Abstract
MicroRNAs (miRNAs) are one class of short, endogenous RNAs which can regulate gene expression at the post-transcriptional level. Previous analysis revealed that mammalian miRNAs tend to cluster on chromosomes. However, the functional consequences of this clustering and conservation property are largely unknown. In this study we present a method to identify signaling pathways targeted by clustered miRNAs. We performed a computational screen for mouse signaling pathways targeted by miRNA clusters. Here, we report that the target genes of 3 miRNA clusters are overrepresented in 15 signaling pathways. We provided experimental evidence that one miRNA cluster, mmu-mir-183–96–182 targets Irs1, Rasa1, and Grb2, all of which are located in the insulin signaling pathway. Theses results suggest that by targeting components with different roles along a signaling pathway, different members of one miRNA cluster can act as a whole to coordinately control the signal transduction process.
Keywords: clustered microRNAs, signaling pathways, insulin pathway
INTRODUCTION
MicroRNAs (miRNAs) are a class of ∼22-nucleotide (nt) endogenous single-stranded RNA which can cleave mRNA or repress the initiation of protein translation by imperfect base pairing with the targeting sequences on 3′-UTR in animals (Bartel 2004). miRNAs are transcribed by RNA polymerase II or III as long primary miRNAs (pri-miRNAs) which range from hundreds to thousands of nucleotides in length. The pri-miRNAs are then processed into mature miRNAs by Drosha and Dicer enzymes sequentially. Studies have shown that miRNAs play crucial roles in a broad range of biological processes including development, differentiation, tissue morphogenesis, and also in several types of diseases such as carcinogenesis and viral infections (Bartel 2004; Kloosterman and Plasterk 2006).
Previous analysis has revealed conserved patterns as clustering of mammalian miRNAs on chromosomes (Altuvia et al. 2005). Megraw et al. (2007) reported that in human, mouse, rat, and chicken, even at a very conservative maximum inter-miRNA distance (MID) of 1 kb, over 30% of all miRNAs are organized into clusters. If the MID extends to 50 kb, over half of all miRNAs are clustered on chromosomes. Expression profiling analysis showed that proximal pairs of miRNAs are generally coexpressed, and an abrupt transition in the correlation between pairs of expressed miRNAs occurs at a distance of 50 kb (Baskerville and Bartel 2005). According to their chromosomal positions, several miRNA clusters have also been experimentally confirmed by RT-PCR or Northern blotting (Cai et al. 2004; Lee et al. 2004; Weston et al. 2006). These evidences strongly suggested that members of a particular miRNA cluster, which are defined by their chromosomal positions, are highly likely to be processed as cotranscribed units. However, the functional consequences of this clustering and conserved property remain unclear.
Analogous to transcriptional regulation (between transcriptional factors and cis-regulatory elements), most miRNAs function to fine tune the expression of hundreds of genes in a combinatorial manner (Bartel 2004). Specifically, different miRNA species have been proposed to regulate a single mRNA target through binding to the same mRNA transcript. Several such cases have also been experimentally verified. For example, Krek et al. (2005) experimentally validated that myotrophin (Mtpn) as the first mammalian gene regulated by miR-375, miR-124, and let-7b coordinately. Other studies also showed that miRNAs 17-5p, 20a, and 106a control monocytopoiesis via binding to the 3′-UTR of transcription factor acute myeloid leukaemia-1 (AML1) mRNA (Fontana et al. 2007).
Dews et al. (2006) recently reported the first evidence of a cluster of miRNAs that could regulate functionally related proteins in cancer. They found that in Myc overexpressing tumors, two anti-angiogenesis genes Tsp1 and Ctgf are downregulated by one Myc activated miRNA cluster miR-17–miR-92. Noticeably, they confirmed that within the cluster, miR-19 is primarily responsible for Tsp1 downregulation and miR-18 for Ctgf downregulation in response to Myc activation. Inspired by their findings, in this paper, we combined the combinatorial regulation concept with the clustering property to screen mouse signaling pathways which are targeted by clustered miRNAs. Especially, we explored another layer of combinatorial regulation, i.e., we hypothesize that each member of a miRNA cluster could regulate one component within the same signaling pathway; thus, the cluster acts coordinately as a whole to control a biological process.
