Abstract
RNA regulators are critical for animal development, especially in the germ line where gene expression is often modulated by changes in mRNA stability, translation, and localization. In this paper, we focus on Caenorhabditis elegans LARP-1, a representative of one La-related protein (Larp) family found broadly among eukaryotes. LARP-1 possesses a signature La motif, which is an ancient RNA-binding domain, plus a second conserved motif, typical of LARP-1 homologs and therefore dubbed the LARP1 domain. LARP-1 appears to bind RNA in vitro via both the La motif and the LARP1 domain. larp-1 null mutants have an oogenesis defect reminiscent of hyperactive Ras-MAPK signaling; this defect is suppressed or enhanced by down- or up-regulating the Ras-MAPK pathway, respectively. Consistent with a role in down-regulating the Ras-MAPK pathway, larp-1 null mutants have higher than normal levels of selected pathway mRNAs and proteins. LARP-1 protein colocalizes with P bodies, which function in RNA degradation. We suggest that LARP-1 functions in P bodies to attenuate the abundance of conserved Ras-MAPK mRNAs. We also propose that the cluster of LARP-1 homologs may function generally to control the expression of key developmental regulators.
Keywords: La motif, Larp, MAPK, P bodies
INTRODUCTION
Post-transcriptional regulation figures prominently in animal development (Colegrove-Otero et al. 2005; Thompson et al. 2007). Controls of mRNA stability, translation, and localization influence when, where, and how much protein is made. Key RNA regulators of animal development include sequence-specific RNA-binding proteins (e.g., PUF proteins), micro-RNAs (e.g., let-7), and RNA modifying enzymes [e.g., cytoplasmic poly(A) polymerase].
The La motif (LM) is an RNA-binding domain first recognized over 20 years ago as the signature motif of the human La autoantigen (Yoo and Wolin 1994). The LM family includes so-called “authentic” La proteins, as defined by their conservation with human La, and the La-related proteins, or Larps (Wolin and Cedervall 2002). Authentic La proteins possess a La motif and an RNA recognition motif (RRM), both located N-terminally; they are predominately nuclear (Yoo and Wolin 1994; Simons et al. 1996; Sobel and Wolin 1999; Marchetti et al. 2000) and associate with nascent RNA polymerase (RNAP) III transcripts (i.e., tRNA, U6 snRNA) (Lerner et al. 1981; Rinke and Steitz 1982; Stefano 1984; Rinke and Steitz 1985; Yoo and Wolin 1994; Kufel et al. 2000; Inada and Guthrie 2004). By contrast, most Larp proteins have a more central La motif and lack a canonical RRM domain. In addition, the La motif sequences of La and Larp proteins cluster into distinct branches of the LM family tree (Sobel and Wolin 1999; Wolin and Cedervall 2002).
Thus far, only a few Larps have been characterized. The ciliate Larps, Euplotes aediculatus p45 and Tetrahymena thermophila p65, bind directly to telomerase RNA and stimulate the assembly and activity of the telomerase holoenzyme (Aigner et al. 2000, 2003; Aigner and Cech 2004; Witkin and Collins 2004; Prathapam et al. 2005). In a similar fashion, human Larp PIP7S binds 7SK snRNA and modulates the integrity of the 7SK snRNP (He et al. 2008). The Saccharomyces cerevisiae Larps, Sro9p and Slf1p, bind RNA homopolymers in vitro, are cytoplasmic, and associate with ribosomes (Sobel and Wolin 1999). Sro9p and Slf1p influence multiple biological processes, including cytoskeletal organization, vesicular transport, and mRNA turnover (Tsukada and Gallwitz 1996; Kagami et al. 1997; Tan et al. 2000; Pan et al. 2006). Finally, a Drosophila Larp has been analyzed, albeit not in great detail. The Drosophila larp gene is controlled by homeotic transcription factors during embryogenesis (Chauvet et al. 2000) and is required for spermatogenesis in adults (Ichihara et al. 2007).
The Caenorhabditis elegans genome encodes two La-related proteins, R144.7/LARP-1 and T12F5.5 (C. elegans Sequencing Consortium 1998). Here we extend previous studies of Larp phylogeny (Sobel and Wolin 1999; Wolin and Cedervall 2002) to show that Larps cluster into two major families based on sequence similarity. LARP-1 is a representative of a broadly conserved Larp family, of which both S. cerevisiae Larps and Drosophila Larp are members, while T12F5.5 is a representative of a second, metazoan-specific Larp family. This work investigates C. elegans LARP-1 and its role in germline development. We find that LARP-1 binds RNA homopolymers via its La motif and a second conserved C-terminal domain. A larp-1 null mutant has germline defects reminiscent of hyperactive Ras-MAPK signaling; genetic interactions indicate that LARP-1 normally attenuates Ras-MAPK signaling in the germ line; and mRNAs encoding certain Ras-MAPK signaling components are elevated in larp-1(0) mutant germ lines. LARP-1 colocalizes with cytoplasmic granules called P bodies, which have been implicated in mRNA degradation. We propose that LARP-1 affects the stability of selected mRNAs via its association with germline P bodies.
RESULTS
C. elegans LARP-1 is representative of the Larp1 family of La motif-containing proteins
To learn how the two C. elegans Larps, R144.7/LARP-1 and T12F5.5, relate to other Larps, we first compared the amino acid sequences of their La motifs (Supplemental Fig, 1A; data not shown) and identified two distinct groups (Fig. 1A). LARP-1 clusters with members found in all eukarya, including both S. cerevisiae Larps, Drosophila Larp, and two human Larps (Larp1 and Larp2), while T12F5.5 belongs to a metazoan-specific branch, which includes human Larp4 and Larp5 (Fig. 1A). We refer to these two Larp clusters as the Larp1 and Larp5 families, respectively; C. elegans T12F5.5 is therefore named LARP-5. The telomerase Larps (E. aediculatus p45 and T. thermophila p65) and the 7SK Larp (human PIP7S) do not cluster with either the Larp1 or Larp5 family (Fig. 1A; data not shown), suggesting the existence of a third Larp family with no clear C. elegans representative.
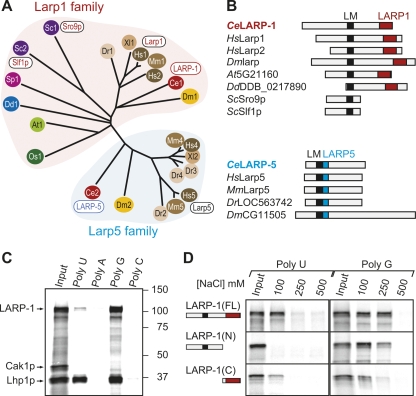
FIGURE 1.
LARP-1 is a conserved Larp that binds RNA. (A) Unrooted phylogenetic tree of Larp proteins, derived by aligning their La motifs with ClustalW (Biology Workbench, San Diego Super Computer Center; http://workbench.sdsc.edu/). La-related proteins cluster into Larp1 (red shaded area) and Larp5 (cyan shaded area) families. Colors denote species: Green, plants (dicot: Arabidopsis thaliana, At; monocot: Oryza sativa, Os); blue, slime mold (Dictyostelium discoideum, Dd); purple, yeasts (Saccharomyces cerevisiae, Sc; Schizosaccaryomyces pombe, Sp); red, nematode (C. elegans, Ce); orange, fruit fly (D. melanogaster, Dm); brown, vertebrates (Danio rerio, Dr; Xenopus laevis, Xl; Mus musculus, Mm; Homo sapiens, Hs). Accession numbers: Ce1 (LARP-1), NP_001040868; Ce2 (LARP-2), NP_491209; Dm1 (larp), AAF35862; Dm2 (CG11505), NP_647793; Hs1 (Larp1), Q6PKGO; Hs2 (Larp2), EAX05185; Hs4 (Larp4), EAW58141; Hs5 (Larp5), NP_055970; Mm1 (mLarp1), Q62Q58; Mm4 (mLarp4), Q8BWW4; Mm5 (mLarp5), NP_766173; Xl1, NP_001089363; Xl2, NP_001091246; Dr1 (Larp1), XP_696560; Dr2, NP_001071030; Dr3, XP_691299; Dr4, XP_696696; Sc1 (Sro9p), P25567; Sc2 (Slf1p), Q12034; Sp1, NP_594554; Dd1 (0,217,890), XP_642918; At1 (5G21160), NP_568409; Os1, EA211350. (B) Schematics of selected Larp1 (top) and Larp5 (bottom) family members with their signature motifs. A La motif (LM; black) is found in all Larps; a C-terminal LARP1 domain (red) is typical of proteins in the Larp1 family; the Larp5 domain (cyan) is immediately adjacent to the La motif in Larp5 family members. (C) Homopolymer binding assay with 35S-labeled proteins, including full-length LARP-1, yeast Lhp1p as a positive control, and yeast Cak1p as a negative control. (Input) Equivalent to 20% of the total protein added to homopolymers; (lanes 2–5) proteins bound to poly(U), poly(A), poly(G), and poly(C) in the presence of 100 mM NaCl. (D) Homopolymer binding assay with 35S-labeled full-length (FL) LARP-1 as well as N-terminal (N) and C-terminal (C) LARP-1 fragments; conventions for La motif and LARP1 domain as in Fig. 1B. (Left) Binding to poly(U); (right) binding to poly(G). (Input) Equivalent to 20% of total protein added to homopolymers; (lanes 2–4) protein bound with increasing NaCl concentrations (100, 250, or 500 mM).
