Abstract
Guanine-rich sequences can adopt intramolecular four-stranded structures, called G-quadruplexes. These motifs have been intensively investigated on the DNA level, but their overall biological relevance remains elusive. Only recently has research concerning the function of G-quadruplexes in RNAs commenced. Here, we demonstrate for the first time, that an RNA G-quadruplex structure inhibits translation in vivo in eukaryotic cells. We investigated the function of a highly conserved, thermodynamically stable RNA G-quadruplex in the 5′-UTR of the mRNA of the human Zic-1 zinc-finger protein. Using dual luciferase reporter assay, we demonstrate that the Zic-1 RNA G-quadruplex represses protein synthesis inside eukaryotic cells. Quantitative RT-PCR assays confirmed that the reduction of protein synthesis is due to regulation of the translation process and not a consequence of reduced transcription. Western blot analysis revealed that expression of Zic-1 is strongly reduced by a 73 nucleotides-long fragment of the UTR containing the G-quadruplex motif. These structures might add to the more recently discovered elements in untranslated regions of mRNAs that regulate their translation.
Keywords: G-quadruplex, RNA structure, Zic-1, translational repression
INTRODUCTION
Guanine-rich nucleic acids have the potential to form unusual secondary structures comprising of four Hoogsteen-paired coplanar guanines, known as G-quadruplexes (GQ) (Gellert et al. 1962). The best characterized G-rich sequences that are able to fold into quadruplex structures are the capping structures at the ends of eukaryotic chromosomes, the telomeres (Mills et al. 2002; Rhodes et al. 2002; Neidle and Parkinson 2003; Cech 2004). Strong evidence for the existence of G-rich quadruplex structures in vivo was observed by staining of telomeres with quadruplex-specific antibodies (Schaffitzel et al. 2001). Furthermore, selective binding of proteins and small molecules to G-rich mRNAs and telomeric DNA (Darnell et al. 2001; Ramos et al. 2003; Granotier et al. 2005; Paeschke et al. 2005) was reported and the formation of DNA GQ structures during transcription was observed (Duquette et al. 2004). Moreover, small molecules that bind to and thereby stabilize DNA GQs have been shown to suppress gene expression of certain proto-oncogenes, hence providing a potential new therapeutic approach (Rangan et al. 2001; Siddiqui-Jain et al. 2002).
Despite the growing body of evidence for the existence of GQs in biological systems (recently reviewed in Patel et al. 2007), their overall functional relevance is still under debate. This is particularly true for the role of DNA GQs, due to the competition between quadruplex formation by one of the two strands and hybridization of the two complementary strands (Li et al. 2003; Risitano and Fox 2003; Kumar and Maiti 2005). Unlike DNA, transcribed RNA is exported to the cytoplasm, where it experiences more changes in the concentration of ions and molecular crowding agents than DNA existing solely in the nucleus. This suggests that RNA GQs can act as tuneable devices depending on cellular conditions.
Research in recent years revealed an unexpected relevance of RNA-mediated regulation of protein synthesis at the post-transcriptional level, e.g., via the formation of secondary structures like hairpin loops (Kozak 1991), riboswitches (Mandal and Breaker 2004), or small RNAs (Rana 2007). A potential regulative role of four-stranded GQ secondary structures has been suggested for alternative splicing of human pre-mRNA (Gomez et al. 2004; Kostadinov et al. 2006). It has also been shown recently that an RNA G-quadruplex in the 5′-UTR of the NRAS mRNA modulates protein synthesis in an artificial in vitro translation system (Kumari et al. 2007). In another study, a G-quadruplex in the fragile X premutation mRNA was described to influence protein synthesis (Khateb et al. 2007). Quadruplex forming G-rich sequences identified by bioinformatic analysis of UTRs of eukaryotic mRNAs were assembled in the GRS_UTR database (Kikin et al. 2008). Furthermore, Wieland and Hartig (2007) introduced tunable RNA GQ motifs to modulate gene expression in bacteria. Here, we demonstrate for the first time that a GQ motif in the 5′-UTR of an mRNA can inhibit translation in living eukaryotic cells. We investigated the effect of an evolutionarily conserved GQ from the mRNA of the zinc-finger protein of the cerebellum 1 (Zic-1). The motif was found to drastically repress translation without affecting the mRNA level in vivo.
