Abstract
Animal genomes contain hundreds of microRNAs (miRNAs), small regulatory RNAs that control gene expression by binding to complementary sites in target mRNAs. Some rules that govern miRNA/target interaction have been elucidated but their general applicability awaits further experimentation on a case-by-case basis. We use here an assay system in transgenic nematodes to analyze the interaction of the Caenorhabditis elegans lsy-6 miRNA with 3′ UTR sequences. In contrast to many previously described assay systems used to analyze miRNA/target interactions, our assay system operates within the cellular context in which lsy-6 normally functions, a single neuron in the nervous system of C. elegans. Through extensive mutational analysis, we define features in the known and experimentally validated target of lsy-6, the 3′ UTR of the cog-1 homeobox gene, that are required for a functional miRNA/target interaction. We describe that both in the context of the cog-1 3′ UTR and in the context of heterologous 3′ UTRs, one or more seed matches are not a reliable predictor for a functional miRNA/target interaction. We rather find that two nonsequence specific contextual features beyond miRNA target sites are critical determinants of miRNA-mediated 3′ UTR regulation. The contextual features reside 3′ of lsy-6 binding sites in the 3′ UTR and act in a combinatorial manner; mutation of each results in limited defects in 3′ UTR regulation, but a combinatorial deletion results in complete loss of 3′ UTR regulation. Together with two lsy-6 sites, these two contextual features are capable of imparting regulation on a heterologous 3′ UTR. Moreover, the contextual features need to be present in a specific configuration relative to miRNA binding sites and could either represent protein binding sites or provide an appropriate structural context. We conclude that a given target site resides in a 3′ UTR context that evolved beyond target site complementarity to support regulation by a specific miRNA. The large number of 3′ UTRs that we analyzed in this study will also be useful to computational biologists in designing the next generation of miRNA/target prediction algorithms.
Keywords: C. elegans, miRNA, lsy-6, cog-1, 3′ UTR
INTRODUCTION
Animal miRNAs are a large class of small regulatory RNAs that control the expression of target genes through binding to complementary target sites in mRNA targets (Lee et al. 1993; Reinhart et al. 2000; Bartel 2004). These sites typically have imperfect complementarity to the miRNA. The ability to target imperfectly complementary target sites together with the large number of miRNAs in genomes (as many as 1000 in the human genome) (Bentwich et al. 2005) has led to the prediction that thousands of genes are miRNA-regulated (Rajewsky 2006b). However, despite the apparent abundance of potential miRNA/3′ UTR target interactions, few animal miRNA/target pairs have been validated to date in a physiologically relevant context (Carthew 2006; Stefani and Slack 2008).
The parameters that govern miRNA/3′ UTR target recognition have grown increasingly complex as more rigorous sequence analyses and experimentation has examined miRNA/target interactions. Initial reports indicated that G:U wobble-free seed matches between position 2-8 of a miRNA and the target site and the ΔG of the miRNA/target heteroduplex were the primary determinants in miRNA target recognition (Doench et al. 2003; Doench and Sharp 2004; Brennecke et al. 2005; Lewis et al. 2005; Grimson et al. 2007). Additional parameters such as the importance of 3′ base-pairing to compensate for weak seed pairing became another factor in identifying potential miRNA targets (Brennecke et al. 2005). However, some other work also gave indications that miRNA/target recognition may be more complex. For example, lin-4 and let-7 binding to some of their targets involves both mismatches within the seed, as well as G:U base pairs (Ha et al. 1996; Vella et al. 2004a). Moreover, it was reported that let-7 requires two unique target sites and that the linker region between them in the lin-41 3′ UTR is required to mediate regulation (Vella et al. 2004a), thereby suggesting that miRNA target sites may require specific 3′ UTR contexts to function.
Robins and Padgett (2005) were the first to explicitly explore the role that mRNA structure may play in influencing miRNA/target recognition. They postulated that the folding of mRNAs may mask some potential miRNA target sites and devised a target prediction algorithm that required that at least three consecutive base pairs in the target site complementary to the seed region of the miRNA (nucleotides 2–8) be unpaired in the mRNA structure. Since this initial report, several groups have further advanced miRNA target prediction by including structural considerations in the form of the difference in free energy between the miRNA bound and unbound states (ΔΔG) as a means to identify potential target 3′ UTRs (Kertesz et al. 2007; Long et al. 2007). Moreover, Zhao et al. (2005, 2007) reported that target sites reside in 3′ UTR regions of high free energy based on the ΔG of the 70-base-pair (bp) 5′ and 3′ of a target site. Another study has added a number of additional considerations that boost site efficacy when attempting to identify potential target 3′ UTRs, including target site location relative to the stop codon, AU-richness around target sites, and preferential base pairing to nucleotides 13–16 of the miRNA (Grimson et al. 2007).
Numerous algorithms now exist that attempt to identify potential miRNA target 3′ UTRs, including PicTar, TargetScan, Miranda, and Mirbase (Stark et al. 2003; John et al. 2004; Krek et al. 2005; Lewis et al. 2005; Robins and Padgett 2005; Griffiths-Jones et al. 2006; Lall et al. 2006). These algorithms vary in their use of conservation, energetics, and structural considerations. The overlap between target sites predicted by different algorithms is, however, still limited (Rajewsky 2006b). The difficulty in target predictions is further underscored by the observation that only a limited number of mRNAs bound to the miRNP/Ago/AIN complexes are predicted to be targeted by known miRNAs (Beitzinger et al. 2007; Zhang et al. 2007). Part of the difficulty in formulating accurate predictions is the lack of data examining the ability of endogenous miRNAs to down-regulate target 3′ UTRs in the cellular context in which such an interaction normally takes place. Another problem is that rules of miRNA/mRNA interactions were built on assay systems that rely on overexpression of the miRNA and its target. Doench and Sharp (2004) reported that endogenously expressed let-7 was incapable of down-regulating some of their 3′ UTR reporter constructs but that some of these same constructs were down-regulated by exogenously supplied let-7. Therefore, these authors warned that overexpression of a miRNA in testing for potential target 3′ UTR regulation may suggest an interaction that may not occur in vivo.
We have developed an assay system that circumvents at least some of these problems (Didiano and Hobert 2006). In this assay system, we analyze the ability of the Caenorhabditis elegans miRNA lsy-6 to regulate a given 3′ UTR. lsy-6 is a miRNA that is expressed in a very small number of neurons, and in one of them, the left ASE (ASEL) neuron, it down-regulates expression of the cog-1 homeobox gene in a 3′ UTR dependent manner (Johnston and Hobert 2003). The assay is based on the use of the ceh-36 promoter to drive the expression of gfp in a pair of bilaterally symmetric neurons, the left and right ASE neurons (ASEL and ASER) (Chang et al. 2003). Control and experimental 3′ UTRs are placed downstream of the gfp coding sequence (Fig. 1A). As lsy-6 is asymmetrically expressed in ASEL, but not in ASER, this constellation provides an assay for evaluating the ability of lsy-6 to down-regulate 3′ UTR sensor constructs in ASEL, but not ASER (Fig. 1B). Notably, this assay system tests the interaction of a miRNA (lsy-6) with its target (cog-1) in exactly the same cell in which the interaction normally takes place. Other possible cell-type specific factors involved in the lsy-6-mediated regulation of the 3′ UTR should therefore be present. Many of the assay systems that have been used in the past to deduce rules of miRNA/target interactions do not display this feature. The second advantage of our assay system is that it performs a direct comparison of gfp expression levels in ASEL with ASER in each individual animal (Fig. 1B; see Material and Methods for more details), thereby providing an internal control that is not provided by many of the population-based transfection systems used to deduce features of miRNA/target interactions. Finally, unlike transfection systems, our system relies on the activity of an endogenous miRNA, rather than relying on an overexpression of the miRNA under investigation. As mentioned above, this is an important point as Doench and Sharp (2004) have previously shown that expression levels have an important impact on the ability of a miRNA to regulate a target 3′ UTR. We note, however, that our assay does rely on the ectopic expression of a sensor construct, thereby raising the issue that a sensor construct may be expressed at too high a level to allow for regulation by lsy-6. We deal with this issue by generating transgenic worms that contain only low copy numbers of the reporter gene, as assessed by gfp expression levels that are just above the level of detection (Fig. 1B). These levels of reporter gene expression easily allow for regulation of the wild-type cog-1 3′ UTR sensor by lsy-6, the control to which we compare our mutated 3′ UTR versions. Moreover, we always generate multiple independent lines for each construct. If we were indeed operating at levels that are close to a saturated threshold, we would expect to observe some variations between individual lines, which we essentially never observe. We are therefore confident that unregulated sensor constructs in our assay system represent true negatives rather than artifacts of excessive transgene expression.
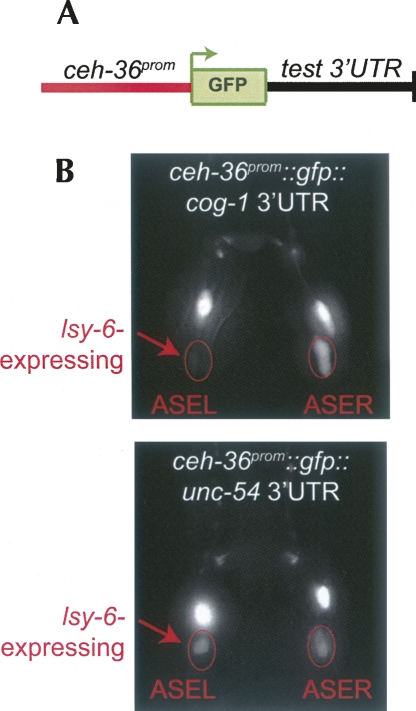
FIGURE 1.
3′ UTR sensor strategy. (A) Schematic representation of 3′ UTR sensor constructs. (B) Example of expression patterns of a regulated (ceh-36∷;gfp-cog-1 3′ UTR) (top) and an unregulated (ceh-36∷;gfp-unc-54 3′ UTR) (bottom) 3′ UTR sensor constructs. These images are the same as shown in Didiano and Hobert (2006) and are shown here again merely as an illustration of our assay system. (Reprinted with permission from Didiano and Hobert 2006; © 1998, Nature Publishing Group [http://www.nature.com/].) lsy-6 is only expressed in ASEL as indicated by arrows. gfp expression is dramatically down-regulated in ASEL but not ASER in the cog-1 3′ UTR sensor construct, but the unc-54 3′ UTR sensor construct displays equal gfp expression in ASEL and ASER. The pair of bright cells above the two ASE cells are the AWCL and AWCR neurons. They serve as internal controls for excluding the scoring of mosaic animals. The degree of regulation can also be quantified using an “asymmetric index” as defined in the Material and Methods and as listed in Supplemental Table 1.