We next compared Larp amino acid sequences outside the La motif. All Larp1 family proteins, except fungal Larps (e.g., ScSro9p, ScSlf1p, SpSPAC1527), bear a C-terminal conserved domain of ∼200 amino acids (Fig. 1B, Supplemental Fig. 1B). This region was recognized previously for mammalian Larp1, nematode R144.7/LARP-1, and fruitfly Larp (Sobel and Wolin 1999; Chauvet et al. 2000; Ichihara et al. 2007); we now report that it is also present in Larps from monocots, dicots, and slime mold. Because this domain is only found in Larp1 family members, we suggest that it be called the LARP1 domain. Proteins in the Larp5 family lack the LARP1 domain but possess a unique conserved region (∼75 amino acids) that resides immediately C-terminal to the La motif (Fig. 1B). Because this domain is only found in Larp5 family members, we suggest that it be called the LARP5 domain. Wolin and Cedervall (2002) report that this LARP5 domain has features of an RRM and suggest that it may contribute to RNA binding. We conclude that LARP-1 represents a family of broadly conserved eukaryotic Larps while LARP-5 defines a second family of metazoan-specific Larps.
LARP-1 appears to bind RNA in vitro
To determine if LARP-1 can bind RNA in vitro, we used a homopolymer-binding assay, as previously described (Swanson and Dreyfuss 1988). Briefly, in vitro-translated 35S-labeled proteins were incubated with homopolymers coupled to beads, washed in varying salt concentrations, eluted, and visualized on protein gels (see Materials and Methods). Full-length LARP-1 was retained by both poly(U) and poly(G), but not by poly(A) or poly(C) (Fig. 1C). The S. cerevisiae La protein, Lhp1p, was similarly retained, but the negative control, Cyclin-associated kinase Cak1p, was not (Fig. 1D). LARP-1 binding is similar to that reported for the yeast Larp Sro9p (Sobel and Wolin 1999). Although we have not excluded the possibility that LARP-1 may bind RNA indirectly via proteins in the in vitro translation extract, the simplest explanation, given the conservation of the LARP-1 La motif, is that LARP-1 binds RNA.
To ask what parts of LARP-1 mediate RNA-binding, we assayed an N-terminal fragment containing the La motif (amino acids 1–831) and a C-terminal fragment containing the LARP1 domain (amino acids 832–1150). Intriguingly, both fragments retained RNA-binding activity, although neither had full activity (Fig. 1D). For example, the N-terminal fragment was retained by poly(G) but not poly(U) while the C-terminal fragment bound weakly to both poly(G) and poly(U). By analogy to the cooperative RNA binding by the LM and RRM of authentic La proteins (Alfano et al. 2004), we suggest that the La motif and LARP1 domain may work together to enhance the RNA-binding activity of Larp1 family members.
First hint of a specific larp-1 biological function
C. elegans larp-1 has been analyzed superficially in large-scale studies. Its mRNA is enriched in the germ line (Reinke et al. 2000), and its promoter drives expression in somatic tissues (McKay et al. 2003). Since germline expression is normally silenced using reporter transgenes (Kelly et al. 1997), the simple conclusion is that larp-1 is expressed in both germ line and soma. Depletion of larp-1 activity by RNA interference (RNAi) causes slow growth and low penetrance embryonic lethality (Gönczy et al. 2000; Kamath et al. 2003; Sonnichsen et al. 2005). Based on these studies, larp-1 appears to have a general biological function that is not essential.
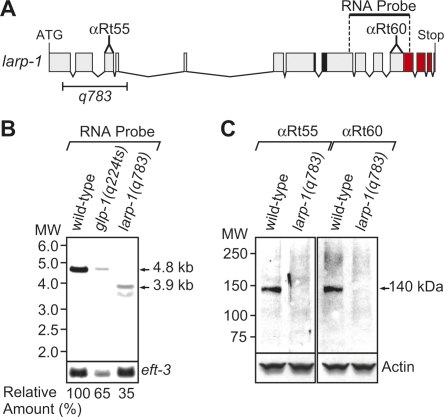
To study LARP-1 and its biological function more rigorously, we generated a larp-1 deletion mutant (Fig. 2A). The larp-1 gene makes a single 4.8-kb transcript in adult hermaphrodites (Fig. 2B, wild-type). Abundance of larp-1 mRNA was reduced, but not absent, in mutants with no germ line [Fig. 2B, glp-1(q224ts)], as would be expected for a transcript expressed in both germ line and soma. The larp-1(q783) deletion removes part of exon 1 and all of exons 2–4 (Fig. 2A) and results in a frame shift and premature termination codon in exon 6, as determined by cDNA sequencing (data not shown). The larp-1(q783) mRNA was smaller and less abundant than wild type [Fig. 2B, larp-1(q783)], and a LARP-1(q783) protein was not detectable on Western blots (Fig. 2C) with antibodies specific to LARP-1 (see Materials and Methods). Therefore, larp-1(q783) is a strong loss-of-function mutation and probably a null allele; henceforth, larp-1(q783) is referred to as larp-1(0).
FIGURE 2.
The larp-1(q783) deletion makes no detectable LARP-1 protein. (A) The larp-1 gene. Predicted start (ATG) and stop codons, plus exons (boxes) and introns (lines) were determined by sequencing cDNAs (see Materials and Methods). The q783 deletion (bracket) removes part of exon 1 and all of exons 2–4. RNA probe was used for Northern blots (B); αRt55 and αRt60 indicate locations of peptides used to generate these LARP-1 antibodies; black region marks La motif; red region denotes LARP1 domain. (B) larp-1 transcript analysis. Northern blots of poly(A) RNA prepared from wild type, glp-1(q224ts) at 25°C, and larp-1(q783). For loading control, we used a probe against eft-3. Relative amount of larp-1 mRNA was calculated as described in Materials and Methods and is presented as a percentage of wild-type amount. (C) LARP-1 protein analysis. Western blots were performed with protein prepared from wild-type and larp-1(q783) adults. A monoclonal antibody directed against actin was used as a loading control.
The larp-1(0) mutant is homozygous viable, but it grows slowly and lays some dead embryos (∼6%); both defects were seen previously by RNAi (Kamath et al. 2003). We also found that larp-1(0) laid fewer embryos than normal [larp-1(0): 207 ± 36, n = 10; wild-type: 272 ± 23, n = 10], and many of the larp-1(0) embryos were smaller than normal (data not shown). These latter defects provided the first clue that larp-1 might influence oogenesis.
