Abstract
In eukaryotes, the canonical process of initiating protein synthesis on an mRNA depends on many large protein factors and the modified nucleotide cap on the 5′ end of the mRNA. However, certain RNA sequences can bypass the need for these proteins and cap, using an RNA structure-based mechanism called internal initiation of translation. These RNAs are called internal ribosome entry sites (IRESes), and the cap-independent initiation pathway they support is critical for successful infection by many viruses of medical and economic importance. In this review, we briefly describe and compare mechanistic and structural groups of viral IRES RNAs, focusing on those IRESes that are capable of direct ribosome recruitment using specific RNA structures. We then discuss in greater detail some recent advances in our understanding of the intergenic region IRESes of the Dicistroviridae, which use the most streamlined ribosome-recruitment mechanism yet discovered. By combining these findings with knowledge of canonical translation and the behavior of other IRESes, mechanistic models of this RNA structure-based process are emerging.
Keywords: viral IRES, ribosome, translation initiation, RNA structure, Dicistroviridae
INTRODUCTION
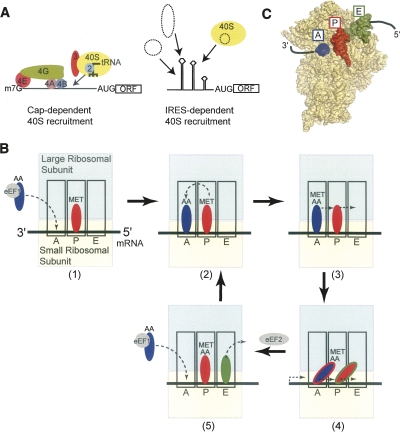
In eukaryotes, the canonical mechanism of initiating translation relies upon many protein factors that recruit the 40S ribosomal subunit to the mRNA, aid in start codon selection, and facilitate the final stages of ribosome assembly (Fig. 1A; Hershey and Merrick 2000; Jackson 2005; Algire and Lorsch 2006). Specifically, the 7-methylguanosine cap on the 5′ end of the mRNA recruits the cap-binding protein, eukaryotic initiation factor (eIF) 4E, which forms the eIF4F complex with eIF4G and eIF4A. This multi-protein·mRNA complex now recruits the 43S complex (40S subunit, eIF3, eIF2, initiator tRNAMet, GTP, eIF1, and eIF1A) to the message, which is placed within the 40S subunit's decoding groove. At this time, the ribosomal subunit scans in a 5′ to 3′ direction until the start codon is located. Recognition of the start codon coupled with GTP hydrolysis, initiation factor release, and 60S subunit recruitment yields an 80S ribosome prepared to enter the elongation phase of protein synthesis (Fig. 1B,C; Pestova et al. 2007).
FIGURE 1.
Mechanisms to recruit the small ribosomal (40S) subunit and ribosomal architecture. (A) Simple diagram comparing canonical cap-dependent translation (left) and IRES driven (right) mechanisms of recruiting the ribosome. In canonical initiation, many factors are needed whereas IRES RNAs use specific RNA sequences either through direct ribosome interaction or with the aid of other factors (dashed ovals). (B) At upper left is a diagram of an 80S ribosome produced by the canonical scanning-dependent initiation mechanism. Shown are the three tRNA-binding sites important for decoding the encrypted message on the mRNA, facilitating the peptidyl transferase mechanism, and maintaining the fidelity of the ribosome. These three sites are the acceptor (A) site (blue), peptidyl (P) site (red), and the exit (E) site (green). The large subunit is shown in cyan, and the small subunit is colored yellow. The elongation cycle is depicted: (1) A tRNA is delivered to the A-site by eEF1, (2) the peptide bond is formed, (3) the A- and P-site tRNAs move on the 40S subunit to form a hybrid state, (4) the hybrid-state tRNA-containing ribosome is a substrate for eEF2-catalyzed translocation (the two-color tRNAs in the figure reflect this hybrid state), and (5) the post-translocation ribosome has tRNAs in the P- and E-sites and can now receive another tRNA in the A-site and eject the deacylated tRNA from the E- site. (C) Crystal structure of a bacterial ribosome bound to three tRNA molecules and mRNA, showing their arrangement in the three sites and with mRNA (Selmer et al. 2006). The 50S subunit is not shown. The pathway of the mRNA into the decoding groove is depicted as a black line. The coloring of the structure is the same as in B. The A-site tRNA (blue) is not complete, as only a portion was visible in the crystal structure.
In addition to this cap-dependent pathway, an alternate cap-independent means of initiating translation exists in which the ribosome is recruited, positioned, and activated on a site other than the 5′ end of the RNA by a specific RNA sequence called an internal ribosome entry site (IRES) (Fig. 1A; Jackson 2005). First discovered in viral mRNAs and since found in cellular messages, IRESes often substantially reduce the need for protein factors (Doudna and Sarnow 2007; Elroy-Stein and Merrick 2007). Although the need for various factors and the interactions with those factors differ, it is clear that the IRES RNA drives translation initiation in these systems (Hellen and Sarnow 2001; Stoneley and Willis 2004). This is most strongly illustrated in IRESes that bind directly to the ribosome. In this review, we introduce the diversity of architecture and mechanisms found in viral IRES RNAs and some common features of IRESes known to bind directly to the ribosome. Focusing on the most mechanistically streamlined IRESes, we then discuss key aspects of this IRES’ structure in regard to direct ribosome recruitment and the implications for function.