We have previously used this assay to test the ability of lsy-6 to regulate mutated versions of the cog-1 3′ UTR as well as heterologous 3′ UTRs that were predicted to be targeted by lsy-6 (Didiano and Hobert 2006). The key findings of this previous analysis were that even though a seed match to the lsy-6 miRNA is required for cog-1 3′ UTR regulation, a seed match is not sufficient to confer regulation. This conclusion was based on (1) an engineered mutation within the cog-1 3′ UTR that did not affect the core lsy-6 seed match, but led to a loss of regulation, (2) the analysis of heterologous 3′ UTRs into which a lsy-6 site was engineered, and (3) the finding that the 3′ UTRs of 13 different genes that contained seed matches to lsy-6 failed to display lsy-6-dependent regulation (Didiano and Hobert 2006). Two independently developed miRNA/target prediction methods accurately predicted the inability of lsy-6 to regulate these 13 genes, therefore lending further credibility to our experimental findings (Long et al. 2007; Zhao et al. 2007). Nevertheless, it was suggested that the 3′ UTR sensors that we tested were expressed at levels too high to allow for regulation by endogenous lsy-6 (Rajewsky 2006a; Bushati and Cohen 2007). However, it appears improbable to us that each of the 13 predicted targets are endogenously expressed at low enough levels to allow for regulation by our lsy-6, and that in our assay system we overexpressed each single one of these 13 different 3′ UTRs, thereby preventing regulation by endogenous lsy-6. This also appears unlikely in light of the fact that the cog-1 gene itself is expressed at low levels in the ASE neurons, a notion derived from reporter gene assays (Palmer et al. 2002). We also tested expression levels of one of the 13 predicted target genes and found it to be not significantly different from that of cog-1 (J. Etchberger and O. Hobert, unpubl.), thereby making the level argument the least parsimonious explanation of our data.
Rajewsky (2006a) and Bushati and Cohen (2007) also raised the possibility that a second lsy-6 target site in the cog-1 3′ UTR, which we had previously reported (Johnston and Hobert 2003) (called site #2 throughout this present paper), may be the explanation for why we do not find a single lsy-6 site to be sufficient to confer regulation. We find this an unlikely explanation since we reported that two of the 13 predicted lsy-6 target 3′ UTRs contain two predicted lsy-6 sites but are nevertheless not regulated by lsy-6 (Didiano and Hobert 2006). Moreover, we had shown that a single lsy-6 site does function in the context of the lin-28 3′ UTR. One of the goals of the present paper is to further address the importance of this second site. The findings that we describe here indicate that this second site cannot explain the lack of sufficiency of seed matches to confer miRNA-mediated 3′ UTR regulation.
Besides testing the importance of both lsy-6 complementary sites in the cog-1 3′ UTR, we describe an extensive mutational analysis of the cog-1 3′ UTR and describe the analysis of a number of engineered, heterologous 3′ UTRs. The findings presented here support the notion that seed matches are not a sufficient predictor for miRNA/target interactions. We show that the sequence context of a 3′ UTR is a critical determinant of target site functionality and propose that entire 3′ UTRs rather than just individual sites evolve to support regulation by a miRNA. We also show that available prediction algorithms fail to provide an accurate assessment of whether a 3′ UTR is regulated by lsy-6. Our large number of experimentally tested 3′ UTRs provide a data set to improve these algorithms.
RESULTS
Assessing the importance of a lsy-6 complementary site in the cog-1 3′ UTR
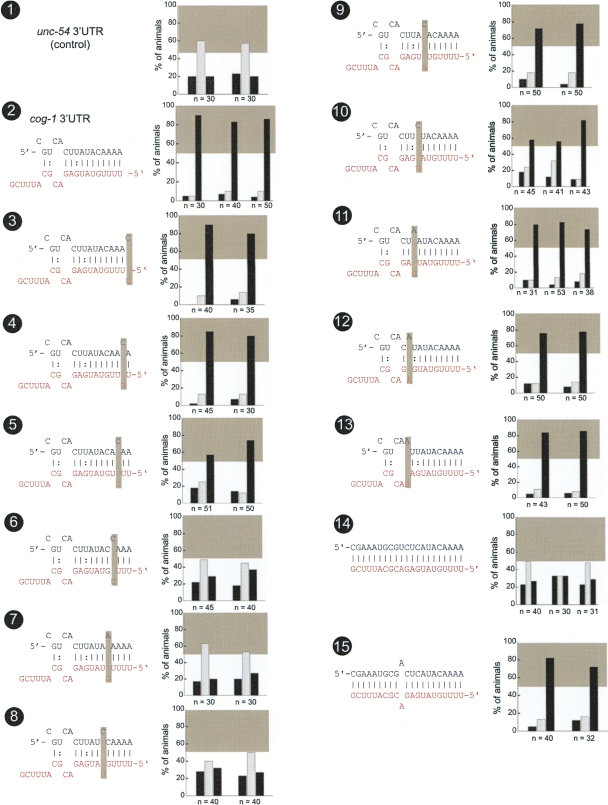
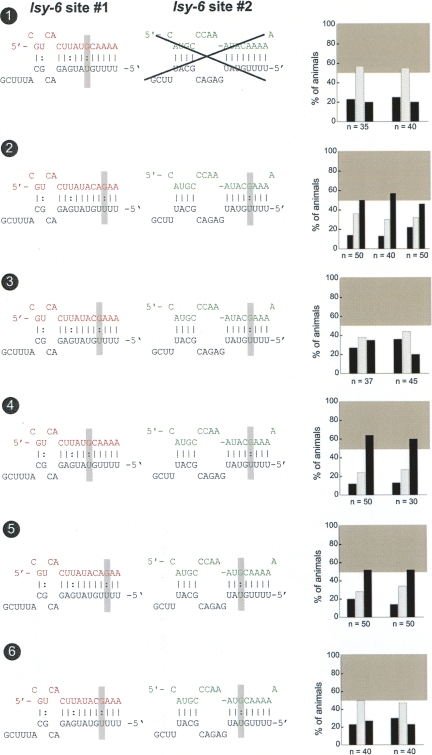
Our previous analysis of the cog-1 3′ UTR has demonstrated an absolute requirement of a lsy-6 complementary site (henceforth called “lsy-6 site #1”) for the correct and differential regulation of the cog-1 3′ UTR in ASEL versus ASER. Deletion of site #1 causes a complete loss of 3′ UTR regulation (Didiano and Hobert 2006). We extended these findings by a systematic point mutational analysis of site #1 (Fig. 2). We find that point mutations in the seed region differentially disrupt 3′ UTR regulation. Point mutations only in seed positions 4, 5, and 6, but not in positions 1–3 or 7–11, disrupt 3′ UTR function significantly. These findings suggest that sequences outside the seed may contribute to lsy-6 function. Indeed, a deletion reducing site #1 from an 11mer seed match to an 8mer seed match results, as we have previously reported, in the complete loss of regulation (shown again in Fig. 4, no. 3, below [note: when “no.” is mentioned, this refers to the numbered black circles in a figure]; Didiano and Hobert 2006). The importance of site #1 can also further be illustrated by its conversion into a site that is perfectly complementary to lsy-6. Such a perfect site is entirely unable to confer regulation (Fig. 2, no. 14). However, regulation can be re-established through reintroduction of a single mismatch (Fig. 2, no. 15). Taken together, various permutations of lsy-6 site #1 illustrate the importance of the overall composition of this site.
FIGURE 2.
Mutational analysis of lsy-6 site #1. Multiple transgenic animals expressing one given sensor construct are scored for gfp expression. Each scored animal is placed into one of three categories, in which relative gfp expression in the two ASE neurons is as follows: ASEL>ASER (left black bar), ASEL=ASER (middle gray bar), or ASEL<ASER (right black bar). See Material and Methods for more details on scoring. Each set of three bars represents an independent transgenic line. The gray box shown in each bar graph is a visual “helper” to indicate what is considered to be a minimal level of regulation for the system; this minimal regulation is arbitrarily defined based on the level of regulation seen with the lsy-6 site #2 deletion, shown in Figure 4, no. 4. The gray box roughly correlates with another quantitative measure for the degree of regulation, called the “asymmetry index,” defined in the Material and Methods and listed in Supplemental Table 1 for each construct analyzed in this paper. Each construct is named based on figure number and number of construct within the figure. The control unc-54 3′ UTR sensor construct displays characteristic unregulated distribution of gfp (no. 1), while the cog-1 3′ UTR sensors display dramatic regulation with the majority of animals falling into the ASER>ASEL category (no. 2). Shaded boxes indicate location of introduced mismatches. Only mismatches in positions 4, 5, and 6 counting from the 5′ end of the miRNA resulted in complete loss of regulation (nos. 6,7,8). Mismatches in positions 1, 2, 7, 9, 10, and 11 resulted in little or no loss of regulation (nos. 3,4,9,11–13). Mismatches in positions 3 and 8 resulted in a moderate loss of regulation (nos. 5,10). If the lsy-6 target site in the cog-1 3′ UTR is converted to have perfect complementary to lsy-6, then regulation is completely lost. However, if a single mismatch is reintroduced into position 12, which is unpaired in the wild type, then regulation is restored (nos. 14,15). Note: Construct nos. 1, 2, and 7 have been previously examined (Didiano and Hobert 2006) and a rescoring of newly generated and scored transgenic lines is shown here (rescored lines show the same results as previously reported).