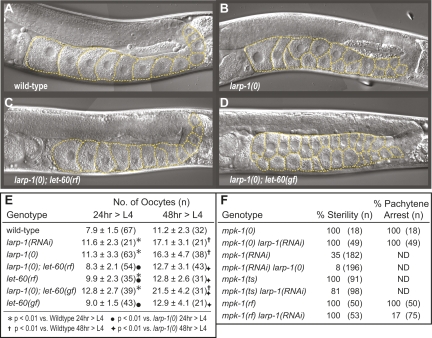
Adult hermaphrodites typically have a single row of 8 to 11 developing oocytes in the proximal arm of each gonad (Fig. 3A). While the overall developmental pattern of the larp-1(0) germ line was similar to wild type (data not shown), the number of oocytes in larp-1(0) gonads was consistently higher than in age-matched, wild-type gonads (Fig. 3B,E). Moreover, most larp-1(0) oocytes were smaller than normal and arranged in multiple rows (Fig. 3, cf. B and A), even though most oocytes were fertilized and produced viable progeny. We conclude that larp-1 is not essential for fertility but is important for proper oocyte development.
FIGURE 3.
LARP-1 attenuates Ras-MAPK signaling during oogenesis. (A–D) Representative differential interference contrast (DIC) images of hermaphrodite germ lines, all at the same stage (48 h after L4). Orange dashed lines outline visible oocytes for each strain. (A) Wild type; (B) larp-1(0); (C) larp-1(0), let-60(rf); and (D) larp-1(0), let-60(gf). (E) Table quantitating the number of visible oocytes in a single proximal arm of animals with the given genotype. Oocyte number per gonad arm was scored at 24 h and at 48 h after the L4 stage. Each number is an average, plus or minus standard deviation, with the total number of gonad arms counted in parentheses. Symbols, where indicated, represent statistical significance with a probability less than 0.01. (F) Table quantitating percentage of sterile animals (left) and percentage of animals with a pachytene arrest phenotype (right). Total number of animals scored (n) shown in parentheses. mpk-1(ts) and mpk-1(ts) larp-1(RNAi) strains were grown at 25°C. All other strains were grown at 20°C.
LARP-1 attenuates Ras-MAPK signaling during oogenesis
The larp-1 oogenesis defects resemble those associated with hyperactive Ras-MAPK signaling; aberrantly increased MAPK activity leads to multiple rows of smaller than normal oocytes in the proximal gonad arm (Hajnal and Berset 2002; Lee et al. 2007b). We hypothesized that LARP-1 might promote normal oogenesis by attenuating Ras-MAPK signaling. Thus, we predicted that the larp-1(0) oocyte defect would be suppressed by lowering Ras-MAPK signaling and enhanced by increasing Ras-MAPK signaling. To test these predictions, we examined double mutants with larp-1(0) and either a reduction-of-function (rf) or a gain-of-function (gf) mutation in the let-60 gene, which encodes the single C. elegans Ras protein (Beitel et al. 1990). The weak let-60(n2021rf) mutation suppressed larp-1(0) oocyte defects (Fig. 3, cf. C and B). Thus, larp-1(0); let-60(rf) germ lines had significantly fewer oocytes than larp-1(0) single mutants (Fig. 3E). In contrast, let-60(n1046gf) enhanced larp-1(0): oocyte number was significantly increased in larp-1(0); let-60(gf) adults compared with either larp-1(0) or let-60(gf) single mutants (Fig. 3, cf. D and B; Fig. 3E). We conclude that hyperactive Ras-MAPK signaling is critical for the larp-1(0) oogenesis defects.
We designed our next set of experiments to test the prediction that removal of larp-1 would suppress the sterility of reduced Ras-MAPK signaling. [We note for these experiments that larp-1(0) does not compromise RNAi efficacy and that larp-1 RNAi affects oogenesis with the same severity as the larp-1(0) mutant (Fig. 3E; data not shown)]. The major C. elegans MAPK, MPK-1, acts downstream of LET-60 in the Ras-MAPK signaling cascade and controls multiple steps of oocyte development, including progression through the pachytene phase of meiosis (Sundaram 2006; Lee et al. 2007b). Strong mpk-1 loss-of-function mutations result in pachytene arrest and render animals sterile (Church et al. 1995; Hsu et al. 2002; Ohmachi et al. 2002). larp-1(RNAi) did not suppress the pachytene arrest of the mpk-1(ga117) null mutant (Fig. 3F), suggesting that larp-1 depletion does not bypass the mpk-1 requirement in oogenesis. However, larp-1 depletion partially suppressed germline defects of reduced MPK-1: mpk-1(RNAi) normally results in 35% sterile animals, but only 8% of mpk-1(RNAi); larp-1(0) animals are sterile (Fig. 3F); mpk-1(ga111ts) is 100% sterile at 25°C (Lackner and Kim 1998) but only 81% sterile after larp-1 RNAi (Fig. 3F); and all mpk-1(oz140rf) germ lines arrest in pachytene at 20°C (Church et al. 1995; Lackner and Kim 1998), but only 17% do so after larp-1 RNAi (Fig. 3F). We conclude that LARP-1 works upstream of, or in parallel to, mpk-1 to attenuate Ras-MAPK signaling during oogenesis.
LARP-1 lowers mpk-1 mRNA and protein levels in the wild-type germ line
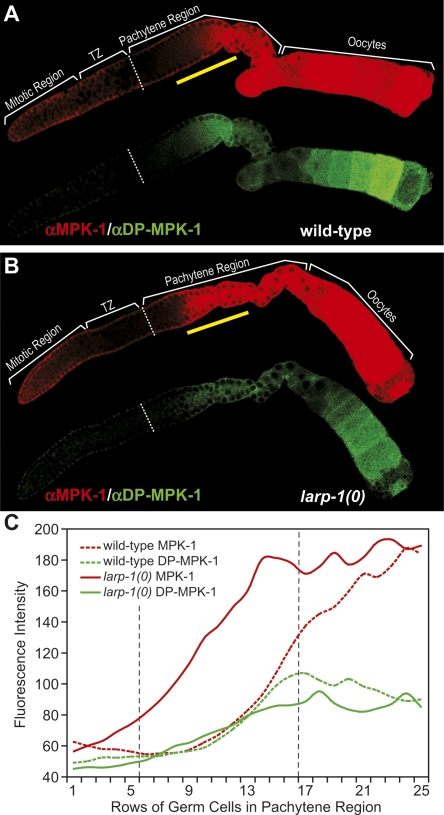
To ask whether LARP-1 normally lowers mpk-1 expression, we compared the abundance of MPK-1 protein and mpk-1 mRNA in wild-type and larp-1(0) germ lines (Figs. 4, 5). To visualize MPK-1 protein, we used the rabbit polyclonal Erk1 (K23) antibody, which detects C. elegans MPK-1 protein (Lackner and Kim 1998; Lee et al. 2007b). Compared with levels in wild-type germ lines (Fig. 4A), MPK-1 was significantly elevated in the larp-1(0) germ line (Fig. 4B), particularly within the mid-pachytene region (Fig. 4A,B, region above the yellow bar) where average MPK-1 levels were twofold higher in larp-1(0) relative to wild type (Fig. 4C, cf. red solid and red dotted lines). We conclude that LARP-1 reduces the expression of MPK-1 in midpachytene germ cells.
FIGURE 4.
LARP-1 lowers abundance of total MPK-1 protein in the pachytene region. (A, B) Dissected germ lines were costained with αErk1 (red), which recognizes C. elegans MPK-1 (Lackner and Kim 1998), and αDP-ERK (green), which recognizes the activated form of MPK-1 (Miller et al. 2001). Germ lines were treated identically and confocal images taken with the same settings at the same magnification. Wild-type (A) and larp-1(0) (B) adult hermaphrodite germ lines, both 24 h after L4. (C) Quantitation of total MPK-1 protein (red lines) and activated MPK-1 (green lines) abundance in wild-type (broken lines) or larp-1(0) (solid lines) pachytene regions. x-axis indicates distance from the transition zone/pachytene region boundary (white broken line in A and B), measured in number of germ cell diameters along the distal to proximal axis of the gonad; y-axis shows average pixel intensity at each germ cell position, averaged across 6 germ lines for wild type and 4 germ lines for larp-1(0). Total MPK-1 abundance is significantly (P < 0.001) higher in larp-1(0) (121 ± 36) than in wild type (66 ± 13) between rows 6–16 of the pachytene region. This region of higher MPK-1 is marked in A and B by a yellow bar, and its boundaries are indicated in C by vertical black dotted lines.
FIGURE 5.