RESULTS
A computational screen framework for mouse signaling pathways targeted by miRNA clusters
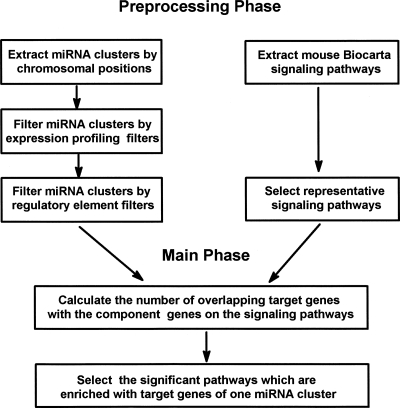
As indicated in Figure 1, to identify mouse signaling pathways targeted by miRNA clusters, we initially extracted mouse miRNA clusters according to mouse chromosomal positions. Although previous studies had indicated that defining clustered miRNA memberships according to chromosomal location is highly consistent with experimental evidences as RT-PCR or Northern blotting, we further filtered miRNA clusters through (1) an expression profiling filter and (2) a regulatory element filter.
FIGURE 1.
Flowchart of the computational screen framework for mouse signaling pathways targeted by the miRNA cluster. The algorithm is divided into two stages. The initial stage is to preprocess both the mouse miRNA clusters and the Biocarta signaling pathways. The second stage involves selecting the significant pathways that are enriched with target genes of one miRNA cluster.
For any particular miRNA cluster that shares the same transcriptional unit, the expression profiles of miRNAs within this cluster should be similar, and thus positively, correlated. We filtered out those miRNA clusters with the mean correlation measures that are less than those of the background correlation in at least two of the three analyzed expression profiles. In addition, we examined for the presence of regulatory elements such as transcription start site (TSS), CpG window, or predicted first exon in the inter-miRNA region of the same cluster. All of these possibly indicated a different transcription initiation. We extensively searched for the elements in the inter-miRNA region of the same cluster, and we filtered out the miRNA clusters in which regulatory elements are found. After filtering signaling pathways, we selected the significant signaling pathways that are enriched with those predicted target genes according to the optimized target prediction data.
Filtering miRNA clusters
Forty-five miRNA clusters which contained 169 miRNAs, were extracted when MID was set to 5 kb. The largest miRNA cluster mmu-mir-669a-3 is located on chromosome 2 with 25 miRNA members (Supplemental Table S1). Among these 45 miRNA clusters, 40 clusters have a correlation value higher than 0.35, the background correlation value of data set published by Thomson et al. (2004). Nineteen out of the 45 clusters have correlation measures significantly higher than the background (nominal P < 0.05) (Supplemental Table S1). Previously, the miRNA members of the mir-17 cluster show consistent upregulation patterns in a Myc-overexpressing Ras-transformed colonocytes (Dews et al. 2006). However, the expressions of these miRNA members are not correlated in two out of three analyzed profiles (Supplemental Table S1), perhaps suggesting that this cluster is only regulated conditionally. In addition, three other miRNA clusters mmu-mir-212, mmu-mir-363, and mmu-mir-382 also do not coexpress in various cellular conditions, indicating that the members within these clusters have independent regulation mechanisms (Supplemental Table S1). Nonetheless, most miRNA clusters defined by chromosomal locations have coexpressed patterns under different cellular conditions, consistent with previous observations (Baskerville and Bartel 2005).
We also filtered the clusters according to regulatory element presence using both FirstEF and Eponine, and found regulatory elements within mmu-mir-212–132 cluster (Supplemental Table S1). Interestingly, the expression profiling filter also shows that the expressions of these two miRNAs are not correlated in all three analyzed expression profiles. Collectively, these evidences strongly suggested that these two miRNAs have independent transcription mechanisms, and they may not function coordinately to regulate signaling pathways.
After these preprocessing phases, we finally obtained 41 miRNA clusters, which show coregulation patterns. Similar analysis were also conducted when MID was set to 50 kb. In all, 55 miRNA clusters were initially selected, and after the preprocessing phases, 45 clusters were used in the following analysis (Supplemental Table S2).