RESULTS AND DISCUSSION
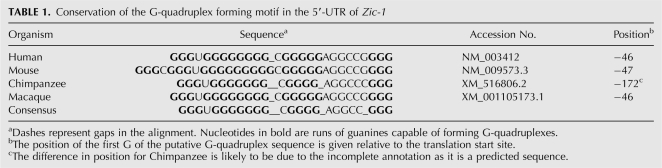
Bioinformatic analyses have shown that up to 376,000 potential DNA GQs exist in the human genome (Huppert and Balasubramanian 2005; Todd et al. 2005). Since we aimed at investigating a possible regulative role in translation, we were interested in motifs being located in the 5′-UTRs of mRNAs. We employed our previously developed computational search algorithm (Scaria et al. 2006) to identify several stable and highly conserved GQ motifs in the 5′-UTRs of mRNAs. As a first candidate, we chose the mRNA of the zinc-finger protein Zic-1. This protein plays a role in cerebellar development (Aruga et al. 1998) and is also considered to be of pathological relevance, as it was found to be overexpressed in medulloblastoma (26/29 cases), while it was not present in any of the other tumors examined (Yokota et al. 1996; Michiels et al. 1999). The 719-nucleotide-(nt) long Zic-1 5′-UTR contains an RNA GQ motif, which starts 46 nt upstream of the translation start site. This motif is evolutionarily conserved across the human, mouse, chimpanzee, and macaque genes orthologous to Zic-1 (Table 1).
TABLE 1.
Conservation of the G-quadruplex forming motif in the 5′-UTR of Zic-1
Thermodynamic and structural characterization of Zic-1 RNA quadruplex
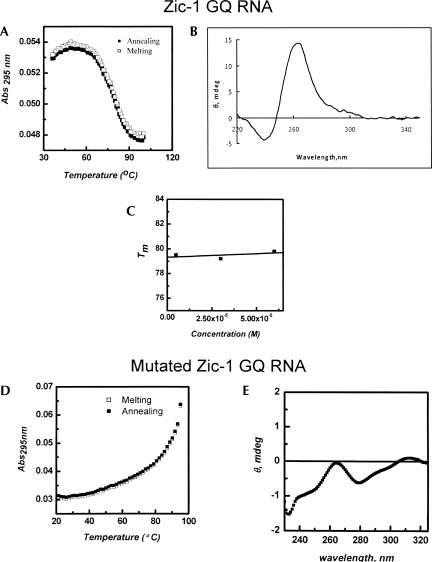
The thermal melting of quadruplex structures can be characterized by an inverse UV transition (Mergny et al. 1998). The UV melting profile of the putative Zic-1 RNA quadruplex sequence in a buffer containing 25 mM KCl shows hypochromic transition with a characteristic sigmoidal curve (Fig. 1A). Curves for melting and annealing were virtually identical. The Tm value was found to be 79°C. At a higher KCl concentration (100 mM), the structure could not be unfolded, even at 95°C, which is indicative of a very stable quadruplex (data not shown). Furthermore, the Tm was found to depend on the nature of the monocation. In the presence of 25 mM NaCl, Tm was 15°C lower than in the presence of KCl at the same concentration (data not shown). This finding confirms the well-known fact that GQ motifs are more stable in the presence of potassium ions than in the presence of any other monocation (Rachwal et al. 2007).
FIGURE 1.
Biophysical analysis of the Zic-1 RNA GQ. (A) Melting and annealing curves were recorded at 295 nm. (B) CD spectrum of Zic-1 RNA GQ. (C) Melting temperature Tm at different concentrations of the RNA GQ. (D) Melting and annealing curve for the mutated Zic-1 GQ RNA. (E) CD spectrum of the mutated Zic-1 GQ RNA. All experiments were carried out at 25 mM KCl.
Circular dichroism (CD) spectroscopy is a standard technique to analyze structural features of GQ motifs (Kumari et al. 2007; Wieland and Hartig 2007). This method was therefore used to characterize the structural conformation of the putative Zic-1 RNA GQ in terms of parallel or antiparallel topology. As shown in Figure 1B, the Zic-1 GQ motif reveals a positive band at around 263 nm and a negative band near 240 nm. These features are typical for a parallel-oriented quadruplex structure. RNA quadruplexes generally disfavor the antiparallel fold, since this would require guanosines to adopt in part the syn-conformation, which is highly unfavorable for RNA due to the ribose C3′-endo conformation (Tang and Shafer 2006).
