Abstract
Human accelerated region 1 (HAR1) is a short DNA region identified recently to have evolved the most rapidly among highly constrained regions since the divergence from our common ancestor with chimpanzee. It is transcribed as part of a noncoding RNA specifically expressed in the developing human neocortex. Employing a panoply of enzymatic and chemical probes, our analysis of HAR1 RNA proposed a secondary structure model differing from that published. Most surprisingly, we discovered that the substitutions between the chimpanzee and human sequences led the human HAR1 RNA to adopt a cloverleaf-like structure instead of an extended and unstable hairpin in the chimpanzee sequence. Thus, the rapid evolutionary changes resulted in a profound rearrangement of HAR1 RNA structure. Altogether, our results provide a structural context for elucidating HAR1 RNA function.
Keywords: brain, chimpanzee, human accelerated region 1 RNA, noncoding RNA, RNA structure
INTRODUCTION
While only about 1.5% of the mammalian genome encodes proteins, recent large-scale cDNA and genomic tiling array transcriptomic analyses revealed that at least 70% is transcribed (The ENCODE Project Consortium 2007; Mehler and Mattick 2007). A large proportion of these transcripts is processed to form large and small regulatory RNAs, and it is legitimate to ask whether these noncoding RNAs, with the exception of the well-characterized species, are functional or just transcriptional noise (Ponjavic et al. 2007). Like all species, Homo sapiens has been shaped by positive selection which, in the genome, can appear as a sudden accelerated change in regions otherwise highly conserved by negative selection. As the search has focused so far on fast evolving protein coding regions only, Pollard et al. (2006a,b) used comparative genomics to predict functional elements in the whole human genome that is 98.5% noncoding. They first scanned regions of the chimpanzee genome with at least 96% identity over 100 base pairs (bp) with the orthologous regions in mouse and rat. In each of the 35,000 such regions, they examined the orthologous segments in all available amniotes, including chicken, opossum, and platypus. In this way, they identified 202 regions (termed human accelerated regions [HAR]) showing a significantly accelerated rate of substitution in humans since the divergence from our common ancestor with the chimpanzee. Genes associated with transcriptional regulation and neurodevelopment were found to be significantly enriched among those adjacent to HAR. The 118 bp HAR1 region possesses the highest rate of acceleration of the HARs: 18 substitutions occurred since the human–chimpanzee ancestor, whereas HAR1 is well conserved across other amniotes with only 2 nucleotide (nt) changes between chicken and chimpanzee (Pollard et al. 2006a,b). Two divergently transcribed genes, HAR1F (forward) and HAR1R (reverse), overlap in a region containing HAR1. None of them has protein coding potential, their transcription products HAR1F and HAR1R RNAs lack homology with any known RNA, and their sequence is poorly conserved across mammals, with the exception of the HAR1 segment. Attempts to elucidate the structure and function of these RNAs were undertaken (Pollard et al. 2006a). Human embryonic brain sections showed strong expression of HAR1F RNA but very little HAR1R RNA in the developing neocortex, a part of the cortex especially well developed in humans, and was not detected in other parts of the forebrain. In situ hybridization of brain sections of the macaque indicated a very similar expression pattern of the HAR1F RNA homolog in the developing neocortex, leading the authors to suggest that the rapid changes that occurred in the human HAR1 could be linked to human brain evolution. However, apart from these findings, these RNAs are still waiting to be attributed a function. Secondary structure models based on sequence alignments were predicted and tested for the human and chimpanzee HAR1 RNA regions, proposing a slightly different structure in the two mammals. However, despite being powerful and indispensable for deriving two-dimensional (2D) structures in divergent sequences, covariation analysis of homologous RNA sequences is fraught with difficulties in this case because of the high sequence conservation of HAR1 regions across nonhuman amniotes. Therefore, in this work, we resorted to probing analysis to investigate the structure of both HAR1 RNAs. With a battery of enzymatic and chemical probes, we found the human and chimpanzee HAR1 RNA secondary structures differed not only from the published models but also dramatically between each other.
RESULTS
The chimpanzee and human HAR1 RNAs adopt distinct secondary structures in vitro
Before undertaking structure probing experiments, both the 118-nt-long human (hHAR1) and chimpanzee (cHAR1) HAR1 RNA transcripts were subjected to native polyacrylamide gels after various denaturation-renaturation protocols in the presence or absence of Mg++. Whatever the conditions, both RNAs were contained in a single band (Fig. 1). However, despite their identical length, the chimpanzee species migrated with a higher electrophoretic mobility, suggesting a more compact conformation.