Selection of significant pathways that are enriched with target genes of one miRNA cluster
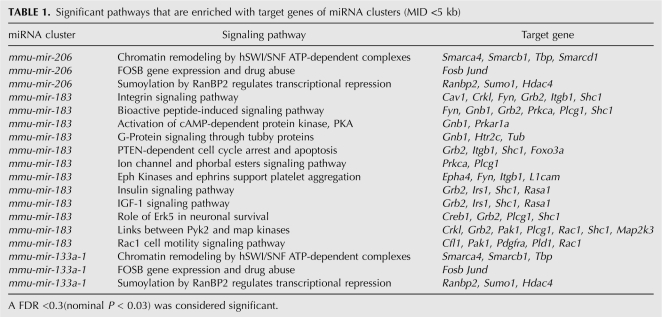
Next, we selected significant pathways that are enriched with target genes of one miRNA cluster among the filtered 97 signaling pathways. As indicated in Table 1, totally 18 cluster–pathway interactions were found by false discovery rate analysis (false discovery rate [FDR] <0.3; nominal P < 0.03), which involved three miRNA clusters and 15 signaling pathways.
TABLE 1.
Significant pathways that are enriched with target genes of miRNA clusters (MID <5 kb)
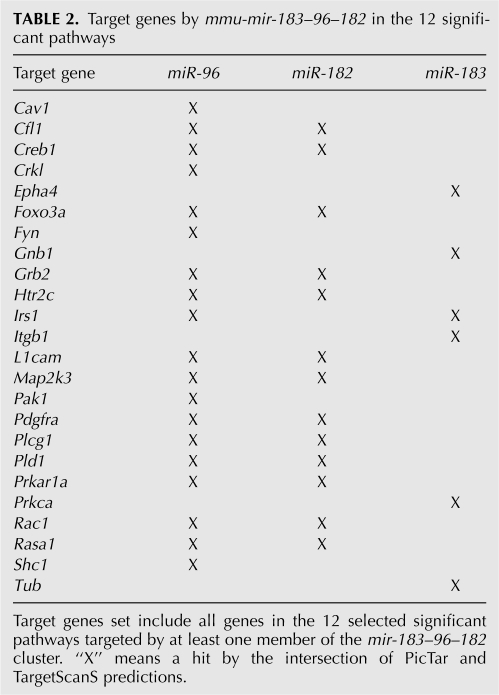
Interestingly, one miRNA cluster mmu-mir-183–96–182 controls 12 signaling pathways. Detailed analysis of the target genes by this cluster in these 12 significant pathways suggested that miR-96 is the major regulator. Other than a few exceptions, miR-96 targets most of the member genes in these 12 signaling pathways (Table 2). Previously, the pri-miRNA polycistronic organization of this cluster has been identified by experimental methods and the coexpression patterns of this cluster were also confirmed (Weston et al. 2006; Xu et al. 2007).
TABLE 2.
Target genes by mmu-mir-183–96–182 in the 12 significant pathways
Consistent with our results, this cluster seems to be involved in at least three biological functions according to the current literature. First, it was reported that one member of this cluster, mir-96, targets protein arginine methyltransferase (PRMT5). Aberrant expression of PRMT5 leads to altered epigenetic modification of chromatin, and consequently impacts on the growth of transformed lymphoid cells (Pal et al. 2007). Second, in colorectal cancer cell lines, all three members of this cluster show consistent upregulations. Forkhead family of transcription factors including CHES 1, FOXF2, FOXK2, FOXO1A, FOXO3A, and FOXQ1, were proposed as putative targets (Bandres et al. 2006). Finally, Xu et al. (2007) reported that Mitf, a transcription factor required for the establishment and maintenance of retinal pigmented epithelium, is a direct target of miR-96 and miR-182, indicating that this miRNA cluster is also important for retinal development and function. We experimentally proved that this cluster is also involved in insulin signaling pathway (see below). Therefore, these evidences suggested that this cluster plays an important role in multiple signaling pathways.