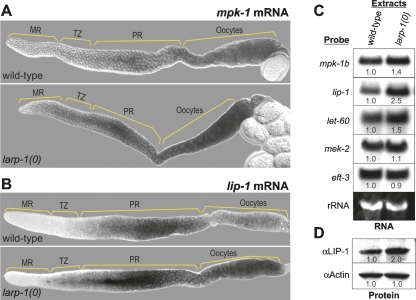
LARP-1 lowers mRNA abundance of selected mRNAs. (A, B) Wild-type (top) or larp-1(0) (bottom) dissected germ lines were examined by in situ hybridization for mpk-1 mRNA (A) or lip-1 mRNA (B). (C) Northern blots of poly(A) RNA from wild-type and larp-1(0) extracts using RNA-specific probes as indicated at left. Ethidium bromide staining of 18S ribosomal RNA is shown at bottom as a loading control. (D) Western blots of protein prepared from wild-type animals (left) and larp-1(0) mutants (right). Actin antibodies were used as a loading control. In C and D, numbers below bands indicate relative abundance in larp-1(0) compared with wild type, with wild-type abundance normalized to 1.0.
To visualize mpk-1 mRNA, we used both in situ hybridization and Northern blots. In both assays, mpk-1 mRNA was more abundant in larp-1(0) than in wild type (Fig. 5A,C). The mpk-1 locus makes two transcripts, mpk-1a and mpk-1b; the latter is the primary germline transcript (Lee et al. 2007a). The in situ probe detected both transcripts, but the Northern probe was specific for the mpk-1b transcript. By in situ hybridization, mpk-1 mRNA was particularly more abundant in the larp-1(0) pachytene region and developing oocytes (Fig. 5A). By Northern blot, mpk-1b was 1.4-fold more abundant in larp-1(0) mutants than in wild type (Fig. 5C, first row). We conclude that LARP-1 normally lowers mpk-1 mRNA and protein levels, at least in mid-pachytene cells, and perhaps more broadly.
LARP-1 lowers mRNA levels of multiple Ras-MAPK signaling components
We next investigated the LARP-1 effect on abundance of the activated form of MPK-1, which relies on dual phosphorylation by a signaling kinase cascade (Sternberg and Han 1998; Sundaram 2006). To this end, we used an antibody that specifically recognizes the dually phosphorylated (DP) form of MPK-1, which we refer to as DP-MPK-1 (Miller et al. 2001). Whereas total MPK-1 protein increased twofold in the middle of the larp-1(0) pachytene region (Fig. 4C, rows 6–16), the active DP-MPK-1 enzyme was not significantly increased in this region (i.e., the average αDP-MPK-1 fluorescence intensity is 66 ± 6 in larp-1(0) versus 58 ± 6 for wild type). However, in the proximal pachytene region (Fig. 4C, rows 13–25), DP-MPK-1 was lower in larp-1(0) than in wild type (i.e., the average αDP-MPK-1 fluorescence intensity is 86 ± 6 in larp-1(0) versus 96 ± 8 for wild type, P < 0.01), a surprising reduction given that total MPK-1 (active plus inactive) was elevated in the same region (i.e., the average MPK-1 fluorescence intensity is 181 ± 9 in larp-1(0) versus 145 ± 35 for wild type, P < 0.01).
A possible explanation is that an inhibitor of MPK-1 activity might also be increased in the larp-1(0) germ line. One such inhibitor is LIP-1, a C. elegans dual specificity phosphatase homolog, which inhibits MPK-1 activity in proximal pachytene nuclei and developing oocytes (Hajnal and Berset 2002). We asked if LARP-1 affects lip-1 expression. By in situ hybridization, lip-1 mRNA appeared more abundant and distributed more broadly in larp-1(0) than in wild-type germ lines (Fig. 5B). Moreover, by Northern blot, lip-1 mRNA was 2.5-fold more abundant (Fig. 5C, second row), and by Western blot, LIP-1 protein was twofold more abundant in larp-1 mutants than in wild type (Fig. 5D). Loading controls were equal for both RNA (i.e., rRNA) and protein blots (i.e., αActin) (Fig. 5C,D). We suggest that increased LIP-1 may lower the activity of MPK-1 and thereby ameliorate the biological consequences of increased MPK-1 expression in the larp-1(0) germ line.
To ask if mpk-1 and lip-1 are the only mRNAs affected by removal of LARP-1, we assayed three other mRNAs, two encoding Ras-MAPK pathway components (i.e., let-60 and mek-2) and one unrelated to the pathway (i.e., elongation factor 3 [eft-3]). By Northern blot, let-60 mRNA was increased (1.5-fold) in larp-1(0) relative to wild type, but the abundance of mek-2 and eft-3 mRNAs (1.1- and 0.9-fold difference, respectively) was not changed in larp-1(0) (Fig. 5C). Thus, LARP-1 specifically affects the abundance of certain mRNAs; it lowers the abundance of three mRNAs encoding MAPK pathway components (i.e., mpk-1, let-60, and lip-1) but does not affect the abundance of all mRNAs, either in the Ras-MAPK pathway or more broadly.
LARP-1 protein is cytoplasmic and punctate but does not localize in P granules
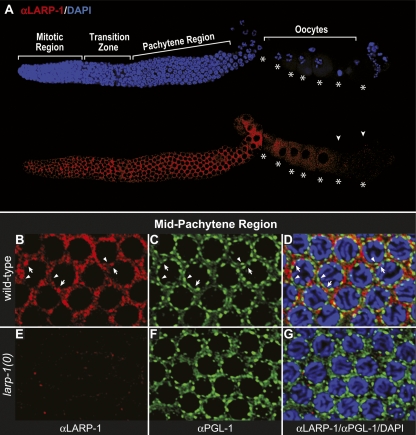
To learn the subcellular location of LARP-1, we stained wild-type and larp-1(0) gonads with affinity-purified LARP-1 antibody (αRt55; see Fig. 2A). LARP-1 expression extended throughout the germ line—from the distal tip into developing oocytes, with a pronounced decline in the most proximal oocytes (Fig. 6A). Moreover, LARP-1 was primarily cytoplasmic (Fig. 6A,B) and concentrated in discrete, irregular-shaped puncta around nuclei (Fig. 6B). LARP-1 puncta were missing in larp-1(0) germ lines, confirming specificity of the LARP-1 antibody (Fig. 6E), and were not detected in the rachis (data not shown).
FIGURE 6.
LARP-1 protein accumulates in cytoplasmic granules but does not colocalize with the PGL-1 marker of P granules. (A) Dissected wild-type germ line costained with αLARP-1 (red) and DAPI (blue). Oocytes are indicated by asterisks. LARP-1 is found throughout the germline cytoplasm of the distal arm but tapers off in maturing oocytes (arrowheads). (B–D) Wild-type germ line costained with αLARP-1 (red) (B, D), αPGL-1 (green) (C, D), and DAPI (blue) (D). (D) The merged image. Confocal images were taken of the mid-pachytene region. Arrows point to LARP-1 perinuclear foci; arrowheads point to PGL-1 perinuclear foci. Virtually no yellow is observed in the merged image. (E–G) larp-1(0) mutant pachytene region treated identically as in B–D and confocal images taken with the same settings at the same magnification.
The LARP-1 puncta were reminiscent of RNA-rich cytoplasmic “germ granules,” also known as P granules in C. elegans (for review, see Strome 2005). To ask if LARP-1 associates with P granules, we costained wild-type germ lines with αLARP-1 and αPGL-1, an antibody that marks P granules (Kawasaki et al. 1998). However, virtually no overlap was seen (Fig. 6B–D), and P granules appeared normal in larp-1 mutant germ lines (Fig. 6F,G). We conclude that LARP-1 is predominately cytoplasmic and enriched in discrete puncta that do not contain PGL-1.
LARP-1 is enriched in P bodies
A major mode of eukaryotic mRNA turnover occurs via three distinct steps: deadenylation, decapping, and 5′ to 3′ degradation (Parker and Song 2004). The enzymes responsible for these events accumulate in discrete cytoplasmic “processing bodies” or P bodies and constitute a “general repression/decay” complex (for review, see Parker and Sheth 2007). In yeast and mammals, the general repression/decay complex consists of the major cytoplasmic deadenylase Ccr4/Pop2/NOT complex, decapping enzyme subunits Dcp1 and Dcp2, decapping activators Dhh1/RCK and Lsm1-7, and the exonuclease Xrn1p (Sheth and Parker 2003; Cougot et al. 2004; Muhlrad and Parker 2005; Barbee et al. 2006; Yang et al. 2006). In C. elegans, decapping enzyme homologs DCAP-1 and DCAP-2 localize to discrete cytoplasmic foci, which are probably analogous to yeast P bodies (Ding et al. 2005; Lall et al. 2005; Squirrell et al. 2006).
