Abstract
The Smaug family of sequence-specific RNA binding proteins regulates mRNA translation and degradation by binding to consensus stem–loop structures in target mRNAs. Vts1p is a member of the Smaug protein family that regulates the stability of target transcripts in Saccharomyces cerevisiae. Here we focus on the mechanism of Vts1p-mediated mRNA decay. Using RNA reporters that recapitulate Vts1p-mediated decay in vivo, we demonstrate that Vts1p stimulates mRNA degradation through deadenylation mediated by the Ccr4p-Pop2p-Not deadenylase complex. We also show that Vts1p interacts with the Ccr4p-Pop2p-Not complex suggesting that Vts1p recruits the Ccr4p-Pop2p-Not deadenylase complex to target mRNAs, resulting in transcript decay. Following deadenylation Vts1p target transcripts are decapped and subsequently degraded by the 5′-to-3′ exonuclease Xrn1p. Decapping and 5′-to-3′ decay is thought to occur in foci known as P-bodies, and we provide evidence that Vts1p function may involve P-bodies. Taken together with previous work, these data suggest that Smaug family members employ a conserved mechanism to induce transcript degradation that involves recruitment of the Ccr4-Pop2-Not deadenylase to target mRNAs.
Keywords: Vts1, mRNA decay, deadenylation, Smaug
INTRODUCTION
The identification of several conserved mRNA degradation pathways in eukaryotes highlights the importance of mRNA turnover as a means of controlling gene expression. In Saccharomyces cerevisiae the major mRNA decay pathway initiates with deadenylation by the Ccr4p-Pop2p-Not deadenylase complex (Decker and Parker 1993; Hsu and Stevens 1993; Muhlrad et al. 1995). Following poly(A) removal the transcript is decapped and then degraded 5′-to-3′ by the exonuclease Xrn1p (Hsu and Stevens 1993; Muhlrad et al. 1994, 1995; Caponigro and Parker 1996). In addition to the general pathway of mRNA decay, sequence-specific RNA binding proteins add an additional level of complexity to the regulation of transcript stability (Garneau et al. 2007). In general these proteins bind target sequences in a given mRNA and can affect transcript stability by interacting with components of the mRNA decay machinery.
The Smaug (Smg) family is a class of proteins, conserved from yeast to humans, that regulate gene expression post-transcriptionally through their ability to bind directly to target mRNAs (Smibert et al. 1996, 1999; Dahanukar et al. 1999; Aviv et al. 2003; Baez and Boccaccio 2005; Semotok et al. 2005). Family members bind RNA with similar specificity through a conserved sterile alpha motif (SAM) domain that is able to interact with stem–loop structures termed Smg recognition elements (SREs) (Smibert et al. 1996, 1999; Dahanukar et al. 1999; Crucs et al. 2000; Aviv et al. 2003; Green et al. 2003). Drosophila Smg, the founding member of this family, regulates mRNA translation and destabilization through separate mechanisms. Smg distinguishes between these roles by recruiting specific trans-acting factors that aid in transcript regulation. Smg represses mRNA translation by recruiting the eIF4E binding protein Cup to target transcripts (Nelson et al. 2004). In contrast, Smg triggers transcript destabilization by recruiting the Ccr4-Pop2-Not deadenylase complex to target mRNAs (Semotok et al. 2005). We have proposed a similar mechanism of mRNA destabilization by Vts1p, the Smg homolog in S. cerevisiae (Aviv et al. 2003). Vts1p can regulate the expression of a reporter mRNA harboring SREs in the 3′UTR of the transcript. This work also showed that reporter expression is regulated at the level of mRNA stability and that both Vts1p and functional SREs are required for this effect. Vts1p-mediated mRNA degradation appears to require the Ccr4p-Pop2p-Not deadenylase complex since reporter mRNA steady-state levels are increased in cells lacking Ccr4p (Aviv et al. 2003), which is the catalytic subunit of this complex (Chen et al. 2002; Tucker et al. 2002; Parker and Song 2004). While potential endogenous targets of Vts1p have been identified by immunoprecipitating Vts1p and identifying copurifying mRNAs via microarray (Aviv et al. 2006), the mechanism that underlies Vts1p-mediated regulation of these transcripts has not been explored.
Here we show that Vts1p induces mRNA decay via the major mRNA decay pathway of deadenylation-dependent decapping and subsequent 5′-to-3′ decay. Using reporter mRNAs we show that Vts1p and Ccr4p are required for the rapid decay and deadenylation of SRE-containing transcripts in vivo and that Vts1p associates with the Ccr4p-Pop2p-Not deadenylase complex, suggesting that Vts1p-mediated decay results from recruitment of this deadenylase to target mRNAs. We demonstrate that deadenylation is the initial step in Vts1p-mediated mRNA decay and that following poly(A) tail removal transcripts are decapped and then degraded in a 5′-to-3′ direction by the exonuclease Xrn1p. Decapping and 5′-to-3′ decay of transcripts is thought to occur in cytoplasmic foci known as P-bodies (Sheth and Parker 2003), and we provide additional evidence that suggests that Vts1p function might involve P-bodies. Together, these data specifically outline the molecular mechanism Vts1p employs to degrade mRNAs. They also provide evidence suggesting that Smg family members employ a conserved mechanism to induce transcript decay, which involves recruitment of the Ccr4-Pop2-Not deadenylase to target mRNAs.