We also generated a set of experimentally testable hypotheses. For example, the epigenetic regulation of chromatin domains by miRNAs has recently been demonstrated through the finding that miR-26a targets the histone methyltransferase enhancer of zeste homolog 2 during myogenesis (Wong and Tellam 2008). Our prediction found that both mmu-mir-206 and mmu-mir-133a-1 clusters target chromatin remodeling hSWI/SNF ATP-dependent complexes, which is a major group of chromatin-modifying complexes. It would be interesting to test whether these miRNA clusters can regulate the chromatin structure via a mechanism involving hSWI/SNF ATP-dependent chromatin remodeling complexes. The same set of cluster–pathway interactions were obtained when the MID set to 50 kb (data not shown), indicating robustness of our prediction results.
Experimental verification of one miRNA cluster targeting the insulin signaling pathway
Insulin signaling pathway is involved in both metabolic and mitogenic aspects of cellular function. In our prediction, one miRNA cluster mmu-mir-183–96–182 targets four members of the insulin signaling pathway. In the intersection of PicTar and TargetScanS prediction, both mir-96 and mir-183 target insulin receptor substrate 1 (irs1), whereas mir-96 and mir-182 target both ras p21 protein activator 1 (rasa1) and growth factor receptor bound protein 2 (grb2).
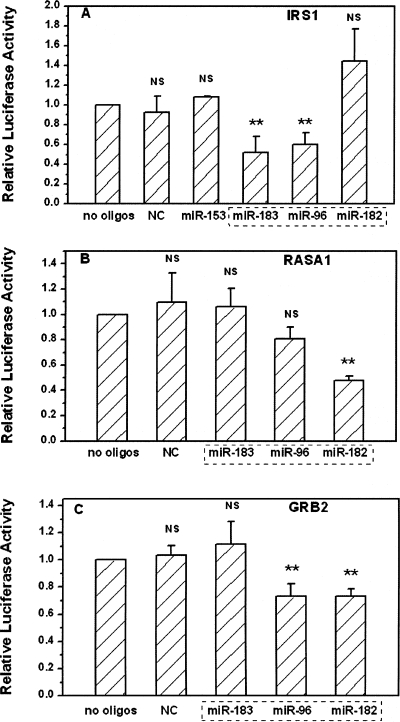
We set up luciferase reporter assays to validate these predicted targets. Fragments of the 3′-UTR of irs1, rasa1, and grb2, including the miRNA target sites, were individually cloned into a firefly luciferase expression vector pGL3. Cells were respectively transfected with these reporters together with a renilla luciferase vector pRL-TK as an internal control. While using mock transfected cells as a control (no oligos), we examined the effects of overexpressing different miRNA oligos on the expressions of these reporters. First, we found that a nonspecific oligo (NC), even at a relatively high concentration, did not alter the expression of all three reporters (Fig. 2A–C). Second, a mir-153 oligo which is predicted to target irs2, did not significantly alter the gene expression of a reporter containing the 3′-UTR of irs1 (Fig. 2A). Third, we experimentally confirmed that both mir-96 and mir-183 repressed the gene expression of this reporter about 40% via targeting the 3′-UTR of irs1 (Fig. 2A). The 3′-UTR of irs1 does not contain a targeting sequence for another member of this cluster mir-182. We did not observe any significant effects of mir-182 oligo on the expression of this reporter, demonstrating the specificity and validity of our reporter system (Fig. 2A).
FIGURE 2.