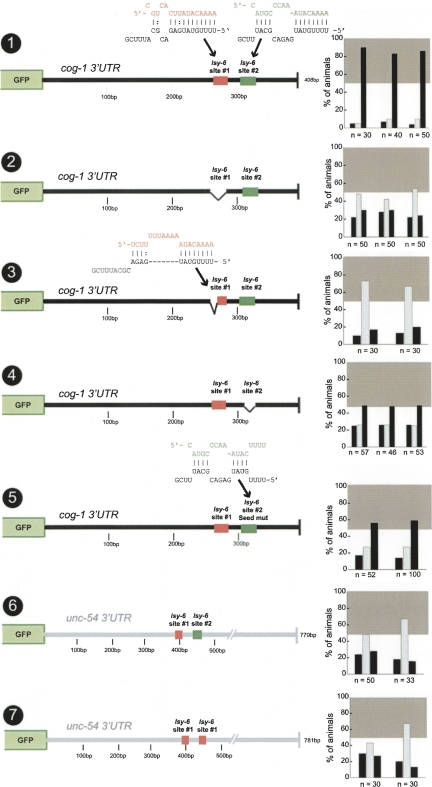
FIGURE 4.
Asymmetric function of two lsy-6 sites in the cog-1 3′ UTR. Construct no. 1 is the same control as shown in Figure 2, no. 2, and is shown for comparison only. A complete deletion (no. 2) or partial deletion resulting in an 8mer seed region of target site #1 (no. 3) each results in a complete loss of regulation, meaning that target site #2 is nonfunctional in isolation. A complete deletion of site #2 (no. 4) or a seed mutant (no. 5) in site #2 each results in a partial loss of regulation. Either two copies of site #1 or a single copy of site #1 in combination with site #2 were inserted into the unc-54 3′ UTR, but neither of these constructs displayed regulation (nos. 6,7). miRNA/target heteroduplexes shown here were generated using RNAhybrid (Rehmsmeier et al. 2004). Construct no. 3 has been previously examined (Didiano and Hobert 2006) and a rescoring of newly generated and scored transgenic lines is shown here (rescored lines show the same results as previously reported). See legend to Fig. 2 and Materials and Methods for explanation of data representation.
Insufficiency of the lsy-6 complementary site #1
We have previously shown that lsy-6 site #1 from the cog-1 3′ UTR can confer regulation when transplanted into the context of the lin-28 3′ UTR but not of the unc-54 3′ UTR (Didiano and Hobert 2006) (shown again, for comparison, in Fig. 3, nos. 2,4). We extended this analysis and find that lsy-6 site #1 fails to confer regulation to the 3′ UTR of the actin gene act-1 (Fig. 3, no. 10), which is, in contrast to the unc-54 3′ UTR (885 bp), a small 3′ UTR (194 bp) that is also low in GC content (35%).
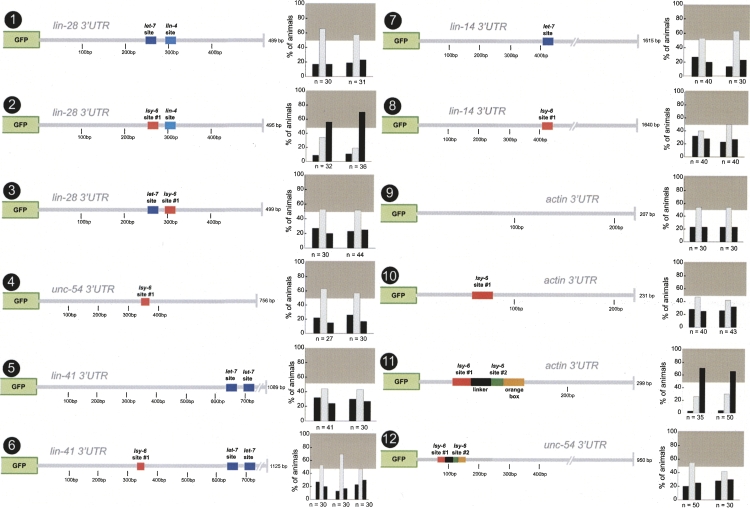
FIGURE 3.
Sufficiency of lsy-6 sites in other 3′ UTR contexts. Single copies of the previously described lsy-6 target site from the cog-1 3′ UTR were inserted into the lin-28, unc-54, lin-41, lin-14, and actin/act-1 3′ UTRs (Didiano and Hobert 2006). None of the heterologous contexts displayed regulation prior to target site insertion (nos. 1,5,7,9). The primary target site was inserted into two position of the lin-28 3′ UTR in place of a presumptive let-7 target site and a lin-4 target site (nos. 2,3). Only a single site insertion into the let-7 site position resulted in regulation (no. 2) but failed to confer regulation in the second position (no. 3). The target site was also inserted into a let-7 position in lin-14 3′ UTR (only let-7 but not lin-4 sites are shown for the lin-14 3′ UTR) but failed to display regulation in these contexts (nos. 6,8). A single site was insufficient to confer regulation to an actin 3′ UTR (no. 10). This context was chosen for its short length and high AU content (66% with target site insertion). Additionally, a cog-1 3′ UTR minimal element, which was defined based on deletion analysis in Figure 8 (nos. 1,2,3), conferred regulation to the actin 3′ UTR but not the unc-54 3′ UTR (nos. 11,12). Construct nos. 1, 2, and 4 have been previously examined (Didiano and Hobert 2006), and a rescoring of newly generated and scored transgenic lines is shown here (rescored lines show the same results as previously reported). See legend to Figure 2 and Materials and Methods for explanation of data representation and Figure 2, nos. 1 and 2, for regulated and unregulated control 3′ UTRs.
The lin-28 3′ UTR is known to be normally regulated by a miRNA, lin-4 (Moss et al. 1997), and also contains additional let-7 target sites (Krek et al. 2005), while neither the actin nor the unc-54 3′ UTR are predicted to be miRNA regulated. We therefore hypothesized that there may be some feature(s) common only to miRNA-regulated 3′ UTRs that enable them to support miRNA regulation. To test this notion, we engineered lsy-6 #1 site into the 3′ UTR of the lin-41 and lin-14 genes, both known miRNA targets (Wightman et al. 1993; Slack et al. 2000). The lsy-6 site was engineered into an AU-rich sequence of the lin-41 3′ UTR and into a let-7 site of the lin-14 3′ UTR. Neither construct is subject to lsy-6 regulation (Fig. 3, nos. 5–8).
Additionally, we find that the lsy-6 site fails to confer regulation when placed in another position in the lin-28 3′ UTR, in which it replaces a lin-4 target site (Fig. 3, no. 3). The lsy-6 site therefore works in one, but not in another, context of the lin-28 3′ UTR (Fig. 3, nos. 2,3). It therefore appears unlikely that the lsy-6 site works in lin-28 because lin-28 contains some cryptic features (e.g., cryptic lsy-6 sites) that permit it to work. If this were the case, the lsy-6 site should have worked in both contexts in the lin-28 3′ UTR, which it does not.
The cog-1 3′ UTR contains two target sites for lsy-6 but these sites are not sufficient in all 3′ UTR contexts
Our previous analysis of the cog-1/lsy-6 interaction focused on only one lsy-6 target site, “site #1,” which contains an 11mer seed region (Didiano and Hobert 2006). However, as we already noted when cloning lsy-6, the cog-1 3′ UTR contains a second target site (Johnston and Hobert 2003), which contains only an 8mer seed (Fig. 3) and which is also less conserved in other species (see below). We call the 8mer seed site “site #2.” How important is site #2 for regulation and could this second site explain why a single lsy-6 site can only work in some, but not all, 3′ UTR contexts? We asked whether lsy-6 site #2 is involved in 3′ UTR regulation to an extent that is comparable to that of site #1 by introducing two distinct mutations. A complete deletion of site #2 or introduction of a 4-bp mutation in the seed results in a reduction, but not a complete loss of regulation, which contrasts the complete loss of regulation observed upon deleting site #1 (Fig. 4, nos. 2,4,5; note that numerical values for the degree of regulation, which allow for comparing different constructs, can be found in Supplemental Table 1).
Could a lack of site #2 explain the lack of sufficiency of a single lsy-6 target site to confer regulation to a 3′ UTR? To address this issue, we engineered a construct in which we inserted site #1 and site #2 into the unc-54 3′ UTR. No regulation was observed (Fig. 4, no. 6). Similarly, placing two copies of site #1 into the unc-54 3′ UTR conferred no regulation (Fig. 4, no. 7; note that the sites are spaced by the same distance as in the cog-1 3′ UTR). We conclude that even though site #2 has some, comparatively minor, role in 3′ UTR regulation, it cannot account for the inability of a lsy-6 site to confer regulation onto heterologous 3′ UTRs. We furthermore note that even within the context of the cog-1 3′ UTR, the mere presence of two 8 bp seed matches to lsy-6, achieved through reduction of the 11mer seed match of site #1 to a 8mer seed (Fig. 4, no. 3), is not sufficient to confer 3′ UTR regulation. The presence of one or several seed matches in a 3′ UTR is therefore not a sufficient criterion to predict regulation of a 3′ UTR by a cognate miRNA.
G:U wobble base pairs in the seed region of the cog-1 target sites result in impaired regulation
We sought to utilize the minor impact of site #2 on 3′ UTR regulation to analyze the issue of the importance of G:U wobble base pairs in miRNA/3′ UTR heteroduplexes. Previous work has shown that in a transfection-based cell culture system, G:U wobbles are detrimental for regulation of a synthetic 3′ UTR by a nonnatural miRNA (Doench and Sharp 2004). In contrast, we previously reported that G:U wobble base pairs were well tolerated in the seed region of lsy-6 site #1, even when two G:U wobbles were introduced (Didiano and Hobert 2006). We continued to address this issue using a “sensitized background,” that is, the cog-1 3′ UTR whose regulation is weakened through a loss in lsy-6 site #2, with the idea being that this may more readily reveal the impact of G:U wobbles on miRNA/target duplex formation. When we combined a G:U wobble base pair in position 6 with a lsy-6 site #2 deletion, we find that this construct displayed no regulation (Fig. 5, no. 1). Next, we introduced G:U wobbles into both lsy-6 site #1 and site #2 simultaneously. Three of the five constructs containing G:U wobbles in both target sites displayed regulation, albeit at levels below wild-type regulation (Fig. 5, nos. 2–6). These results indicate that G:U wobbles can be detrimental to lsy-6-mediated regulation in specific circumstances; however, G:U wobbles should not be used as a criterion by which to eliminate potential miRNA/target interactions.
FIGURE 5.