RESULTS
Ccr4p is required for Vts1p-mediated mRNA decay
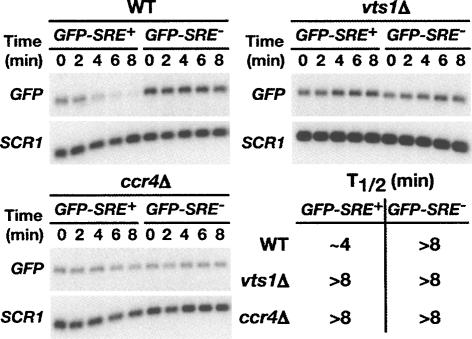
We previously proposed that Vts1p regulates mRNA stability through a mechanism that requires the Ccr4p-Pop2p-Not deadenylase complex (Aviv et al. 2003). This model was based on experiments that showed that the steady-state levels of a Vts1p-regulated reporter mRNA increase in a ccr4Δ strain. To further test this model we first compared the stability of a reporter mRNA in wild-type cells to cells carrying a deletion in either the VTS1 gene or CCR4 gene, which encodes the catalytic subunit of the deadenylase (Chen et al. 2002; Tucker et al. 2002; Parker and Song 2004). This reporter, which has been previously shown to recapitulate Vts1p-mediated decay in vivo (Aviv et al. 2003), encodes green fluorescent protein (GFP) under the control of a galactose-inducible promoter and has three SREs in its 3′UTR (GFP-SRE+). A similar reporter bearing SREs in which the loop sequences are mutated to block Vts1p binding (GFP-SRE−) serves as a control.
To measure the stability of the reporter mRNA we used a transcriptional pulse-chase approach where reporter gene transcription was induced by the treatment of cells with galactose for 16 min and then inhibited by the addition of glucose. The level of the GFP reporter mRNA was then measured at 2-min intervals by Northern blotting with the zero time point corresponding to the glucose addition. As we have previously reported the GFP-SRE+ mRNA is rapidly degraded (half-life of ∼4 min) in a manner that is dependent on both Vts1p and wild-type SREs (Fig. 1, with quantification of these data shown in Supplemental Fig. 1; Aviv et al. 2003). Here we show that rapid degradation of GFP-SRE+ mRNA requires Ccr4p as it is significantly stabilized in the ccr4Δ strain. These data confirm our initial hypothesis that Ccr4p is required for Vts1p-mediated mRNA degradation.
FIGURE 1.
Vts1p stimulates the decay of target mRNAs. GFP reporter transcription was induced with galactose and then shut off with glucose and GFP mRNA levels were assayed at the times indicated after transcriptional shutoff by Northern blot. Levels of GFP mRNA were measured in wild-type (WT), vts1Δ, and ccr4Δ cells. SCR1 RNA serves as a loading control. Half-lives (T1/2) for each strain are as indicated.
Vts1p target mRNAs undergo Ccr4p-dependent deadenylation
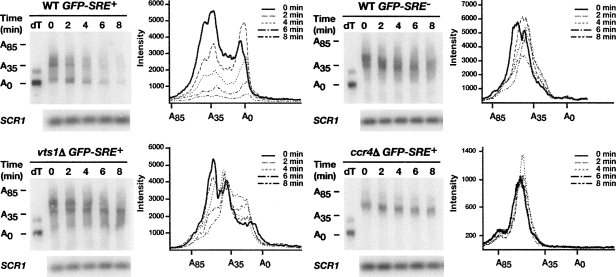
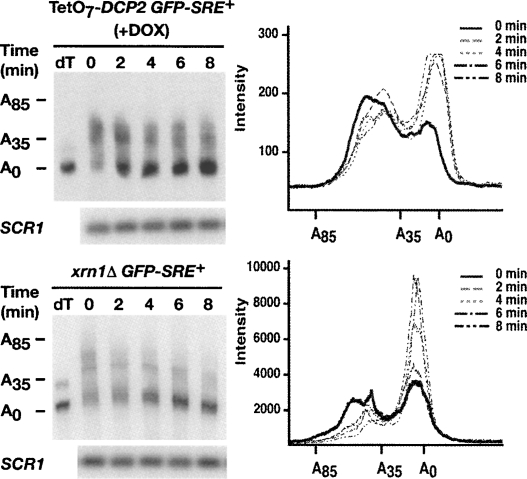
To determine if degradation of the GFP-SRE+ transcript involved deadenylation by Ccr4p, we analyzed the poly(A) tail length of each GFP reporter in wild-type, vts1Δ, and ccr4Δ cells. We used an RNase H cleavage method to measure the poly(A) tail length of the GFP reporter mRNAs over an 8-min transcriptional pulse-chase time course. A specific antisense oligonucleotide that hybridizes to the GFP open reading frame was added to total RNA from each time point and cleaved with RNase H to produce a short 3′-end fragment of the GFP reporter mRNA. The poly(A) tail length of this fragment was measured by performing high resolution polyacrylamide electrophoresis and Northern blotting. In one sample reporter specific oligonucleotide and oligo(dT) was included and, as such, treatment with RNase H produced a 3′-end fragment without a poly(A) tail serving as a marker for the deadenylated 3′-end fragment.
