Abstract
Maintenance of the intestinal epithelium is based on well-balanced molecular mechanisms that confer the stable and continuous supply of specialized epithelial cell lineages from multipotent progenitors. Lineage commitment decisions in the intestinal epithelium system involve multiple regulatory systems that interplay with each other to establish the cellular identities. Here, we demonstrate that the microRNA system could be involved in intestinal epithelial cell differentiation, and that microRNA-194 (miR-194) is highly induced during this process. To investigate this inducible expression mechanism, we identified the genomic structure of the miR-194-2, -192 gene, one of the inducible class of miR-194 parental genes. Furthermore, we identified its transcriptional regulatory region that contains a consensus-binding motif for hepatocyte nuclear factor-1α (HNF-1α), which is well known as a transcription factor to regulate gene expression in intestinal epithelial cells. By chromatin immunoprecipitation assay and luciferase reporter analysis, we revealed that pri-miR-194-2 expression is controlled by HNF-1α, and its consensus binding region is required for the transcription of pri-miR-194-2 in vivo in an intestinal epithelial cell line, Caco-2. Our observations indicate that microRNA genes could be targets of lineage-specific transcription factors and that microRNAs are regulated by a tissue-specific manner in the intestinal epithelium. Therefore, our work suggests that induced expression of these microRNAs have important roles in intestinal epithelium maturation.
Keywords: microRNA, miR-194, Caco-2; differentiation, HNF-1α, intestine
INTRODUCTION
The intestinal epithelial system is a paradigm for the production of distinct cell lineages from multipotent progenitors (Crosnier et al. 2006). The molecular mechanism of balanced and continuous generation of intestinal epithelial cells has been extensively investigated, as it would be involved in the pathogenesis of gastrointestinal disorders such as intestinal epithelial tumors. Recent studies highlighted the critical roles of specific signaling pathways directing activation of certain transcription factors, such as Notch and Wnt pathways, in the development of intestinal epithelium, clearly indicating that its developmental program is under the control of dynamic gene regulatory networks (Sancho et al. 2004; Fre et al. 2005; Gregorieff and Clevers 2005; Stanger et al. 2005; Clarke 2006; Crosnier et al. 2006).
microRNAs (miRNAs) are 21–23-nucleotide (nt) noncoding RNAs that function as post-transcriptional regulators of gene expression in various species (Ambros 2004; Bartel 2004; Zamore and Haley 2005). miRNAs recognize their target(s) with the partially complementary sequences and repress their translation or modify their stability (Olsen and Ambros 1999; Jing et al. 2005). miRNAs play essential roles in diverse events, including control of developmental timing (Wightman et al. 1993), differentiation (Chen et al. 2004, 2006; Esau et al. 2004; Kim et al. 2006), apoptosis, cell proliferation (Brennecke et al. 2003; Cheng et al. 2005; Cimmino et al. 2005), and organ development (Giraldez et al. 2005).
miRNAs initially appear as relatively long transcripts called pri-miRNA, and it is now widely accepted that many pri-miRNAs are transcribed by RNA polymerase II (Cai et al. 2004; Lee et al. 2004), suggesting that tissue- and time-specific expression of miRNAs is probably specified at the level of pri-miRNA transcription. Although hundreds of miRNAs were cloned in various species, the regulatory mechanisms of specific pri-miRNAs are still largely unknown. Studies regarding regulation of each pri-miRNA would therefore provide clues to the further understanding of miRNA biology in general or of particular functions in specific cell lineages.
In this study, we focused on the miRNA system as novel molecular machinery in intestinal epithelial cell differentiation. Our high-throughput miRNA expression profiling revealed a dynamic change of the miRNA expression pattern during Caco-2 differentiation, which is one of the most widely known intestinal epithelial differentiation models. In the model, we found that dozens of miRNAs were up- or down-regulated. Among such miRNAs, miR-194 was found to be highly up-regulated with tight tissue specificity. To know the precise regulatory mechanism, we determined the genomic structure of pri-miR-194-2. Given the genomic structure, we also identified a highly conserved genomic region close to the transcription start site of pri-miR-194-2 that contains a consensus binding motif for hepatocyte nuclear factor-1α (HNF-1α). Chromatin immunoprecipitation (ChIP) assay confirmed physical binding of HNF-1α to this conserved region in vivo, and other functional assays further showed the transcriptional role of HNF-1α for the pri-miR-194-2 transcription, suggesting that the conserved genomic region is the core promoter element for pri-miR-194-2 and that HNF-1α is the key transcription factor for the expression of this miRNA.