G:U wobble base pairing in the seed region impairs but does not necessarily abolish regulation. Construct no. 1 consists of a G:U wobble base pair in position 6 of site #1 in the background of a site #2 deletion and fails to display regulation. Construct nos. 2–6 were engineered to contain G:U wobble base pairs in both lsy-6 site #1 and site #2. Three of five of these double G:U wobble constructs display intermediate regulation (nos. 2,4,5). Shaded boxes indicate location of G:U wobbles introduced. See legend to Fig. 2 and Materials and Methods for explanation of data representation and Figure 2, nos. 1 and 2, for regulated and unregulated control 3′ UTRs.
Extra-target site sequences in the cog-1 3′ UTR influence lsy-6 mediated regulation
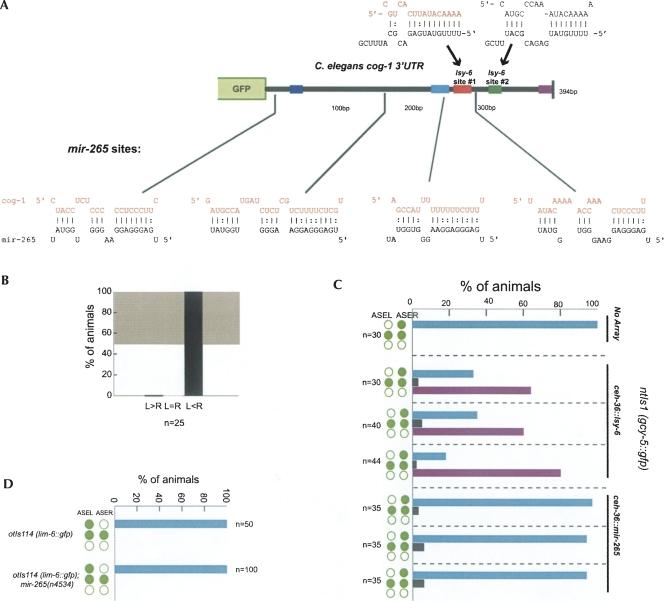
Our analysis has shown that even though lsy-6 complementary site #1 is an essential feature for cog-1 3′ UTR regulation, neither single nor multiple lsy-6 sites are sufficient to confer regulation. An obvious explanation for these observations is that additional features in the cog-1 3′ UTR are required for miRNA regulation, which, together with the lsy-6 sites, may also confer regulation on heterologous 3′ UTRs. What other features in the cog-1 3′ UTR could be involved in regulation? One attractive candidate would simply be another miRNA binding site in the cog-1 3′ UTR. There are 4 potential binding sites for the functionally uncharacterized miRNA mir-265 in the cog-1 3′ UTR (Fig. 6). To ask whether mir-265 is involved in the ASEL-specific down-regulating of the cog-1 gene, we undertook three separate experiments. First, we assayed the regulation of the wild-type cog-1 3′ UTR sensor in mir-265 null mutants (Miska et al. 2007) and found no effect on sensor gene expression (Fig. 6B). Second, we ectopically expressed mir-265 in ASEL and ASER. The same ectopic expression, when done with lsy-6, results in a down-regulation of the cog-1 target and an ensuing loss of ASER cell fate (Fig. 6C; Johnston and Hobert 2003). However, such an effect cannot be observed upon ectopic expression of mir-265 (Fig. 6C). Third, we tested whether cog-1 regulation is disrupted in a mir-265 null mutant strain by assaying the regulation of a cog-1 effector gene, lim-6. While loss of lsy-6 leads to a completely penetrant loss of lim-6 expression, caused by loss of cog-1 repression (Johnston and Hobert 2003), no such effect can be observed in mir-265 null mutants (Fig. 6D). We conclude that mir-265 plays no significant role in regulating the cog-1 3′ UTR.
FIGURE 6.
mir-265 does not affect cog-1 activity. (A) Potential target sites for mir-265 in the cog-1 3′ UTR shown below 3′ UTR schematic, with lsy-6 target sites shown above. (B) The cog-1 3′ UTR sensor otIs185 (Sarin et al. 2007) is down-regulated normally in the ASEL neuron in mir-265(n4534) null mutant animals. The degree of regulation is the same as shown for the same sensor in a wild-type background (Sarin et al. 2007). (C) The ASE bilateral ceh-36 promoter was used to drive expression of lsy-6 and mir-265 hairpin precursors. ntIs1(gcy-5::gfp) was used as a cell-fate marker to test the ability of lsy-6 and mir-265 to down-regulate cog-1, thereby driving ASEL cell fate in ASER (green circles represent gcy-5::gfp expression pattern as ASER-specific [top], ASE bilateral [middle], or loss of expression [bottom]. All three lines misexpressing lsy-6 results in a “two ASEL phenotype,” as previously reported (Johnston and Hobert 2003). None of the three lines misexpressing mir-265 results in a phenotype, indicating that mir-265 is incapable of downregulating cog-1 via its 3′ UTR in ASER. (D) The mir-265 null mutant n4534 shows no lsy phenotype using the lim-6::gfp ASEL specific reporter (otIs114). Green circles indicate reporter expression in ASEL and ASER.
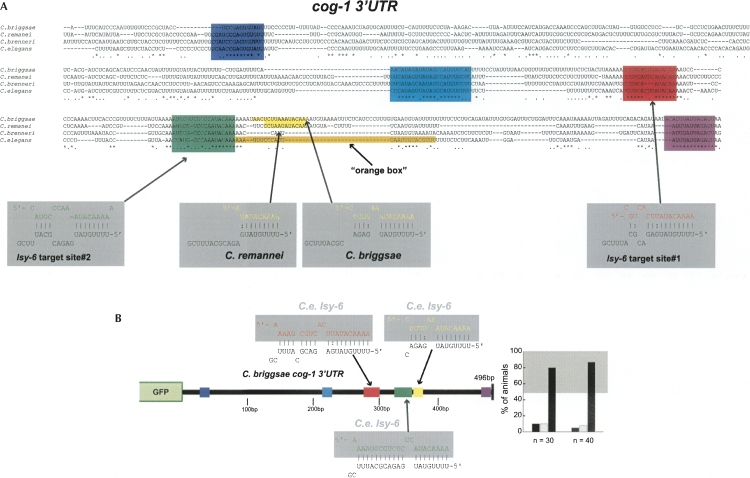
As an alternative means to identify potentially relevant sequences in the cog-1 3′ UTR, we identified phylogenetically conserved sequences in the 3′ UTR, utilizing 4 different nematode genomes (Fig. 7A). We confirmed that regulation is conserved across phylogeny by generating a sensor construct in which we test the C. briggsae cog-1 3′ UTR for ASEL-specific down-regulation and find this 3′ UTR to indeed be regulated as well (Fig. 7B).
FIGURE 7.
Evolutionary conservation of the cog-1 3′ UTR. (A) Alignment of cog-1 3′ UTR between C. elegans and three related nematode species. Asterisk (*) indicates perfect conservation; period (.) indicates conservation across any three of the four species. Conserved motifs shown in various colors (dark blue, light blue, and purple). We arbitrarily define conserved motifs as being at least 10-bp long and displaying >66% conservation across all four species. C. elegans lsy-6 heteroduplexes with target sites from the C. elegans cog-1 3′ UTR are shown in red for site #1 and green for site #2. Two of the four species, C. briggsae and C. remanei, also contain a third potential lsy-6 target site shown in yellow. The “orange box” is a sequence in the C. elegans 3′ UTR that we find to contribute to 3′ UTR regulation (Fig. 8). (B) cog-1 3′ UTR regulation is conserved across phylogeny. The C. briggsae cog-1 3′ UTR confers down-regulation in ASEL when expressed in C. elegans. See Figure 2, nos. 1 and 2, for controls. See the legend to Figure 2 and Materials and Methods for explanation of data representation.
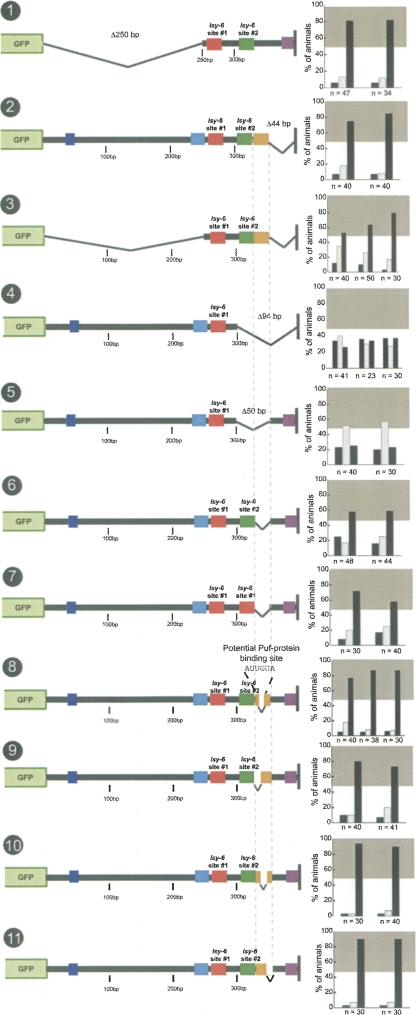
To test the importance of the phylogenetically conserved regions in the 3′ UTR, we undertook a deletion analysis of the C. elegans cog-1 3′ UTR. We find that the first 250 bases of the 394-bp cog-1 3′ UTR, which contain two phylogenetically conserved sequence motifs (Fig. 7A, dark and light blue boxes), are not required for regulation (Fig. 8, no. 1). Deletion of the last 44 bp (351–394), which contained another, strongly conserved motif (Fig. 7A, purple box), also showed no effect on regulation (Fig. 8, no. 2). Combining the 5′ 250-bp deletion with the 3′ 44-bp deletion, which removes all phylogenetically conserved motifs aside from the conserved lsy-6 sites, leads to a modest decrease in regulation, but the 3′ UTR still remains well regulated (Fig. 8, no. 3). The remaining 92 bp (258–350) therefore define a minimal regulatable fragment from the cog-1 3′ UTR, that is not only sufficient to confer regulation in isolation (Fig. 8, no. 3), but also when placed into the context of the actin 3′ UTR (Fig. 3, no. 11). Notably, though, this minimal element can still not function in the unc-54 3′ UTR (Fig. 3, no. 12).
FIGURE 8.