In wild-type cells, at time zero there were two distinct populations of the GFP-SRE+ mRNA with one having poly(A) tail lengths ranging from 30 to 50 nucleotides (nt) and another population having very short poly(A) tails (Fig. 2). This latter population presumably represents transcripts that accumulated and were deadenylated during the 16-min induction with galactose, suggesting rapid deadenylation of the transcript. If the GFP-SRE+ mRNA is subject to rapid deadenylation followed by decay, this would predict that during a transcriptional pulse-chase experiment the longer poly(A) tail species would be converted to the shorter tail species, which at subsequent time points would disappear. However, the short half-life of the GFP-SRE+ transcript might make it difficult to observe these intermediates. Nonetheless, soon after the addition of glucose we reproducibly detected a decrease in the amount of the longer poly(A) tail species and an increase in the amount of the shorter tail population (compare the distribution of the poly(A) tail lengths of transcripts at time equals zero to the 2-min time point) while at later time points the shorter tail population disappears.
FIGURE 2.
Vts1p induces the deadenylation of target mRNAs. GFP reporter transcription was induced with galactose and then shut off with glucose and an RNase H cleavage assay was used to measure the poly(A) tail length of GFP reporter mRNAs in wild-type (WT), vts1Δ, and ccr4Δ strains at the times indicated after transcriptional shutoff. Where indicated oligo(dT) was added to remove the poly(A) tail by RNase H treatment, providing a marker for deadenylated mRNA (dT). The distribution of poly(A) tail lengths at the indicated times after transcriptional shutoff are displayed on the graphs. SCR1 RNA serves as a loading control.
In contrast, the majority of the GFP-SRE+ mRNA in vts1Δ cells and ccr4Δ cells and the GFP-SRE− mRNA in wild-type cells retained a long poly(A) tail distribution of 30–50 nt that exhibited little or no accumulation of short poly(A) tail transcripts during the 8-min time course (Fig. 2). Taken together these data suggest that recruitment of Vts1p to GFP mRNA via SREs stimulates rapid Ccr4p-dependent deadenylation. In addition, they demonstrate a strong correlation between defects in deadenylation (observed in either the vts1Δ or ccr4Δ strains or when the SREs are mutated) and stabilization of GFP-SRE+ mRNA, indicating that deadenylation plays a key role in Vts1p-mediated transcript decay. Additional data described below lend further support to the central role of deadenylation in Vts1p-mediated transcript decay.
Vts1p interacts with the Ccr4p-Pop2p-Not deadenylase complex
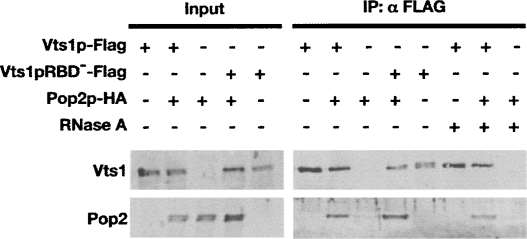
Our pervious work suggests that Drosophila Smg induces mRNA degradation by physically recruiting the Ccr4-Pop2-Not deadenylase complex to target transcripts (Semotok et al. 2005). Given the requirement of Ccr4p for Vts1p-mediated mRNA decay in S. cerevisiae, we sought to determine whether Vts1p physically interacts with a component of the Ccr4p-Pop2p-Not deadenylase complex in vivo, Pop2p. We performed coimmunoprecipitation experiments using whole-cell lysates from cells expressing Vts1p tagged with a Flag epitope and Pop2p tagged with an HA epitope. Lysates from cells expressing only one of the epitope-tagged proteins served as negative controls. Vts1p-Flag was immunoprecipitated using anti-Flag resin, and the immunoprecipitates were analyzed by Western blot. Pop2p-HA was present in the anti-Flag immunoprecipitate when the Vts1p-Flag protein was present (Fig. 3). As a control, Pop2p-HA was not immunoprecipitated from an extract lacking Vts1p-Flag. The coimmunoprecipitation of Vts1p with Pop2p was RNA independent, as it was observed in the presence of RNase A and when Vts1p harbored an amino acid change (A498Q) that blocks its ability to bind RNA binding (Vts1pRBD−) (Aviv et al. 2003). This suggests that Vts1p physically associates with a component of the Ccr4p-Pop2p-Not deadenylase complex and supports a model in which Vts1p recruits the deadenylase to a target transcript to initiate deadenylation-dependent mRNA degradation.
FIGURE 3.
Vts1p interacts with Pop2p. Vts1p-Flag and Pop2p-HA are expressed in yeast cells individually or in combination as indicated, and crude extracts were subjected to immunoprecipitation using an anti-Flag resin. Starting crude extracts (input) and the resulting immunoprecipitates (IP:αFLAG) were analyzed by Western blot. Input samples represent 5% for Vts1p blots and 0.5% for Pop2p blots of material used in the immunoprecipitations. The addition of RNase A or use of the Vts1p RNA binding mutant (Vts1pRBD −) are as indicated.
