Abstract
Noncoding RNAs constitute a huge repertoire of gene regulatory molecules. Our previous, fine-resolution characterization of a mouse meiotic recombination hotspot from chromosome 8 resulted in identification of 2.4-kb unspliced and polyadenylated noncoding mrhl RNA. The gene is expressed in multiple tissues and is also present in rat but absent in humans. Here we report that the mrhl RNA gets processed to a small 80-nucleotide (nt) RNA species and is mediated by the Drosha complex. We also observe that the 80-nt Drosha product could be processed further to a 22-nt small RNA by Dicer in an in vitro reaction. However, this 22-nt product was not detected in vivo. The 80-nt as well as the 2.4-kb full-length RNA are nuclear-localized, showing distinct punctate nuclear signal. The colocalization of the noncoding RNA with Drosha and Nucleolin suggests the nucleolus as the site of processing of the 2.4-kb primary transcript. Additional foci of the processed 80-nt RNA were also observed outside the nucleolus, suggesting its role in some specific chromatin domain(s). Thus, this study reports a novel noncoding mrhl RNA that is processed and restricted within the cell nucleus.
Keywords: noncoding RNA, Drosha, mouse
INTRODUCTION
The analysis of the draft sequence of the human genome has revealed that coding sequences account for <2% of its total transcriptome (The ENCODE Project Consortium 2007). Several noncoding RNAs (ncRNAs) have been identified whose functions are yet to be determined. By definition, “noncoding RNA” refers to the RNA that is transcribed but not translated to protein (Mattick and Makunin 2005). The complexity of the eukaryotic genome and the presence of a huge repertoire of different classes of RNA indirectly show that these noncoding transcripts are, indeed, functional molecules that are involved in cellular regulation at various levels (Eddy 2001; Huttenhofer et al. 2005). There are a variety of RNA molecules in the cell that display a remarkable range of functions far beyond those already known for messenger, ribosomal, and transfer RNA (Szymanski et al. 2003). Recent evidence is giving newer insights into the complexity and varied functions of the noncoding RNA in cellular physiology.
The eukaryotic genome encodes two distinct categories of other ncRNAs, referred to as “small ncRNAs” and “long mRNA-like ncRNAs” (Kim 2005). Much of the recent attention has been focused on small RNAs that are 22–33 nucleotides (nt) in length, which include miRNAs, siRNAs, piRNAs, and Rasi RNAs. The central theme of these small RNAs is their functional role in gene silencing by interaction with chromatin or by base-pairing with complementary mRNA or DNA (Fire et al. 1998; Mattick and Makunin 2005; Girard et al. 2006). The miRNA, which has been a widely studied small RNA class, is involved in silencing of many messenger RNAs, regulating gene function at the level of post-transcription (Lee et al. 2002; Ambros et al. 2003a; Kim 2005).
The long ncRNAs, many of which are transcribed by RNA polymerase II and spliced and polyadenylated, like Xist, Tsix, Air, H19, and roX, are implicated in a variety of regulatory processes, such as imprinting (Constancia et al. 2000), X-chromosome inactivation (Clemson et al. 1996; Gilbert et al. 2000), DNA methylation, transcription, RNA interference, chromatin-structure dynamics, and antisense regulation (Bernstein and Allis 2005; Prasanth and Spector 2007). In addition, ncRNAs such as MALAT-1, BC-1, and BC-200 serve as prognostic markers for cancer, while the prion-associated RNAs (SCA-8, etc.) are implicated in a number of neurological disorders (Costa 2005). Apart from this, there are RNAs such as IPW, involved in Prader-Willi syndrome (Wevrick et al. 1994), and LIT-1, which is implicated in Beckwith–Wiedemann syndrome (BWS) (Niemitz et al. 2004). A recent addition to this group of noncoding RNAs is the natural antisense transcript (NAT), which act in cis and regulate sense/antisense transcripts (Lapidot and Pilpel 2006). They are generally coexpressed with their targets and involved in competitive transcriptional interference. Although the mechanism of many regulatory processes mediated by RNAs of different lengths still remains a mystery, it is certainly becoming clear that RNA is an important cellular regulatory molecule.
We have recently identified a noncoding RNA that was encoded in a mouse genomic locus possessing recombination hotspot activity (Nishant et al. 2004). This 2.4-kb transcript was shown to be an unspliced polyadenylated transcript expressed in multiple mouse tissues. The transcript does not possess a significant open reading frame (ORF) but has a considerable propensity to form a stable secondary structure. We subsequently started to probe the function of this noncoding RNA. We demonstrate here that this noncoding RNA is processed by Drosha to an ∼80-nt intermediate RNA that is nuclear-restricted. However, the 22-nt mature miRNA is not detected in vivo, although the 80-nt intermediate RNA can serve as a substrate for Dicer in vitro. This noncoding RNA gene is present in the mouse and rat genomes but is absent in human and also shows tissue-specific expression in mouse. The structural properties of the RNA, ability to get processed by RNase III-type enzymes, and the unique nuclear localization pattern suggest a potential regulatory role within the nucleus.
RESULTS
Characterization of mrhl RNA
An earlier study from our laboratory involved a detailed characterization of a mouse meiotic recombination hotspot locus mapping to mouse chromosome 8 (Nishant et al. 2004). Transcriptional analysis of this region as probed by a Northern analysis for the total RNA from testis and liver revealed a positive signal to one of the contigs at the 2.4-kb region. The in silico analysis of this transcript did not reveal any open reading frame that was >100 base pairs (bp). The largest ORF also did not seem to have any Kozak sequences at its immediate 5′ proximal region. Thus it was concluded that this RNA could be a noncoding transcript (Fig. 1A). The RNA was then subjected to secondary-structure prediction analysis by the Mfold program and found to have considerable propensity to form stable secondary structure at 37°C at physiological ionic strength. Expression of this RNA was also studied by Northern analysis on multiple tissues, which showed that the mrhl RNA is expressed only in testis, liver, kidney, and spleen and is absent in brain, lungs, heart, and skeletal muscles (Nishant et al. 2004). When we looked at the conservation of the RNA across many different species by performing BLAST against the different genome databases, we could find significant homology of mrhl only in the rat genome. In rat, the gene is located on Chromosome 19, and the gene shares ∼80% homology with the mouse gene, and most interestingly was absent in the human genome (Fig. 1B). We have also carried out an extensive bioinformatics search to identify the homologs of mrhl RNA in other species by looking at the syntenic region across different organisms. In mouse, the mrhl RNA is adjacent to the phosphorylase kinase β (phkb) gene on chromosome 8. We analyzed the up-to-100-kb region flanking the phkb gene in various species using low-stringency BLAST and also by using BLAST two-sequence options. The extensive search also did not identify any significant homologs in any other species except in the case of the rat genome.
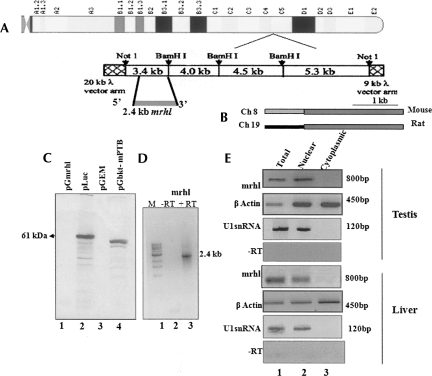
FIGURE 1.