DIFFERENT APPROACHES TO CAP-INDEPENDENT TRANSLATION INITIATION
Viral IRES groups
The viral IRESes are divided into groups based on related secondary structures and functional requirements for protein factors. At one end of the spectrum are certain IRESes from the picornaviruses, such as encephalomyocarditis virus (EMCV) (Jang et al. 1988), poliovirus (Pelletier et al. 1988), and hepatitis A virus (HAV) (Glass et al. 1993). These IRESes are unable to bind directly to the ribosome, and to date there is no definitive evidence that they directly contact the ribosome (except at and adjacent to the start codon). These IRESes require a subset of the canonical initiation factors to function (Thompson and Sarnow 2000). For example, EMCV requires eIF2, eIF3, eIF4A, ATP, and the cleaved C-terminal portion of eIF4G (Pestova et al. 1996a,b), and other picornavirus IRESes rely on a similar set of factors (Pestova et al. 1996a; Kolupaeva et al. 1998; Pilipenko et al. 2000). These IRESes can be subdivided into two groups (Jackson and Kaminski 1995; Jackson 2005), each with related secondary structures (Pilipenko et al. 1989a,b). In one group are those that function efficiently in rabbit reticulocyte lysate (RRL) and that initiate translation at the 3′ end of the IRES; members of this group include EMCV, foot-and-mouth disease virus (FMDV), and Theiler's murine encephalomyelitis virus (TMEV). In another group are those that initiate downstream of the IRES RNA and that usually require supplementation of RRL with a source of additional protein factors (such as HeLa cell extract) called IRES trans-activating factors (ITAFs). Members of this group include poliovirus, rhinovirus, and coxsackievirus. Though the requirements for canonical factors are similar among these IRESes, the need for and identity of ITAFs varies (Hellen and Sarnow 2001; Jackson 2005).
Midway in the spectrum are viral IRESes that bind directly to the ribosome and need fewer canonical initiation protein factors to begin translation. In this group the IRES RNA prefolds into an extended structure with local areas of compact folded RNA (Kieft et al. 1999); some of these areas involve an RNA pseudoknot (Wang et al. 1995). Members of this group include the hepatitis C virus (HCV) (Tsukiyama-Kohara et al. 1992) and classical swine fever virus (CSFV) (Rijnbrand et al. 1997). Recent evidence suggests that a subset of picornaviruses, including porcine teschovirus 1 (PTV-1), possess IRESes that fall into this group (Pisarev et al. 2004; Chard et al. 2006). Through direct interactions with the small ribosomal subunit and eIF3, these IRESes place the start codon in the decoding site, eliminating ribosomal scanning (Reynolds et al. 1996; Pestova et al. 1998; Kolupaeva et al. 2000a,b). These IRESes also use the eIF2·tRNA·GTP ternary complex and upon GTP hydrolysis, codon–anticodon dependent release of GDP/phosphate, and the action of eIF5B, the 60S ribosomal subunit binds to form the 80S ribosome–IRES complex, followed by the first peptide bond formation (Pestova et al. 1998; Unbehaun et al. 2004; Locker et al. 2007). Various IRES RNA secondary structure elements, including a pseudoknot near the 3′ end of the IRES (Wang et al. 1995), have been shown to influence the 40S subunit, eIF3, and ternary complex recruitment, as well as eIF2 release and 80S ribosome formation (Wang et al. 1995; Kieft et al. 2001; Ji et al. 2004; Otto and Puglisi 2004; Locker et al. 2007).
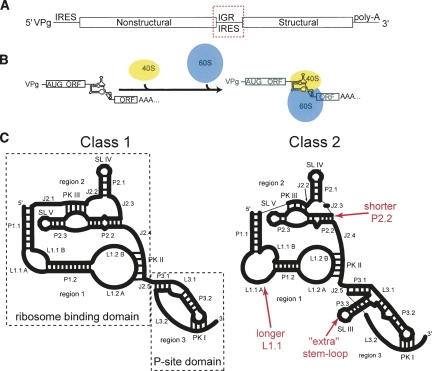
At the very end of the spectrum reside viral IRESes that require no protein factors to directly interact with and recruit both the 40S and 60S subunits and initiate translation without initiator tRNAMet (for review, see Jan 2006). The IRES RNAs of this group form a more intricate fold than the other groups that includes two independently folded domains (Costantino and Kieft 2005). The two domains are a compact double-nested pseudoknot structure that provides the affinity for the 40S subunit (Jan and Sarnow 2002; Nishiyama et al. 2003) and an additional pseudoknot element at the 3′ end of the IRES that is placed into the P-site in the small subunit's decoding groove (Wilson et al. 2000a; Kanamori and Nakashima 2001). These IRESes are found within the intergenic region of the viral RNA genomes of the Dicistroviridae (Fig. 2A), which include Plautia stali intestine virus (PSIV) (Sasaki and Nakashima 1999), cricket paralysis virus (CrPV) (Wilson et al. 2000b), and Taura syndrome virus (TSV) (Mari et al. 2002; Hatakeyama et al. 2004). Using an all RNA-based mechanism, these IRESes position the 80S ribosome at the 5′ end of the second cistron without initiator tRNAMet, ATP, GTP, or any protein factors (Fig. 2B; Sasaki and Nakashima 1999, 2000; Wilson et al. 2000a,b; Jan and Sarnow 2002; Nishiyama et al. 2003; Cevallos and Sarnow 2005; Pisarev et al. 2005). Once on the IRES, delivery of a tRNA to the A-site by eIF1A induces the ribosome to translocate prior to peptide bond formation and then to move to elongation (Jan et al. 2003; Pestova and Hellen 2003). Thus, translation for these IRESes begins from the A-site and from a non-AUG codon. The first decoded codon used in this mechanism can in fact be any codon, though some are more efficient than others (Shibuya et al. 2003).