Vts1p-mediated transcript decay requires mRNA decapping and 5′-to-3′ degradation
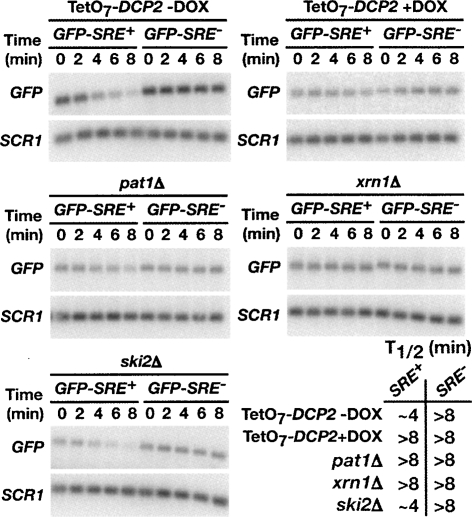
In yeast mRNA deadenylation can trigger decapping and subsequent 5′-to-3′ exonucleolytic decay (Caponigro and Parker 1996). Our data suggest that Vts1p mediates transcript degradation by recruiting the Ccr4p-Pop2p-Not deadenylase complex to initiate mRNA deadenylation. To determine if Vts1p target transcripts are decapped and degraded 5′-to-3′, we measured the stability of our GFP reporters in strains defective for decapping or 5′-to-3′ decay. To test whether Vts1p target transcripts are decapped, we examined the stability of the GFP reporters in a strain deficient for DCP2, the catalytic component of the decapping enzyme (Steiger et al. 2003). As DCP2 is essential in our strain background, we employed a strain expressing DCP2 under a tetracycline (tet)-regulatable promoter (TetO7-DCP2) (Mnaimneh et al. 2004). With this promoter-shutoff system, repression of DCP2 transcription is achieved by the addition of doxycycline (DOX), a tetracycline analog, to the growth medium. In the absence of DOX, TetO7-DCP2 is expressed and the GFP-SRE+ transcript decays at a rate similar to wild-type (Fig. 4, with results quantitated in Supplemental Fig. 2). When TetO7-DCP2 transcription is repressed by the addition of DOX, the stability of GFP-SRE+ is significantly increased, indicating that decapping is required for rapid turnover of the GFP-SRE+ mRNA. Also, consistent with the role of decapping in Vts1p-mediated decay, we find that GFP-SRE+ mRNA is stabilized in a strain deleted for the decapping activator PAT1 (Hatfield et al. 1996; Zhang et al. 1999; Bonnerot et al. 2000; Bouveret et al. 2000; Tharun et al. 2000). Interestingly, degradation of the GFP-SRE+ mRNA does not require another decapping activator Dhh1p (Supplemental Fig. 2), suggesting that Dhh1p is not required for the decapping of all mRNAs.
FIGURE 4.
Vts1p-mediated mRNA decay requires components of the major mRNA decay pathway. GFP reporter transcription was induced in the indicated deletion strains with galactose and then shut off with glucose and GFP mRNA levels were assayed at the times indicated after transcriptional shutoff by Northern blot. Transcriptional pulse-chase was performed with TetO7-DCP2 in the absence or presence of DOX as indicated. SCR1 RNA serves as a loading control. Half-lives (T1/2) for each strain are as indicated.
Following deadenylation-dependent decapping, the body of a transcript typically is degraded in a 5′-to-3′ direction by the exonuclease Xrn1p (Hsu and Stevens 1993; Muhlrad et al. 1994, 1995). To determine whether Vts1p-mediated mRNA degradation requires 5′-to-3′ decay, we measured the stability of the GFP reporters in an xrn1Δ strain. We found that GFP-SRE+ mRNA was significantly stabilized in the xrn1Δ strain over the 8-min time course (Fig. 4; Supplemental Fig. 2). We conclude that 5′-to-3′ decay by the exonuclease Xrn1p is required for Vts1p-mediated mRNA decay.
Thus far we have demonstrated that deadenylation, decapping, and 5′-to-3′ decay are important for Vts1p-mediated mRNA degradation. However after deadenylation a transcript can also be degraded by a complex of 3′-to-5′ exonucleases known as the exosome (Anderson and Parker 1998). To determine whether 3′-to-5′ decay is required for Vts1p-mediated decay, we examined the stability of the GFP reporters in a ski2Δ strain that is defective for exosome function. The half-life of the GFP-SRE+ reporter in ski2Δ cells was similar to wild type (Fig. 4; Supplemental Fig. 2) indicating that the exosome does not play a significant role in Vts1p-mediated decay.
Our data have shown that deadenylation, decapping, and 5′-to-3′ decay are all required for Vts1p-mediated mRNA degradation, leading us to hypothesize that Vts1p targets transcripts for deadenylation, which triggers decapping and subsequent 5′-to-3′ decay. To illustrate that deadenylation is the first step in Vts1p-mediated decay, which in turn triggers decapping and subsequent 5′-to-3′ decay of the message, we examined the poly(A) tail length of the GFP-SRE+ mRNA in TetO7-DCP2 and xrn1Δ cells. We find that inhibition of DCP2 expression and deletion of the XRN1 gene results in the accumulation of a stable deadenylated form of GFP-SRE+ mRNA (Fig. 5) indicating that deadenylation precedes decapping and 5′-to-3′ exonucleolytic decay. These data therefore are consistent with Vts1p triggering decay via deadenylase recruitment.