Noncoding RNA encoded on mouse Chromosome 8 is nuclear-restricted. (A) Chromosomal position of 17.2-kb mouse meiotic hotspot locus from mouse Chromosome 8 and the contig map (Nishant et al. 2004). (B) Schematic map of homology between the mouse and the rat mrhl gene. (C, lane 1) In vitro transcription-coupled translation assay of pGmrhl (2.4-kb RNA gene cloned into pGEM3Z vector downstream from T7 promoter); (lane 2) positive control luciferase encoding 61-kDa protein; (lane 3) empty vector negative control; (lane 4) pGbktT7-mPTB encoding 58-kDa mouse poly-pyrimidine tract-binding protein. (D) RT-PCR of RNA isolated from the in vitro transcription-coupled translation of pGmrhl in the presence and absence of RT. (M) The 1-kb DNA ladder. (E) RT-PCR of the mrhl RNA from testis and liver total, nuclear, and cytoplasmic RNA. β-Actin was used as the positive control. U1snRNA was also used as a control to assess the purity of fractions. A no-RT reaction was used as a negative control to eliminate amplification from DNA.
In order to ensure that the RNA does not code for any protein, we carried out an in vitro coupled transcription and translation assay. The 2.4-kb noncoding RNA gene that was cloned under a T7 promoter was used for the reaction in the presence of 35S methionine using the rabbit reticulolysate system. As shown in Figure 1C, there was no protein product detected in the lane of pGmrhl, whereas the control plasmids that harbor luciferase and the mouse poly-pyrimidine tract-binding protein gave respective protein products. The negative control that contained only empty vector also did not give any protein product. In order to verify that the gene is, indeed, transcribed full length in the in vitro transcription reaction, we also did an RT-PCR amplification of the mrhl transcript by isolating RNA from the same reaction. Figure 1D shows the PCR amplification of the gene product, which shows that the 2.4-kb RNA gene is, indeed, transcribed but not translated in the in vitro coupled transcription and translation experiment. A no-RT reaction did not give any amplification, eliminating the possibility of the plasmid getting amplified in the reaction.
The 2.4-kb mrhl RNA is nuclear-localized
We next investigated the subcellular localization pattern of the mrhl RNA. The testis and liver tissues from the mouse were fractionated to the nucleus and cytoplasm by differential centrifugation, and the RNA was isolated. RT-PCR was done subsequently using the gene-specific primers for the noncoding RNA. Figure 1E shows the result, where we see the amplification of mrhl RNA in total RNA as well as in the nuclear fraction but not in the cytoplasmic fraction. The β-actin mRNA was amplified in all the fractions as probed by single exonic primers. The U1 snRNA was used to assess the purity of the nuclear and cytoplasmic fractions. It was amplified only in the nuclear fraction but not in the cytoplasm. A no-RT control was also performed in order to eliminate any false amplification that might arise due to DNA contamination. Thus the mrhl RNA appears to be nuclear-restricted and does not get transported into the cytoplasm. The nuclear localization of mrhl RNA was not surprising, as many noncoding RNAs have been shown earlier to exhibit definite nuclear-localization patterns mediating potent regulatory events such as regulation of coding genes (CTN) (Prasanth et al. 2005), imprinting (H19, Air) (Sleutels et al. 2002; Gabory et al. 2006), and dosage compensation (Xist, roX) (Gilbert et al. 2000; Smith et al. 2000; Reik and Walter 2001).
The 2.4-kb mrhl RNA is processed further to an intermediate RNA species
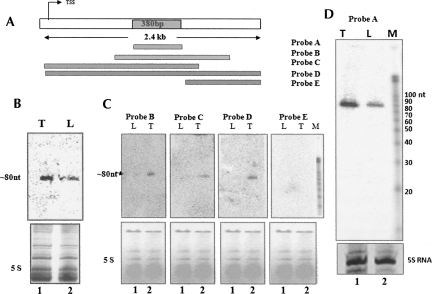
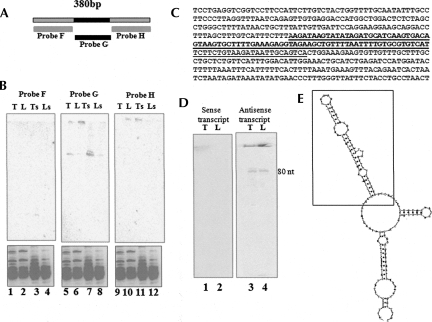
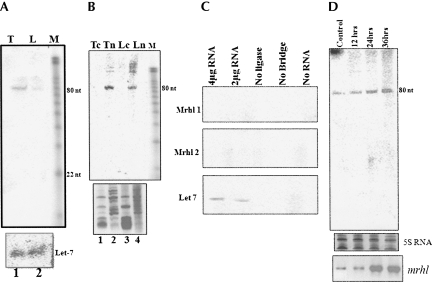
The 2.4-kb mrhl RNA showed an extensive secondary structure comprising of a stem–loop and hairpin loops (Nishant et al. 2004). It is now fairly established that many of the microRNAs are transcribed as a primary transcript by polymerase II. This primary transcript possesses stem–loop structure that will be recognized and processed by the nuclear RNase III enzyme Drosha in the nucleus and subsequently to a 22-nt miRNA by Dicer in the cytoplasm (Lee et al. 2002, 2003; Gregory et al. 2004). Hence, we wanted to examine whether the 2.4-kb noncoding RNA also gets processed further. For this purpose, total RNA was separated in a 15% polyacrylamide urea gel, and a Northern blot was carried out on both testis and liver RNA. The blot was hybridized using a body-labeled probe against the purified 2.4-kb PCR product. Interestingly, we observed a signal from both the testis as well as the liver RNA at the ∼80-nt position (Fig. 2B). In order to narrow down the region that actually generates the 80-nt product, we amplified different regions of the noncoding RNA gene, and Northern analysis was carried out using each of them as a probe against total RNA extracted from liver and testis. Figure 2A shows the different primer positions used in generating the amplicons of different regions of the transcript. Figure 2C shows the autoradiogram of such a Northern analysis. The blots showing the hybridization signal are derived from the amplicons that shared the 380-bp common region that has been highlighted in Figure 2A. Hybridization with probe E failed to show any signal, as it does not overlap with the small region identified and represented at the far end of the gene. The signal that arises due to the presence of the 380-bp common region was further confirmed by using that region (Fig. 2D, probe A) as a probe. Thus, the region that gave signal was mapped to 380 bp from the original 2.4 kb. We further narrowed down the region corresponding to 80 nt by carrying out Northern blot analysis using amplicons of a 100-bp fragment each. The shorter regions from the 380-bp fragment used for the Northern are shown in Figure 3A and the sequences in Figure 3C. The specific region of 96 bp was identified as the one giving rise to the signal in the Northern analysis (Fig. 3B). However, one cannot rule out the possibility of few molecules of the 80-nt processed RNA originating from other regions of the primary transcript that fall below the level of detection by hybridization. Additional Northern hybridization experiments were carried out using the identified small region by using a radiolabeled RNA probe transcribed either in the sense or antisense directions. This would ensure that the signal in the Northern was truly arising from the same strand as the primary transcript. Figure 3D clearly shows that the hybridization occurs with the antisense transcript as a probe but not with the sense probe, confirming that in vivo the 80-nt RNA is of the same strand and is the processed product of the primary transcript.