FIGURE 2.
The Dicistroviridae IGR IRESes. (A) The single-stranded positive sense RNA genome of Dicistroviridae is diagrammed. The mRNA is linked to a 5′-terminal VPg peptide and has a poly-A tail. The IRES is located in the intergenic region (IGR) between two open reading frames and is shown in the red box. (B) The mechanism of initiation on the Dicistroviridae IGR IRES is shown. These IRESes directly interact with both ribosomal subunits to assemble the 80S ribosome without the use of any other protein factors. (C) The IGR IRESes can be divided into two types, and cartoon secondary structures of the two types are shown. At left, the class 1 are represented by the PSIV IGR IRES; at right, the class 2 are represented by the TSV IGR IRES. The secondary structures presented are to scale with respect to loop length, and numbers of base pairs are shown in the helices. The differences between the two classes are illustrated by red text. With regard to the labels, P stands for helix (paired), L means loop, J stands for junction, PK denotes pseudoknot, and SL means stem–loop. The three regions of the IRES are labeled, and the two functional and structural domains are boxed. Note that we use the term “region” to refer to a group or collection of logically connected secondary structural elements or interactions, but “domain” is reserved for entities experimentally shown to fold independently. Hence, regions 1 and 2 fold together to form the “ribosome-binding domain,” and region 3 is itself also a domain, the “P-site domain” (also often referred to as just “domain 3”).
Similarities between two of the IRES groups
Members of the last two IRES groups described above bind directly to the ribosome, and in both cases specific RNA structure drives this process. Despite differences in RNA structures and factor requirements, similarities are emerging between these IRESes in terms of their interactions with the ribosome. Because none of the members of the other groups of IRESes directly binds the ribosome and there is no direct structural data regarding their three-dimensional conformations, we cannot yet draw parallels between those IRESes and the IRESes that directly bind the ribosome.
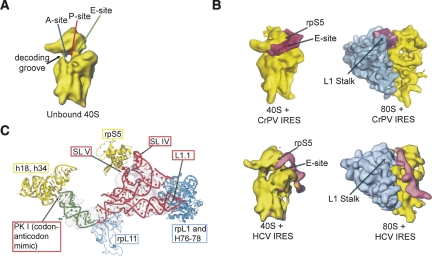
Similarities between the Dicistroviridae IGR IRESes and the HCV IRES are evident in cryo-electron microscopy (cryo-EM) reconstructions of the HCV and CrPV IRESes bound to the 40S subunit and 80S ribosome (Fig. 3A,B; Spahn et al. 2001b, 2004; Boehringer et al. 2005; Schuler et al. 2006). Both the CrPV IGR IRES and HCV IRES make direct contact with the E-site of the small ribosomal subunit, and both change the 40S subunit's conformation, resulting in the “head” rotating relative to the “body.” Because both IRESes contact the back or solvent side of the head, this interaction might be necessary for IRES-induced conformational changes in the small ribosomal subunit that are critical for function. Indeed, when the domain of the HCV IRES RNA (domain II) making this contact is removed, the 40S subunit conformational changes no longer occurred (Spahn et al. 2001b), and mutations to this domain inhibit the formation of the 80S ribosome on the HCV IRES (Ji et al. 2004; Otto and Puglisi 2004; Locker et al. 2007). Furthermore, similar 40S subunit conformational changes are seen in response to eIF1 and eIF1A (factors involved in scanning and start codon selection) binding to the subunit (Passmore et al. 2007). These changes are interpreted as switching the subunit from an “open” scanning conformation to a “closed” conformation (ribosome is clamped down on the mRNA), suggesting parallels between the scanning- and IRES-dependent mechanisms. In the context of the 80S ribosome, both the CrPV IRES and the HCV IRES interact with a feature of the large subunit called the L1 stalk (also known as the L1 protuberance) (Fig. 3; Nishiyama et al. 2003; Spahn et al. 2004; Boehringer et al. 2005; Pfingsten et al. 2006; Schuler et al. 2006). Hence, this interaction could also be important in IRES-driven translation, although the location of the interaction differs between the two IRESes. It should also be noted that the conformations of the 80S ribosome when complexed with each of the two IRESes are different from one another (Boehringer et al. 2005; Schuler et al. 2006), suggesting that, although both IRESes contact the L1 stalk, the effects of this interaction differ. More details regarding these interactions and the Dicistroviridae IGR IRESes are contained later in this review.
FIGURE 3.