Repression of irs1, rasa1, and grb2 3′-UTR by mmu-mir-183–96–182 cluster. 3′-UTR of target genes were inserted into the 3′ end of the luciferase gene in the pGL3 control vector. Cells were transfected with or without 10 nM miRNA oligos indicated, pGL3 vectors containing the 3′-UTR of target genes, and an internal control Renilla luciferase vector. The luciferase activity levels were determined after 24 h. Data were generated from triplicate in at least three independent experiments. The ratios of Firefly luciferase to Renilla luciferase expression were normalized to no oligos transfection. **P < 0.01 for no oligos versus various oligos treatments, NS: no significant difference. (A) pGL-3-Irs1 plasmid cotransfected with miR-153, miR-183, miR-96, miR-182, and nonspecific oligos (NC). NC, miR-153, and miR-182 were employed as negative controls as predicted by both Pictar and TargetScanS algorithms. (B) pGL-3-Rasa1 plasmid cotransfected with miR-183, miR-96, miR-182, and nonspecific oligos (NC). Both NC and mir-183 were employed as negative controls as predicted by both Pictar and TargetScanS algorithms. (C) pGL-3–Grb2 plasmid cotransfected with miR-183, miR-96, miR-182, and nonspecific oligos (NC). Both NC and mir-183 were employed as negative controls as predicted by both Pictar and TargetScanS algorithms. A dotted rectangle indicates the miRNA cluster mmu-mir-183–96–182.
We also observed strong repression of a luciferase reporter through presumed binding of mir-182 to the 3′-UTR of rasa1 (Fig. 2B). Moderate but significant repression of a luciferase reporter was also observed in pGL-3-grb2 plasmid cotransfected with mir-96 or mir-182 oligos (Fig. 2C). Although rasa1 was also a predicted target of mir-96 by optimized prediction method, there was no significant difference between cotransfections with or without mir-96 oligos, which possibly indicated that this was a false positive prediction.
DISCUSSION
One principle of gene regulation by miRNA is coordinate gene expression. In this model, individual miRNA may reduce the expression of target genes moderately while a collection of miRNAs may act in a combinatorial fashion to exert significant effects (Bartel 2004; Stark et al. 2005). Bioinformatics analysis agrees with this principle as well, since it is common to find multiple and independent binding sites on most, if not all, target genes. Different groups had also reported in experimental instances that miRNAs can act cooperatively through multiple target sites on the same gene. Recent studies have widened the scope of this combinatorial regulation such that a few miRNAs can regulate a group of functionally related genes instead of a single gene (Hua et al. 2006). These are convenient and efficient strategies to regulate cellular processes, but the key issue is that these regulatory miRNAs should themselves be regulated coordinately. Therefore, certain mechanisms must exist to guarantee the coordinate expression of these miRNAs.
Combining the coregulation concept with the observation that mammalian miRNAs tend to cluster on chromosomes, we proposed that the clustering and conserved patterns provide a natural mechanism to explain the principle of coregulation by miRNAs. Since those clustered miRNAs are largely regulated under the same control, upon stimulation, members within the same cluster could express and function in a cooperative manner. Thus, the downstream effect, for example, to completely shut down a biological process, is ensured. However, the above regulation model does not rule out the possibility that a single miRNA could greatly alter a cellular process. For instance, a miRNA may target an essential regulator of a signaling pathway which leads to alterations in multiple downstream effectors.
From the discovery of miRNAs, plenty of evidences have demonstrated the increasingly important role of these tiny molecules in developmental processes and carcinogenesis. However, relatively few are known for their regulation on cellular metabolic processes. Here, we provided the first evidence of an interplay between one miRNAs cluster mmu-mir-96–182–183 and the insulin signaling pathway.
The IRS proteins are major players for the metabolic aspect of the insulin signaling cascade because they mediate the activations of intracellular signaling molecules, such as PI 3-kinase (PI3k), Akt, and molecular target of rapamycin (mTOR). On the other hand, the proliferative and mitogenic effects of insulin are mediated largely via the activation of Ras signaling pathway (Draznin 2006). Grb2 is a classical adapter molecule that controls Ras and MAPK activations, which are important for growth factor signaling. Therefore, Grb2 is a potential target of antitumor therapy (Dharmawardana et al. 2006). Rasa1, which transforms active GTP-bound Ras into inactive GDP-bound form, links the insulin signaling with Ras signaling pathway. Loss of Rasa1 could lead to increased activation of Ras, which has been linked to various tumors (Downward 2003).