In a mutated variant of the Zic-1 GQ RNA various guanosines were replaced by adenosine. To confirm that this RNA does not form a GQ structure, a melting curve (Fig. 1D) at 295 nm and a CD spectrum (Fig. 1E) were recorded. Neither a distinct hypochromic transition in the melting curve at 295 nm nor characteristic bands in the CD spectrum were observed, indicating that this sequence is a suitable control that does not form a GQ structure.
To further investigate whether the GQ of the original Zic-1 sequence is formed intermolecularly or intramolecularly, we determined the melting temperature Tm at different concentrations of the RNA GQ oligonucleotides (5–60 μM) in buffer containing 25 mM KCl. As can be seen in Figure 1C, Tm remains virtually unchanged at 79°C in the concentration range analyzed. This finding is indicative for the formation of an intramolecular GQ motif. Together, the UV and CD profiles reflect that the G-rich Zic-1 RNA folds into a very stable, parallel intramolecular GQ structure under near physiological pH and salt conditions.
Inhibition of translation in living human cells
Our next aim was to investigate the influence of the RNA GQ motif on translation in living eukaryotic cells. Previous studies have shown that GQ motifs in the 5′-UTR of an mRNA can reduce translation in a rabbit reticulocyte lysate in vitro (Kumari et al. 2007). Furthermore, artificial GQ motifs that mask the Shine–Dalgarno sequences were found to suppress gene expression in bacteria (Wieland and Hartig 2007). However, to the best of our knowledge, the question of whether or not RNA GQ structures influence translation in vivo in eukaryotic cells has not yet been addressed.
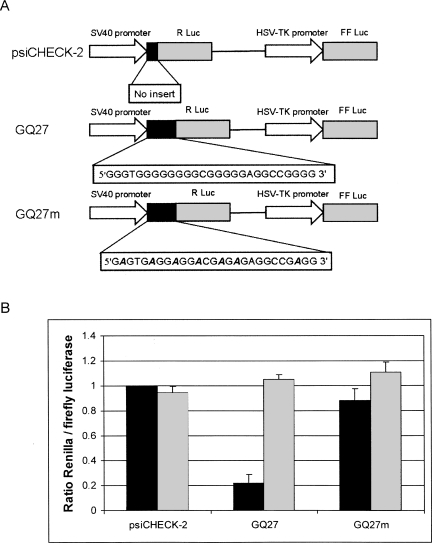
To clarify this important point, we performed dual luciferase assays in HeLa cells. The psiCHECK-2 vector from Promega allows simultaneous expression of Renilla and firefly luciferase from a single plasmid (Fig. 2A). To evaluate the influence of the Zic-1 RNA GQ on the efficiency of translation, we cloned a 27-base pair-long DNA sequence encoding the GQ motif upstream of the Renilla luciferase. This plasmid was named GQ27 (Fig. 2A). As a control, a mutated version of the GQ was used, in which several guanosines of the original sequence were replaced by adenosines preventing the formation of the GQ structure (Fig. 2A, GQ27m).
FIGURE 2.
(A) Plasmids for the investigation of the influence of the GQ motif on translation. The original psiCHECK-2 vector (Promega) expresses Renilla and firefly luciferase under control of the SV40 and HSV-TK promoter, respectively. The plasmid GQ27 contains the 27-nt-long GQ-motif of the Zic-1 5′-UTR upstream of the start codon. In the control plasmid GQ27m several guanosines were replaced by adenosines (bold and italics). (B) Relative levels of Renilla and firefly luciferase. Protein activity was determined by dual-luciferase reporter assays (black bars). Plasmids were transfected into HeLa cells. Twenty-four hours after transfection cells were harvested and dual-luciferase assays were carried out. The ratios of Renilla and firefly luciferase activity were calculated. The ratio of the control plasmid psiCHECK-2 was set as 1 and the other values were normalized accordingly. Average values and standard deviations obtained from four independent experiments are shown. Relative mRNA levels were determined by real-time PCR (gray bars). Again, HeLa cells were transfected with either of the plasmids and total RNA was isolated after 24 h. CT values of Renilla and firefly luciferase mRNAs were determined by quantitative RT-PCR. Ratios of Renilla and firefly mRNA levels were calculated as CT (Renilla)/CT (firefly). Averages and standard deviations of three independent experiments are shown.