Viral IRES interactions with the ribosome. (A) Cryo-EM reconstruction of a naked 40S subunit, with the approximate locations of the three tRNA binding sites (Spahn et al. 2001b). (B) Cryo-EM reconstructions of viral IRES RNAs bound to the ribosome. At upper left is CrPV IGR IRES bound to the small subunit with a resolution of ∼20 Å; at upper right the IRES is bound to the 80S ribosome with a resolution at ∼17.3 Å (Spahn et al. 2004; Schuler et al. 2006). At lower left is the HCV IRES bound to the 40S subunit with a resolution at ∼20 Å (Spahn et al. 2001b); at lower right is the HCV IRES bound to the 80S ribosome at ∼15 Å (Boehringer et al. 2005). The IRESes are colored magenta, the small ribosomal subunit is colored yellow, and the large ribosomal subunit is colored blue. Key ribosome structural elements and ribosomal protein locations are labeled. (C) Crystal structures of the two IGR IRES domains are shown docked into the cryo-EM density of the CrPV IRES bound to an 80S ribosome at ∼7.3 Å (Schuler et al. 2006). The ribosome-binding domain from PSIV is in red, and the P-site domain is in green (Pfingsten et al. 2006; Costantino et al. 2008). The modeled positions of ribosome elements that are likely to interact with the IRES are shown and labeled with 40S subunit elements in yellow and 60S elements in blue, and the relevant IRES structures are labeled. The positions of these proteins and rRNA are based on the model of the evolutionarily conserved core of the yeast 80S ribosome (Spahn et al. 2001a; Schuler et al. 2006).
In addition to ribosome conformational changes, the conformation of both the CrPV and HCV IRES RNAs change upon movement from the 40S subunit-bound to 80S ribosome-bound forms (Spahn et al. 2004; Boehringer et al. 2005). Thus, these representatives of the two IRES groups make similar ribosomal contacts and both possess a combination of static and dynamic IRES structural elements.
THE DICISTROVIRIDAE IGR IRESES AND DIRECT RIBOSOME RECRUITMENT
The Dicistroviridae IGR IRESes (hereafter referred to as the “IGR IRESes”) provide a starting point for studying RNA-structure–based ribosome recruitment without the added layer of complexity generated by the need for proteins. The remainder of this review focuses on IGR IRES structure and direct interactions with the ribosome.
Diversity and similarity within the IGR IRES group
Diversity exists within the Dicistroviridae IGR IRESes in terms of their primary sequence, but all are predicted to adopt a similar secondary structure consisting of two highly conserved stem–loops that interact with the small ribosome subunit (SL IV, SL V), multiple helices, as well as three pseudoknot tertiary interactions (Kanamori and Nakashima 2001; Nishiyama et al. 2003; Jan 2006). However, secondary structure differences in all three regions of the IGR IRES RNA warrant division into two classes (Fig. 2C; Kanamori and Nakashima 2001; Nishiyama et al. 2003; Jan 2006). Examples of class 1 IGR IRESes are CrPV and PSIV, and an example of a class 2 is TSV. Compared to the class 1 IGR IRESes, the class 2 contain additional structural elements, possess a longer but conserved sequence in loop L1.1, and some sequences found in the crystal structure of the PSIV IGR IRES ribosome-binding domain (discussed in more detail in the next section) to be responsible for maintaining specific intramolecular contacts are altered in class 2 relative to class 1 (Nishiyama et al. 2003; Hatakeyama et al. 2004). However, the class 2 IGR IRESes still adopt a three-dimensional fold similar to that found in the class 1 that contains two independently folded domains and which presents the strongly conserved apical loops of SL IV and SL V for binding to the 40S subunit (Pfingsten et al. 2006, 2007). Although the mechanism of 40S subunit binding is conserved in both classes, evidence suggests that the interactions and manipulations of the large ribosomal subunit are different due to the presence of SL III found in the class 2 IGR IRESes (Pfingsten et al. 2007). Because domain 3 (P-site domain) structurally and functionally mimics a tRNA (see the next section) (Jan et al. 2003; Costantino et al. 2008), it might be that the additional stem–loop of the class 2 IGR IRESes mimics tRNAs with a long variable arm, but this remains to be tested.
Crystal structures of the IGR IRES
Within the last two years, crystal structures of both independently folded domains of the IGR IRES have been solved (Pfingsten et al. 2006; Costantino et al. 2008). Specifically, the ribosome-binding domain of the PSIV IRES (Fig. 2C) and the P-site domain of the CrPV IRES were solved to 3.1 and 2.4 Å, respectively (Fig. 2C). When these two structures are combined, they provide the first complete high-resolution structural picture of an IRES RNA (Fig. 3C). Although a detailed description of these structures is beyond the scope of this Mini-Review, we discuss several features important for IRES-ribosome interactions.
The structure of the larger, double-pseudoknotted ribosome-binding domain of the PSIV IGR IRES shows that the two stem–loops (SL IV and SLV) that interact directly with the 40S subunit are positioned adjacent to one another by underwound helix P2.2 (Pfingsten et al. 2006). This helix is involved in many noncanonical intramolecular interactions with the “core” of the fold that involve highly conserved nucleotides (Kanamori and Nakashima 2001). Within this folded structure, regions 1 and 2 lie side-by-side, and this arrangement places loop L1.1 of the IRES in position to interact with the L1 stalk of the large ribosomal subunit (Pfingsten et al. 2006). The structure suggests that the ribosome-binding surfaces are prepositioned when the IRES folds, providing an explanation for direct recruitment of both subunits. In addition, the structure reveals likely areas of stable structure and dynamic elements that might work together within the IRES recruitment process (Pfingsten et al. 2006). This structure agrees very well with models based on cryo-EM reconstructions (Spahn et al. 2004; Schuler et al. 2006).