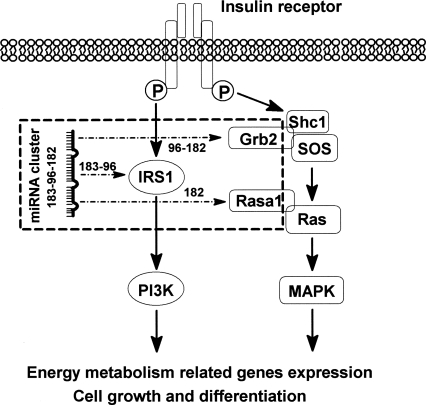
Importantly, we showed that three members of mmu-mir-96–182–183 have distinct but coordinate roles in controlling insulin signaling. Mir-96 and mir-183 are mainly responsible for regulating Irs1, which is the key mediator of metabolic aspect of insulin pathway, whereas mir-182 targets Rasa1 and Grb2, both of which play important roles in linking insulin signaling with the Ras pathway. Thus we proved that by targeting components with different roles along the insulin signaling pathway, mmu-mir-183–96–182 can coordinately control the insulin signal transduction. This regulation model is illustrated in Figure 3. Due to the potential bias of a reporter assay, further in vivo experiments are required to confirm the regulation and to establish the physiological consequences.
FIGURE 3.
Model of the regulation of the insulin signaling pathway by miRNA cluster mmu–mir-183–96–182. Mir-96 and mir-183 are mainly responsible for regulating Irs1, which is the key mediator of the metabolic aspect of the insulin pathway, whereas mir-182 targets Rasa1 and Grb2, both of which play important roles in linking insulin signaling with the Ras pathway. A dotted arrow indicates the experimentally verified miRNA and target gene interactions via luciferase activity assay.
Recently, several groups reported about the global analysis of predicted miRNA targets with sets of genes (i.e., pathway or gene ontology [GO] term). Gusev et al. (2007) reported GO terms as well as biological functions targeted by five coexpressed miRNAs sets. Cui et al. (2006) analyzed distribution of the predicted miRNA targets in the cellular signaling network. But no one explicitly explored the functional consequence of clustered miRNAs with signaling pathways. Our approach can be extended forward to GO and other pathway system (i.e., KEGG) analysis as well as human pathway analysis. It is also important to identify signaling pathways that are conditionally and tissue specifically regulated by miRNA clusters. This would require further fine tuning the mining methods and obtaining additional miRNA expression profiles under specific conditions. However, this direction of research is beyond the scope of our current study. Such systems biological approaches will provide insight into the principles of the regulation of signaling networks by miRNAs and facilitate experimentally identifying interactions between miRNAs and target mRNAs.
MATERIALS AND METHODS
Data sources
Mouse miRNA clusters data
According to chromosome positional relationships between miRNAs, any two miRNAs with maximum distance less than defined distance were considered to be in the same cluster. We derived mouse miRNAs clusters from miRGen database (Megraw et al. 2007). Forty-five miRNA clusters which contains 169 miRNAs were extracted when MID was set to 5 kb. Fifty-five miRNA clusters which contains 202 miRNAs were found when MID was set to 50 kb.
Optimized targets prediction data
Sethupathy et al. (2006) compared various miRNA target-prediction programs both individually and in combination with other programs. They suggested that the intersection of PicTar and TargetScanS prediction could achieve both high sensitivity and specificity compared to other programs or program combinations. This optimized target prediction data was used in this study. It includes 21,225 miRNA–target gene pairs, which totally involved 112 miRNAs and 3586 target genes.
Mouse Biocarta pathway data
We use the compiled pathway data by Tian et al. (2005). It is consisted of 277 Mouse Biocarta pathway data, and could be downloaded from http://compbio.med.harvard.edu/Supplements/PNAS05.html.
miRNA expression data
We used three mouse miRNA expression profile data sets. Thomson data: the investigators used a dual-channel microarray to monitor expression levels of 124 mouse miRNAs in seven mouse tissues and four different embryonic stages, embryonic stem cells, and embryoid bodies (Thomson et al. 2004). Beuvink data: chemically modified 2′-O-(2-methoxyethyl)-(MOE) oligoribonucleotide probes were arrayed onto Evanescent Resonance microchips to detect 200 miRNAs expression in eight mouse tissues (Beuvink et al. 2007). Betel data: miRNA expression profiles derived from a comprehensive cloning and sequencing effort of 64 mouse small RNA libraries extracted from major organs and cell types. Expression values were reported as normalized clone counts for 400 miRNAs (Betel et al. 2008).