Mutational dissection of the cog-1 3′ UTR. The first 250 bp and last 44 bp of the cog-1 3′ UTR are not required for regulation (nos. 1,2). and base pairs 258–350 of the cog-1 3′ UTR alone are sufficient for regulation (no. 3). However, this minimal element is not sufficient in all heterologous contexts (nos. 11,12). A deletion of the last 94 bp of the cog-1 3′ UTR results in a complete loss of regulation (no. 4). A deletion of 300–350 bp results in a complete loss of regulation (no. 5). Deletion of 325–350 bp results in a partial loss of regulation (no. 6). This region does not affect lsy-6 target site #2. Replacing site #2 with a second copy of site #1 in the background of a 325- to 350-bp deletion resulted in slightly increased, but sub-wildtype regulation (no. 7). This 25-bp motif contains a potential PUF-protein binding site, but a deletion of this site had no affect on regulation (no. 8). Scanning deletion analysis of the 25-bp motif by two 8-bp and a 9-bp deletion did not recapitulate the 25-bp deletion effect on regulation (nos. 9,10,11). See legend to Figure 2 and Materials and Methods for explanation of data representation and Figure 2, nos. 1 and 2, for regulated and unregulated control 3′ UTRs.
Deletions into this 92-bp minimal region lead to a loss of 3′ UTR regulation. Specifically, a deletion that removes the last 94 bp (301–394) of the complete cog-1 3′ UTR (eliminating 50 bp of the 92-bp minimal region) results in the complete loss of regulation (Fig. 8, no. 4). The in vivo importance of this 3′ terminal region is underscored by our identification of a mutant allele of the cog-1 locus in a genetic screen; this mutant allele produces a cog-1 gain-of-function phenotype, that is, derepression of cog-1 in ASEL, and harbors a similar deletion at the end of the 3′ UTR (105-bp deletion) (Sarin et al. 2007). The last 94 bp of the cog-1 3′ UTR contain the lsy-6 site #2; however, the 94-bp deletion has a larger effect on the regulation than a mutation of the lsy-6 site #2 alone (cf. Fig. 8, no. 4, and Fig. 4, nos. 4,5). This indicates that some sequence in addition to site #2 in the last 94 bp contributes to the regulation. A 50-bp deletion from 301 to 350 bp, encompassing lsy-6 site #2 (309–324 bp) plus adjacent 25 bp of sequences still resulted in a complete loss of regulation (Fig. 8, no. 5). We term these adjacent 25 bp the “orange box” (see sequence in Fig. 7A). Nineteen out of 25 (76%) base pairs in the orange box are AU base pairs, which is not much above the overall AU content of the entire cog-1 3′ UTR (65%). A deletion of the 25-bp orange box alone (326–350) results in a modest but significant loss of regulation (Fig. 8, no. 6). Taken together, lsy-6 site #2 plus an adjacent, modestly conserved 25-bp region, the orange box, are jointly required for cog-1 regulation; deletion of either sequence results in modest defects, deleting both sites together causes a complete loss of 3′ UTR regulation, comparable only to the loss of lsy-6 site #1. The orange box region does not appear to merely increase the efficacy of the lsy-6 site #2, as the replacement of site #2 with site #1 in the absence of the orange box does not restore normal regulation (Fig. 8, no. 7).
The orange box acts independently of position and can be substituted with AU- but not GC-rich sequences
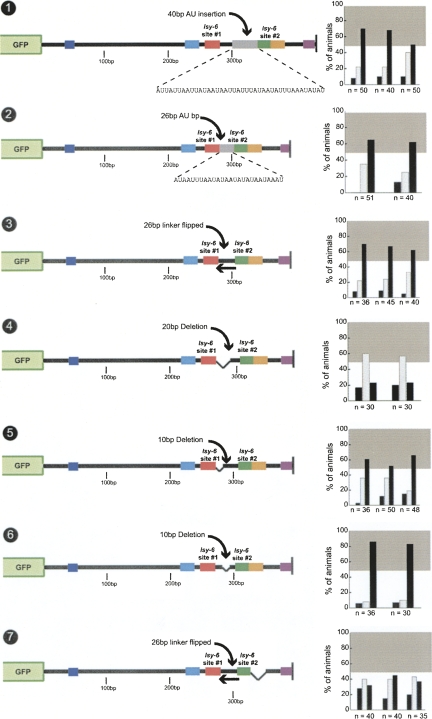
One potential function of the orange box could be its binding to a cofactor, either another regulatory RNA or a protein. With the limitation of miRNA target predictions in mind (Rajewsky 2006b), we find that no miRNA is predicted to pair with the orange box. A PUF-type protein was recently shown to be involved in the regulation of a miRNA-controlled 3′ UTR (Nolde et al. 2007), and the orange box contains a AUUGUA motif that may constitute a potential PUF-protein binding site (Wharton et al. 1998). However, deleting the AUUGUA motif has no affect on 3′ UTR regulation (Fig. 8, no. 8). Next, we deleted chunks of the 25-bp orange box by removing three different 8- to 9-bp regions. None of the smaller deletions recapitulate the effect of the full 25-bp deletion (Fig. 8, nos. 9–11).
The addition of a 20-bp insert consisting of only A's or of A's and U's between lsy-6 site #2 and the orange box does not impair regulation (Fig. 9, nos. 1,2). Intriguingly, the introduction of the AU-insert abrogates the importance of the orange box, that is, deletion of the orange box has no effect on the regulation of the 3′ UTR when the heterologous AU-insert is present (Fig. 9, no. 3). In other words, the function of the 25-bp orange box (which contains 25% GC and is therefore not any more or less AU- or GC-rich than the whole cog-1 3′ UTR) and an arbitrary 20-bp AU-rich sequence are equivalent.
FIGURE 9.
Functional analysis of the orange box. Distancing the 25-bp orange box from lsy-6 site #2 by a 20-bp A or a AU spacer does not affect regulation (nos. 1,2). Deleting the orange box in the presence of the 20-bp AU spacer does not result in a reduction of regulation (no. 3). Distancing the 25-bp orange box from site #2 with a 20-bp GC spacer also does not affect regulation (no. 4), but deleting the orange box in this context results in a complete loss of regulation (no. 5). See legend to Fig. 2 and Materials and Methods for explanation of data representation and Figure 2, nos. 1 and 2, for regulated and unregulated control 3′ UTRs.
The insertion of a 20-bp GC-rich sequence between lsy-6 site #2 and the orange box also does not impair regulation (Fig. 9, no. 4). However, in contrast to the inserted AU sequences, the GC sequences cannot substitute for the orange box. That is, a deletion of the orange box in the GC-inserted construct completely abolishes regulation (Fig. 9, no. 5).
At first sight, one interpretation of these findings may be that the 25-bp orange box provides an energetically favorable context for lsy-6 site #2 to function in. This explanation would be in accordance with the recently deduced nonstructured and AU-rich nature of sequences that flank miRNA binding sites (Zhao et al. 2005; Grimson et al. 2007; Zhao et al. 2007). However, we note that this explanation is rendered unlikely by our finding that the orange box and lsy-6 site #2 function independently, that is, the effect of the loss of lsy-6 site #2 is enhanced by loss of the orange box (cf. Fig. 4, no. 4, and Fig. 8, nos. 5,6). Moreover, in the context of the GC insertion (which cannot substitute for the orange box), the orange box can still function from the distance, thereby ruling out that the orange box provides a proximal, local context for site #2 to function.
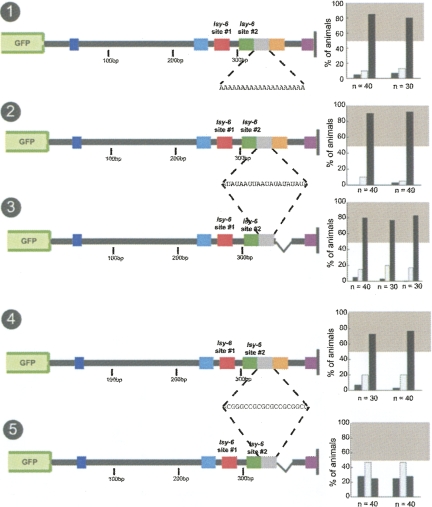
The linker region between the two target sites is required for wild-type regulation
Are there other regions besides the orange box that are important for cog-1 3′ UTR function? A previous study on let-7-mediated 3′ UTR regulation has found that both the composition and the distance between two let-7 target sites in the lin-41 3′ UTR can influence miRNA-mediated regulation (Vella et al. 2004a). We tested whether the linker region between the two lsy-6 target sites in the cog-1 3′ UTR influences 3′ UTR regulation in a number of manners (Fig. 10). To test whether the base sequence composition of the linker affected regulation, we replaced the 26-bp linker between the two sites (35% GC content) with a 26-bp linker consisting entirely of A and U base pairs. This mutation results in only a reduction, but not elimination, of regulation (Fig. 10, no. 2). Similarly, an inversion of the linker sequence only slightly affects regulation (Fig. 10, no. 3). A 40-bp extension of the linker also has only minimal effects on regulation (Fig. 10, no. 1) This indicates that the sequence composition and length of the linker region have only a minor impact on regulation, which is in contrast to the let-7/lin-41 3′ UTR case, in which the linker region was shown to be absolutely required for regulation (Vella et al. 2004a).
FIGURE 10.
A 26-bp linker between lsy-6 site #1 and #2 provides contextual information that is length independent. Insertion of a 40-bp AU spacer 3′ to the endogenous 26-bp spacer between the two target sites in the cog-1 3′ UTR results in a partial loss of regulation (no. 1). Replacing the endogenous linker with a 26-bp AU linker (no. 2) or inverting the linker also resulted in a partial loss of regulation (no. 3). A 20-bp deletion that shortens the linker to 6 bp results in a complete loss of regulation (no. 4). Only one of two separate 10-bp deletions of the linker region resulted in a reduction of regulation (no. 5), indicating that the deletion that had an effect was due to contextual changes rather than in a reduction of the linker regions length below an ideal length (nos. 5,6). This may be due to contextual changes or reduction of the linker below a critical length or a combination of both. Combining the linker inversion mutation with a deletion of the orange box, examined in Figures 8 and 9, renders the cog-1 3′ UTR virtually nonfunctional (no. 7). See legend to Figure 2 and Materials and Methods for explanation of data representation and Fig. 2, nos. 1 and 2, for regulated and unregulated control 3′ UTRs.