FIGURE 1.

The chimpanzee and human HAR1 RNAs have distinct electrophoretic mobilities on native polyacrylamide gels. RNAs (C, chimpanzee; H, human) were stained with ethidium bromide.
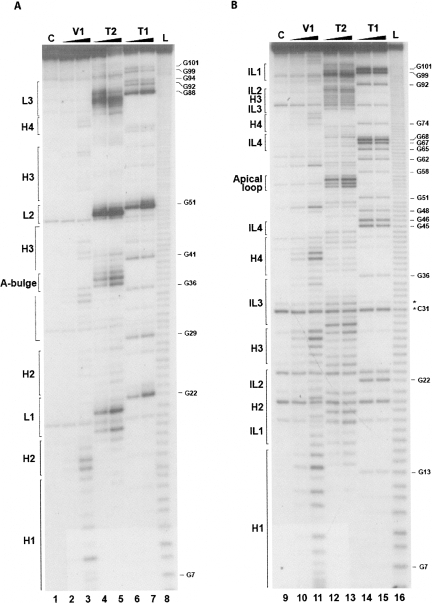
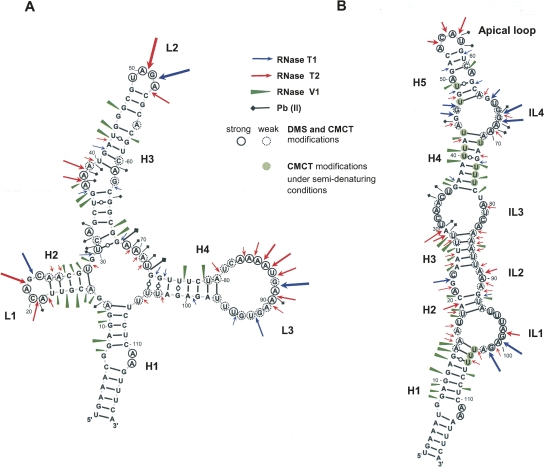
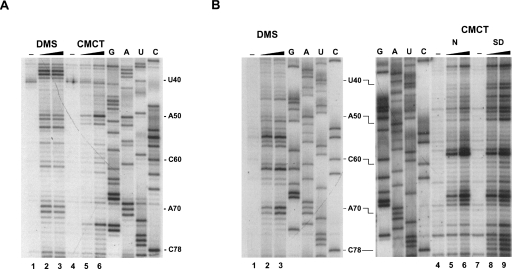
A panoply of nucleases and chemical reagents was employed to probe the hHAR1 and cHAR1 RNA structures, according to Walczak et al. (1996). Representative gels of enzymatic probings are shown in Figure 2. RNase T1 cleaves after G, RNase T2 after any nucleotide with a preference for A, and RNase V1 cleaves in helical and stacked regions without base specificity. High intensity RNase T2 cleavage sites occurred in hHAR1 RNA at U18-A21, A37-A39, A50-G51, and A83-A90 (Fig. 2A, lanes 4,5), and after G22, G51, and G88 by RNase T1 (Fig. 2, lanes 6,7). Phosphodiester bonds cut by RNase V1 are mostly G7-U16, U35-A37, G41-G44, C67-G68, U76-U77, and C78-U79 (Fig. 2A, lanes 2,3). Based on these results, we manually built a 2D model compatible with all the high intensity cleavages (Fig. 3A). Alternative structures predicted by Mfold (Zuker 2003) were discarded because of their incompatibility with the experimental results. Considerable support for manual refinement of the model was provided by lead-induced cleavages. Pb++ cations, which catalyze phosphodiester bond hydrolysis in single strands, yielded a discrete pattern (Supplemental Fig. 1A, lanes 2–4) indicating that A32 is bulged out, confirming it for A37-A39, and substantiating that G68-U72 are single-stranded (Fig. 3A). To further establish the validity of the model, hHAR1 RNA was submitted to chemical modifications. Dimethysulfate (DMS) methylates positions N1 of adenines and N3 of cytosines while carbodiimide (CMCT) reacts at N3 of uracils and N1 of guanines, if they are not engaged in hydrogen bonding. Reactivities toward DMS and CMCT are shown in Figure 4A (Supplemental Fig. 2A, full-size gels). Data on the entire molecule, displayed in Figure 3A, correlate well with enzymatic accessibilities and Pb++ induced cleavages: for example, loops 1, 2, 3, A36, and A37 were highly reactive to chemicals. Combined together, the whole set of experimental results support the proposed human HAR1 2D model.