Because HNF-1α is one of the critical factors of intestinal epithelial gene expression during differentiation and maturation, miR-194 might constitute a certain part of its regulatory network, thereby contributing to the regulation of gene expression program in intestinal epithelial cells. Therefore, our work suggests that induced expression of miRNAs has an important role in intestinal epithelium maturation.
RESULTS
miR-194 is highly induced during intestinal epithelial cell differentiation
Adopting the differentiation system of intestinal epithelial cell line Caco-2, possible alteration of miRNA expression pattern during intestinal epithelial differentiation was examined. Induction of differentiation in Caco-2 cells was performed by conventional long-term confluent culture method. Following confirmation of maturation status by the differentiation markers such as lactase-phlorizin hydrolase (LPH) and sucrase-isomaltase (SI) (Fig. 1A), 156 mature miRNAs were quantified in differentiated and proliferative cells by the TaqMan-based high-throughput profiling method. As shown in Figure 1, B and C, several miRNAs were significantly up- or down-regulated during Caco-2 differentiation as judged by Ct values. Among such genes, expression of miR-133a and miR-133b has been previously detected in tissues other than intestine (Chen et al. 2006). Also, miR-146 has been reported to control Toll-like receptor and cytokine signaling (Taganov et al. 2006), whereas the miR-34 family has been shown to be involved in the p53 network (He et al. 2007). Inhibition of miR-148 and 210 increases the level of apoptosis, while inhibition of miR-152 decreases cell growth (Cheng et al. 2005). Consequently, differentially expressed miRNAs observed in the present experiment suggested that miRNA machineries that control general physiological events are involved in epithelial cell differentiation.
FIGURE 1.
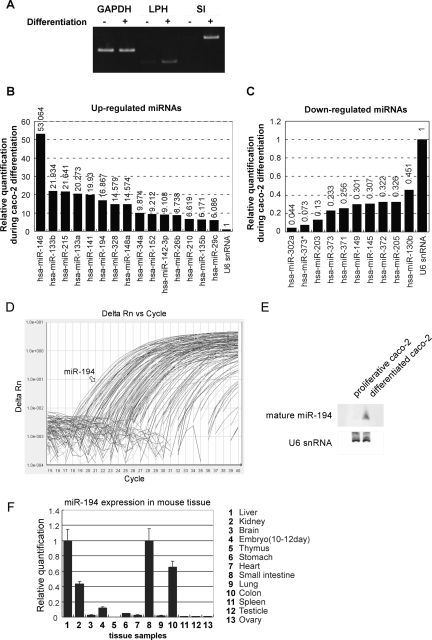
Altered expression profile of miRNA during Caco-2 differentiation. (A) Detection for differentiation markers by RT-PCR. RT-PCR was performed on total RNAs extracted from proliferative or differentiated Caco-2 cells. PCR products were separated by 5% acrylamide gel electrophoresis and stained by EtBr. (B) Up-regulated miRNAs during Caco-2 differentiation. A total of 156 miRNAs were quantified by TaqMan miRNA assays Human Panel–Early Access Kit. Relative Quantification (RQ) was normalized by U6 RNA endogenous control. Quantification results were arranged by RQ and cut-off detectors whose Ct values were more than 30 cycles in differentiated Caco-2 cells. (C) Down-regulated miRNAs during Caco-2 differentiation, observed in the quantification performed in B. (D) Amplification plot by TaqMan miRNA assay in differentiated Caco-2 cells. Arrow indicates amplification plot of miR-194. (E) Northern blot of mature miR-194. Total RNAs (30 μg) were separated in 8 M urea 12% PAGE and transferred to Nybond N+. After UV cross-linking, the membrane was hybridized with DIG-labeled RNA probe. Hybridized probes were detected by AP-conjugated anti-DIG antibody and visualized by CDP-star. (F) miR-194 expression in various mouse tissues. Expression of miR-194 was quantified by TaqMan miRNA assay. U6 snRNA was used as endogenous control. The expression level in small intestine was set to 1.