FIGURE 2.
mrhl RNA is processed to ∼80-nt intermediate RNA. (A) Scheme of different regions of mrhl used for probe generation and its position. The highlighted region represents the sequence shared between probes B, C, and D giving a positive signal. (B) Small RNA Northern blot of total RNA separated in 15% polyacrylamide-urea gel probed with 2.4-kb full-length gene (probe D) by Klenow labeling using random primers on (lane 1) mouse testis and (lane 2) liver total RNA. (Bottom panel) Ethidium bromide-stained pattern for loading control. (C) Northern blot on (lane 1) liver and (lane 2) testis RNA separated on 15% polyacrylamide-urea gel using each of the probes B, C, D, and E. (Bottom panel) The ethidium bromide-stained pattern for the loading control. (D) Small RNA Northern blot using probe A spanning the 380-bp region that corresponds to the hybridization signal. (Lane 1) Testis RNA; (lane 2) liver RNA. (Bottom panel) The ethidium bromide-stained pattern. (M) The RNA ladder (Ambion).
FIGURE 3.
Characterization of region giving rise to ∼80-nt signal. (A) Probes used for identification of ∼80-nt region within the narrowed down 380-bp region (Fig. 2A). (B) Small RNA Northern blot of total RNA from (lanes 1,5,9) testis, (lanes 2,6,10) liver, and small RNAs enriched (Ambion miRNA isolation kit) from (lanes 3,7,11) testis and (lanes 4,8,12) liver using the probes F, G, and H. (Bottom panel) Ethidium bromide-stained pattern. (C) Sequence of the 380-bp region that was narrowed down from the 2.4-kb region is shown, and highlighted is the 90-nt region that gave the signal in Northern blots. (D) Northern blot on mouse testis and liver RNA by using (lane 1,2) sense probe and (lane 3,4) antisense probe derived from T7 and SP6 promoters, respectively. (E) Secondary structure of the 380-nt region showing stem–loop structures predicted by Mfold that maps to the region corresponding to hybridization signal. The inset is the ∼80-nt region that possess the stem–loop structure.
Drosha mediates processing of 2.4-kb mrhl RNA
The microRNAs that are 22-nt RNA molecules regulating mRNA expression post-transcriptionally are derived from a pre-miRNA that is ∼60–80 nt in length by the action of Dicer (Lee et al. 2002; Kim 2005). The pre-miRNA itself is a product of cleavage of a larger transcript by the enzyme Drosha (Cai et al. 2004). The basic requirements in a particular RNA species to be processed by Drosha have been identified. The RNAs to be processed by Drosha often fold back to give a stem–loop structure with a large stem, an internal bulge, and a large loop region. Apart from this, Drosha also requires flanking nonstructured RNA for its cleavage (Han et al. 2006). Interestingly, the sequence of 2.4 kb when folded using an Mfold program also generated a stem–loop structure with a large stem, an internal bulge, and a loop region along with flanking unstructured RNA (Fig. 3E). This region of the sequence also corresponded to the ∼80-nt fragment that we detected in vivo as shown in Figure 3B, suggesting that the 2.4-kb transcript may serve as a substrate for the Drosha machinery. We subsequently examined this possibility, whether the 2.4-kb transcript can generate the 80-nt fragment, in an in vitro assay using a cell-free system. For this purpose, we performed the in vitro processing assay according to the method described for miRNA processing by Han et al. (2004). The gene cloned between the T7 and SP6 promoters in pGmrhl was transcribed in both sense and antisense orientations and was incubated with whole cell extract from testis. The processed RNA was separated on a 15% polyacrylamide gel containing 7 M urea. The results presented in Figure 4A show an ∼80-nt processed RNA band that was seen only from the sense transcript but not from the antisense transcript.
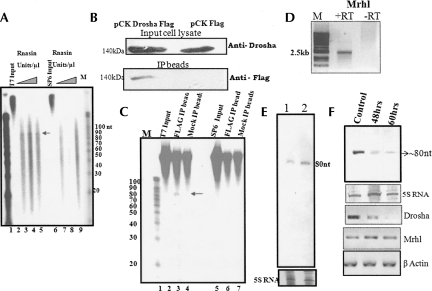
FIGURE 4.
Drosha-mediated processing of the mrhl RNA. RNA processing assay of the 2.4-kb transcript derived from T7 (sense) and SP6 (antisense) promoters using total testicular cell lysate. (A) The primary 2.4-kb transcript obtained from (lane 2) T7 or (lane 6) SP6 polymerase-mediated transcription. A processing assay was carried out with (lanes 3,7) 2 units, (lanes 4,8) 3 units, and (lanes 5,9) 4 units of RNasin/μL. The processed ∼80-nt RNA was seen only in lanes 3, 4, and 5 containing the sense transcripts. (M) The molecular size marker. (B) Flag IP was performed from the HEK 293T cells transfected with the pCK-Drosha–Flag construct. Mock transfection contains the empty vector pCK Flag without the insert. (Upper panel) The Western blot of the input cell lysates using Drosha antibody; (bottom panel) Western blot of Flag IP beads using Flag antibody. Flag Drosha fusion protein at 140 kDa is seen only in the IP beads from pCK Drosha Flag-transfected cell lysate but not from pCK Flag-transfected cell lysate. (C) Processing assay using the Flag immunoprecipitated beads containing Drosha complex. (Lanes 2,5) The input T7 and SP6 polymerase-derived RNA from the 2.4-kb primary transcript. (Mock IP, lanes 4,7) The processing reaction carried out using mock-transfected beads. (Lanes 3,6) The reaction carried out with Flag IP beads. The products were separated on 15% urea PAGE and autoradiographed. (M) The 10-nt RNA ladder. (D) RT-PCR of mrhl RNA from the mouse Gc1-Spg cell line in the presence and absence of RT. (E) Small RNA Northern blot on (lane 1) mouse Gc1-spg cell line and (lane 2) RNA isolated from 10-d-old mouse testis probed with probe G (Fig. 3A). (Bottom panel) The ethidium bromide-stained pattern. (F) Small RNA Northern blot on Gc1 Spg total RNA probed for 80-nt RNA after Drosha down-regulation. The cells were harvested 48 h and 60 h post-transfection with a combination of two Drosha siRNAs (QIAGEN) and compared with lipofectamine-transfected control. (Bottom panel, 5S RNA) The ethidium bromide-stained gel pattern that was used as the loading control. (Bottom three panels) The RT-PCR analysis of Drosha, mrhl, and β-actin using specific primers.