FIGURE 5.
Deadenylation is the first step in Vts1p-mediated mRNA decay. GFP reporter transcription was induced with galactose and then shut off with glucose and an RNase H cleavage assay was used to measure the poly(A) tail length of the GFP-SRE+ reporter mRNA in TetO7-DCP2 in the presence of DOX (+DOX) and in an xrn1Δ strain at the times indicated after shutoff. Where indicated oligo(dT) was added to remove the poly(A) tail by RNase H treatment, providing a marker for deadenylated mRNA (dT). The distribution of poly(A) tail lengths at the indicated times after transcriptional shutoff are displayed on the graphs. SCR1 RNA serves as a loading control.
Vts1p-mediated decay of an endogenous target mRNA requires Ccr4p and Xrn1p
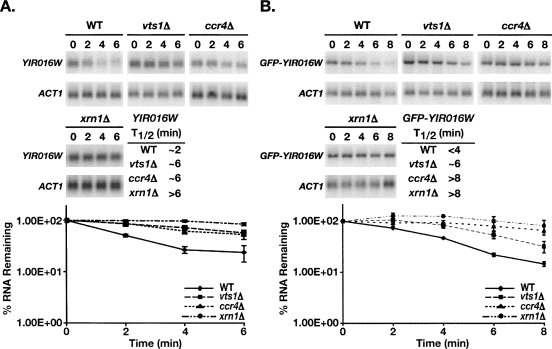
We have shown that Vts1p induces the degradation of artificial reporter transcripts via deadenylation followed by decapping and 5′-to-3′ decay. To validate our model we set out to determine if Vts1p uses this mechanism to degrade a bona fide endogenous mRNA target. We previously identified 79 potential Vts1 target transcripts through a pulldown/microarray approach and validated the interaction of Vts1p with a subset of these mRNAs by showing that we could detect these transcripts in Vts1p immunoprecipitates via Northern blot (Aviv et al. 2006). Of these validated transcripts a subset showed an increase in steady-state levels in vts1Δ cells compared to wild type, making them potential targets of Vts1p-medaited decay. YIR016W mRNA exhibited the greatest increase, and thus we examined the effect of Vts1p on YIR016W mRNA stability by measuring the half-life of YIR016W mRNA after treatment of cells with the transcriptional inhibitor thiolutin. Northern blotting revealed that the transcript had a half-life of ∼2 min in wild-type cells while in the vts1Δ strain the transcript was significantly stabilized with a half-life of ∼6 min (Fig. 6A). To rule out the possibility that thiolutin treatment was having unanticipated effects on YIR016W mRNA we also generated a construct in which GFP is fused to the YIR016W ORF under the control of the GAL1 promoter (GFP-YIR016W) and measured the stability of this reporter mRNA after inducing reporter gene transcription with galactose and then shutting off transcription with glucose. In these experiments GFP-YIR016W mRNA had a half-life of <4 min in wild-type cells, and its stability was increased in the vts1Δ strain with a half-life of ∼6 min (Fig. 6B). Taken together these data indicate that Vts1p is required for the rapid decay of YIR016W mRNA in vivo.
FIGURE 6.
Stability of YIR016W mRNA is regulated by Vts1p. (A) Transcription was inhibited by the addition of thiolutin at time zero. Endogenous YIR016W mRNA levels were measured in wild-type (WT), vts1Δ, ccr4Δ, and xrn1Δ cells by Northern blot. ACT1 mRNA serves as a loading control. The results of four independent experiments were quantitated and normalized using the levels of ACT1 mRNA and graphed with error bars representing standard deviation. The half-life (T1/2) of the mRNA in the different strains is as indicated. (B) Transcription of the GFP-YIR016W reporter was induced with galactose and then shut off with glucose. Reporter mRNA levels were measured in wild-type (WT), vts1Δ, ccr4Δ, and xrn1Δ cells by Northern blot. ACT1 mRNA serves as a loading control. The results of at least two independent experiments were quantitated and normalized using the levels of ACT1 mRNA and graphed with error bars representing standard deviation. The half-life (T1/2) of the mRNA in the different strains is as indicated.
Having confirmed that Vts1p is required to stimulate rapid degradation of YIR016W mRNA, we examined the mechanisms involved by assaying the stability of both endogenous YIR016W mRNA using thiolutin and GFP-YIR016W mRNA via transcriptional pulse-chase in ccr4Δ and xrn1Δ cells. We found that both endogenous YIR016W mRNA and GFP-YIR016W mRNA exhibited increased stability in these strains, indicating that Ccr4p and Xrn1p are required for rapid decay of these transcripts (Fig. 6A,B). Since transcripts must be decapped to be susceptible to 5′-to-3′ decay by Xrn1p (Hsu and Stevens 1993; Muhlrad et al. 1994, 1995; Caponigro and Parker 1996), we can infer that decapping is likely also required. Thus we have confirmed that the mechanism of Vts1p-mediated mRNA decay outlined using an artificial reporter transcript holds true for an in vivo mRNA target.