The three vector constructs (psiCHECK-2, GQ27, or GQ27m) were transfected into HeLa cells, and 24 h after transfection, cells were harvested for dual luciferase assays. The ratio of Renilla and firefly luciferase activity in the original psiCHECK-2 control vector was ∼30. Figure 2B (black bars) shows the ratios of Renilla and firefly luciferase activity normalized to the value obtained for the unmodified psiCHECK-2 vector. Strikingly, insertion of the Zic-1 RNA GQ27 motif upstream of the Renilla luciferase start codon drastically reduces protein synthesis by ∼80%. In contrast, the mutated version GQ27m, which does not form a stable quadruplex structure (Fig. 1D,E), does not significantly influence translation. The extent of translational repression found in living cells is comparable to what was reported for an in vitro translation system before. Balasubramanian and coworkers reported an approximately fourfold decrease in protein synthesis in rabbit reticulocyte lysate caused by a GQ motif in the 5′-UTR of the NRAS mRNA (Kumari et al. 2007).
We next wanted to know whether the GQ motif represses gene expression on the transcriptional or translational level. To this end, we performed real-time PCR assays with lysates from those that were transfected with the empty psiCHECK-2, GQ27 and GQ27m plasmid, respectively. Twenty-four hours after transfection, total RNA was isolated and quantitative RT-PCR assays were performed. As can be seen in Figure 2B (gray bars) almost identical levels were observed for all three constructs, i.e., the GQ motif does not reduce the mRNA level of the Renilla luciferase. We thus conclude that the decrease of protein synthesis is due to repression of translation rather than a consequence of reduced transcription.
Suppresion of Zic-1 protein synthesis in the presence of G-quadruplex motif
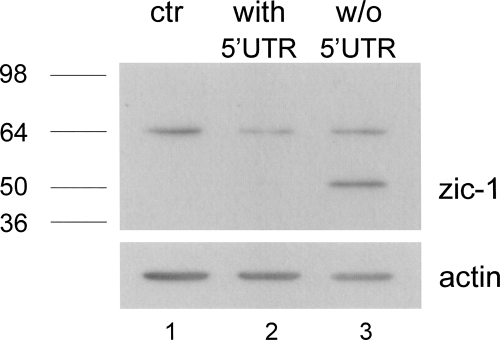
In order to investigate the role of the RNA GQ on the Zic-1 expression, we analyzed synthesis of the Zic-1 protein in the presence or absence of the guanosine-rich sequence upstream of the start codon. Eukaryotic HeLa cells were transfected with a plasmid encoding Zic-1 either in the presence or absence of a 73-nt-long fragment of the natural 5′-UTR containing the GQ-forming sequence. Twenty-four hours after transfection, cells were harvested and lysates for Western blots were prepared. An unspecific band of ∼64 kDa is detected in all samples, but no endogenous expression of Zic-1 was observed in untransfected HeLa cells (Fig. 3, lane 1). Similarly, virtually no Zic-1 expression could be detected from plasmids containing the fragment of the 5′-UTR (Fig. 3, lane 2). In sharp contrast, a strong band at the expected size of ∼50 kDa was observed in the lysate of cells which were transfected with the plasmid encoding Zic-1 in the absence of the 5′-UTR (Fig. 3, lane 3). These results clearly confirm that the GQ motif strongly represses protein synthesis.
FIGURE 3.
Expression of Zic-1 in the presence and absence of the partial 5′-UTR, containing the GQ motif. HeLa cells were transfected with 0.8 μg of the pcDNA3.1 plasmid expressing Zic-1 with (lane 2) or without (lane 3) partial 5′-UTR. Cells were lysed 24 h after transfection. Western blot with Zic-1 specific antibodies was performed as described in the Materials and Methods section. Lane 1 shows the lysate of untransfected cell; an actin blot was performed as a loading control.
The functional relevance of G-quadruplex structures in living cells is still under investigation. In our present study, we demonstrate that an RNA GQ in the 5′-UTR strongly represses protein synthesis. Since the GQ motif does not influence the mRNA level, we conclude that this effect is due to inhibition of translation. GQ motifs occurring in numerous genes might be a new class of elements in untranslated regions that add to the recently discovered repertoire of regulatory elements in mRNAs like riboswitches and aptazymes. Since a number of low molecular weight substances have been developed that specifically bind to G quadruplexes, these motifs can be considered as an attractive target for small molecular therapeutics.
MATERIALS AND METHODS
Oligonucleotides
The following RNA oligonucleotides for spectroscopic studies was purchased from Chemgenes Corp.:
Zic-1 5′-GGGUGGGGGGGGCGGGGGAGGCCGGGG-3′; and
Mutated Zic-1 5′-GAGUGAGGAGGACGAGAGAGGCCGAGG-3′.