Although identification of sets of miRNA as regulators of general events is important, here, we rather sought to find some miRNAs that are regulated specifically in intestinal epithelial cells. Among differentially expressed miRNAs, miR-194 was one of the highly induced miRNAs during differentiation and showed the highest intensity in differentiated Caco-2 cells (Fig. 1D). Induction of mature miR-194 upon Caco-2 differentiation was also confirmed by Northern blot (Fig. 1E). Also, tissue distribution of mature miRNA-194 in an adult mouse was identified to be relatively specific in intestinal tracts (Fig. 1F), consistent with some previous results in other organisms such as zebrafish embryo (Wienholds et al. 2005).
These data collectively suggested that, among numerous regulated miRNAs, miRNA-194 is highly and specifically induced in intestinal epithelial cells during its differentiation.
Genomic structures of miR-194 primary transcripts
Given the high and specific expression of miR-194 in intestinal epithelial cells, we further sought to characterize this miRNA to know its regulation. According to miRBase, mature miR-194 can be derived from two separate loci on human genome that are registered as miR-194-1 and miR-194-2 (Fig. 2A). The miR-194-1 and miR-194-2 are encoded on human chromosomes 1 and 11, respectively. As like many other miRNAs, both miR-194-1 and miR-194-2 are so-called clustered miRNA and have cluster partners miR-215 and miR-192, respectively.
FIGURE 2.
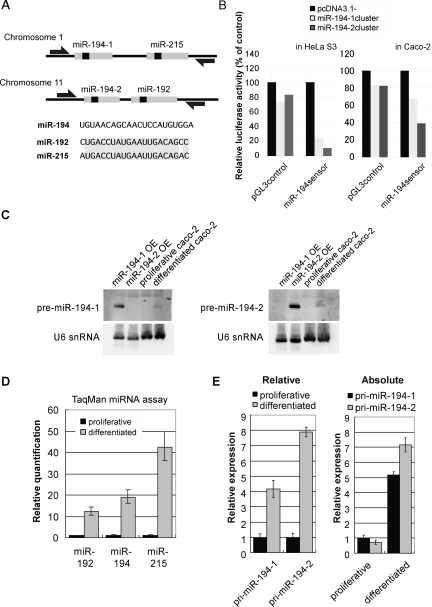
Expression of miR-194-2, -192 cluster was induced during Caco-2 differentiation. (A) Genomic organization of miR-194 clusters. miR-194-1 and miR-215 are located within an ∼400-bp region in chromosome 1. miR-194-2 and miR-192 are located within an ∼300-bp region in chromosome 11. Open arrows represent pri-miRNA detection primers used in E. (B) Both miR-194 loci potentially express mature miR-194. miR-194 expression vector and pGL3miR-194sensor were cotransfected into HeLa S3 cells (left) or Caco-2 cells (right). As pGL3miR-194sensor contained a sequence completely complementary to mature miR-194 in 3′UTR of luciferase, expression of mature miR-194 is detected by reduced firefly luciferase activity by RNAi. Firefly luciferase reporter activities were normalized by Renilla luciferase. The mean value of cells cotransfected with pcDNA3.1 mock vector was set to 100. (C) Northern blot of the pre-miR-194s, each arising from distinct loci. Total RNA (30 μg) extracted from proliferative or differentiated Caco-2 cells that were pretreated with Dicer siRNA was analyzed. Positive control (10 μg total RNA) was obtained from Dicer knockdowned 293 cells transfected with miR-194 cluster expression vectors (miR-194-1 OE and miR-194-2, OE, respectively). Probes were designed to hybridize around the loop sequence of each transcript. (D) Inducible expression of miRNAs from miR-194 clusters during Caco-2 differentiation. Expression of miR-192, miR-194, and miR-215 were quantified by TaqMan miRNA assay. U6 snRNA was used as endogenous control to normalize expression of miRNAs. The expression level of each miRNA in proliferative Caco-2 cells was set to 1. (E) Two distinct miR-194 loci contribute to its induced expression during Caco-2 cell differentiation. RT-PCR was performed with primers indicated in A, and GAPDH was used as endogenous control. For relative quantification, expression of each pri-miR-194 within proliferative Caco-2 was used as a standard (left). For absolute quantification, genomic DNA was used as a standard. Bar graph is drawn so that pri-miR-194-1 in proliferative Caco-2 cells is set to 1 (right).
