. Sci. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D., Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem. Sci. 1990;15:186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J., Siddell S.G. An “elaborated” pseudoknot is required for high frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res. 1993;1:5838–5842. doi: 10.1093/nar/21.25.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H.E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani H.D., Masiarz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Jovine L., Hainz T., Oubridge C., Nagai K. Crystallization and preliminary X-ray analysis of the conserved domain IV of Escherichia coli 4.5S RNA. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1033–1037. doi: 10.1107/s0907444900006910. [DOI] [PubMed] [Google Scholar]
- Ke A., Doudna J.A. Crystallization of RNA and RNA–protein complexes. Methods. 2004;34:408–414. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Kim Y.G., Su L., Maas S., O'Neill A., Rich A. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc. Natl. Acad. Sci. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.G., Maas S., Wang S.C., Rich A. Mutational study reveals that tertiary interactions are conserved in ribosomal frameshifting pseudoknots of two luteoviruses. RNA. 2000;8:1157–1165. doi: 10.1017/s1355838200000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.G., Maas S., Rich A. Comparative mutational analysis of cis-acting RNA signals for translational frameshifting in HIV-1 and HTLV-2. Nucleic Acids Res. 2001;29:1125–1131. doi: 10.1093/nar/29.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Shiffeldrim N., Gan H.H., Schlick T. Candidates for novel RNA topologies. J. Mol. Biol. 2004;341:1129–1144. doi: 10.1016/j.jmb.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Kunkel T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.Y., Shapiro B.A., Chen J.H., Nussinov R., Maizel J.V. RNA pseudoknots downstream of the frameshift sites of retroviruses. Genet. Anal. Tech. Appl. 1991;8:191–205. doi: 10.1016/1050-3862(91)90013-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt J., Napthine S., Kontos H., Brierley I. Evidence for an RNA pseudoknot loop-helix interaction essential for efficient −1 ribosomal frameshifting. J. Mol. Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Cavanagh D., Green P., Inglis S.C. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology. 1991;184:531–544. doi: 10.1016/0042-6822(91)90423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manktelow E., Shigemoto K., Brierley I. Characterization of the frameshift signal of Edr, a mammalian example of programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2005;33:1553–1563. doi: 10.1093/nar/gki299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczinke B., Fisher R., Vidakovic M., Bloys A.J., Brierley I. Secondary structure and mutational analysis of the ribosomal frameshift signal of Rous sarcoma virus. J. Mol. Biol. 1998;284:205–225. doi: 10.1006/jmbi.1998.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels P.J., Versleijen A.A., Verlaan P.W., Pleij C.W., Hilbers C.W., Heus H.A. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O., Moran S.J., Stuart D.I., Gilbert R.J.C., Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon P.L., Cornish P.V., Suram S.V., Giedroc D.P. Thermodynamic analysis of conserved loop-stem interactions in P1-P2 frameshifting RNA pseudoknots from plant Luteoviridae. Biochemistry. 2002a;41:10665–10674. doi: 10.1021/bi025843c. [DOI] [PubMed] [Google Scholar]
- Nixon P.L., Rangan A., Kim Y.G., Rich A., Hoffman D.W., Hennig M., Giedroc D.P. Solution structure of a luteoviral P1-P2 frameshifting mRNA pseudoknot. J. Mol. Biol. 2002b;322:621–633. doi: 10.1016/s0022-2836(02)00779-9. [DOI] [PubMed] [Google Scholar]
- Pallan P.S., Marshall W.S., Harp J., Jewett F.C., 3rd, Wawrzak Z., Brown B.A., 2nd, Rich A., Egli M. Crystal structure of a luteoviral RNA pseudoknot and model for a minimal ribosomal frameshifting motif. Biochemistry. 2005;44:11315–11322. doi: 10.1021/bi051061i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin N.T., Chamorro M., Varmus H.E. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: Demonstration by expression in vivo. J. Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotto M., Saudek V., Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Plant E.P., Jacobs K.L., Harger J.W., Meskauskas A., Jacobs J.L., Baxter J.L., Petrov A.N., Dinman J.D. The 9 Å solution: How mRNA pseudoknots promote efficient programmed −1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C.W.A., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reil H., Kollmus H., Weidle U.H., Hauser H. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J. Virol. 1993;67:5579–5584. doi: 10.1128/jvi.67.9.5579-5584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah C., Böni J., Huder J.B., Vogt H.R., Mühlherr J., Zanoni R., Miserez R., Lutz H., Schüpbach J. Phylogenetic analysis and reclassification of caprine and ovine lentiviruses based on 104 new isolates: Evidence for regular sheep-to-goat transmission and worldwide propagation through livestock trade. Virology. 2004;319:12–26. doi: 10.1016/j.virol.2003.09.047. [DOI] [PubMed] [Google Scholar]
- Shen L.X., Tinoco I. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J. Mol. Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- Staple D.W., Butcher S.E. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Su L., Chen L., Egli M., Berger J.M., Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat. Struct. Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S., Hickerson R.P., Noller H.F. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- ten Dam E.B., Pleij C.W.A., Bosch L. RNA pseudoknots: Translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E.B., Verlaan P.W., Pleij C.W. Analysis of the role of the pseudoknot component in the SRV-1 gag-pro ribosomal frameshift signal: Loop lengths and stability of the stem regions. RNA. 1995;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- Turner D.H., Sugimoto N., Freier S.M. RNA structure prediction. Annu. Rev. Biophys. Biophys. Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- van Batenburg F.H., Gultyaev A.P., Pleij C.W. PseudoBase: Structural information on RNA pseudoknots. Nucleic Acids Res. 2001;29:194–195. doi: 10.1093/nar/29.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G., Wimberly B., Tinoco I., Jr Conformation and dynamics of an RNA internal loop. Biochemistry. 1989;28:7760–7772. doi: 10.1021/bi00445a036. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wills N.M., Du Z., Rangan A., Atkins J.F., Gesteland R.F., Hoffman D.W. Comparative studies of frameshifting and nonframeshifting RNA pseudoknots: A mutational and NMR investigation of pseudoknots derived from the bacteriophage T2 gene 32 mRNA and the retroviral gag-pro frameshift site. RNA. 2002;8:981–996. doi: 10.1017/s1355838202024044. [DOI] [PMC free article] [PubMed] [Google Scholar]