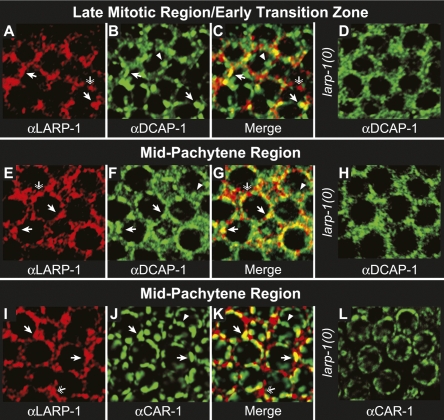
To test whether LARP-1 accumulates in P bodies, we costained wild-type and larp-1(0) germ lines with αLARP-1 and a polyclonal antibody directed against DCAP-1. Consistent with the staining of DCAP-1 in the embryo (Squirrell et al. 2006), DCAP-1 was predominately cytoplasmic and accumulated in irregularly shaped puncta. DCAP-1 was most abundant in the mitotic region, midpachytene region, and distal oocytes, and least abundant in the transition zone, early pachytene region, and most proximal oocytes (Supplemental Fig. 2A). LARP-1 and DCAP-1 overlapped throughout the distal germ line and colocalized in perinuclear foci (Fig. 7A–C; Supplemental Fig. 2). However, not all DCAP-1 foci stained positively for αLARP-1 and not all LARP-1 foci were positive for αDCAP-1 (Fig. 7C). These results indicate that LARP-1/DCAP-1 foci define a unique subset of P bodies in the germ line.
FIGURE 7.
LARP-1 colocalizes with P body proteins in the distal germ line. Wild-type (A–C, E–G) and larp-1(0) (D, H) germ lines costained with αLARP-1 (red) and αDCAP-1 (green). Confocal images were taken of the late mitotic region/early transition zone (A–D) and mid-pachytene region (E–H). Arrows point to foci that stain with both αLARP-1 and αDCAP-1, which are yellow in the merged images. Arrowheads point to foci specific for αDCAP-1 staining; double arrows point to foci specific for αLARP-1 staining. Wild-type (I–K) and larp-1(0) (L) germ lines costained with αLARP-1 (red) and αCAR-1 (green). Arrows point to foci that stain with both αLARP-1 and αCAR-1, which are yellow in the merged image. Arrowheads point to foci specific for αCAR-1 staining; double arrows point to foci specific for αLARP-1 staining. In all cases, wild-type and larp-1(0) germ lines were treated identically and confocal images taken with the same settings at the same magnification.
We also costained germ lines for LARP-1 and CAR-1, an Scd6 homolog (Albrecht and Lengauer 2004; Anantharaman and Aravind 2004). Scd6 homologs are thought to act as translational repressors that reside in a variety of RNA regulatory particles, including P bodies, P granules, Drosophila neuronal granules, and mammalian stress granules (Lieb et al. 1998; Audhya et al. 2005; Boag et al. 2005; Wilhelm et al. 2005; Barbee et al. 2006; Decker and Parker 2006; Squirrell et al. 2006; Yang et al. 2006). In C. elegans, primarily two types of CAR-1 foci exist: those with P body proteins, such as DCAP-1, and those with P granule protein PGL-1 (Lieb et al. 1998; Audhya et al. 2005; Boag et al. 2005; Wilhelm et al. 2005; Barbee et al. 2006; Decker and Parker 2006; Squirrell et al. 2006; Yang et al. 2006). We observed significant overlap of LARP-1 and CAR-1 in the mid-pachytene region (see Supplemental Fig. 2B). Since LARP-1 is excluded from P granules (Fig. 6), we reasoned that the LARP-1/CAR-1 foci were P bodies. Moreover, as with DCAP-1 and LARP-1 colocalization, only a subset of CAR-1 and LARP-1 foci colocalized (Fig. 7I–K). These results confirm that LARP-1 accumulates in a subset of germline P bodies. Intriguingly, in larp-1(0) mutant germ lines, the DCAP-1 and CAR-1 foci were less prominent and distinct (Fig. 7D,H,L). The size and number of P bodies depends on the amount of mRNA available for decapping. When mRNA decay is blocked prior to the decapping step, such as during deadenylation, P bodies fail to form (Sheth and Parker 2003). Alternatively, P bodies grow in size and number when mRNA specific binding proteins, or miRNAs, actively target specific mRNAs to them for decapping and degradation (Liu et al. 2005; Franks and Lykke-Andersen 2007). One simple possibility is that LARP-1 normally localizes to P bodies and enhances their formation by promoting degradation of specific mRNAs, such as mpk-1 and lip-1.
DISCUSSION
In this work, we show that LARP-1 is a likely RNA-binding protein that reduces the levels of Ras-MAPK mRNAs and promotes oogenesis. Moreover, LARP-1 colocalizes in cytoplasmic granules with two P-body components. The following discussion places this work in context with previous Larp studies and proposes a mechanism for how Larp1 family members may affect cellular functions and development.
LARP-1, a conserved La-related protein, harbors two RNA-binding domains
Previous studies placed La motif-containing proteins into two broad categories, the authentic La proteins and the La-related proteins, or Larps (Sobel and Wolin 1999; Wolin and Cedervall 2002). We show that Larps themselves fall into at least two major families, a more widely conserved Larp1 family and a metazoan-specific Larp5 family. The C. elegans genome encodes a single Larp1 family member, LARP-1, and a single Larp5 family member, LARP-5. This work focuses on LARP-1, as a representative of the Larp1 family. C. elegans LARP-1 possesses a central La motif and a C-terminal ∼200 amino acid sequence that is strongly conserved among most Larp1 family members. We dub this sequence the LARP1 domain (this paper). Within the LARP1 domain is a repeated sequence, the DM15 tandem repeat, which had been previously noted (Marchler-Bauer et al. 2007); the significance of the DM15 repeat is not known. The RNA-binding characteristics and structural properties of the La motif have been studied extensively for authentic La proteins (Ohndorf et al. 2001; Alfano et al. 2004; Dong et al. 2004; Teplova et al. 2006). As expected, the full-length LARP-1 protein binds RNA, and an N-terminal LARP-1 fragment harboring its La motif also binds RNA homopolymers in vitro. Surprisingly, a C-terminal LARP-1 fragment, which lacks the La motif but harbors the LARP1 domain, can also bind RNA homopolymers in vitro. Therefore, LARP-1 likely possesses two RNA-binding domains, the La motif and the LARP1 domain. This LARP1 domain is present in most Larp1 family members, including those from Dictyostelium, monocots, dicots, and animals. Exceptions include the yeast Larps, which cluster with LARP-1 based on their La motif sequences. A challenge for the future is to dissect the individual and cooperative functions of the La motif and LARP1 domain. One possibility is that the La motif and LARP1 domain cooperate to enhance RNA-binding specificity of LARP-1 for its natural substrate. This idea is based on our in vitro RNA-binding studies (this paper) and the well-established cooperative RNA-binding by the La motif and RRM of authentic La proteins (Ohndorf et al. 2001; Alfano et al. 2004; Teplova et al. 2006). However, alternative scenarios are also possible. The LARP1 domain could be critical for the overall tertiary structure of LARP-1 or the LARP1 domain could interact with other proteins. For example, LARP-1 accumulates in P bodies (see below), and the LARP1 domain could have a conserved role in that association.
C. elegans LARP-1 attenuates the abundance of specific mRNAs that control oogenesis
Two previous studies suggested that Larps influence metazoan development (Chauvet et al. 2000; Ichihara et al. 2007). Both focused on Drosophila Larp, which like C. elegans LARP-1, is in the Larp1 family. Chauvet and colleagues (2000) found that the larp gene is a target of homeobox transcription factors during embryogenesis, and Ichihara and colleagues (2007) reported that larp is required for male meiosis. However, neither study addressed how larp achieves its effect on development.