The structure of the CrPV IRES domain of the IRES that docks into the small subunit's P-site reveals that this pseudoknot element closely mimics the mRNA codon–tRNA anticodon interaction in a novel form of structural mimicry (Costantino et al. 2008). In the IGR IRES this domain can be removed without affecting 80S ribosome recruitment, but it must be present for translation initiation (Jan and Sarnow 2002; Nishiyama et al. 2003; Costantino and Kieft 2005). When placed in the P-site, this domain likely mimics interactions normally associated with mRNA and tRNA, and this might put the ribosome into a pretranslocation state. When this structure is combined with cryo-EM reconstructions and other crystal structures, its docked position, overall location of the IRES on the ribosome (over the E-site and in the P-site), and functional assays studying the actions of elongation factors lead to the hypothesis that the IGR IRESes mimic a P/E hybrid state tRNA (Costantino et al. 2008; Spahn et al. 2004; Schuler et al. 2006; Yamamoto et al. 2007).
IGR IRES contacts to the 40S subunit
Combining the crystal structure with cryo-EM reconstructions and traditional biochemical approaches reveals details of the interactions between the IGR IRESes and the 40S subunit that are necessary to recruit the subunit (Fig. 3C). SL IV and SL V contact the back of the “head” of the 40S subunit, in position to interact with two ribosomal proteins (rp): rpS5 (in position to interact with SL IV) and an unidentified protein called rpSX (in position to interact with SL V) (Schuler et al. 2006). SL IV cross-links to rpS25 but not rpS5, suggesting that rpS5 might interact with SL V (Nishiyama et al. 2007). Because a prokaryotic homolog for rpS25 does not exist, it has been suggested this could be why the IGR IRESes do not function in bacteria (Nishiyama et al. 2007). Further investigation is required to sort out the details of stem–loop interactions with proteins at the IRES–40S interface; however, some interesting observations can be made by comparing the HCV IRES with the IGR IRESes. The HCV IRES RNA domain II cross-links to rpS5 (Fukushi et al. 2001); this is the HCV IRES domain previously mentioned as contacting the back side of the “head” and changing the conformation of the 40S subunit (Spahn et al. 2001b). If the conserved sequences within the apical loop of this domain are mutated, the IRES’ ability to form the 80S–IRES complex is substantially reduced (Reynolds et al. 1996; Odreman-Macchioli et al. 2001; Kalliampakou et al. 2002; Otto and Puglisi 2004; Locker et al. 2007), establishing a correlation between the HCV IRES’ interaction with rpS5, 40S subunit conformational change, eIF2 release, and 80S ribosome formation. This indicates that an interaction with rpS5 is potentially necessary for function, raising the possibility that the same is true of the IGR IRESes.
The proposed interactions between the IGR IRES and rpS5 and rpS25 are critical for affinity and proper placement of the IGR IRESes; it is interesting to consider additional implications of these interactions based on their bacterial counterpart's known functions in translation. Besides binding tRNA located in the E-site of the ribosome, the bacterial counterpart of rpS5 (rpS7), interacts with bacterial rpS11 to help form the exit channel around the mRNA (Wower et al. 1993; Yusupov et al. 2001). When this interaction is mutated, the large ribosomal subunit's ability to interact with the small subunit-containing preinitiation complex is reduced (Robert and Brakier-Gingras 2003). In addition, this mutation increases the small ribosomal subunit's capacity to bind mRNA but the translational fidelity of the ribosome is decreased, attributable to increased flexibility in the ribosome's “head” and a loss in communication between conformational changes in the A- and E-sites (Robert and Brakier-Gingras 2003). In eukaryotes, a rpS5–rpS14 is positioned to take the place of the bacterial rpS7–rpS11 (Passmore et al. 2007). Hence, it possible that rpS25 aids the rpS5–rpS14 interaction to promote the conformational changes in the 40S ribosomal subunit. Recently, human rpS5 was substituted for yeast rpS5, resulting in a viable yeast strain that nonetheless showed decreased growth (Galkin et al. 2007). These ribosomes appeared to have elongation or termination defects, reduced association of eEF3 and eEF1A, increased frameshifting activity, and hyperaccurate recognition of the UAA stop codon. Perhaps most interesting in this Mini-Review, these hybrid ribosomes have increased ability to bind to the CrPV IGR IRES and to translate messages driven by this IRES, suggesting that rpS5 is indeed a primary determinant of small ribosome recruitment by the IGR IRESes.
In the case of rpS25, the protein's exact function is unknown, but it is necessary for translation (Ferreira-Cerca et al. 2005) and it cross-links not only to rpS5 (Tolan and Traut 1981; Gross et al. 1983) but also to mRNA when in the 80S ribosome (Takahashi et al. 2005). Hence, these elements are in close enough proximity for an interaction to occur and the putative similarities between rpS7/rpS11 interaction in bacteria and rpS5/rpS14 interaction in eukaryotes suggest the hypothesis that the IRES’ interactions with rpS5 and rpS25 promote 40S subunit conformational changes and subsequent 60S subunit association.