After establishing that the 2.4-kb transcript can be processed to generate an ∼80-nt fragment in a cell-free extract, we wanted to examine whether this can be demonstrated with Drosha (RNasen in mammals). For this purpose, we carried out an in vitro Drosha assay in which the plasmid pCK harboring Drosha Flag was transfected into HEK 293T and the Drosha flag complex was pulled down using anti-Flag agarose beads. It was essential that we used Drosha complexed with another RNA-binding protein, DGCR8, since the primary miRNA cleavage is brought about by a Drosha–DGCR8 complex also known as the “microprocessor complex.” The beads containing the complex were directly used for the Drosha assay using in vitro transcribed sense or antisense mrhl 2.4-kb RNA as the substrate. A Western blot analysis using anti-Drosha antibody confirmed the presence of Drosha in the input cell lysates of the Flag Drosha transfected cell line (Fig. 4B). The mock transfected cell line also showed the presence of endogenous Drosha. Western blot analysis on the immunoprecipitated beads using anti-Flag antibody detected the Drosha Flag fusion protein at 140 kDa, only in the immuno-pull-down of the pCK Drosha Flag transfected cell line but not in the mock pCK Flag transfected cell line. Figure 4C shows the autoradiogram of the processing assay, and it can be seen that only the sense transcript is cleaved by the Drosha complex to generate the ∼80-nt fragment but not the antisense transcript. As a control, we also used mock-transfected cells, which did not show any processed ∼80-nt RNA product. In order to unequivocally demonstrate that the Drosha machinery is involved in the processing of the 2.4-kb mrhl RNA primary transcript in vivo, we used the siRNA approach to down-regulate Drosha and look for the ∼80-nt processed intermediate. This experiment was carried out on a Gc1-Spg cell line of mouse spermatogonial cells. Figure 4D shows the RT-PCR analysis of the 2.4-kb RNA from the Gc1-Spg cell line in the presence and absence of RT, where the signal was seen only in the RT lane, demonstrating that this cell line does express the 2.4-kb noncoding RNA. A Northern blot of RNA from the same cell line was also done in order to confirm the presence of the processed ∼80-nt intermediate RNA. As can be seen in Figure 4E, the 80-nt RNA is detected in mouse Gc1-Spg cells (Fig. 4E, lane 1). The ∼80-nt processed RNA intermediate was also observed in the testicular cells from 10-d-old mouse containing predominantly spermatogonial cells (Fig. 4E, lane 2). The results of the siRNA experiment using the Gc1-Spg cell line are shown in Figure 4F. The densitometric analysis showed that Drosha was down-regulated to almost 70% after 60 h post-transfection. At the same time, the ∼80-nt processed intermediate was also reduced to ∼66% that of the control. We have also found that under these conditions of Drosha silencing there is no change in the 2.4-kb transcript level. Thus, this experiment demonstrates Drosha's involvement in the processing of the 2.4-kb mrhl RNA to the ∼80-nt intermediate RNA.
The above experiments have shown that the 2.4-kb mrhl RNA is processed into an ∼80-nt intermediate by the Drosha enzyme machinery. As mentioned above, in the miRNA processing pathway the 60–80-nt intermediate RNA is further processed into small 20–22-nt miRNA species by the Dicer machinery. It is interesting to note that we did not observe any small 22-nt RNA being generated in the small RNA Northern blot described in previous experiments, indicating that in vivo, this RNA may not be processed toward miRNA species. As a positive control, we have probed the presence of Let-7 miRNA in the RNA samples, and the result shown in Figure 5A also confirms that there is no technical problem in detecting the mature microRNA in our Northern blot experiments. The same blot was stripped and reprobed using an mrhl probe, where again only the ∼80-nt intermediate RNA was detected but not the 22-nt mature miRNA. As mentioned above, the entire process of miRNA maturation happens partially in the nucleus, where Drosha is present, and the precursor to the mature miRNA takes place with the aid of cytoplasmic Dicer. We then examined the localization of the ∼80-nt RNA in subcellular fractions. We obtained the nuclear and the cytoplasmic fractions from testis and liver tissues as described above and performed a small RNA Northern analysis by the same probe that was used to detect the ∼80-nt RNA. Again we could detect the 80-nt RNA in the nuclear compartment but not in the cytoplasm (Fig. 5B). Some additional bands were also observed in the 150-nt size region in the nuclear fraction, which we presume to be the partially processed primary transcript in the nucleus. Thus the 80-nt intermediate RNA is also nuclear-restricted and not being transported to cytoplasm. In order to confirm that the 22 nt has not been generated in vivo, we resorted to the recently described sensitive technique of the splinted ligation method (Maroney et al. 2007). The results of this experiment described in Figure 5C indicate that the positive control Let 7 is detected by the splinted ligation method with 2 μg of total RNA, whereas the mrhl RNA was not detected even at 4 μg of RNA. We have used two bridge oligos spanning each of the two complementary strands of the ∼80-nt mrhl small RNA as we were not sure of the mature miRNA strand that could be generated in vivo. The negative control experiment that did not have ligase, RNA, or the bridge oligo did not give rise to any signal. Furthermore, we also overexpressed the 2.4-kb mrhl RNA in the Gc1-Spg cell line under the CMV promoter and tried to detect the 22-nt RNA, so that the primary transcript is available in abundance to generate a detectable quantity of the mature miRNA. We harvested the cells at 12 h, 24 h, and 36 h post-transfection with a pCDNA mrhl construct and carried out the Northern blot using an ∼80-nt region-specific probe (probe G). As seen from Figure 5D, although there is a marginal increase in the 80-nt precursor form by 24-h and 36-h post-transfection, we could not see the 22-nt mature RNA as monitored by Northern analysis, which also confirms our conclusion that the 22 nt may not be formed in the in vivo scenario. The expression pattern of the 2.4-kb mrhl primary transcript under the overexpressed conditions was also monitored by RT-PCR (Fig. 5D, bottom panel).
FIGURE 5.
Twenty-two-nucleotide mature microRNA is not generated in vivo. (A) Small RNA Northern blot using probe that corresponds to the 80-nt intermediate region. (Lane 1) Testis RNA; (lane 2) liver RNA. (M) The RNA ladder (Ambion). (Bottom panel) The positive control, where the blot was used for hybridizing to Let-7 miRNA. (B) Northern blot on the RNA isolated from nuclei and cytoplasm of (Tn, Tc) testis and (Ln, Lc) liver tissues with probe G showing signal only in the nuclear lanes but not in the cytoplasm. (Bottom panel) The ethidium bromide-stained gel pattern. (C) Splinted ligation experiment on total testis RNA for detection of microRNAs (Maroney et al. 2007). Let7, the positive control, is detected at 4 and 2 μg of RNA concentration, whereas the mrhl bridge oligos 1 and 2 representing each strand of the 80-nt intermediate RNA failed to give a positive signal. The negative controls with no ligase, or no bridge oligo, or no RNA did not give any signal. (D) Overexpression of the 2.4-kb mrhl RNA in the mouse Gc1 Spg cell line using the pCDNA mrhl construct. Small RNA Northern blot using the 80-nt region as probe on total RNA isolated from cells 12 h, 24 h, and 36 h post-transfection. (Bottom panel) The RT-PCR of 2.4-kb RNA overexpression at various time points. The ethidium bromide-stained gel showing 5S RNA represents the loading control.