In this study, we show that LARP-1 affects C. elegans oogenesis by attenuating the abundance of mRNAs in the Ras-MAPK pathway. Three lines of evidence support this idea. First, oocyte development is abnormal in larp-1 null mutants in a manner reminiscent of mutants with hyperactive Ras-MAPK signaling. Second, larp-1 interacts genetically with genes in the Ras-MAPK pathway as predicted. Third, the abundance of three mRNAs in the Ras-MAPK pathway (let-60/Ras, mpk-1/MAPK, and lip-1/MKP) is increased in larp-1 null mutants relative to wild type. Importantly, LARP-1 does not attenuate all germline mRNAs. Therefore, LARP-1 functions selectively to control mRNA abundance.
Although LARP-1 controls the Ras-MAPK pathway in the germ line, it does not appear to have a similar effect in the vulva (K. Nykamp, unpubl.). We do not yet understand why LARP-1 has tissue-specific effects on Ras-MAPK signaling, since larp-1 mRNA is expressed in both germline and somatic tissues (Reinke et al. 2000; McKay et al. 2003; this paper). One simple explanation might be that LARP-1 protein is not expressed in the vulva. Alternatively, LARP-1 may work with germline-specific factors to attenuate Ras-MAPK signaling. Regardless, the role of LARP-1 in the germ line can provide a model for analyzing its effect on development more broadly. Based on our findings with C. elegans LARP-1, we suggest that the Larp1 family of proteins may function to control the expression of key mRNAs during development.
LARP-1 function may be linked to P bodies
The germline cytoplasm contains many perinuclear and cytoplasmic granules. Some granules harbor PGL-1 and have been termed P granules or germ granules (Kawasaki et al. 1998); some harbor components of P bodies (Audhya et al. 2005; Boag et al. 2005), which in yeast and mammals are sites of mRNA turnover (Parker and Sheth 2007). LARP-1 protein does not colocalize with PGL-1 but does colocalize with two P body markers: DCAP-1, a predicted decapping enzyme (Cohen et al. 2005; Lall et al. 2005), and CAR-1, an RNA-binding protein and likely translational repressor (for review, see Decker and Parker 2006). The colocalization of LARP-1 with P body proteins is intriguing given our finding that the levels of specific mRNAs and proteins are reduced by LARP-1.
The most prominent colocalization of DCAP-1 and LARP-1 is in the late mitotic region and early transition zone of the germ line, an area where mpk-1 and lip-1 mRNAs are normally not detected (Lee et al. 2006, 2007a). Yet, in larp-1(0) germ lines, both mpk-1 and lip-1 mRNAs are seen in the mitotic region. Cleavage of the 5′ cap (i.e., decapping) is a critical step in mRNA repression: it permits 5′ to 3′ exonucleolytic digestion and blocks translational initiation (Coller and Parker 2004, 2005). In yeast and mammals, the Dcp1/Dcp2 heterodimer executes decapping (Lykke-Andersen 2002; Steiger et al. 2003) in P bodies (Sheth and Parker 2003). In C. elegans, the likely decapping enzyme is DCAP-1/DCAP-2 (Cohen et al. 2005; Lall et al. 2005), which colocalizes in granules with conserved P body components (Ding et al. 2005; Lall et al. 2005; Squirrell et al. 2006). An intriguing model is that LARP-1 accumulation with DCAP-1 in P bodies may be critical for the degradation of mpk-1 and lip-1 mRNAs.
Larps are widely conserved but poorly understood, especially in multicellular organisms. This study of C. elegans LARP-1 uncovers characteristics that may be applicable to Larp1 family members more generally: LARP-1 possesses two RNA-binding domains, it controls the abundance of selected mRNAs, and it is associated with cytoplasmic granules specialized for mRNA turnover and mRNA repression. But of course, these results raise many additional questions. How does LARP-1 control mRNA abundance? What is the role of LARP-1 in P bodies? Is the LARP-1 control of Ras-MAPK signaling conserved? Answering these questions is beyond the scope of this work. However, we suggest that their analysis in the C. elegans germ line will shed light on conserved functions of Larps.
MATERIALS AND METHODS
Nematode strains and RNAi
All strains were maintained at 20°C as described (Brenner 1974), unless noted otherwise. We used the wild-type Bristol N2 strain and the following mutants: LGIII: glp-1(q224ts) (Austin and Kimble 1987), larp-1(q783) null (this paper), mpk-1(ga111ts) (Lackner and Kim 1998), mpk-1(ga117) null (Lackner et al. 1994), mpk-1(oz140rf) (Church et al. 1995); the mpk-1(ga117, oz140) mutations were balanced by qC1 (Austin and Kimble 1989); LGIV: let-60(n1046gf, n2021rf) (Beitel et al. 1990). The larp-1(q783) deletion was generated as described (Kraemer et al. 1999) and outcrossed seven times. The larp-1(q783) deletion breakpoints are at genomic positions 5,017,363/5,017,364 and 5,016,193/5,016,194 (1170 bp). The mpk-1(ts) strain was maintained at 20°C and examined for sterility after shifting to 25°C. RNAi was performed by feeding L1s bacteria expressing double-stranded RNAs corresponding to the gene of interest (Kamath et al. 2003). Fertility, pachytene arrest, average number of oocytes, and embryonic lethality were all scored by standard methods. Unless stated otherwise, phenotype scoring was performed at 24 h after mid-L4 stage.
RNA-binding assay
Full-length larp-1 was amplified from a mixed-stage cDNA library, λACT-RB2 (gift from Robert Barstead, Oklahoma Medical Research Foundation), using oligos KN158 (5′-GGATCCGCCGAGAAGCAGCCCATGC-3′) and KN166 (5′-GAATTCATCTTACTTCTTGGTTGATTGTAGAAG-3′). Full-length larp-1 cDNA was sequenced, and intron/exon boundaries are shown in Figure 2A. The N-terminal and C-terminal larp-1 fragments were generated using KN158/KN173 (5′-GAATTCTTCTTCATTGAACTCTCCTTGATTTTG-3′) and KN172 (5′-GGATCCGTTGAAATTGGATTGCGCAGATACG-3′)/KN166 primer pairs, respectively. S. cerevisiae LHP1 and CAK1 were amplified from genomic DNA (gift from Craig Stumpf, Wickens Laboratory, University of Wisconsin–Madison) using primer pairs KN183 (5′-GGATCCCCACAACAAGAGGAGCAAGAGAAACC-3′)/KN184 (5′-CTCGAGGAATCACTCCTTGTGCTCCTCAGCG-3′) and KN185 (5′-GGATCCCTGGATAGTATAGACATTACACACTG-3′)/KN186 (5′-GAATTCTTATGGCTTTTCTAATTCTTGCAAGATTC-3′), respectively. Fragments were digested with BamHI and EcoRI (or BamHI/XhoI for LHP1) and cloned into pCITE4a (Novagen).
Coupled transcription/translation (TNT T7 Quick Coupled System, Promega) reactions with each pCITE4a expression plasmid were performed separately in the presence of [35S]methionine as instructed by the manufacturer. Similar amounts of LARP-1, Cak1p, and Lhp1p proteins were combined and incubated for 10 min at 4°C in binding buffer (10 mM Tris-Cl at pH 7.4, 2.5 mM MgCl2, 100 mM NaCl, 0.5% Triton-X 100) with 50 μg of the indicated homopolymer immobilized on sepharose [GE Healthcare, poly(U) and poly(A)] or agarose [Sigma, poly(C) and poly(G)]. Separate binding reactions with poly(U) and poly(G) were performed for LARP-1(FL), LARP-1(N), and LARP-1(C) fragments in binding buffer (10 mM Tris-Cl at pH 7.4, 2.5 mM MgCl2, 0.5% Triton-X 100) plus the indicated NaCl concentration for 10 min at 4°C. In all cases, beads were washed four times, and protein was eluted by boiling beads in Laemmli cocktail (10 mM Tris-HCl at pH 6.8, 4% SDS, 20% glycerol, 1.3 M β-mercaptoethanol, 4 mg/mL Bromphenol blue). Eluted proteins were separated on 4%–12% gradient gels (Cambrex). The gels were exposed to a phosphor screen (Molecular Dynamics) for 2–4 h and analyzed using a Typhoon Scanner (GE Healthcare).