FIGURE 2.
Enzymatic probing of the human (A) and chimpanzee (B) HAR1 RNAs. Samples were incubated for 3 min (lanes 2,10) and 10 min (lanes 3,11) with RNase V1, 1 min (lanes 4,12) and 3 min (lanes 5,13) with RNase T2, and 1 min (lanes 6,14) and 3 min (lanes 7,15) with RNase T1. L: alkaline ladder (lanes 8,16). Lanes 1,9: controls without enzyme. Digests were run on 10% denaturing polyacrylamide gels. Guanine positions and the structural features shown in Figure 3 are indicated. In chimpanzee HAR1 RNA (panel B), band doublets occurred (marked, e.g., by an asterisk at C31), arising from 5′ addition of untemplated residues by T7 RNA polymerase (Pleiss et al. 1998). However, this event did not prohibit reliable assignment of the cleaved positions. H, helix; IL, internal loop; L, loop, referring to the 2D structures in Figure 3.
FIGURE 3.
Two distinct experimentally supported secondary structure models for HAR1 RNAs. (A) The cloverleaf-like model of the human HAR1 RNA. (B) The chimpanzee HAR1 RNA adopts a hairpin structure. The length and thickness of the symbols represent the intensity of the cleavages. Bases reactive to DMS or CMCT under native conditions are circled; weak reactivities are depicted by dotted circles. Bases modified by CMCT under semidenaturing conditions only are displayed with a green background. H, helix; IL, internal loop; L, loop.
FIGURE 4.
Chemical probing of HAR1 RNAs with dimethylsulfate (DMS) and carbodiimide (CMCT). Only a selected area is shown; full-size gels are displayed in Supplemental Figure 2. After reaction, primer extension products of human (A) and chimpanzee (B) HAR1 RNAs were run on 8% denaturing polyacryamide gels. Treatment was for 5 min with 1 μL of DMS diluted 1/10 (lanes 2) or 1/2 (lanes 3); CMCT (60 mg/mL) was for 10 min with 1 μL (lanes 5,8) or 4 μL (lanes 6,9). CMCT reactions were performed in (B) under native (N, lanes 5,6) or semidenaturing (SD, lanes 8,9) conditions. Lanes 1,4,7: controls without reagent. G, A, U, C: sequencing lanes. Numbering is as in Figure 3. Reverse transcriptase stops immediately 3′ to the modified base.
Figure 2B shows the pattern of enzymatic accessibilities in the cHAR1 RNA. Immediately apparent are the differences in the digestion profiles between cHAR1 and hHAR1 RNAs (Fig. 2, cf. A and B). Besides, weak RNase T2 cleavages were also observed in regions cut by RNase V1, particularly in helices H3 and H4 (Fig. 2B), rendering less straightforward the identification of paired and loop regions. Nevertheless, major RNase T1 cleavages appeared after G45, G46, G67, G68, G99, and G101, whereas A29-U30, A54-A56, and A98-A100 were mostly accessible to RNase T2. Regions susceptible to RNase V1 resided between G7-U18, A24-U28, U35-U40, and U75-C78. Complementary chemical probing analyses were required to propose a 2D model. They are shown in Figure 4B and Supplemental Figures 1B and 2B. Pb++ induced hydrolyzes were found in internal loop 4 and the apical loop; bases in the apical loop and internal loops 1–4 were reactive to DMS and CMCT; U75-U77 became reactive to CMCT under semidenaturing conditions only (i.e., in the presence of EDTA and the absence of salt), indicative of base-pairing. The most satisfying 2D model (Fig. 3B) accounting for the enzymatic and chemical probings was built using Mfold (Zuker 2003) and manually refined. The simultaneous presence of RNases T2 and V1 cleavages in some areas is suggestive of a weakly stable hairpin, with the transitory formation of alternative base pairs especially at helices H2, H3, and to a lesser extent at helix H4, probably owing to the high AU content. This was consistently obtained regardless of the presence or absence of Mg++, whatever the concentration of Na+ and K+ salts (in the range 50–300 mM), and under various denaturing-renaturing steps prior to probing the structure (data not shown).
Hence, in contrast to the human HAR1 RNA stable cloverleaf-like structure, the chimpanzee counterpart folds into a weakly stable extended hairpin.