Although the composition of the linker does not appear to have a strong impact on 3′ UTR regulation, its mere presence does. Deleting 20 bp from the 26-bp linker region, results in a complete loss of regulation (Fig. 10, no. 4). However, two smaller separate 10-bp deletions that cover the essential 20-bp region have small or no effect on regulation (Fig. 10, nos. 5,6). Together with the results of the linker inversion, these results argue that in contrast to the lin-41/let-7 linker region, which cannot be replaced by heterologous sequences (Vella et al. 2004a), the linker region between the two lsy-6 sites tolerates significant sequence alterations.
Taken together, we have shown that two regions that flank the two lsy-6 binding sites, the orange box 3′ of lsy-6 site #2 and the linker region 3′ of lsy-6 site #1, have an impact on 3′ UTR regulation. Both flanking regions function independently and together are essential for 3′ UTR regulation, as we find that combining the 26-bp linker inversion mutation, which alone has only a minimal effect on regulation (Fig. 10, no. 3), with a deletion of the orange box, which alone has small effects on regulation (Fig. 8, no. 6), results in an almost complete loss of regulation (Fig. 10, no. 7). It is notable that this construct has two intact lsy-6 target sites #1 and #2, yet it is not regulated. These results indicate that the lsy-6 target sites are only functional when present within an appropriate 3′ UTR context.
lsy-6 target sites are only fully functional in their endogenous positions within the cog-1 3′ UTR
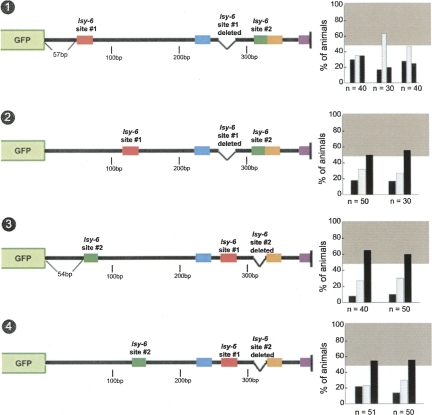
We further addressed the importance of position and context of the lsy-6 sites by moving site #1 and site #2 into different positions with the otherwise wild-type cog-1 3′ UTR. We find that neither site #1 nor site #2 is functional when placed at a position in which the 3′ end of the target site is 70 bp 3′ of the stop codon in the cog-1 3′ UTR. This distance from the stop is much above the recently proposed minimal distance of a miRNA target site to a stop codon and also is located in an “off-center position,” which was also suggested to be favorable for miRNA target site function (Grimson et al. 2007). In spite of these apparently favorable conditions, moving site #1 in this position leads to a complete loss of regulation (Fig. 11, no. 1). Moving site #2 leads to the same moderate loss of regulation observed upon entirely deleting site #2 (cf. Fig. 11, no. 3, and Fig. 4, no. 4). When moved to another location in the cog-1 3′ UTR, site #1 confers some limited, but clearly impaired level of regulation (Fig. 11, no. 2), while another location for site #2 has the same effects as having no site #2 at all (Fig. 11, no. 4).
FIGURE 11.
lsy-6 target sites in the cog-1 3′ UTR are only fully functional in endogenous positions in the 3′ UTR. lsy-6 target site #1 displays no regulation when moved to one position but displays weak regulation when moved to a second position within the cog-1 3′ UTR (nos. 1,2). Similarly, moving site #2 to the same positions within the cog-1 3′ UTR results in a level of regulation similar to those displayed by a site #2 deletion (nos. 3,4, see also Fig. 4, no. 4). This indicates that site #1 and site #2 are only capable of achieving wild-type levels of regulation when they are in their endogenous positions. See legend to Figure 2 and Materials and Methods for explanation of data representation.
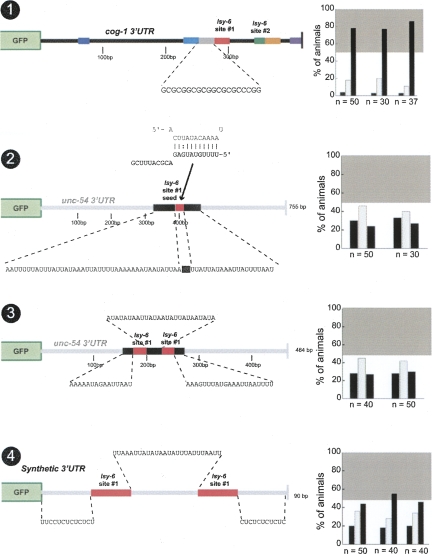
The base-pair composition surrounding the target sites does not influence regulation in a manner correlating with AU or GC richness
What is it exactly about the lsy-6 target site context that is so important for regulation? Several reports have indicated that miRNA target sites reside in AU-rich regions of 3′ UTRs (Robins and Padgett 2005; Grimson et al. 2007). High ΔG values, favored by high AU content, of sequences around the target site have also been proposed to be important for functionality (Zhao et al. 2007). The primary lsy-6 target site #1 normally resides in a location with 83% AU content (over 30 bp) 5′ to the site, while the 26-bp linker sequence 3′ to site #1 is only 58% AU in content, which is comparable to the overall AU content of the cog-1 3′ UTR (65%) (Fig. 7A). When we alter site #1 context by adding 20 bp of GCs 5′ to the site #1, thereby dramatically changing local AU content, we observe no significant effect on regulation (Fig. 12, no. 1). As mentioned above in the context of studying the orange box, the introduction of GC sequences 3′ of lsy-6 site #2 also has little effect on regulation (Fig. 9, no. 4).
FIGURE 12.
The AU-richness of the sequence flanking target sites does not correlate with functionality. Inserting 20 bp of GCs 5′ to target site #1 in the cog-1 3′ UTR, distancing the site from a conserved AU-rich sequence, has no effect on the level of regulation (no. 1). Inserting the seed region of target site #1 into an AU-rich region in the unc-54 3′ UTR fails to confer regulation (no. 2). Construct no. 3 is a shortened unc-54 3′ UTR in which two copies of site #1 are inserted into an AU-rich region with a 26-bp AU linker region between the sites; this construct also fails to display regulation. Construct no. 4 is a completely synthetic 3′ UTR consisting of two copies of site #1 also with a 26-bp AU linker surrounded by CU base pairs. This construct displays weak levels of regulation. Note that all engineered linker regions are 26 bp long, which is the length of the linker region between the two target sites in the cog-1 3′ UTR. Construct no. 4 consists of 16 bp from the cog-1 3′ UTR that contains the entire target site #1, whereas all other site #1 insertions are 25-bp sequences from the cog-1 3′ UTR. See legend to Figure 2 and Materials and Methods for explanation of data representation and Fig. 2, nos. 1 and 2, for regulated and unregulated control 3′ UTRs.
We addressed this issue further by attempting to “activate” the nonfunctional lsy-6 target sites in the unc-54 3′ UTR context (described above in Figs. 3, no. 4; Figs. 4, no. 7), by surrounding a single lsy-6 site #1 seed region in the context of the unc-54 3′ UTR with >20-bp AU-rich sequences. This manipulation does not confer regulation on the 3′ UTR (cf. Fig. 3, no. 4, and Fig. 12, no. 2). We also changed the construct described above with two lsy-6 site #1, placed into the unc-54 3′ UTR, which conferred no regulation (Fig. 4, no. 7). If both sites are embedded into an AU-rich context, regulation is still not observed (Fig. 12, no. 3). Taken together, these results indicate that in the case of the lsy-6/cog-1 3′ UTR interaction, no simple trend involving the base pair composition around the target sites correlates with the extent of regulation of the 3′ UTR.
Finally, we generated a synthetic 3′ UTR construct consisting of two copies of site #1 with a 26-bp AU linker and CU bp 5′ and 3′ to the target sites (Fig. 12, no. 4). The CU flanking sequences were designed to prevent secondary structure around the target sites. This construct displays a small level of regulation, which is not comparable to the level of regulation observed with the intact cog-1 3′ UTR. Based on these incomplete levels of regulation, we conclude that in those cases in which lsy-6 sites do not confer regulation (e.g., in unc-54, actin, lin-41, lin-14 3′ UTR context) this is not because these 3′ UTR sequences inhibit lsy-6 sites from functioning. Rather, in those contexts where lsy-6 site(s) are functional (i.e., the cog-1 3′ UTR context), contextual features enable the lsy-6 site(s) to function.
Predicting experimental results
Based on theoretical considerations and/or cell culture based transfection assays, researchers in several different laboratories have developed means to assess whether a miRNA may exert regulation on a putative target sequence. Zhao et al. (2005, 2007) have proposed that the energetic features (ΔG of folding) of sequences surrounding a miRNA target site are a predictor for miRNA/target interactions. Long et al. (2007) and Kertesz et al. (2007) utilized overall folding properties of the mRNA target to predict miRNA/target interaction. Grimson et al. (2007) reported that local AU content provides a means to assess successful miRNA/target regulation. We do not find any strong correlation between any of these parameters and the presence and strength of regulation of the many constructs we have generated in this study (Supplemental Table 1).
We also tested each 3′ UTR sequence for predicted folding patterns and observed no notable correlation between regulation and predicted secondary structure (Supplemental Table 1). That is, we found that in some regulated 3′ UTR, the lsy-6 sites lie within secondary structure elements while in several nonregulated 3′ UTR, the sites are exposed (Supplemental Table 1). In sum, the ability to predict whether a 3′ UTR is regulated by a miRNA remains limited.
DISCUSSION
We have described here 156 transgenic lines that express 71 different 3′ UTR sensor constructs and assessed the ability of an endogenously expressed miRNA, acting within its normal cellular context, to regulate these sensor constructs. We draw the following conclusions from our studies.