RNA concentration was determined by UV measurement using an extinction coefficient of 26.63 × 104 M−1 cm−1 and 28.82 × 104 M−1 cm−1 for Zic-1 and mutated Zic-1, respectively.
DNA oligonucleotides for PCR and cloning were purchased from TIB MOLBIOL.
Spectroscopy
For spectroscopic studies, we prepared RNA samples at 5 μM concentration in a buffer containing 10 mM sodium cacodylate at pH 7.4, and 25 or 100 mM KCl as indicated in the Results and Discussion section. Samples were annealed by heating at 90°C for 10 min and subsequent cooling down to 5°C. CD experiments were performed at 20°C using a Jasco J-810 spectropolarimeter (Jasco Hachioji, Tokyo, Japan) equipped with a Peltier temperature controller. CD scans were taken in triplicates and their average was calculated. A CD spectrum of the buffer was recorded and subtracted from the spectrum obtained for the RNA-containing solution. Data were zero-corrected at 350 nm.
UV-melting studies were carried out on a Cary 100 UV-visible spectrophotometer (Varian) equipped with a Peltier temperature controller. Samples were heated to 90°C and cooled down to 5°C at a 0.2°C min−1 temperature gradient, and absorption data recorded at 295 nm were collected every 0.1 min on both annealing and melting steps.
Construction of plasmids
To construct the plasmid GQ27, a synthetic DNA duplex encoding the G-rich sequence of the Zic-1 5′UTR was inserted into the unique NheI restriction site of the psiCHECK-2 plasmid (Promega) upstream of the Renilla luciferase start codon. The following DNA oligonucleotides were used:
GQ27 sense: CTAGCGGGTGGGGGGGGCGGGGGAGGCCGGGGA; and
GQ27 antisense: CTAGTCCCCGGCCTCCCCCGCCCCCCCCACCCG.
The cloning procedure results in the following mRNA sequence (the GQ motif is underlined; the start codon is given in italics):
5′-AUAGCUAGCGGGUGGGGGGGGCGGGGGAGGCCGGGGAGCUAGCCACCAUG-3′.
The control vector GQ27m containing the GQ motif with several G to A substitutions was generated with the following oligonucleotides:
GQ27m sense: CTAGCGAGTGAGGAGGACGAGAGAGGCCGAGGA; and
GQ27m antisense: CTAGTCCTCGGCCTCTCTCGTCCTCCTCACTCG.
A Zic-1 cDNA clone with a partial 5′-UTR containing the GQ forming region was obtained from Bio Cat as the pCR4-TOPO vector. The Zic-1 cDNA with or without the partial 5′UTR was subcloned into the eukaryotic expression vector pcDNA3.1.
Cell culture
HeLa cells were grown at 37°C in a humidified atmosphere containing 5% CO2 in Dulbecco's modified Eagles medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM glutamine, nonessential amino acids, and the antibiotics penicillin and streptomycin.
Transient transfection and dual luciferase reporter assay
Transfection was carried out in 24-well plates using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions. Twenty-four hours after transfection, cells were resuspended in passive lysis buffer (Promega). The firefly and Renilla luciferase activities are measured using the Dual-luciferase Reporter Assay kit (Promega) according to the manufacturer's instructions with a luminometer (TD 20/20 Luminometer, Turner BioSystems).
Quantitative real-time PCR assay
For quantitative analysis of the mRNA levels, total cellular RNA was isolated from transfected cells using the RNeasy mini kit along with RNase-free DNase to remove any traces of DNA contamination (QIAGEN). The first-strand cDNA was synthesized with M-MLV reverse transcriptase (Promega). cDNA was then subjected to quantitative real-time PCR, using a SYBR-Green PCR Master Mix (Applied Biosystems) and the ABI Prism 7300 Detection system (Applied Biosystems).
Western blot
Zic-1 protein was immunoblotted with an anti-Zic-1 primary antibody (ab24204, Abcam). Actin was subsequently probed as an internal loading control (MAB1501R, Chemicon). Goat antirabbit and goat antimouse secondary antibodies conjugated with horse-radish peroxidase (HRP) were used for Zic-1 and actin detection, respectively. Antigen–antibody complexes were visualized by the ECL system (Pierce).