Vts1p localizes to P-bodies in xrn1Δ cells
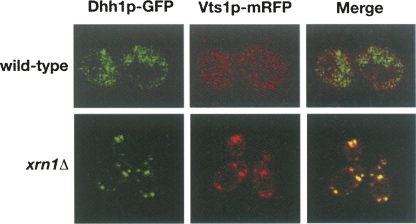
In yeast, mRNAs undergoing degradation can be localized along with the mRNA decay machinery, including the decapping enzymes and the 5′-to-3′ exonuclease Xrn1p, to distinct cytoplasmic foci known as processing or P-bodies (Sheth and Parker 2003). We have shown that deadenylation-dependent decapping and 5′-to-3′ decay are required for Vts1p-mediated mRNA degradation, suggesting that Vts1p may associate with these sites during transcript degradation. We constructed a monomeric red fluorescent protein (mRFP) C-terminal fusion of Vts1p and examined its localization in live cells. To visualize P-bodies we used a GFP C-terminal fusion of Dhh1p, a known P-body component (Sheth and Parker 2003). In wild-type cells Vts1p-mRFP does not colocalize with Dhh1p (Fig. 7). Given the apparent rapid rate of Vts1p-mediated decay, we theorized that the association of Vts1p with P-bodies may be too transient to observe in wild-type cells under standard conditions. We therefore examined the localization of Vts1p-mRFP in an xrn1Δ strain that is blocked for 5′-to-3′ decay and therefore retains degrading transcripts in P-bodies (Sheth and Parker 2003). In xrn1Δ cells, Vts1p-mRFP was concentrated in foci that colocalize extensively with Dhh1p-GFP (Fig. 7). These results indicate that Vts1p-mRFP localizes to P-bodies in an xrn1Δ strain and suggest that Vts1p-mediated mRNA degradation may take place in P-bodies.
FIGURE 7.
Vts1p localizes to P-bodies in xrn1Δ cells. Dhh1p-GFP (left panel) and Vts1p-mRFP (middle panel) were coexpressed in wild-type or xrn1Δ cells and visualized by confocal microscopy. The merged images are shown in the right panel.
DISCUSSION
Here we detail the molecular mechanism of Vts1p-mediated mRNA decay using both an in vivo reporter mRNA and a bona fide target of Vts1p. Our data are consistent with a model whereby Vts1p induces their degradation via Ccr4p-mediated deadenylation. Vts1p coimmunoprecipitates with Pop2p in an RNA-independent manner, suggesting that Vts1p interacts with the Ccr4p-Pop2p-Not deadenylase complex. Whether Vts1p interacts directly with one or more components of the deadenylase or indirectly through another protein remains to be determined. While our data do not rule out other possibilities, they are consistent with a model whereby Vts1p bound to an mRNA facilitates deadenylase recruitment to that transcript, which in turn triggers mRNA destabilization.
Deadenylation-dependent decapping followed by 5′-to-3′ decay by the exonuclease Xrn1p has been identified as the major mRNA decay pathway in yeast, and our work demonstrates that Vts1p functions within this pathway. However, microarray experiments suggest that <20% of yeast mRNAs are significantly up-regulated in dcp1Δ or xrn1Δ strains, suggesting other mechanisms play significant roles (He et al. 2003). Clearly additional work is required to determine the extent to which different mechanisms contribute to global regulation of mRNA degradation.
Smg family members employ a conserved mechanism to induce transcript degradation
Previous work suggests that Drosophila Smg employs a similar mechanism, involving deadenylation mediated by the recruitment of the Ccr4-Pop2-Not complex, to initiate the degradation of target mRNAs (Semotok et al. 2005). While Smg initiates transcript decay through a similar mechanism to that of Vts1p, the precise mechanism that underlies decay after deadenylation is not known. Nonetheless, the mechanistic similarities raise the strong possibility that regions conserved between Vts1p and Smg are responsible for deadenylase binding. Both Vts1p and Smg carry an as yet uncharacterized region designated Smg similarity region 1 as well as a SAM domain, either of which could be responsible for deadenylase binding. As these regions are also present in Smg homologs in other species, this would support a model whereby destabilization of mRNAs by Ccr4-Pop2-Not recruitment is evolutionarily conserved among the Smg family. However, human Smg 1 represses the translation of a cotransfected artificial reporter gene without inducing transcript degradation (Baez and Boccaccio 2005). Either this protein has lost the ability to induce transcript degradation or the cell type used in these experiments lacks a factor required to destabilize mRNAs.
Several other RNA binding proteins have been suggested to induce transcript decay via recruitment of the Ccr4-Pop2-Not deadenylase complex. These include Puf family members, AU-rich element RNA binding proteins TTP and BRF-1, the cold-shock domain containing protein UNR, and the Drosophila proteins Nanos and Bicaudal C (Chang et al. 2004; Lykke-Andersen and Wagner 2005; Goldstrohm et al. 2006, 2007; Chicoine et al. 2007; Hook et al. 2007; Kadyrova et al. 2007). Thus, deadenylase recruitment may be a common mechanism employed to regulate the stability of specific mRNAs.