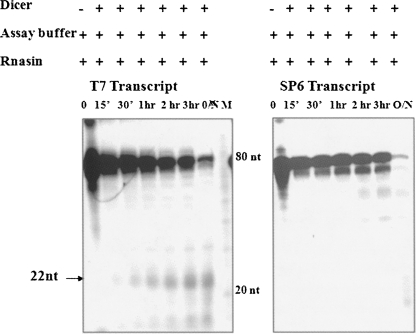
However, we were still curious to know whether this ∼80-nt RNA species can be cleaved in vitro by Dicer to generate the 22-nt miRNA. Toward this direction, we amplified the region that corresponds to the 80-nt region using T7 and SP6 adapter primers for generating sense and antisense transcripts, respectively. These in vitro transcribed RNAs were used as substrates for an in vitro Dicer assay using recombinant human Dicer. As can be seen from Figure 6, a 22-nt RNA fragment was generated from only the sense transcript but not from the antisense transcript. Thus, the ∼80-nt RNA that we have identified does have the propensity to act as a substrate for Dicer in an in vitro scenario.
FIGURE 6.
Processing of ∼80-nt RNA to 22-nt small RNA by Dicer in vitro. Dicer assay for in vitro transcribed 80-nt RNA from T7 (sense) and SP6 (antisense) promoters using recombinant human Dicer. The reaction was carried out for different time periods as indicated. The sequence corresponding to the 80-nt processed RNA was amplified using T7 and SP6 adapter primers, and the RNA was in vitro transcribed using T7 (sense) or SP6 (antisense) polymerase and used as substrates for Dicer. (M) The 10-nt RNA ladder.
The mrhl RNA localizes to specific nuclear compartment
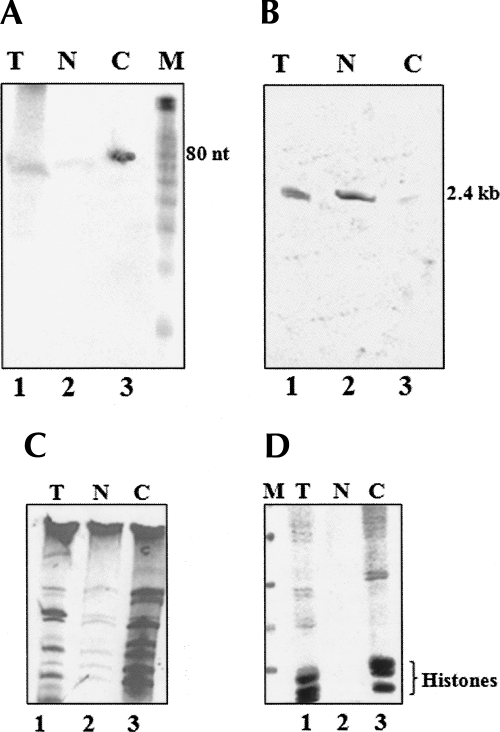
Since the experiments described above showed that both the 2.4-kb primary transcript and the processed ∼80-nt fragment were nuclear-restricted, we wanted to examine their intranuclear localization pattern. Initially, we fractionated nuclear extracts into nucleoplasmic and chromatin fractions, and the isolated RNAs were separated on a 15% and 5% polyacrylamide containing 7 M urea and hybridized with probe G (Fig. 3A), which encompassed the ∼80-nt region with a flanking nucleotide stretch that will hybridize both to the ∼80-nt as well as the 2.4-kb primary transcript. As can be seen in Figure 7, A and B, the 2.4-kb primary transcript was enriched in the nucleoplasmic fraction, while the ∼80-nt fragment was enriched in the chromatin fraction. The integrity of the RNA was also checked in an ethidium bromide-stained gel as shown in Figure 7C. We could authenticate the subnuclear fractions by analyzing their protein composition on a 10% SDS-PAGE. The protein pattern showed in Figure 7D clearly reveals the presence of histones only in the chromatin fraction and absent in nucleoplasm.
FIGURE 7.
Distribution of precursor and the processed RNA between nucleoplasm and chromatin. Testicular nuclei were processed to isolate the nucleoplasmic and chromatin fractions, and the total RNA was hybridized with probes against the 2.4-kb primary transcript and the 80-nt processed product. (A) Northern blot using probe G of RNA from (lane 1) total (T), (lane 2) nucleoplasm (N), and (lane 3) chromatin (C) of testis separated on 15% PAGE containing 7 M urea showing bands at the 80-nt position. (M) The 10-nt RNA ladder. (B) Northern blot using probe G of RNA from (lane 1) total (T), (lane 2) nucleoplasm (N), and (lane 3) chromatin (C) of testis separated on 5% PAGE containing 7 M urea showing bands at the 2.4-kb position. (C, lanes 1–3) RNA profile of total (T), nucleoplasm (N), and chromatin (C) of mouse testis on a 15% PAGE containing 7 M urea. (D) Protein profile by 15% SDS-PAGE of the fractionated (lane 1) total nucleus, (lane 2) nucleoplasm, and (lane 3) chromatin showing enrichment of histones in the chromatin fraction.
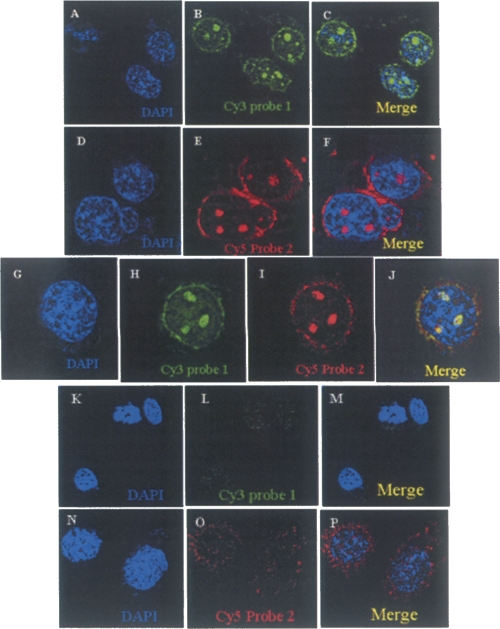
We further examined the in vivo localization of this noncoding RNA within the nucleus, by a fluorescence RNA in situ hybridization experiment. We used two probes spanning different regions of the primary transcript, one falling within the ∼80-nt region and another away from that region to score for the full-length 2.4-kb transcript. Each of the probes was synthesized using LNA technology for better hybridization stability and specificity (Elmqn et al. 2005). They were labeled with Cy3 for the probe 1 falling within the ∼80-nt region and with Cy5 for the probe 2 falling outside the region but within the 2.4-kb transcript. This experiment was carried out on the Gc1-Spg cell line of mouse spermatogonial cells, which expresses both the 2.4-kb primary transcript as well as the processed ∼80-nt RNA. The in situ hybridizations were performed in the presence of 50% formamide concentration and high stringency washes. The in situ fluorescence images are shown in Figure 8, A–F. Most interestingly, we observed clear, large 3-4 punctate signals hybridizing to both the probes. When the cells were hybridized simultaneously, both the probes showed colocalization patterns (Fig. 8G–J). The signal observed in the large punctuate foci using the ∼80-nt probe 1 does not distinguish between hybridization of the probe to either the 2.4-kb primary transcript or the processed ∼80-nt intermediate per se. However, there were some additional signals from probe 1 that correspond to the processed ∼80-nt RNA that did not colocalize. In a control experiment, we treated the cells with RNase A prior to hybridization, in which case no fluorescence signal was seen (Fig. 8K–P).