Seed matches are not a sufficient predictor for miRNA/target interactions
This notion stems from the following results. First, through mutating the 5′ half (3′ in reference to the miRNA) of the first lsy-6 site, we have generated a 3′ UTR that contains two perfect 8mer seed matches to lsy-6 but is not regulated (Didiano and Hobert 2006). Second, we have shown that 13 3′ UTRs with seed matches to lsy-6 are not regulated by lsy-6 (Didiano and Hobert 2006). We have extended this finding here by showing that the two lsy-6 sites do not function in a variety of different contexts, when surrounding sequences are mutated, when transplanted into different, heterologous 3′ UTRs, or when moved within the cog-1 3′ UTR. Our results are consistent with a previous, frequently overlooked study by Vella et al. (2004a) that demonstrated that an engineered 3′ UTR with multiple seed matches to the let-7 miRNA fails to display down-regulation in a natural cellular context.
lsy-6 requires imperfectly complementary target sites to mediate down-regulation of the cog-1 3′ UTR
A cog-1 3′ UTR sensor with a perfectly complementary lsy-6 target site displays no down-regulation, indicating that lsy-6 is primarily entering the ALG-1/ALG-2 miRNA processing pathway and that the bulged base pairs in the miRNA/target heteroduplex are required by the ALG-1/ALG-2 complex for efficient recognition. These in vivo observations bolster recent biochemical work that has shown that depending on the complementarity of the hairpin precursor structure miRNA or siRNAs get sorted into distinct silencing complexes (Forstemann et al. 2007; Steiner et al. 2007; Tomari et al. 2007). This sorting is not 100% discriminative as we observe that within a heterologous context (unc-54 3′ UTR), three repeated, perfectly complementary lsy-6 sites confer a rde-1-dependent, though modest, down-regulation of the sensor in ASEL (L. Cochella and O. Hobert, unpubl.). In light of these issues, the previously reported use of “perfect sensors” to detect sites of endogenous miRNA expression (Mansfield et al. 2004) needs to be viewed with a substantial degree of caution.
G:U wobbles can be tolerated in the lsy-6/cog-1 heteroduplex
We have shown that even though G:U wobble base-pairing can weaken miRNA/target interaction, G:U base-pairing does not abrogate regulation. Even if G:U wobbles are engineered into both lsy-6 sites, regulation is still preserved. The presence of G:U base pairs should therefore not be categorically used to exclude miRNA/target interactions. This notion is also supported by G:U wobbles being present and functionally required in let-7/lin-41 target sites (Vella et al. 2004b).
Asymmetry of multiple target sites
The let-7/lin-41 miRNA target interaction was the first experimentally validated example of a synergistic interaction of two miRNA sites (Vella et al. 2004a). Deletion of either of two let-7 site in the lin-41 target 3′ UTR causes a loss of 3′ UTR regulation. More recent studies further underscored the potency with which two miRNA sites can cooperate to induce 3′ UTR regulation (Grimson et al. 2007; Saetrom et al. 2007). The cog-1 3′ UTR also contains two lsy-6 sites but their functional importance is highly asymmetric. Only deletion of one site strongly affects 3′ UTR regulation; loss of the other site only has a limited effect. Moreover, the spacing of the two sites does not conform with the optimal spacing of cooperating sites; not only are the two lsy-6 sites more distantly spaced than previously reported examples (Saetrom et al. 2007), but their spacing can be even further increased without significant loss of 3′ UTR regulation. Also, in one other 3′ UTR context, that of lin-28, lsy-6 site #1 can also function alone. lsy-6 site #2 likely works via lsy-6 binding, rather than by merely providing some structural context, as mutating the seed disrupts its function. One conceivable function of lsy-6 site #2 is to provide a recruitment platform for lsy-6. Such a recruitment model has been proposed to be important in opening secondary structural elements and has proven powerful in correctly predicting several in vivo miRNA/target interactions (Long et al. 2007).
Another difference between the multiple target site configuration of the lin-41 3′ UTR and the cog-1 3′ UTR is that the linker sequence between the two let-7 sites needs to be of specific length and sequence content (Vella et al. 2004a). In contrast, we can lengthen or invert the linker sequence between the two lsy-6 sites with little effect on regulation.
AU richness or unstructuredness is not a sufficient predictor for lsy-6 target site functionality
Recent bioinformatics analysis combined with transfection studies argues that miRNA target site efficacy depends on surrounding AU-rich sequences (Grimson et al. 2007). We find that this is not the case for lsy-6. Decreasing the AU richness of the lsy-6 site #1 does not impair 3′ UTR regulation, and inclusion of AU-rich flanking sequences to lsy-6 sites in a 3′ UTR that is normally refractory to regulation does not enable regulation. Moreover, an analysis of all constructs generated in this study does not show any strong correlation between AU-richness and regulation (Supplemental Table 1).
Contextual features that promote miRNA activity
A number of constructs that we have generated in this study argue for the overall importance of 3′ UTR features beyond miRNA target sites, which act in conjunction with miRNA target sites and need to be present in a specific configuration relative to miRNA binding sites. We have defined two contextual features that are critical for cog-1 3′ UTR function, both of which are located 3′ to the two lsy-6 sites, namely, a linker region between lsy-6 site #1 and #2 and a region located downstream of lsy-6 site #2, the “orange box.” Both sequences are AU-rich, but not more AU-rich than the whole cog-1 3′ UTR. Neither sequence appears to carry overtly specific sequence information, as they each can be replaced by AU-only sequences (or, in the case of the linker, can be inverted). Also, neither sequence is absolutely required for 3′ UTR regulation in isolation, but when affected in combination (inversion of the linker+deletion of the orange box), a complete loss of 3′ UTR regulation is observed.
The two lsy-6 sites and the linker region and orange box are alone sufficient to impart robust regulation on a reporter gene and can also convert the normally nonregulated 3′ UTR of the actin gene into a miRNA-regulated 3′ UTR (Fig. 3). Notably, the two lsy-6 sites and the linker region and orange box need to be present in a specific configuration, as moving either of the lsy-6 sites out of their normal context results in defects comparable to the deletion of the respective site. However, at least the orange box (and perhaps also the linker region, which we did not test) need not be in absolute proximity to the lsy-6 site #2 as the insertion of either AU-only or GC-only sequences between site #2 and the orange box does not affect orange box function.
Given their similar features and relative placement it is conceivable that the linker and orange box regions may fulfill similar functions. As those functions may not be entirely sequence specific and as many RNA-binding proteins display limited specificity in RNA binding (Fierro-Monti and Mathews 2000), it is conceivable that each region may provide a binding platform for an RNA binding protein. Such protein binding is unlikely to function to improve the recruitment of lsy-6 to its cognate sites, as illustrated by the independent action of site #2 and the orange box. If the orange box were to merely serve to recruit lsy-6 to site #2, then the function of the orange box would be obsolete if site #2 were removed. To the contrary, we find that the effect of a deletion of site #2 is enhanced by loss of the orange box, arguing for an independent function of the orange box. The binding of miRNAs and other factors may thus be independent events and, in analogy to transcriptional enhancesosomes (Merika and Thanos 2001), a joint assembly of multiple trans-acting factors, miRNAs, and proteins may be required to exert regulatory control over a 3′ UTR. Alternatively, the linker region and orange box may cooperate to provide a structural context that allows for miRNA-mediated regulation.
miRNA target predictions
We find a limited correlation with target prediction algorithms that allow the user to provide 3′ UTR sequences (StarMir, Pita). Target genes predicted by other prediction programs also did not show regulation in our assay system (Didiano and Hobert 2006). Other features with proposed predictive values such as target-side surrounding AU-richness or target-side surrounding ΔG values did also not accurately predict our experimental results. Current prediction algorithms calculate target efficacy based on interactions between the mRNA with itself and the mRNA with a miRNA. However, these interactions are occurring in a complex cellular environment in which mRNAs and miRNAs are likely bound by cellular RNA-binding proteins, which is currently impossible to account for in silico. Genomes across phylogeny encode multitudes of proteins of unknown function with predicted RNA-binding motifs. One member of a large family (>100 members in C. elegans) of RNA binding proteins was in fact recently shown to control miRNA binding efficacy to its target mRNA (Kedde et al. 2007).
Is lsy-6 an exemplary miRNA?
At this point it is not clear whether lessons learned from the lsy-6/cog-1 target interaction are generally applicable to other miRNA/target interactions. However, if there is one lesson to be learned from the history of the miRNA field, it is that one should not discard individual case studies as mere oddities (Ruvkun et al. 2004). As our analysis revealed a striking reliance on some presently rather ill-defined contextual features of the 3′ UTR, we consider it of paramount importance to analyze miRNA/target interaction in a cellular environment in which the miRNA and target normally interact. Such cellular environment may impart on the miRNA/target interaction by providing the correct set of cofactors, such as mRNA-binding proteins. Studies that place miRNAs and their target into heterologous context and also often rely on overexpression of the two reactants provide, in our opinion, no more generalizable results than the single miRNA/target interaction that we have dissected here.
CONCLUSIONS
Our study indicates that miRNA/target interactions rely on features beyond the miRNA complementary site in a target gene, a situation that is reminiscent of transcription factor/target recognition. Transcription factor binding sites abound in genome sequences, yet only a limited set are occupied and utilized in vivo, apparently due to relatively poorly understood contextual features of genomic DNA sequence (see for example, Etchberger et al. 2007). Our results therefore raise the possibility that the critical role of 3′ UTR context may limit the number 3′ UTRs that may be regulated by a given miRNA and therefore that less genes in a genome are regulated by miRNAs than currently anticipated.
MATERIAL AND METHODS
DNA constructs
All 3′ UTR sensor constructs are based on the same vector platform generated by cloning 1.8 kb of the ceh-36 promoter (1883 bp to 36 bp upstream of the ATG start codon) into the Hind III/BamH1 sites of the canonical pPD95.75 gfp vector, kindly provided by Andy Fire (Stanford University). All tested 3′ UTRs were generated by cloning into the EcoRI/EagI restriction sites. This process removes the unc-54 3′ UTR and replaces it with experimental 3′ UTRs. All mutant variants of the cog-1 3′ UTR derivatives of a cloned PCR product from genomic DNA isolated from the N2 wild-type strain isolate. All heterologous 3′ UTRs, with the exception of unc-54, which is already present in pPD95.75, were also amplified and cloned from N2 genomic DNA. For the lin-28 3′ UTR constructs, we did not include the first 50 bp of the 3′ UTR since it contained a cryptic lsy-6 site with a poor seed match (5mer) but relatively favorable energetic pairing. With the exception of those constructs described below, mutations were introduced by PCR fusion. The 3′ UTR is amplified as two individual PCR fragments using internal primers, which are overlapping and contain the mutation of interest. These PCR fragments are used as a template in a second round of PCR using the same outside primers containing the restrictions sites (Hobert 2002). PCR fusion products were subcloned into the EcoRI/EagI sites mentioned above. Some mutations near the 3′ end of the 3′ UTR were introduced using long primers containing the mutation of interest and the Eag1 restriction site (e.g., Fig. 8, nos. 5,7,11). In Figure 3, construct no. 12 has the cog-1 3′ UTR minimal element inserted into a vector linker region 3′ of gfp and 5′ of the unc-54 3′ UTR in pPD95.75. In Figure 12, construct no. 3 is a shortened unc-54 3′ UTR; this construct was made by PCR fusion with the internal primers annealing to AU-rich sequences in the unc-54 3′ UTR, thereby removing the intervening sequence and replacing it with the target site inserts. In Figure 12, construct no. 4 was made by cloning annealed complementary primers containing restriction sites. Constructs involving multiple mutations were made using a cloned 3′ UTR containing one of the mutations as a template in a second round of PCR fusion-based mutagenesis. All constructs were sequenced to confirm introduction of mutation. The sequence of all constructs can be found in the Supplementary Information.