Northern blots
Total RNA was extracted from staged adults (24 h after L4) by Trizol Reagent (Invitrogen) and enriched for poly(A) RNA using the poly(A) Purist Kit (Ambion). For each lane, 5 μg of poly(A) RNA was run on a 1% agarose gel under denaturing conditions, transferred, and UV-crosslinked to a positively charged nylon membrane (BrightStar-Plus, Ambion). In vitro-transcribed 32P-radiolabeled antisense RNA probes were prepared for each mRNA indicated as instructed by the Strip-EZ RNA T7 kit (Ambion). Hybridization and washes were carried out at 68°C using NorthernMax (Ambion) reagents. Hybridized membranes were exposed for 2 h to a phosphor screen (Molecular Dynamics). We analyzed screens using a Typhoon Scanner (GE Healthcare) and quantitated bands with ImageQuant Software (Molecular Dynamics).
Antibody generation and protein analyses
To generate LARP-1-specific antibodies, rats were injected with synthetic keyhole-limpit-hemocyanin (KLH)-conjugated peptides (Genemed Synthesis, Inc.) corresponding to the N terminus (amino acids 228–242, αRt55) or C terminus (amino acids 971–989, αRt60). Whole worm lysates were prepared from staged adults (24 h after mid-L4 stage) by grinding worms (∼500,000) suspended in HB(A) buffer (50 mM HEPES at pH 7.6, 10 mM KCL, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM EGTA, 1× Halt Protease Inhibitor [Pierce] cocktail) with a mortar and pestle in liquid N2. Lysates were homogenized (Wheaton Dounce Tissue Grinder, Fisher Scientific) and cleared of cell debris by low-speed centrifugation (3000g). Extracts were stored at −80°C in HB(D) buffer (50 mM HEPES at pH 7.6, 10 mM KCL, 1.5 mM MgCl2, 100 mM NaCl, 0.2 mM EDTA, 20% glycerol, 1 mM DTT). For Western analysis, 10 μg of total protein per lane was separated on 4%–12% gradient gels (Cambrex) and the following antibodies were used: αRt55 serum (1:500 dilution), αRt60 serum (1:500 dilution), αLIP-1 (Lee et al. 2006; 1:500 dilution), and anti-actin C4 (MP Biomedicals; 1:40,000 dilution). Quantitation of protein bands was done using a Gel Logic 100 Imaging System (Kodak).
In situ mRNA hybridization and immunofluorescence
In situ hybridization was performed as described (Lee et al. 2007a) using antisense digoxigenin-labeled DNA probes for lip-1 (cDNA nucleotides 42–559) and mpk-1 (cDNA nucleotides 246–748). For antibody staining, adult germ lines were extruded, fixed with 1.5% paraformaldehyde for 2 h at room temperature followed by methanol for 5 min at −20°C, and then treated with 0.1% Triton-X for 5 min at room temperature. After blocking for 1 h with 0.5% BSA in PBS, fixed germ lines were incubated overnight at room temperature with primary antibodies followed by 2 h at 4°C with secondary antibodies. LARP-1 was detected using purified αRt55 (1:25 dilution) and Cy3-labeled anti-Rat secondary (Jackson ImmunoResearch, 1:400 dilution). αCAR-1 (gift from J. Squirrell and J. White, University of Wisconsin–Madison; 1:50 dilution) and αDCAP-1 (gift from P. Anderson, University of Wisconsin–Madison; 1:1500 dilution) primary antibodies were used as described (Squirrell et al. 2006) and detected by Alexa Fluor 488-labeled anti-rabbit secondary (Molecular Probes, 1:200 dilution). DAPI was used to visualize DNA. MPK-1 and DP-MPK-1 levels were analyzed using rabbit polyclonal ERK1 (K23) (Santa Cruz Biotechnology, 1:400 dilution) and mouse monoclonal Anti-DP-ERK (Sigma-Aldrich, 1:100 dilution) antibodies, respectively. Secondary antibodies used were Cy3-labeled anti-rabbit (Jackson ImmunoResearch, 1:400 dilution) and Alexa Fluor 488-labeled anti-mouse (Molecular Probes, 1:200 dilution). Images were captured using a Zeiss LSM510 Meta confocal microscope and processed using ImageJ and Adobe Photoshop CS2. Anti-ERK1 (K23) and Anti-DP-ERK signals were quantitated in ImageJ by obtaining the average pixel intensity for each row of germ cells and then averaging this value among the number of germ lines indicated. In all cases, wild-type and mutant germ lines were treated identically.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Laura Vanderploeg for help with figure preparation and members of the Kimble Laboratory for their comments and suggestions. This work was supported by NIH grant GM069454 to J.K.; J.K. is an investigator at the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1066008.
REFERENCES
- Aigner S., Cech T.R. The Euplotes telomerase subunit p43 stimulates enzymatic activity and processivity in vitro. RNA. 2004;10:1108–1118. doi: 10.1261/rna.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner S., Lingner J., Goodrich K.J., Grosshans C.A., Shevchenko A., Mann M., Cech T.R. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 2000;19:6230–6239. doi: 10.1093/emboj/19.22.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner S., Postberg J., Lipps H.J., Cech T.R. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry. 2003;42:5736–5747. doi: 10.1021/bi034121y. [DOI] [PubMed] [Google Scholar]
- Albrecht M., Lengauer T. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 2004;569:18–26. doi: 10.1016/j.febslet.2004.03.126. [DOI] [PubMed] [Google Scholar]
- Alfano C., Sanfelice D., Babon J., Kelly G., Jacks A., Curry S., Conte M.R. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004;11:323–329. doi: 10.1038/nsmb747. [DOI] [PubMed] [Google Scholar]
- Anantharaman V., Aravind L. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC Genomics. 2004;5:45. doi: 10.1186/1471-2164-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., Hyndman F., McLeod I.X., Maddox A.S., Yates J.R., 3rd, Desai A., Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans . J. Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J., Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Austin J., Kimble J. Transcript analysis of glp-1 and lin-12, homologous genes required for cell interactions during development of C. elegans. Cell. 1989;58:565–571. doi: 10.1016/0092-8674(89)90437-6. [DOI] [PubMed] [Google Scholar]
- Barbee S.A., Estes P.S., Cziko A.M., Hillebrand J., Luedeman R.A., Coller J.M., Johnson N., Howlett I.C., Geng C., Ueda R., et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel G.J., Clark S.G., Horvitz H.R. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature. 1990;348:503–509. doi: 10.1038/348503a0. [DOI] [PubMed] [Google Scholar]
- Boag P.R., Nakamura A., Blackwell T.K. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans . Development. 2005;132:4975–4986. doi: 10.1242/dev.02060. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chauvet S., Maurel-Zaffran C., Miassod R., Jullien N., Pradel J., Aragnol D. dlarp, a new candidate Hox target in Drosophila whose orthologue in mouse is expressed at sites of epithelium/mesenchymal interactions. Dev. Dyn. 2000;218:401–413. doi: 10.1002/1097-0177(200007)218:3<401::AID-DVDY1009>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Church D.L., Guan K.-L., Lambie E.J. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans . Development. 1995;121:2525–2535. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- Cohen L.S., Mikhli C., Jiao X., Kiledjian M., Kunkel G., Davis R.E. Dcp2 Decaps m2,2,7GpppN-capped RNAs, and its activity is sequence and context dependent. Mol. Cell. Biol. 2005;25:8779–8791. doi: 10.1128/MCB.25.20.8779-8791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrove-Otero L.J., Minshall N., Standart N. RNA-binding proteins in early development. Crit. Rev. Biochem. Mol. Biol. 2005;40:21–73. doi: 10.1080/10409230590918612. [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Parker R. CAR-1 and Trailer hitch: Driving mRNP granule function at the ER? J. Cell Biol. 2006;173:159–163. doi: 10.1083/jcb.200601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans . Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Dong G., Chakshusmathi G., Wolin S.L., Reinisch K. Structure of the La motif: A winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 2004;23:1000–1007. doi: 10.1038/sj.emboj.7600115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T.M., Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes & Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Echeverri C., Oegema K., Coulson A., Jones S.J.M., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Hajnal A., Berset T. The C. elegans MAPK phosphatase LIP-1 is required for the G2/M meiotic arrest of developing oocytes. EMBO J. 2002;21:4317–4326. doi: 10.1093/emboj/cdf430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N., Jahchan N.S., Hong E., Li Q., Bayfield M.A., Maraia R.J., Luo K., Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu V., Zobel C.L., Lambie E.J., Schedl T., Kornfeld K. Caenorhabditis elegans lin-45 raf is essential for larval viability, fertility and the induction of vulval cell fates. Genetics. 2002;160:481–492. doi: 10.1093/genetics/160.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara K., Shimizu H., Taguchi O., Yamaguchi M., Inoue Y. A Drosophila orthologue of larp protein family is required for multiple processes in male meiosis. Cell Struct. Funct. 2007;32:89–100. doi: 10.1247/csf.07027. [DOI] [PubMed] [Google Scholar]