IGR IRES contacts to the 60S subunit
The CrPV IRES can recruit the 80S ribosome by two mechanisms: initial 40S subunit binding followed by 60S subunit binding, or direct IRES binding to preformed 80S ribosomes (Pestova et al. 2004). Cryo-EM reconstructions and docked X-ray structures show how the IRES contacts the 60S ribosomal subunit within the 80S ribosome and also reveal that the CrPV IRES changes structure upon subunit joining (Spahn et al. 2004; Pfingsten et al. 2006; Schuler et al. 2006). Specifically, when compared to the 40S subunit bound form, the 80S ribosome-bound IRES seems to retract slightly from the A-site toward the E-site, and some of the contacts with the 40S subunit are altered (Spahn et al. 2004). In addition, although the overall global conformation of the IRES is similar between the 40S subunit and 80S ribosome-bound forms, there are some parts of the IRES’ density that appear as a more-pronounced globular shape and others that appear thinner. Higher-resolution cryo-EM structures and docking of the crystal structure of the PSIV IGR IRES’ ribosome-binding domain into the cryo-EM structures are revealing what parts of the IRES are involved in these conformational changes (Pfingsten et al. 2006; Schuler et al. 2006), but the precise roles of these changes in initiation are not yet clear.
The IGR IRES uses a highly conserved loop (L1.1) to interact with a feature of the 60S subunit called the L1 stalk. Mutating this loop impairs its ability to interact with the 60S subunit and form the 80S ribosome–IRES complex (Pfingsten et al. 2006), and chemical probing found that a nucleotide within this loop of the PSIV IGR IRES is protected upon formation of the 80S ribosome (Nishiyama et al. 2003). The L1 stalk contains rpL1 (bacterial homolog is rpL10) and rRNA helix H77, suggesting the IRES contacts one or both of these elements. Hypotheses regarding the significance of the IRES’ interaction with the L1 stalk can be developed by examining the known functions of the stalk in translation. During translation, the L1 stalk interacts with the T-loop of E-site–bound tRNA (Agrawal et al. 2000; Yusupov et al. 2001), thus the IGR IRES might locally mimic this interaction. In bacterial ribosomes, the dynamic L1 stalk not only moves during the translocation process but might facilitate the movement of the deacylated tRNA from the P-site to the E-site (Valle et al. 2003) and is believed to aid in the removal of the E-site tRNA (Gomez-Lorenzo et al. 2000; Harms et al. 2001; Yusupov et al. 2001). Hence, it is possible that the L1 stalk aids in the movement of the IRES during translocation before peptide bond formation and also in removal of the IRES from the ribosome. Cryo-EM reconstructions suggest that the portion of the IRES contacting the L1 stalk undergoes structural changes as it goes from the 40S subunit bound to the 80S subunit bound state (Spahn et al. 2001b). This change is propagated through the IRES to alter how the tRNA-mimicking domain is positioned in relation to the P-site of the decoding groove. Hence, functionally important allosteric changes in both the IRES and the L1 stalk appear to be induced by this interaction.
INSIGHTS INTO THE MECHANISM OF TRANSLATION INITIATION ON THE IGR IRESES
To function, the IGR IRES must position and prime an 80S ribosome to accept an aminoacylated-tRNA brought to the A-site by eEF1A and then undergo a translocation event facilitated by eEF2. By interacting with proteins whose prokaryotic homologs bind the E-site tRNA (Wower et al. 1993; Harms et al. 2001; Yusupov et al. 2001), the IRES induces conformational changes in the 80S ribosome and appears to manipulate it into adopting a pretranslocation state. In bacterial ribosomes programmed to adopt the pretranslocation state, the tRNAs are in a hybrid state and EF-G interacts with the ribosome and drives translocation (Dorner et al. 2006; Spiegel et al. 2007). Likewise, in eukaryotes eEF2 has an affinity for the 80S ribosome–IGR IRES complex, and this interaction is important for the translocation event (Yamamoto et al. 2007). In IGR IRES–80S ribosome complexes, stable binding of the first aminoacylated-tRNA to the first codon of the ribosome depends on the presence of both eEF2 and eEF1A, suggesting that translocation from the A- to the P-site (before a peptide bond is made) must occur for stable binding of the tRNA (Yamamoto et al. 2007). It is only when eEF2 promotes translocation that tRNA becomes more stably bound in the P-site. This behavior could be explained by the hypothesis that the IRES mimics a P/E hybrid state of tRNA and moves the ribosome into a pretranslocation state (Yamamoto et al. 2007). The recently solved crystal structure of the P-site domain of CrPV and the biochemical results illustrating the dynamic nature of this domain also support this hypothesis (Costantino et al. 2008).
Events after the initial IGR IRES/ribosome translocation event remain poorly understood, as this state has not been studied in great detail structurally or biochemically. However, one might hypothesize that after the eEF2-triggered translocation event, the IRES’ P-site domain (domain/region 3) could move to the E-site like a deacylated tRNA (Valle et al. 2003). Although speculative at this point, the IGR IRES’ structure could undergo conformational changes and allow the IRES’ structures to interact with the E-site and maintain contact with rpS5, rpS25, and the L1 stalk. When the time comes for the 80S ribosome to release the IRES, the IRES’ interaction with the L1 stalk could play a pivotal role in the process, similar to the role of the L1 stalk in bacterial ribosomes (Harms et al. 2001; Yusupov et al. 2001).