For expression of lsy-6 and mir-265 in both ASEL and ASER, two constructs, ceh-36∷;lsy-6 and ceh-36∷;mir-265, were generated. Both microRNAs hairpins were amplified from N2 genomic DNA and either directly fused to the ceh-36 promoter by PCR fusion or first cloned into pPD95.75 containing the ceh-36 promoter and then PCR-amplified before injection.
Transgenic lines
Transgenic lines were generated by injecting the 3′ UTR sensor constructs at 5 ng/μL with rol-6(d) at 100 ng/μL as an injection marker into the gonad of wild-type N2 animals. Stable transgenic lines were selected in the F2 generation.
ceh-36∷;lsy-6 and ceh-36∷;mir-265 were injected as PCR products at 40 ng/μl with rol-6 (100 ng/μl) into ntIs1(gcy-5∷;gfp) animals.
Scoring 3′ UTR regulation
All strains were maintained at 20°C prior to scoring. All lines were scored blind to genotype under a Zeiss Axioplan 2 microscope. To minimize the inclusion of mosaic animals that may have lost the reporter in either ASEL or ASER or both lineages, only those animals were scored in which expression in the AWCL and AWCR neurons was observed (see gfp image in Fig. 1). The fluorescence intensity of the 3′ UTR sensor constructs is compared between ASEL and ASER in each individual animal. This assay is quantitative in that it quantifies the percentage of animals whose gfp expression pattern falls into one of three categories: ASEL>ASER, ASEL=ASER, and ASEL<ASER. Therefore, our assay quantifies regulation levels over a population based on qualitative differences in expression levels in individual animals. We express our sensor constructs at a low concentration resulting in dim expression levels that are ideal for discerning regulation. We have spot-checked our qualitative assessment of gfp expression using AxioVision imaging software (Zeiss, Inc.) that measures the pixilation intensity of ASEL versus ASER. We found that our visual qualitative scoring of each individual animal is sensitive to 25% differences in gfp expression. Importantly, this 25% limit of sensitivity is well below the inherent noise levels of the in vivo system, in which variation of gfp expression intensity from an extrachromosomal array varies from transgenic line to transgenic line and from animal to animal within the same transgenic line. This noise is apparent in nonregulated control unc-54 3′ UTR; ∼15%–30% of animals show ASEL>ASER gfp expression but about the same number of animals also shows ASEL<ASEL gfp expression. This noise in the system makes the assay extremely sensitive to regulation. Whenever a 3′ UTR is lsy-6-regulated, these “noise categories” are transformed into striking levels of differences in gfp intensity in ASEL versus ASER; i.e., there are still wild-type levels of fluorescence intensity in ASER, but fluorescence is dramatically down-regulated in ASEL. Additionally, those constructs that display regulation do so consistently across transgenic lines, despite variation of gfp expression intensity between independent lines.
Due to variability in DNA copy number on transgenic arrays, the levels of noise vary from line to line and construct to construct, thereby preventing a comparison of absolute numbers of animals in each category in different lines. For example, despite tightly controlled injection concentrations, expression levels may vary from one array to another by severalfold. Assume that copy number of DNA on array #1 expression is 2× that of array #2 and assume that both arrays contain DNA that is regulated by lsy-6. Scoring by our method (which does not compare array to array, but compares the ASEL>ASER and ASER>ASEL category within a transgenic line) will reveal that both DNAs are regulated. In contrast, if one were to compare absolute fluorescence intensity from different lines carrying varying copy numbers, one would reach the inaccurate conclusion that the construct carrying more copies is not regulated as well as the one carrying fewer copies.
We have also developed an “asymmetry index” as a metric of regulation. This index is calculated as the total across lines of (ASER>ASEL)–(ASER<ASEL)/Total, and has a range of –1.0 to +1.0, with +1.0 representing absolute asymmetry and perfect regulation. The indices for all examined constructs are shown in Supplemental Table 1. We consider a score of 0.2 as an arbitrary cutoff for regulated versus nonregulated, as roughly indicated by the gray boxes shown in the graphs of the individual figures.
Bioinformatics tools
Alignments of cog-1 3′ UTRs in Figure 7 were made using T-COFFEE with minor corrections by hand (http://tcoffee.vital-it.ch/cgi-bin/Tcoffee/tcoffee_cgi/index.cgi) (Notredame et al. 2000). All microRNA/mRNA target heteroduplexes shown in the paper are based on RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html) (Rehmsmeier et al. 2004), with the exception of those in Figure 2, which display single base pair mismatches based on the wild-type seed #1, and Figure 5, which displays G:U wobbles. Other tools used to generate the data in Supplemental Table 1 are described in the Supplemental Table 1 legend.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Q. Chen for generating transgenic lines; E. Miska, E. Alvarez-Saavedra, and R. Horvitz for providing a mir-265 null allele prior to publication; A. Goldsmith for providing the ceh-36∷;lsy-6 construct; L. Cochella and J. Etchberger for sharing unpublished data; and the Hobert laboratory members for discussions and comments on the manuscript. This work was funded by NIH R01 NS050266-01 and 2R01NS039996-05 to O.H. and by an NIH predoctoral fellowship to D.D.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1082708.
REFERENCES
- Bartel D.P. MicroRNAs. Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Beitzinger M., Peters L., Zhu J.Y., Kremmer E., Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Carthew R.W. Gene regulation by microRNAs. Curr. Opin. Genet. Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Chang S., Johnston R.J., Jr, Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans . Genes & Dev. 2003;17:2123–2137. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D., Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen C.P., Sharp P.A. siRNAs can function as miRNAs. Genes & Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger J.F., Lorch A., Sleumer M.C., Zapf R., Jones S.J., Marra M.A., Holt R.A., Moerman D.G., Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes & Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro-Monti I., Mathews M.B. Proteins binding to duplexed RNA: One motif, multiple functions. Trends Biochem. Sci. 2000;25:241–246. doi: 10.1016/s0968-0004(00)01580-2. [DOI] [PubMed] [Google Scholar]
- Forstemann K., Horwich M.D., Wee L., Tomari Y., Zamore P.D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha I., Wightman B., Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans . Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Kedde M., Strasser M.J., Boldajipour B., Vrielink J.A., Slanchev K., le Sage C., Nagel R., Voorhoeve P.M., van Duijse J., Orom U.A., et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lall S., Grun D., Krek A., Chen K., Wang Y.L., Dewey C.N., Sood P., Colombo T., Bray N., Macmenamin P., et al. A genome-wide map of conserved microRNA targets in C. elegans . Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 . Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Long D., Lee R., Williams P., Chan C.Y., Ambros V., Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- Mansfield J.H., Harfe B.D., Nissen R., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., et al. MicroRNA-responsive “sensor” transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- Merika M., Thanos D. Enhanceosomes. Curr. Opin. Genet. Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- Miska E.A., Alvarez-Saavedra E., Abbott A.L., Lau N.C., Hellman A.B., McGonagle S.M., Bartel D.P., Ambros V.R., Horvitz H.R. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss E.G., Lee R.C., Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Nolde M.J., Saka N., Reinert K.L., Slack F.J. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′ UTR-mediated repression of the let-7 microRNA target gene, hbl-1 . Dev. Biol. 2007;305:551–563. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Palmer R.E., Inoue T., Sherwood D.R., Jiang L.I., Sternberg P.W. Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6.1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev. Biol. 2002;252:202–213. doi: 10.1006/dbio.2002.0850. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. L(ou)sy miRNA targets? Nat. Struct. Mol. Biol. 2006a;13:754–755. doi: 10.1038/nsmb0906-754. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006b;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Robins H., Li Y., Padgett R.W. Incorporating structure to predict microRNA targets. Proc. Natl. Acad. Sci. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G., Wightman B., Ha I. The 20 years it took to recognize the importance of tiny RNAs. Cell. 2004;116:S93–S96. doi: 10.1016/s0092-8674(04)00034-0. [DOI] [PubMed] [Google Scholar]
- Saetrom P., Heale B.S., Snove O., Jr, Aagaard L., Alluin J., Rossi J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., O'Meara M.M., Flowers E.B., Antonio C., Poole R.J., Didiano D., Johnston R.J., Jr, Chang S., Narula S., Hobert O. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics. 2007;176:2109–2130. doi: 10.1534/genetics.107.075648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F.J., Basson M., Liu Z., Ambros V., Horvitz H.R., Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Russell R.B., Cohen S.M. Identification of Drosphila microRNA targets. PLoS Biol. 2003;1:1–13. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G., Slack F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell. Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Steiner F.A., Hoogstrate S.W., Okihara K.L., Thijssen K.L., Ketting R.F., Plasterk R.H., Sijen T. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans . Nat. Struct. Mol. Biol. 2007;14:927–933. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Du T., Zamore P.D. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella M.C., Choi E.Y., Lin S.Y., Reinert K., Slack F.J. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′ UTR. Genes & Dev. 2004a;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella M.C., Reinert K., Slack F.J. Architecture of a validated microRNA∷;target interaction. Chem. Biol. 2004b;11:1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Wharton R.P., Sonoda J., Lee T., Patterson M., Murata Y. The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Zhang L., Ding L., Cheung T.H., Dong M.Q., Chen J., Sewell A.K., Liu X., Yates J.R., 3rd, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N., Tsuchihashi T., McManus M.T., Schwartz R.J., Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]