The role of foci in the function of Smg family members
Vts1p does not localize to P-bodies in wild-type cells but can be found in these structures in an xrn1Δ strain. While these data could suggest that Vts1p-mediated mRNA degradation takes place in P-bodies, it is also possible that P-bodies play no role in Vts1p function and more work will be required to address this issue. Nonetheless, the localization of Vts1p to cytoplasmic foci is not a unique phenomenon among the Smg protein family. The intracellular distribution of Drosophila Smg is highly punctate with foci of varying size that show a modest colocalization with P-bodies (Zaessinger et al. 2006). Neither the composition of these Smg foci nor their functional significance is currently known. When expressed in fibroblasts, human Smg 1 forms cytoplasmic foci that lack GW182, a P-body marker, but contain several proteins that are found in stress granules (Baez and Boccaccio 2005). Stress granules are induced by environmental stress and contain nontranslating mRNAs, a subset of translation initiation factors, 40S ribosomal subunits, and a number of RNA binding proteins (Anderson and Kedersha 2006). While P-bodies and stress granules are distinct structures, they share various components and under some circumstances physically interact with one another. Whether the human Smg foci represent a novel function for this protein or are in some way related to the function of the yeast and Drosophila family members awaits further investigation.
MATERIALS AND METHODS
Yeast strains and media
Yeast strains used in this study were derivatives of the wild-type BY4741 (Brachmann et al. 1998). All deletion strains are as described by Winzeler et al. (1999), with the exception of ccr4Δ strain, which is described by Woolstencroft et al. (2006). The TetO7-DCP2 strain is described by Mnaimneh et al. (2004). The strains were transformed by standard techniques, and plasmids were maintained by growth in selective media.
mRNA analysis
Transcriptional pulse-chase experiments employed GFP-SRE+ and GFP-SRE− reporters described by Aviv et al. (2003). Both reporters express the GFP open reading frame in p413GAL1 with ∼160 nt of the CYC1 3′UTR containing the 3xSRE+ or 3xSRE− sequence, which is described by Smibert et al. (1996). For these experiments cells were grown at 30°C to mid-log phase in selective medium containing 2% raffinose and cooled to 20°C, with the exception of Figure 6, in which transcriptional shut-off was performed at 30°C. An accurate measure of reporter mRNA stability at 30°C was not possible due to the short half-life of the GFP-SRE+ reporter in wild-type cells. Cooling to 20°C slowed the decay of GFP-SRE+ mRNA to allow for measurement of reporter mRNA half-lives. GFP reporter transcription was initiated by the addition of galactose to a final concentration of 2%. After 16 min, transcription was repressed by the addition of glucose to a final concentration of 4%. Total RNA was isolated by glass bead lysis in LET buffer (100 mM LiCl, 20 mM EDTA, 25 mM Tris-HCl at pH 8.0) and LET-equilibrated phenol at the indicated time points and analyzed by Northern blot. SCR1 or ACT1 RNA was used for normalization of reporter transcript levels as indicated. Where indicated, thiolutin (Carbosynth Limited) was added to cultures to a final concentration of 20 μg/mL at time 0 of the time course. RNase H cleavage assays were preformed as described by Decker and Parker (1993), using an oligonucleotide that hybridized ∼330 nt upstream of the GFP reporter poly(A) site. To control for differences in RNA concentration in these experiments an amount of each RNA sample was analyzed via a standard Northern blot and probed for SCR1 RNA. Northern blots were exposed to PhosphorImager screens and analyzed with ImageQuant Software with the exception of TetO7-DCP2 in Figure 5, which was exposed to X-ray film. The GFP-YIR016W reporter was under the control of the GAL1 promoter and expressed a GFP N-terminal fusion to the YIR016W ORF plus 597 nt of YIR016W downstream sequence.
Immunoprecipitations
The plasmid expressing Vts1p-Flag was generated in pRS316 with 574 bases of genomic sequence upstream and 396 nt downstream of the VTS1 ORF. Vts1p-Flag also harbors a VSV epitope that was used for protein detection by Western blot. The Pop2p-HA plasmid was created in pRS313 with a C-terminal 3X HA tag and 549 nt of genomic sequence upstream and 530 nt downstream of the POP2 ORF. Cells were harvested and lysed in KHT buffer (150 mM KCl, 30 mM HEPES at pH 7.4, 0.1% Tween) by glass bead lysis and clarified at 15,000 rpm for 10 min. Anti-Flag M2 affinity gel (Sigma) was added to the supernatant and bound for 3 h at 4°C. Beads were washed four times at 4°C with KHT buffer and then twice in 100 mM KCl, 100 mM HEPES (pH 7.4) at room temperature for 10 min. Vts1p-VSV-Flag and associated proteins were eluted with Flag-peptide (Sigma) for 10 min at room temperature. Where indicated 0.35 μg/μL RNase A (Fermentas) was added during 3 h incubation at 4°C. VSV antibodies (Bethyl Laboratories) and HA antibodies (Abcam) were used to detect VSV-tagged and HA-tagged proteins, respectively.