FIGURE 8.
The mrhl RNA localizes to the nucleus. RNA in situ on mouse Gc1 Spg cells using LNA probes. (A–C) RNA in situ performed using probe 1 (encompassing the sequence within the 80-nt region) labeled with Cy3. (B) Punctate nuclear localization of the RNA. (C) The merge image. (D–F) RNA in situ hybridization using probe 2 labeled with Cy5 encompassing the region outside the 80-nt region but within the 2.4-kb mrhl RNA. (G–J) The colocalization of (H) probe 1 and (I) probe 2 showing a (J) clear overlapping pattern. Cy3 shows additional diffused signal apart from the colocalized spots. (K–P) RNA in situ of cells using (K–M) probe 1 and (N–P) probe 2, where cells are treated with RNase A prior to hybridization showing no signal. The nucleus is stained with DAPI.
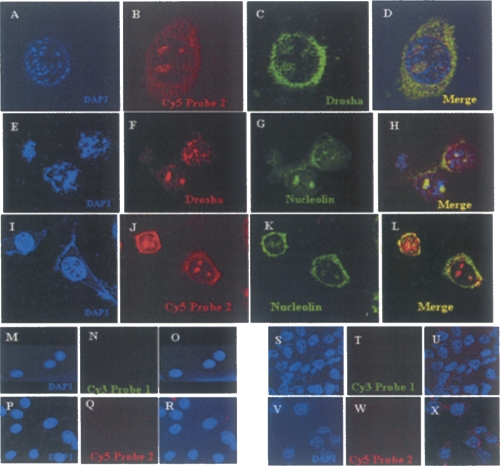
We extended this observation further to look at the localization of this 2.4-kb mrhl transcript with any of the known nuclear protein(s). Initially, we did the experiment with Drosha to see the colocalization of Drosha with the RNA, and we did, indeed, find that Drosha and the 2.4-kb primary transcript colocalize to each other (Fig. 9A–D), strengthening our earlier findings of Drosha's involvement in the processing of the 2.4-kb mrhl RNA. Some reports have shown the presence of Drosha in the nucleolus (Fukuda et al. 2007). Hence, we carried out colocalization of Drosha and Nucleolin, a nucleolus-specific protein. Figure 9, E–H, shows the colocalization of Drosha and Nucleolin. As we observed colocalization of Drosha and Nucleolin, we wanted to examine whether we could colocalize the RNA also to Nucleolin. Thus we did a colocalization analysis of the RNA with Nucleolin. Interestingly, the RNA and Nucleolin also colocalized, showing the presence of the RNA in the nucleolus (Fig. 9I–L). A similar experiment was performed in the RAG1 cell line (data not shown), which is a mouse renal adeno-carcinoma cell line, and we could demonstrate the presence of this RNA in a similar pattern in those cell lines also with predominant localization in the nucleolus, showing that the result presented so far is a general theme and not an artifactual signal due to some property of the cell line itself. As mentioned above, the 2.4-kb RNA coding gene is absent in humans, and our earlier experiment showed that this gene is not expressed in brain tissue in mouse. As an additional negative control, we carried out similar RNA in situ in HeLa (human) as well as C6 rat glioma (brain) cell lines. As can be seen in Figure 9, M–R and S–X, we did not detect any signal in either of these cell lines.
FIGURE 9.
The nucleolus is the site of processing of mrhl RNA. Immunolocalization combined with RNA in situ on the mouse cells. (A–D) RNA in situ on the mouse Gc1 Spg cells using LNA probe 2 labeled with (B) Cy5 and (C) immunofluorescence using anti-Drosha followed by anti-rabbit Alexa 488. (D) The colocalization of both the signals showing overlapping regions in the nucleus. Immunolocalization of (F) Drosha and (G) nucleolin in the nucleus using anti-rabbit Alexa 568 and anti-mouse Alexa 488 secondary antibodies. (H) The merge of the two images showing colocalization. Colocalization of mrhl RNA hybridized to (J) cy5-labeled LNA probe 2 with (K) nucleolin showing (L) overlapping signals. (M–X) Negative controls using (M–R) HeLa and (S–X) rat C6 glioma cells that do not express the RNA, where no hybridization of the LNA probes could be visualized. The nucleus is stained with DAPI.
DISCUSSION
We had earlier identified a 2.4-kb transcript from mouse chromosome 8D that is an unspliced, polyadenylated, noncoding RNA (Nishant et al. 2004). Here we have provided unequivocal experimental evidence that it is a noncoding RNA species by carrying out an in vitro transcription-coupled translation assay. The gene encoding this noncoding RNA transcript is also found in the rat genome on chromosome 19 with 80% identity. It is interesting to note that we did not find this gene to be present in the human genome database by a bioinformatics search. This is not completely surprising since recent evidence shows that noncoding RNA genes may not be conserved across species (Pollard et al. 2006). Even for the miRNAs for which evolutionary conservation was considered to be a major factor for classifying them as miRNAs, recent reports suggest that there are several authentic miRNAs that fail to satisfy this criterion and that these RNAs are, in fact, believed to play some role in the evolution of that species as well (Ambros et al. 2003b).
The extensive secondary structure of the 2.4-kb mrhl RNA prompted us to look for its putative RNA substrate in the miRNA pathway. We did observe a processing of this large RNA into a small intermediate RNA of ∼80 nt in length. The ∼80-nt small intermediate mouse RNA is also found to have 95% identity to the rat RNA sequences. The ∼80-nt RNA region from the long transcript was further delineated, and we did not find any short ORF within this region, eliminating the possibility of any small peptides having been generated. Rather, we did find a foldback structure like a stem–loop and speculated that the RNA might be a potential pre-miRNA. The Northern blot on fractionated cellular compartments also showed the presence of the ∼80-nt RNA in the nucleus. This was very interesting because the pre-miRNA generated in the nucleus would have to be transported to the cytoplasm for its processing by Dicer, but our experiment did not find any cytoplasmic pre-miRNA. This in vivo identification of an ∼80-nt processed species was also supported by in vitro experiments in which the 2.4-kb transcript was cleaved to an ∼80-nt small RNA by total cell-free lysate as well as by purified Drosha complex. Interestingly, we also found that the 80-nt small RNA species can be cleaved by Dicer in vitro to a 22-nt microRNA species. The lack of detection of the 22-nt mature RNA processed product in vivo can be explained in two scenarios, where, in one, the RNA being export-defective cannot bind to exportin 5 for its nuclear cytoplasmic transport, as the majority of pre-miRNAs require exportin 5 for transport to the cytoplasm (Bohnsack et al. 2004). The other reason may be that it is transiently expressed at some developmental or differentiation time point, which we are not able to score for at present. It is becoming very clear that there exist multiple levels of regulation operating within an miRNA pathway. For example, the Caenorhabditis elegans miR38 is expressed only in the embryo, whereas the precursor is ubiquitously detected in all stages of development, indicating that the maturation might be temporally regulated (Ambros et al. 2003a). Since some of the miRNAs are known to determine major cell fate decisions, it has become vital for the system to tightly regulate their expression in a highly temporal and spatial manner (Lagos-Quintana et al. 2002; He et al. 2005). It is still plausible that under certain physiological or pathological conditions, this 22-nt species may be generated in vivo and found in cytoplasm targeting specific mRNAs.