DISCUSSION
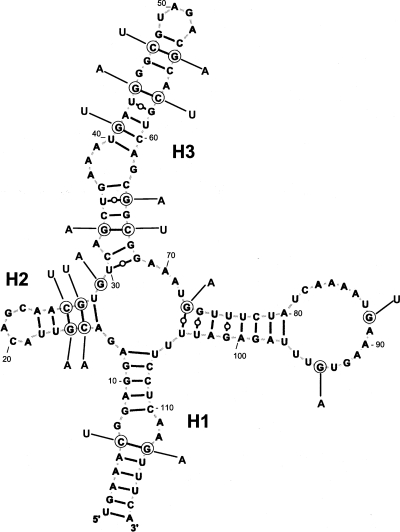
The identification of human accelerated region 1 (HAR1) in the human genome is a recent discovery (Pollard et al. 2006a,b). The noncoding HAR1F and HAR1R RNAs, which contain the HAR1 region, are localized in the neocortex but nothing is known about their function. The salient finding of our work is the unequivocal derivation of two distinct, experimentally derived secondary structure models for human and chimpanzee HAR1 RNAs. The differences between the published structures (Pollard et al. 2006a), and those resulting from the present study, are twofold. First, major differences appear in human HAR1 RNA at helices H3 and H4 that are formed in our model by base pairs involving nucleotides that differ from those published. Thus, H4 has no real homolog in the Pollard et al. model. Second, and very importantly, we established a 2D structure for the chimpanzee HAR1 RNA that varies considerably from the human model. Only helix H1 is common to both species. It is remarkable that a cloverleaf-like model can be proposed for the chimpanzee HAR1 RNA (Fig. 5), six base-pair covariations and two U•G to U-A base pair changes supporting the existence of helices H1, H2, and H3. However, almost all of the 18 changes that appeared in humans do not support the chimpanzee hairpin model. In addition, other lines of evidence argue in favor of the structural dichotomy (human cloverleaf-like versus chimpanzee hairpin) that we observed in vitro. First, the hairpin structure of the chimpanzee HAR1 RNA was observed under a variety of experimental conditions. Besides, this RNA has an electrophoretic mobility differing from its human counterpart in native polyacrylamide gels. Second, Mfold calculated that the hairpin is more thermodynamically stable than the cloverleaf-like structure in the chimpanzee RNA.
FIGURE 5.
The chimpanzee HAR1 RNA can adopt the cloverleaf-like structure of the human HAR1 RNA. The 18 nt changes in the chimpanzee HAR1 RNA are shown at positions circled in the displayed human HAR1 RNA. Six compensatory changes maintain Watson–Crick base pairs in helices H1, H2, and H3.
The central question that remains to be addressed is the elucidation of the function of the HAR1 RNAs. Interestingly, HAR1F RNA, which harbors the 118-nt-long HAR1 region, was shown to be coexpressed with the cortical patterning protein Reelin in early developing human and macaque brains (Pollard et al. 2006a). This protein is a marker of the Cajal–Retzius neurons localized in the neocortex that is particularly well developed in humans and often associated with higher cognitive functions. Therefore, in a review article, Ponting and Lunter (2006) proposed the human HAR1 RNA to be a strong candidate for the emergence of innovative function in the human neocortex. That the HAR1 RNA region might represent one functional domain of HAR1F RNA is highly likely because (1) the strong sequence conservation among nonhuman amniotes clearly indicates that HAR1 is under functional constraint; and (2) HAR1F RNA sequences, upstream of and downstream from the HAR1 region, do not align in species other than primates because of the low degree of conservation. Figure 5 shows that the chimpanzee HAR1 RNA has the potential to fold according to the cloverleaf-like model, yet it did not under our experimental conditions. If the hairpin structure needs to undergo a conformational change in vivo into the cloverleaf-like structure to exert its function, it may require a cofactor such as an RNA-binding protein. By contrast, the already-acquired cloverleaf-like structure of the human HAR1 RNA may enable it to achieve an identical function without a conformational change and/or a stabilizing factor. Finding such a cofactor is challenging and will constitute an important early step in understanding more about HAR1 RNA function. The computer search for noncoding RNAs is notoriously difficult owing to their lack of sequence signature (Hammann and Westhof 2007). Interestingly, it has been shown that the Human Accelerated Regions result from GC-biased gene conversion, a neutral process resulting from recombination events (Pollard et al. 2006b; Galtier and Duret 2007). The present results offer a structural basis for the contributions of biased gene conversion to the molecular evolution of noncoding RNAs.