Computational framework
miRNA profiling filter
We computed the Pearson coefficient for each pair of miRNAs within the same cluster. The mean correlation coefficients were used as a correlation measure for a miRNA cluster. For each miRNA profile, we also randomly sampled 1000 pairs of miRNAs and computed the pearson coefficient. The average value of positive correlation measures was used as a background correlation. Finally, we filtered out such clusters that the mean correlation measure is less than those of the background correlation in at least two of the three miRNA profiles. If there are <2 miRNAs within the same cluster measured in the three expression profiles, the miRNA cluster passes the filter automatically as currently there are not enough evidences to show this cluster does not coexpressed. To assess the statistical significance of the correlation measure for each miRNA cluster, the fraction of 1000 random sampling pairs of miRNAs having equal to or larger than the correlation measure was reported as nominal P value.
Regulatory elements filter
We used FirstEF and Eponin programs to search for possible regulatory elements among the inter-miRNA region within the same cluster. FirstEF is a decision tree based promoter detection programs to predict exon, promoter, and CpG window of sequences (Davuluri et al. 2001). The Eponine program provides a probabilistic method for detecting transcription start sites (TSS) in the mammalian genomic sequence (Down and Hubbard 2002). For the analysis, we used the UCSC human multiz28way conservation track to get the corresponding mouse regions. All the sequence analyses were done at Galaxy platform (Giardine et al. 2005; Karolchik et al. 2008). We filtered out such clusters that the regulatory elements were found by any one of the two detection programs. miRNA cluster that passes both miRNA profiling filter and regulatory elements filter were used in the following analysis.
Filtering pathways
For the 277 Biocarta signaling pathways, there are some pathways that only a few or even none of its component genes are predicted as miRNA targets in the optimized target prediction data. Only those signaling pathways with at least 50% of their component genes listed in the optimized target prediction data as target genes were selected for further analysis. After this selection, there remained 97 signaling pathways.
Finding significant interactions between clusters and pathways
For each cluster and each signaling pathway, we first found predicted target genes of all the miRNA members within this cluster. Then, we calculated the number of irredundant overlapping target genes with the component genes on the signaling pathway. One thousand simulated miRNA clusters of the same size were also randomly sampled from the 112 miRNAs with target prediction and then the corresponding number of overlapping targets was recorded. We set the P value of the cluster–pathway pair as the fraction of 1000 random sampling having equal to or larger than the number of overlapping targets of the real miRNA cluster. The P value based on such randomizations was used to assess whether the observed number of overlapping genes for a cluster–pathway pair was achieved by chance. Multiple statistical tests were controlled by false discovery rate (FDR) (Benjamini and Hochberg 1995). After applying the same procedure to all the cluster–pathway pairs, we selected the cluster–pathway pair at FDR <0.3 (nominal P < 0.03).
Experimental verification
The siRNA duplexes mir-96, mir-153, mir-182, and mir-183 were purchased from Shanghai Gene-Pharma Co. siRNA duplexes with nonspecific sequences were used as a negative control (NC). Mouse genes 3′-UTR sequences were downloaded from Pictar website (http://pictar.bio.nyu.edu/cgi-bin/PicTar_vertebrate.cgi). The fragments of irs1 (mm_NM_005544.0) 3′-UTR (nucleotides 33–1001), grb2 (mm_NM_203506.0) 3′-UTR (nucleotides 82-598), and rasa1 (NM_002890.0) 3′-UTR (nucleotides 9-936) were PCR-amplified and subcloned into the XbaI site in pGL3-Control vector (Promega Corp.). Constructs were all sequence verified (Guangzhou Invitrogen Biotechnology Co., Ltd.). In addition to nonspecific sequences as a negative control, specific negative controls were employed (both mir-153 and mir-182 for irs1, and mir-183 for rasa1, and grb2) because no target sites were predicted for the corresponding miRNA oligos by both Pictar and TargetScanS algorithms.