CONCLUSION AND SUMMARY
The Dicistroviridae IGR IRESes use the most streamlined mechanism of all known IRESes. In the last few years, a combination of structural, biophysical, biochemical, and genetic approaches has provided insight into how these IRES RNAs fold, interact with the ribosome, and manipulate the translation machinery. Through a specific conserved RNA fold, these IRESes contact specific parts of both the large and small subunits and in so doing change the conformation of the ribosome. The nature of these manipulations and the position of the IGR IRES suggest that they mimic tRNA–ribosome interactions to induce the ribosome into a pretranslocation conformation. In this way, the IGR IRESes initiate translation by “fast-forwarding” the ribosome to the canonical elongation cycle. Parts of this model remain speculative, but it provides the basis for the next round of experiments aimed at fully understanding the function of this remarkable RNA.
ACKNOWLEDGMENTS
The authors thank Christian Spahn for providing us with cryo-EM density files and Professors Elan Eisenmesser and Christopher Hellen for critical reading of this review. IRES RNA studies in the Kieft Laboratory are supported by grants R01 GM072560 and R03 AI072187 from the National Institutes of Health and by Research Scholar Grant 0805801GMC from the American Cancer Society.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.987808.
REFERENCES
- Agrawal R.K., Spahn C.M., Penczek P., Grassucci R.A., Nierhaus K.H., Frank J. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J. Cell Biol. 2000;150:447–460. doi: 10.1083/jcb.150.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire M.A., Lorsch J.R. Where to begin? The mechanism of translation initiation codon selection in eukaryotes. Curr. Opin. Chem. Biol. 2006;10:480–486. doi: 10.1016/j.cbpa.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Boehringer D., Thermann R., Ostareck-Lederer A., Lewis J.D., Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: Remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Cevallos R.C., Sarnow P. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J. Virol. 2005;79:677–683. doi: 10.1128/JVI.79.2.677-683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chard L.S., Bordeleau M.E., Pelletier J., Tanaka J., Belsham G.J. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J. Gen. Virol. 2006;87:927–936. doi: 10.1099/vir.0.81546-0. [DOI] [PubMed] [Google Scholar]
- Costantino D., Kieft J.S. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino D.A., Pfingsten J.S., Rambo R.P., Kieft J.S. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner S., Brunelle J.L., Sharma D., Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat. Struct. Mol. Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A., Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews M.B., Sonenberg N., Hershey J.W.B., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 129–153. [Google Scholar]
- Elroy-Stein O., Merrick W.C. Translation initiation via cellular internal ribosome entry sites. In: Mathews M.B., Sonenberg N., Hershey J.W.B., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 155–172. [Google Scholar]
- Ferreira-Cerca S., Poll G., Gleizes P.E., Tschochner H., Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Fukushi S., Okada M., Stahl J., Kageyama T., Hoshino F.B., Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- Galkin O., Bentley A.A., Gupta S., Compton B.A., Mazumder B., Kinzy T.G., Merrick W.C., Hatzoglou M., Pestova T.V., Hellen C.U., et al. Roles of the negatively charged N-terminal extension of Saccharomyces cerevisiae ribosomal protein S5 revealed by characterization of a yeast strain containing human ribosomal protein S5. RNA. 2007;13:2116–2128. doi: 10.1261/rna.688207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass M.J., Jia X.Y., Summers D.F. Identification of the hepatitis A virus internal ribosome entry site: In vivo and in vitro analysis of bicistronic RNAs containing the HAV 5′ noncoding region. Virology. 1993;193:842–852. doi: 10.1006/viro.1993.1193. [DOI] [PubMed] [Google Scholar]
- Gomez-Lorenzo M.G., Spahn C.M., Agrawal R.K., Grassucci R.A., Penczek P., Chakraburtty K., Ballesta J.P., Lavandera J.L., Garcia-Bustos J.F., Frank J. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 Å resolution. EMBO J. 2000;19:2710–2718. doi: 10.1093/emboj/19.11.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B., Westermann P., Bielka H. Spatial arrangement of proteins within the small subunit of rat liver ribosomes studied by cross-linking. EMBO J. 1983;2:255–260. doi: 10.1002/j.1460-2075.1983.tb01414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. High-resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Hatakeyama Y., Shibuya N., Nishiyama T., Nakashima N. Structural variant of the intergenic internal ribosome entry site elements in dicistroviruses and computational search for their counterparts. RNA. 2004;10:779–786. doi: 10.1261/rna.5208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen C.U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B., Merrick W.C. Pathway and mechanism of initiation of protein synthesis. In: Mathews M.B., editor. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 33–88. [Google Scholar]
- Jackson R.J. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- Jackson R.J., Kaminski A. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Jan E., Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- Jan E., Kinzy T.G., Sarnow P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S.K., Krausslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Fraser C.S., Yu Y., Leary J., Doudna J.A. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl. Acad. Sci. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliampakou K.I., Psaridi-Linardaki L., Mavromara P. Mutational analysis of the apical region of domain II of the HCV IRES. FEBS Lett. 2002;511:79–84. doi: 10.1016/s0014-5793(01)03300-2. [DOI] [PubMed] [Google Scholar]
- Kanamori Y., Nakashima N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7:266–274. doi: 10.1017/s1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft J.S., Zhou K., Jubin R., Murray M.G., Lau J.Y., Doudna J.A. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- Kieft J.S., Grech A., Adams P., Doudna J.A. Mechanisms of internal ribosome entry in translation initiation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:277–283. doi: 10.1101/sqb.2001.66.277. [DOI] [PubMed] [Google Scholar]