ACKNOWLEDGMENTS
The authors thank Diana Rothe and Denise Werk for their help, and Volker A. Erdmann for fruitful discussions. A.A. is a fellow of the German Academic Exchange Service (DAAD) and a Senior Research Fellow (SRF) of the University Grants Commission (UGC), India. M.D. is a fellow of the German Academic Exchange Service (DAAD). V.S. and M.H. thank the Council for Scientific and Industrial Research (CSIR), India, for a Senior Research Fellowship. Financial support from the Free University Berlin, the Fonds der Chemischen Industrie, and Council for Scientific and Industrial Research (CSIR), India, is gratefully acknowledged.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1001708.
REFERENCES
- Aruga J., Minowa O., Yaginuma H., Kuno J., Nagai T., Noda T., Mikoshiba K. Mouse Zic1 is involved in cerebellar development. J. Neurosci. 1998;18:284–293. doi: 10.1523/JNEUROSCI.18-01-00284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T.R. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes & Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez D., Lemarteleur T., Lacroix L., Mailliet P., Mergny J.L., Riou J.F. Telomerase down-regulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granotier C., Pennarun G., Riou L., Hoffschir F., Gauthier L.R., De Cian A., Gomez D., Mandine E., Riou J.F., Mergny J.L., et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005;33:4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khateb S., Weisman-Shomer P., Hershco-Shani I., Ludwig A.L., Fry M. The tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance the in vivo translation of fragile X premutation mRNA. Nucleic Acids Res. 2007;35:5775–5788. doi: 10.1093/nar/gkm636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikin O., Zappala Z., D'Antonio L., Bagga P.S. GRSDB2 and GRS_UTRdb: Databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008;36:D141–D148. doi: 10.1093/nar/gkm982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostadinov R., Malhotra N., Viotti M., Shine R., D'Antonio L., Bagga P. GRSDB: A database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Kumar N., Maiti S. The effect of osmolytes and small molecule on Quadruplex-WC duplex equilibrium: A fluorescence resonance energy transfer study. Nucleic Acids Res. 2005;33:6723–6732. doi: 10.1093/nar/gki961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., Bugaut A., Huppert J.L., Balasubramanian S. An RNA G-quadruplex in the 5′-UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Miyoshi D., Nakano S., Sugimoto N. Structural competition involving G-quadruplex DNA and its complement. Biochemistry. 2003;42:11736–11744. doi: 10.1021/bi034168j. [DOI] [PubMed] [Google Scholar]
- Mandal M., Breaker R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- Michiels E.M., Oussoren E., Van Groenigen M., Pauws E., Bossuyt P.M., Voute P.A., Baas F. Genes differentially expressed in medulloblastoma and fetal brain. Physiol. Genomics. 1999;1:83–91. doi: 10.1152/physiolgenomics.1999.1.2.83. [DOI] [PubMed] [Google Scholar]
- Mills M., Lacroix L., Arimondo P.B., Leroy J.L., Francois J.C., Klump H., Mergny J.L. Unusual DNA conformations: Implications for telomeres. Curr. Med. Chem. Anticancer Agents. 2002;2:627–644. doi: 10.2174/1568011023353877. [DOI] [PubMed] [Google Scholar]
- Neidle S., Parkinson G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- Paeschke K., Simonsson T., Postberg J., Rhodes D., Lipps H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- Patel D.J., Phan A.T., Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachwal P.A., Findlow I.S., Werner J.M., Brown T., Fox K.R. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007;35:4214–4222. doi: 10.1093/nar/gkm316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A., Hollingworth D., Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9:1198–1207. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana T.M. Illuminating the silence: Understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- Rangan A., Fedoroff O.Y., Hurley L.H. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Fairall L., Simonsson T., Court R., Chapman L. Telomere architecture. EMBO Rep. 2002;3:1139–1145. doi: 10.1093/embo-reports/kvf246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risitano A., Fox K.R. Stability of intramolecular DNA quadruplexes: Comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- Scaria V., Hariharan M., Arora A., Maiti S. Quadfinder: Server for identification and analysis of quadruplex-forming motifs in nucleotide sequences. Nucleic Acids Res. 2006;34:W683–W685. doi: 10.1093/nar/gkl299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J., Pluckthun A. In vitro generated antibodies specific for telomeric guanine–quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.F., Shafer R.H. Engineering the quadruplex fold: Nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland M., Hartig J.S. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Yokota N., Aruga J., Takai S., Yamada K., Hamazaki M., Iwase T., Sugimura H., Mikoshiba K. Predominant expression of human zic in cerebellar granule cell lineage and medulloblastoma. Cancer Res. 1996;56:377–383. [PubMed] [Google Scholar]