MATERIALS AND METHODS
Plasmid constructs
HAR1 DNAs were assembled from oligodeoxynucleotides H1-H8 for human, H1 and C9-C15 for the chimpanzee counterpart, and H1 incorporating the T7 promoter followed by three consecutive Gs for higher transcription yield (Supplemental Table 1). The 5′-phosphorylated oligos were heated to 90°C, annealed by slow cooling to room temperature, and ligated overnight. An aliquot of each ligation reaction was PCR amplified with the H1/H8 and H1/C15 couples, respectively. Products were cloned into pGEM T-Easy to yield pGhHAR1 and pGcHAR1 (human and chimpanzee HAR1) which were next PCR amplified with H16/H17 and H16/C18, respectively, to remove the endogenous pGEM T-Easy T7 promoter and the undesired 5′ and 3′ sequences flanking the inserts. H16 incorporated a BamHI site upstream of the T7 promoter, H17 and C18 introduced EcoRI and SmaI sites 3′ to the HAR1 sequences. BamHI-EcoRI digests of the PCR products were ligated to BamHI-EcoRI cleaved pT7BckX vector (Fagegaltier et al. 2000), generating pT7hHAR1 and pT7cHAR1. pT7BckX contains a sequence complementary to the universal primer between SmaI and a downstream XhoI site.
T7 transcription
pT7hHAR1 and pT7cHAR1 were linearized with SmaI or XhoI. RNAs originating from SmaI-linearized templates were used for native gel assays, and enzymatic and Pb(II) cleavages. RNAs transcribed from XhoI linearized vectors contained an extra sequence at the 3′ end for hybridization to the universal primer and were used for chemical probing. Transcription by T7 RNA polymerase was conducted as in Fagegaltier et al. (2000).
Native polyacrylamide gel electrophoresis
Prior to loading, RNA samples were denatured at 90°C for 2 min, snap cooled on ice or slowly annealed to 4°C in the appropriate buffer, then run on 12% nondenaturing polyacrylamide gels for 2–3 h at 4°C in 90 mM Tris-borate buffer pH 8.3 with or without 2 mM MgCl2.
Enzymatic and Pb (II) cleavages
Dephosphorylated RNAs were 5′-end labeled with 32P. RNase and Pb(II) cleavages were performed as in Fagegaltier et al. (2000). Enzymatic digestions were performed at 20°C for 1–10 min in 12 μL containing 50 mM HEPES pH 7.5, 100 mM K acetate, 0–20 mM Mg acetate, with 0.2 U/μg RNase V1, 0.01 U/μg RNase T2, 0.1 U/μg RNase T1. Alkaline ladders were obtained by incubation in 50 mM carbonate buffer pH 8.9 for 5–10 min at 90°C. Pb(II) cleavages occurred at 20°C for 5 min in the same buffer with 1–8 mM Pb(II) acetate.
Chemical probing
Chemical modifications with dimethylsulfate (DMS) or carbodiimide (CMCT) were performed under native or semidenaturing conditions at 20°C, essentially according to Walczak et al. (1996) and Fagegaltier et al. (2000). Modified bases were detected on 8% denaturing polyacrylamide gels after reverse transcription of the 32P-labeled universal primer.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank C. Allmang, P. Carbon, B. Masquida, M. Pheasant, and P. Romby for careful reading of the manuscript and advice. We thank A. Schweigert for skillful technical assistance. This work was supported by grants from the CNRS.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1054608.
REFERENCES
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D., Lescure A., Walczak R., Carbon P., Krol A. Structural analysis of new local features in SECIS RNA hairpins. Nucleic Acids Res. 2000;28:2679–2689. doi: 10.1093/nar/28.14.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Hammann C., Westhof E. Searching genomes for ribozymes and riboswitches. Genome Biol. 2007;8:210.1–210.11. doi: 10.1186/gb-2007-8-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler M.F., Mattick J.S. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol. Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- Pleiss J.A., Derrick M.L., Uhlenbeck O.C. T7 RNA polymerase produces 5′-end heterogeneity during in vitro transcription from certain templates. RNA. 1998;4:1313–1317. doi: 10.1017/s135583829800106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.S., Salama S.R., Lambert N., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006a;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Pollard K.S., Salama S.R., King B., Kern A.D., Dreszer T., Katzman S., Siepel A., Pedersen J.S., Bejerano R., Baertsch R., et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006b;2:1599–1611. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J., Ponting C.P., Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., Lunter G. Human brain gene wins genome race. Nature. 2006;443:149–150. doi: 10.1038/nature05154. [DOI] [PubMed] [Google Scholar]
- Walczak R., Westhof E., Carbon P., Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]