For luciferase activity assay, HeLa cells were seeded 24 h prior to transfection in 96-well tissue culture plates. The next day, cells were cotransfected with various constructs and internal control pRL-TK and 10 nM of each miRNA duplex using Lipofectamine 2000 in accordance with the manufacturer's instructions. Cell lysate was collected 24 h after transfection. Firefly and Renilla luciferase activities were measured using a Dual Luciferase Reporter Assay System (Promega Corp.). All of the assays were performed in triplicate in at least three independent experiments. We assessed statistical significance using two-tailed Student's t-test.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank the UCSC Genome Bioinformatics Group for helpful discussion on genome analysis for this work, and Molly Megraw at the University of Pennsylvania for kindly providing us with the optimized target prediction data. We also thank Zhanfang Kang for his assistance on graphic preparation. The research is supported by grants from the National Science Foundation of China #30672463, the Chinese National Science and Technology Department #2006CB50390, the Chinese Academy of Sciences #KSCX2-YW-R-085, and a start-up grant from GIBH.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.997708.
REFERENCES
- Altuvia Y., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres E., Cubedo E., Agirre X., Malumbres R., Zarate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzo M., et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuvink I., Kolb F.A., Budach W., Garnier A., Lange J., Natt F., Dengler U., Hall J., Filipowicz W., Weiler J. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucleic Acids. 2007;35:e52. doi: 10.1093/nar/gkl1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Yu Z., Purisima E.O., Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol. Syst. Biol. 2006;2:46. doi: 10.1038/msb41000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R.V., Grosse I., Zhang M.Q. Computational identification of promoters and first exons in the human genome. Nat. Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- Dews M., Homayouni A., Yu D., Murphy D., Sevignani C., Wentzel E., Furth E.E., Lee W.M., Enders G.H., Mendell J.T., et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardana P.G., Peruzzi B., Giubellino A., Burke T.R., Jr, Bottaro D.P. Molecular targeting of growth factor receptor-bound 2 (Grb2) as an anti-cancer strategy. Anticancer Drugs. 2006;17:13–20. doi: 10.1097/01.cad.0000185180.72604.ac. [DOI] [PubMed] [Google Scholar]
- Down T.A., Hubbard T.J. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Draznin B. Molecular mechanisms of insulin resistance: Serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: The two sides of a coin. Diabetes. 2006;55:2392–2397. doi: 10.2337/db06-0391. [DOI] [PubMed] [Google Scholar]
- Fontana L., Pelosi E., Greco P., Racanicchi S., Testa U., Liuzzi F., Croce C.M., Brunetti E., Grignani F., Peschle C. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev Y., Schmittgen T.D., Lerner M., Postier R., Brackett D. Computational analysis of biological functions and pathways collectively targeted by co-expressed microRNAs in cancer. BMC Bioinformatics. 2007;8(Suppl. 7):S16. doi: 10.1186/1471-2105-8-S7-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Lv Q., Ye W., Wong C.K., Cai G., Gu D., Ji Y., Zhao C., Wang J., Yang B.B., et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Kuhn R.M., Baertsch R., Barber G.P., Clawson H., Diekhans M., Giardine B., Harte R.A., Hinrichs A.S., Hsu F., et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M., Sethupathy P., Corda B., Hatzigeorgiou A.G. miRGen: A database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Baiocchi R.A., Byrd J.C., Grever M.R., Jacob S.T., Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26:3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P., Megraw M., Hatzigeorgiou A.G. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′-UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Thomson J.M., Parker J., Perou C.M., Hammond S.M. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Tian L., Greenberg S.A., Kong S.W., Altschuler J., Kohane I.S., Park P.J. Discovering statistically significant pathways in expression profiling studies. Proc. Natl. Acad. Sci. 2005;102:13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston M.D., Pierce M.L., Rocha-Sanchez S., Beisel K.W., Soukup G.A. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Wong C.F., Tellam R.L. microRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J. Biol. Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- Xu S., Witmer P.D., Lumayag S., Kovacs B., Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]