The next major question we raised was a possible role of the ∼80-nt RNA within the nucleus itself. There are numerous RNAs reported in the literature that elicit its function at the nuclear level. The well-studied regulatory RNAs such as Xist in mammals bring about long-range chromatin interactions and are indispensable for the establishment of dosage compensation by forming inactive X-chromosomes (Constancia et al. 2000; Heard 2004). Prasanth et al. (2005) have also reported CTN RNA, which is activated upon stress conditions and regulates its counterpart, the cation-activated channel protein mRNA, by alternate promoter and poly(A) site usage. The 8-kb CTN RNAs are localized specifically in subnuclear compartments like nuclear speckles and are post-transcriptionally cleaved to produce the protein-coding mRNA. Similarly, a screen for nuclear transcripts has identified two noncoding RNAs that are linked and are associated with S35 splicing domains in the nucleus (Hutchinson et al. 2007). These results suggest that RNAs that are involved in a multitude of functions are actually targeted to specific compartments in the cells and do not show a generalized uniform localization pattern (Pagano et al. 2007).
The fluorescent RNA in situ experiments using LNA probes also show a specific localization pattern of both the 2.4-kb transcript and the processed ∼80-nt RNA species to the nucleolus. There was also additional extra-nucleolar signal observed with probe 1 labeled with Cy3 apart from the one that was colocalizing with Nucleolin and Drosha. Furthermore, we have also shown biochemically that the processed ∼80-nt RNA species is associated with chromatin fraction. Analysis of these extra-nucleolar chromatin domain(s) harboring the processed ∼80-nt RNA species will give valuable insights into the possible function of the 2.4-kb mrhl RNA.
MATERIALS AND METHODS
Animals and reagents
Male BALB/c mice and CD1 mice were obtained from the Central Animal Facility at the Indian Institute of Science, Bangalore, India, and the Animal facility at JNCASR. [α-32P]dATP and other radioactive isotopes were obtained from the Board of Radiation and Isotope Technology (BRIT; India).
RNA isolation and RT-PCR
Total RNA from mouse testis and other tissues was isolated by using TRIzol reagent (Invitrogen) as per the manufacturer's protocol. Approximately 2–3 μg of RNA were taken and reverse-transcribed by using oligo(dT) reverse primers or random primers. About 1/20 of the RT product was used for PCR with gene-specific primers for the noncoding transcript.
RNA was isolated from the nucleus and cytoplasm from liver and testis by the following protocol. The tissue was homogenized in buffer A (10 mM Tris-Cl at pH 7.4, 5 mM MgCl2, 1 mM CaCl2, 40 mM NaHSO3, 0.1 mM PMSF) containing 0.34 M sucrose. The homogenate was spun at 1000g to separate nuclear and cytoplasmic fractions. The nuclear fraction was suspended in buffer A containing 2.3 M sucrose and spun at 100,000g for 1 h. The nuclear pellet was washed once in buffer A containing 0.34 M sucrose, and the RNA was isolated using TRIzol reagent. For cytoplasmic RNA isolation, the supernatant after separating the nuclear pellet was taken and the RNA was extracted by using phenol:chloroform and by ethanol precipitation. RNA obtained from fractionated tissues was used for RT-PCR or Northern analysis.
Subnuclear fractionation
The nuclei isolated from the tissues as described previously were resuspended in buffer A and sonicated for 5 sec at 25 amplitude in a sonicator (Vibra sonics) and layered on a 30% sucrose cushion and centrifuged at 1000g for 20 min to fractionate it into chromatin and nucleoplasm. RNA was isolated from both the fractions after digestion with RNase free DNAse (Sigma). An aliquot of the input was also used for profiling the protein composition to check the purity of the preparation.
In vitro coupled transcription/translation
The in vitro coupled transcription/translation was done using the T7 rabbit reticulolysate system (Promega). Briefly, the gene cloned downstream from the T7 promoter in the pGem3Zf+ vector, was added to rabbit reticulolysate along with 35S-methionine, and incubated for 90 min at 30°C. The in vitro translated products were separated in 15% SDS-PAGE. The gel was dried and exposed to a PhosphorImager screen. RNA was also isolated from the same mixture after DNase treatment, and RT-PCR was performed for the specific genes of interest.
Small RNA Northern hybridization
The small RNA PAGE was carried out as described previously (Lagos-Quintana et al. 2002). Approximately 30 μg of RNA in loading buffer were denatured for 15 min at 65°C and loaded on the 15% PAGE containing 7 M urea gel. The EtBr-stained gel was used to check the integrity and the quantity of RNA. The small RNAs were transferred to a Nylon membrane and immobilized under UV. The membrane was prehybridized for 2 h at 42°C in prehybridization buffer (7% SDS, 5× SSC, 2× Denhart's reagent, 0.2 M sodium phosphate, 100 μg/mL denatured single-stranded DNA. The amplified PCR product representing full-length or part of the mrhl gene was gel purified and was used for probe generation by Klenow labeling with random primers. Alternatively, RNA probes derived from in vitro transcription in the presence of [α-32P]UTP were also used for strand-specific Northern analysis. Hybridization was carried out overnight at 42°C. The membrane was then washed twice in 2× SSC/0.5% SDS at room temperature and once each in 1× SSC/0.1% SDS at 42°C and 0.1× SSC/0.1% SDS at 42°C and exposed to a PhosphorImager screen. The Let-7 probe (Sigma) was used as a positive control.
Splinted ligation
Splinted ligation was carried out as described by Maroney et al. (2007). The sequence of the oligos used in the experiment is given below. The Linker oligo was purchased from IDT, while the Bridge oligos were from Sigma:
Linker Oligo: 5′-CGCTTATGACATTddC-3′;
Bridge Mrhl 1: 5′-GAATGTCATAAGCGAAGCACTTACTGTCACTTGATGCA-3′;
Bridge Mrhl 2: 5′-GAATGTCATAAGCGCAAAATTAAAACAGCTTCTACCTCT-3′; and
Bridge Let 7: 5′-GAATGTCATAAGCGAACTATACAACCTACTACCTCA-3′.
miRNA processing assay
The miRNA processing assay was carried out using mouse testicular total cell lysates. Briefly, the testis was decapsulated and treated with collagenase IV (Sigma). The cell pellet was resuspended in buffer D (20 mM HEPES at pH 7.9, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, 5% glycerol) and sonicated to lyse the cells. The processing reaction was carried out as described previously (Han et al. 2004).
In vitro Drosha processing assay
HEK293T cells were transfected with pCK Drosha Flag or pCK Flag. The cells were harvested 48 h post-transfection, and the pellet was suspended in buffer D. The cells were sonicated to lyse them and the supernatant after clearing was immunoprecipitated using anti-Flag agarose beads (Sigma). The beads after washing with buffer D were directly used for the processing assay as described previously.
In vitro Dicer assay
The in vitro transcribed sense (T7) or antisense (SP6) transcript of the purified ∼80-nt region was used for a Dicer assay as per the manufacturer's protocol (Ambion). An aliquot of the reaction was taken at different time points and resolved on a 15% urea-PAGE gel. The gel was dried and autoradiographed.