Confocal microscopy
The Vts1p-mRFP plasmid was generated by C-terminally tagging Vts1p with monomeric RFP in pRS316 with 574 nt of genomic sequence upstream and 396 nt downstream of the VTS1 ORF. The Dhh1p-GFP plasmid was generated by C-terminally tagging Dhh1p with GFP in pRS315 with 517 nt of genomic sequence upstream and 430 nt downstream of the DHH1 ORF. Cells expressing fluorescent proteins were grown to mid-log phase in selective media and visualized with a Zeiss LSM 510 confocal microscope using the 100× objective. Images shown are of a single focal plane and were generated with Zeiss LSM browser software.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Heli Vari for her excellent technical expertise and Tim Hughes, Dan Durocher, and Mike Tyers for reagents. L.M.R. is supported by an Ontario Graduate Scholarship and M.A.B is supported by a National Sciences and Engineering Research Council Scholarship. This work was supported by an operating grant from the National Cancer Institute of Canada with funds from the Terry Fox Run.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.955508.
REFERENCES
- Anderson P., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.S., Parker R.P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv T., Lin Z., Lau S., Rendl L.M., Sicheri F., Smibert C.A. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat. Struct. Biol. 2003;10:614–621. doi: 10.1038/nsb956. [DOI] [PubMed] [Google Scholar]
- Aviv T., Lin Z., Ben-Ari G., Smibert C.A., Sicheri F. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat. Struct. Mol. Biol. 2006;13:168–176. doi: 10.1038/nsmb1053. [DOI] [PubMed] [Google Scholar]
- Baez M.V., Boccaccio G.L. Mammalian Smaug is a translational repressor that forms cytoplasmic foci similar to stress granules. J. Biol. Chem. 2005;280:43131–43140. doi: 10.1074/jbc.M508374200. [DOI] [PubMed] [Google Scholar]
- Bonnerot C., Boeck R., Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E., Rigaut G., Shevchenko A., Wilm M., Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Caponigro G., Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae . Microbiol. Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Yamashita A., Chen C.Y., Yamashita Y., Zhu W., Durdan S., Kahvejian A., Sonenberg N., Shyu A.B. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes & Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Chiang Y.C., Denis C.L. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicoine J., Benoit P., Gamberi C., Paliouras M., Simonelig M., Lasko P. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev. Cell. 2007;13:691–704. doi: 10.1016/j.devcel.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Crucs S., Chatterjee S., Gavis E.R. Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol. Cell. 2000;5:457–467. doi: 10.1016/s1097-2765(00)80440-2. [DOI] [PubMed] [Google Scholar]
- Dahanukar A., Walker J.A., Wharton R.P. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila . Mol. Cell. 1999;4:209–218. doi: 10.1016/s1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- Decker C.J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A.C., Seay D.J., Hook B.A., Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- Green J.B., Gardner C.D., Wharton R.P., Aggarwal A.K. RNA recognition via the SAM domain of Smaug. Mol. Cell. 2003;11:1537–1548. doi: 10.1016/s1097-2765(03)00178-3. [DOI] [PubMed] [Google Scholar]
- Hatfield L., Beelman C.A., Stevens A., Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae . Mol. Cell. Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Li X., Spatrick P., Casillo R., Dong S., Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- Hook B.A., Goldstrohm A.C., Seay D.J., Wickens M. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 2007;282:15430–15438. doi: 10.1074/jbc.M611253200. [DOI] [PubMed] [Google Scholar]
- Hsu C.L., Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova L.Y., Habara Y., Lee T.H., Wharton R.P. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes & Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S., Davierwala A.P., Haynes J., Moffat J., Peng W.T., Zhang W., Yang X., Pootoolal J., Chua G., Lopez A., et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C.J., Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes & Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C.J., Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.R., Leidal A.M., Smibert C.A. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004;23:150–159. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Semotok J.L., Cooperstock R.L., Pinder B.D., Vari H.K., Lipshitz H.D., Smibert C.A. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert C.A., Wilson J.E., Kerr K., Macdonald P.M. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes & Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- Smibert C.A., Lie Y.S., Shillinglaw W., Henzel W.J., Macdonald P.M. Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. RNA. 1999;5:1535–1547. doi: 10.1017/s1355838299991392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M., Carr-Schmid A., Schwartz D.C., Kiledjian M., Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., He W., Mayes A.E., Lennertz P., Beggs J.D., Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tucker M., Staples R.R., Valencia-Sanchez M.A., Muhlrad D., Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae . EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Woolstencroft R.N., Beilharz T.H., Cook M.A., Preiss T., Durocher D., Tyers M. Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. J. Cell Sci. 2006;119:5178–5192. doi: 10.1242/jcs.03221. [DOI] [PubMed] [Google Scholar]
- Zaessinger S., Busseau I., Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- Zhang S., Williams C.J., Wormington M., Stevens A., Peltz S.W. Monitoring mRNA decapping activity. Methods: A Companion to Methods in Enzymology. 1999;17:46–51. doi: 10.1006/meth.1998.0706. [DOI] [PubMed] [Google Scholar]