- Inada M., Guthrie C. Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc. Natl. Acad. Sci. 2004;101:434–439. doi: 10.1073/pnas.0307425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M., Toh-e A., Matsui Y. SRO9, a multicopy suppressor of the bud growth defect in the Saccharomyces cerevisiae rho3-deficient cells, shows strong genetic interactions with tropomyosin genes, suggesting its role in organization of the actin cytoskeleton. Genetics. 1997;147:1003–1016. doi: 10.1093/genetics/147.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kawasaki I., Shim Y.H., Kirchner J., Kaminker J., Wood W.B., Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans . Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Kelly W.G., Xu S., Montgomery M.K., Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B., Crittenden S.L., Gallegos M., Moulder G., Barstead R., Kimble J., Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans . Curr. Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- Kufel J., Allmang C., Chanfreau G., Petfalski E., Lafontaine D.L., Tollervey D. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 2000;20:5415–5424. doi: 10.1128/mcb.20.15.5415-5424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M.R., Kim S.K. Genetic analysis of the Caenorhabditis elegans MAP kinase gene mpk-1 . Genetics. 1998;150:103–117. doi: 10.1093/genetics/150.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M.R., Kornfeld K., Miller L.M., Horvitz H.R., Kim S.K. A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans . Genes & Dev. 1994;8:160–173. doi: 10.1101/gad.8.2.160. [DOI] [PubMed] [Google Scholar]
- Lall S., Piano F., Davis R.E. Caenorhabditis elegans decapping proteins: Localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol. Biol. Cell. 2005;16:5880–5890. doi: 10.1091/mbc.E05-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Hook B., Lamont L.B., Wickens M., Kimble J. LIP-1 phosphatase controls the extent of germline proliferation in Caenorhabditis elegans . EMBO J. 2006;25:88–96. doi: 10.1038/sj.emboj.7600901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Hook B., Pan G., Kershner A.M., Merritt C., Seydoux G., Thomson J.A., Wickens M., Kimble J. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007a;3:e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Ohmachi M., Arur S., Nayak S., Francis R., Church D., Lambie E., Schedl T. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics. 2007b;177:2039–2062. doi: 10.1534/genetics.107.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M.R., Boyle J.A., Hardin J.A., Steitz J.A. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. 1981;211:400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- Lieb B., Carl M., Hock R., Gebauer D., Scheer U. Identification of a novel mRNA-associated protein in oocytes of Pleurodeles waltl and Xenopus laevis . Exp. Cell Res. 1998;245:272–281. doi: 10.1006/excr.1998.4249. [DOI] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti M.A., Tschudi C., Kwon H., Wolin S.L., Ullu E. Import of proteins into the trypanosome nucleus and their distribution at karyokinesis. J. Cell Sci. 2000;113:899–906. doi: 10.1242/jcs.113.5.899. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J.B., Derbyshire M.K., DeWeese-Scott C., Gonzales N.R., Gwadz M., Hao L., He S., Hurwitz D.I., Jackson J.D., et al. CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay S.J., Johnsen R., Khattra J., Asano J., Baillie D.L., Chan S., Dube N., Fang L., Goszczynski B., Ha E., et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans . Cold Spring Harb. Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- Miller M.A., Nguyen V.Q., Lee M.-H., Kosinski M., Schedl T., Caprioli R.M., Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Parker R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmachi M., Rocheleau C.E., Church D., Lambie E., Schedl T., Sundaram M.V. C. elegans ksr-1 and ksr-2 have both unique and redundant functions and are required for MPK-1 ERK phosphorylation. Curr. Biol. 2002;12:427–433. doi: 10.1016/s0960-9822(02)00690-5. [DOI] [PubMed] [Google Scholar]
- Ohndorf U.M., Steegborn C., Knijff R., Sondermann P. Contributions of the individual domains in human La protein to its RNA 3′-end binding activity. J. Biol. Chem. 2001;276:27188–27196. doi: 10.1074/jbc.M102891200. [DOI] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae . Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Prathapam R., Witkin K.L., O'Connor C.M., Collins K. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2005;12:252–257. doi: 10.1038/nsmb900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V., Smith H.E., Nance J., Wang J., Van Doren C., Begley R., Jones S.J., Davis E.B., Scherer S., Ward S., et al. A global profile of germline gene expression in C. elegans . Mol. Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Rinke J., Steitz J.A. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1982;29:149–159. doi: 10.1016/0092-8674(82)90099-x. [DOI] [PubMed] [Google Scholar]
- Rinke J., Steitz J.A. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 1985;13:2617–2629. doi: 10.1093/nar/13.7.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons F.H., Broers F.J., Van Venrooij W.J., Pruijn G.J. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp. Cell Res. 1996;224:224–236. doi: 10.1006/excr.1996.0132. [DOI] [PubMed] [Google Scholar]
- Sobel S.G., Wolin S.L. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol. Biol. Cell. 1999;10:3849–3862. doi: 10.1091/mbc.10.11.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B., Koski L.B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A.M., Artelt J., Bettencourt P., Cassin E., et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans . Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Squirrell J.M., Eggers Z.T., Luedke N., Saari B., Grimson A., Lyons G.E., Anderson P., White J.G. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol. Biol. Cell. 2006;17:336–344. doi: 10.1091/mbc.E05-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano J.E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- Steiger M., Carr-Schmid A., Schwartz D.C., Kiledjian M., Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P.W., Han M. Genetics of RAS signaling in C. elegans . Trends Genet. 1998;14:466–472. doi: 10.1016/s0168-9525(98)01592-3. [DOI] [PubMed] [Google Scholar]
- Strome S. Specification of the germ line. WormBook. 2005 doi: 10.1895/wormbook.1.9.1. www.wormbook.org (ed. The C. elegans Research Community). WormBook, [DOI] [PMC free article] [PubMed]
- Sundaram M.V. RTK/Ras/MAPK signaling. WormBook. 2006 doi: 10.1895/wormbook.1.80.1. www.wormbook.org (ed. The C. elegans Research Community). WormBook, [DOI]
- Swanson M.S., Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q., Li X., Sadhale P.P., Miyao T., Woychik N.A. Multiple mechanisms of suppression circumvent transcription defects in an RNA polymerase mutant. Mol. Cell. Biol. 2000;20:8124–8133. doi: 10.1128/mcb.20.21.8124-8133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M., Yuan Y.R., Phan A.T., Malinina L., Ilin S., Teplov A., Patel D.J. Structural basis for recognition and sequestration of UUUOH 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B., Wickens M., Kimble J. Translational control in development. In: Mathews M.B., et al., editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 507–544. [Google Scholar]
- Tsukada M., Gallwitz D. Isolation and characterization of SYS genes from yeast, multicopy suppressors of the functional loss of the transport GTPase Ypt6p. J. Cell Sci. 1996;109:2471–2481. doi: 10.1242/jcs.109.10.2471. [DOI] [PubMed] [Google Scholar]
- Wilhelm J.E., Buszczak M., Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila . Dev. Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Witkin K.L., Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes & Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin S.L., Cedervall T. The La Protein. Annu. Rev. Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- Yang W.H., Yu J.H., Gulick T., Bloch K.D., Bloch D.B. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.J., Wolin S.L. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: A yeast homolog of the La autoantigen is dispensable for growth. Mol. Cell. Biol. 1994;14:5412–5424. doi: 10.1128/mcb.14.8.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]