- Kolupaeva V.G., Pestova T.V., Hellen C.U., Shatsky I.N. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- Kolupaeva V.G., Pestova T.V., Hellen C.U. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000a;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva V.G., Pestova T.V., Hellen C.U. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA. 2000b;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N., Easton L.E., Lukavsky P.J. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari J., Poulos B.T., Lightner D.V., Bonami J.R. Shrimp Taura syndrome virus: Genomic characterization and similarity with members of the genus cricket paralysis-like viruses. J. Gen. Virol. 2002;83:915–926. doi: 10.1099/0022-1317-83-4-915. [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Yamamoto H., Shibuya N., Hatakeyama Y., Hachimori A., Uchiumi T., Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T., Yamamoto H., Uchiumi T., Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007;35:1514–1521. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odreman-Macchioli F., Baralle F.E., Buratti E. Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J. Biol. Chem. 2001;276:41648–41655. doi: 10.1074/jbc.M104128200. [DOI] [PubMed] [Google Scholar]
- Otto G.A., Puglisi J.D. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- Passmore L.A., Schmeing T.M., Maag D., Applefield D.J., Acker M.G., Algire M.A., Lorsch J.R., Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Kaplan G., Racaniello V.R., Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell. Biol. 1988;8:1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Hellen C.U. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Hellen C.U., Shatsky I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996a;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky I.N., Hellen C.U. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996b;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Lomakin I.B., Hellen C.U. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Lorsch J.R., Hellen C.U. The mechanism of translation initiation in eukaryotes. In: Mathews M.B., Sonenberg N., Hershey J.W.B., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- Pfingsten J.S., Costantino D.A., Kieft J.S. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten J.S., Costantino D.A., Kieft J.S. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J. Mol. Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E.V., Blinov V.M., Chernov B.K., Dmitrieva T.M., Agol V.I. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 1989a;17:5701–5711. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E.V., Blinov V.M., Romanova L.I., Sinyakov A.N., Maslova S.V., Agol V.I. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: An analysis of the segment controlling translation and neurovirulence. Virology. 1989b;168:201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- Pilipenko E.V., Pestova T.V., Kolupaeva V.G., Khitrina E.V., Poperechnaya A.N., Agol V.I., Hellen C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes & Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- Pisarev A.V., Chard L.S., Kaku Y., Johns H.L., Shatsky I.N., Belsham G.J. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev A.V., Shirokikh N.E., Hellen C.U. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C. R. Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Reynolds J.E., Kaminski A., Carroll A.R., Clarke B.E., Rowlands D.J., Jackson R.J. Internal initiation of translation of hepatitis C virus RNA: The ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- Rijnbrand R., van der Straaten T., van Rijn P.A., Spaan W.J., Bredenbeek P.J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J. Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F., Brakier-Gingras L. A functional interaction between ribosomal proteins S7 and S11 within the bacterial ribosome. J. Biol. Chem. 2003;278:44913–44920. doi: 10.1074/jbc.M306534200. [DOI] [PubMed] [Google Scholar]
- Sasaki J., Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J., Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M., Connell S.R., Lescoute A., Giesebrecht J., Dabrowski M., Schroeer B., Mielke T., Penczek P.A., Westhof E., Spahn C.M. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- Selmer M., Dunham C.M., Murphy F.Vt., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Shibuya N., Nishiyama T., Kanamori Y., Saito H., Nakashima N. Conditional rather than absolute requirements of the capsid coding sequence for initiation of methionine-independent translation in Plautia stali intestine virus. J. Virol. 2003;77:12002–12010. doi: 10.1128/JVI.77.22.12002-12010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C.M., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA-ribosome and subunit–subunit interactions. Cell. 2001a;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Spahn C.M., Kieft J.S., Grassucci R.A., Penczek P.A., Zhou K., Doudna J.A., Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001b;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Spahn C.M., Jan E., Mulder A., Grassucci R.A., Sarnow P., Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Spiegel P.C., Ermolenko D.N., Noller H.F. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M., Willis A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors, and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Hirayama S., Odani S. Ribosomal proteins cross-linked to the initiator AUG codon of a mRNA in the translation initiation complex by UV-irradiation. J. Biochem. 2005;138:41–46. doi: 10.1093/jb/mvi096. [DOI] [PubMed] [Google Scholar]
- Thompson S.R., Sarnow P. Regulation of host cell translation by viruses and effects on cell function. Curr. Opin. Microbiol. 2000;3:366–370. doi: 10.1016/s1369-5274(00)00106-5. [DOI] [PubMed] [Google Scholar]
- Tolan D.R., Traut R.R. Protein topography of the 40 S ribosomal subunit from rabbit reticulocytes shown by cross-linking with 2-iminothiolane. J. Biol. Chem. 1981;256:10129–10136. [PubMed] [Google Scholar]
- Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbehaun A., Borukhov S.I., Hellen C.U., Pestova T.V. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes & Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Wang C., Le S.Y., Ali N., Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- Wilson J.E., Pestova T.V., Hellen C.U., Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000a;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- Wilson J.E., Powell M.J., Hoover S.E., Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000b;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower J., Scheffer P., Sylvers L.A., Wintermeyer W., Zimmermann R.A. Topography of the E site on the Escherichia coli ribosome. EMBO J. 1993;12:617–623. doi: 10.1002/j.1460-2075.1993.tb05694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Nakashima N., Ikeda Y., Uchiumi T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J. Biol. Chem. 2007;282:7770–7776. doi: 10.1074/jbc.M610887200. [DOI] [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]