Overexpression and silencing studies
mrhl cloned in a pCDNA vector under the CMV promoter was transfected into 80% confluent Gc1-Spg cells using Lipofectamine 2000 as per the manufacturer's instructions. The cells were harvested at various time points post-transfection, and RNA was extracted using TRIzol reagent. For the silencing experiment, a combination of two Drosha siRNAs (QIAGEN) was transfected in similar fashion, and total RNA and protein were used for various experiments.
In situ RNA hybridization with LNA probes
The LNA probes specific for the mrhl RNA and the processed transcript were labeled with Cy3 and Cy5 (Exiqon) and hybridized in situ on mouse cell lines as described by Prasanth et al. (2005) with some modifications. Briefly, the cells grown on the coverslips were washed once with PBS and fixed with 4% formaldehyde and 10% acetic acid for 10 min at room temperature. The cells were dehydrated in 70% ethanol, subsequently rehydrated in PBS, and permeablized in PBS with 0.01% Triton X-100. After washing twice in PBS, the cells were prehybridized for ∼1 h in solution containing 10% dextran sulfate, 2 mM vanadyl ribonucleotide complex, 0.2% BSA, 40 μg of Escherichia coli tRNA, 2× SSC, 50% formamide, and 200 μg/mL sheared single-stranded salmon sperm DNA. The prehybridization was followed by hybridization with 10 nM concentration of the probe in the same buffer for 8 h at 50°C in a moist chamber. The cells were then washed twice in buffer containing 50% formamide and 2× SSC followed by once in 50% formamide in 0.5% SSC and 0.1% SDS. The coverslips were mounted subsequently on 60% glycerol in PBS containing 10 μg/μL DAPI.
For immunofluorescence, the cells after hybridization washes were once again washed with PBS, and appropriate dilutions of the primary antibody (mouse monoclonal anti Nucleolin [Santa Cruz] and rabbit polyclonal anti-Drosha [a gift from Narry Kim, Seoul National University]) were added and incubated for 1 h at room temperature in a moist chamber followed by three washes in PBS containing 0.1% Tween 20. This was followed by incubation with appropriate secondary antibody conjugated with Alexa Fluor for 1 h at room temperature in a moist chamber. The cells were washed thrice in PBS containing 0.1% Tween 20 and dried. The coverslips were mounted on 60% glycerol in PBS containing DAPI. The images were acquired in an LSM 10 Meta confocal microscope (Carl Zeiss), and images were analyzed by image analysis software provided by Carl Zeiss.
ACKNOWLEDGMENTS
We thank Professor Narry V. Kim for the Drosha–Flag construct and Drosha antibodies used in this work. We thank B.S. Suma for help in confocal microscopy and V. Ramesh for the mPTB construct. G.G. is a Senior Research Fellow of the Council of Scientific and Industrial Research. M.R.S.R. thanks the Department of Science and Technology for the J.C. Bose Fellowship. Financial support from the Department of Science and Technology and the Department of Biotechnology is gratefully acknowledged.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.838308.
REFERENCES
- Ambros V., Lee R.C., Lavanway A., Williams P.T., Jewell D. MicroRNAs and other tiny endogenous RNA from C. elegans . Curr. Biol. 2003a;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., et al. A uniform system of micro RNA annotation. RNA. 2003b;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Allis C.D. RNA meets Chromatin. Genes & Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bohnsack M.T., Czaplinski K., Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export to pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C.M., McNeil J.A., Willard H.F., Lawrence J.B. Xist RNA paints the inactive X chromosome at interphase: Evidence for novel RNA involved in nuclear chromosome structure. J. Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia M., Dean W., Lopes S., Moore T., Kelsey G., Reik W. Deletion of silencer element in IgF2 results in loss of imprinting dependent of H19. Nat. Genet. 2000;26:203–206. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- Costa F.F. Noncoding RNAs: New players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. Noncoding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Elmqn J., Thonberg H., Ljungberg K., Frieden M., Westergaard M., Xu Y., Wahren B., Liang Z., Prum H., Koch T., et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mellow C.C. Potent and specific genetic interference by double stranded RNA in C. elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fukuda T., Yamagata K., Fujiyama S., Matsumoto T., Koshida I., Yoshimura K., Mihara M., Naitou M., Endoh H., Nakamura T., et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- Gabory A., Ripoche M.A., Yoshimizu T., Dandolo L. H19 gene regulation and function of non coding RNA. Cytogenet. Genome Res. 2006;113:188–193. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- Gilbert S.L., Pehrson J.R., Sharp P.A. Xist RNA association with specific regions of the inactive x chromatin. J. Biol. Chem. 2000;275:36491–36494. doi: 10.1074/jbc.C000409200. [DOI] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Nam J.W., Kim Y.K., Jin H., Kim V.N. The Drosha–DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell. 2006;125:887–902. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 2004;16:247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A., Schattner P., Polacek N. Noncoding RNAs: Hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Kim V.N. Small RNAs: Classification, biogenesis, and function. Mol. Cell. 2005;19:1–15. [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lapidot M., Pilpel Y. Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Maroney P.A., Chamnongpol S., Souret F., Nilsen T.W. A rapid, quantitative assay for direct detection of microRNA and other small RNA sing splinted ligation. RNA. 2007;13:930–936. doi: 10.1261/rna.518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J.S., Makunin I.V. Small regulatory RNAs in mammals. Hum. Mol. Genet. 2005;14:R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- Niemitz E.L., DeBaun M.R., Fallon J., Murakami K., Kugoh H., Oshimura M., Feinberg A.P. Microdeletion of LIT1 in Familial Beckwith–Wiedemann Syndrome. Am. J. Hum. Genet. 2004;75:844–849. doi: 10.1086/425343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant K.T., Ravishankar H., Rao M.R.S. Characterization of a mouse recombination hot spot locus encoding a novel nonprotein coding RNA. Mol. Cell. Biol. 2004;24:5620–5634. doi: 10.1128/MCB.24.12.5620-5634.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano A., Castelnuovo M., Tortelli F., Ferrari R., Dieci G. New small nuclear RNA gene-like transcriptional units as sources of regulatory transcripts. PLoS Genet. 2007;3:0174–0184. doi: 10.1371/journal.pgen.0030001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.S., Salama S.R., Lambert N., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Prasanth K.V., Spector D.L. Eukaryotic regulatory RNAs: An answer to the “genome complexity” conundrum. Genes & Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Prasanth K.V., Prasanth S., Xuan Z., Hearn S., Freier S., Bennett C., Zhang M., Spector D. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Reik W., Walter J. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Sleutels F., Zwartand R., Barlow D.P. The noncoding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:68–73. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Smith E.R., Pannuti A., Gu W., Steurnagel A., Cook R.G., Allis C.D., Lucchesi J.C. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M., Barciszewska M.Z., Zywicki M., Barciszewski J. Noncoding RNA transcripts. J. Appl. Genet. 2003;44:1–19. [PubMed] [Google Scholar]
- Wevrick R., Kerns J.A., Francke U. Identification of a novel paternally expressed gene in the Prader–Willi syndrome region. Hum. Mol. Genet. 1994;3:1877–1882. doi: 10.1093/hmg/3.10.1877. [DOI] [PubMed] [Google Scholar]