To examine whether either of the two loci can generate mature miR-194, we constructed miRNA expression vectors in which miR-194 loci were inserted downstream of the CMV promoter, and observed mature miR-194 generation using luciferase sensor reporter assay. As luciferase sensor mRNA have perfect complementary sequence against mature miR-194 in 3′UTR, if mature miR-194 is generated from the miR-194 loci, firefly sensor mRNA is cleaved by RNAi effect. Thus, miRNA generation could be observed as reduction of luciferase activity. Forced expression of each locus showed marked decrease of the sensor luciferase activity in both Hela S3 and Caco-2 cells (Fig. 2B). As the results of this assay showed that both loci are able to generate mature miR-194, we sought to examine which locus is the origin of miR-194 that is induced upon differentiation of intestinal epithelial cells. Northern blot analysis of pre-miRNAs showed up-regulation of both pre-miRNAs (Fig. 2C), whereas TaqMan miRNA assay showed up-regulation of miR-194, miR-192, and miR-215 upon Caco-2 differentiation (Fig. 2D). Quantitative RT-PCR using a reverse-transcribed sample as a standard template also showed induction of pri-miRNA from both loci upon differentiation, but a relatively higher induction of pri-miR-194-2 was observed, compared with pri-miR-194-1 (Fig. 2E, left). Absolute quantification using genomic DNA as a standard template, which enables quantitative comparison between pri-miR-194-1 and pri-miR-194-2, also demonstrated that expression of pri-miR-194-2 was more abundant in differentiated Caco-2 cells, compared with pri-miR-194-1 (Fig. 2E, right).
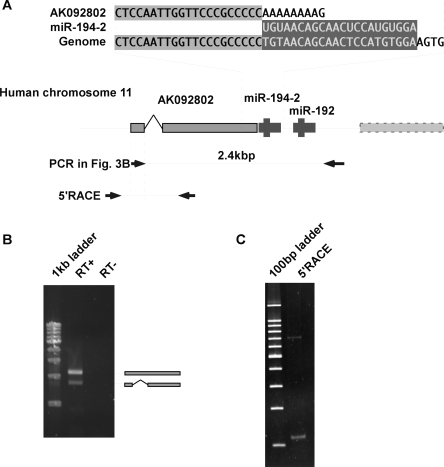
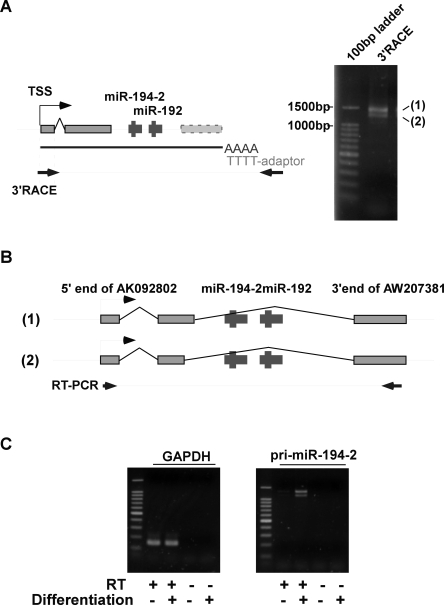
Having these observations, we focused on miR-194-2 cluster and further characterized this locus. Upon the database search, a registered cDNA, AK092802, was found as a putative 5′ part of the pri-miR-194-2 transcript (Fig. 3A). As shown by RT-PCR, the registered transcript AK092802 and the region encoding pre-miR-194-2 were transcribed primarily as a single RNA (Fig. 3B). Also, the 5′ region of the transcript including the transcription start site of pri-miR-194-2 was determined by 5′ RACE. Although 5′ RACE presented two fragments, sequence analysis revealed that these two fragments arise from the same transcription start site, while the difference in length was due to splicing (Fig. 3C). Furthermore, we performed 3′ RACE to identify the structure of pri-miR-194-2, -192 (Fig. 4A). The identified pri-miRNA shared the same 3′ terminal with that of another registered transcript, AW207381 (Fig. 4B), and induced expression of the entire transcript was confirmed during Caco-2 cell differentiation (Fig. 4C). Therefore, we examined the genomic structure of pri-miR-194-2 that gives rise to a part of the inducible miR-194 during intestinal epithelial cell differentiation.
FIGURE 3.
Identification of the transcription start site of pri-miR-194-2. (A) Genomic sequence and EST information surrounding miR-194-2. 3′ End of AK092802 is adjacent to 5′end of miR-194. Arrow bars represent predicted PCR products. (B) AK092802 and miR-194-2, -192 cluster are linked in a single transcript. RT-PCR was performed by total RNA extracted from differentiated Caco-2 cells, using primers indicated in A. PCR products were separated by 1% agarose gel electrophoresis and stained by EtBr. Sequence analysis of the amplified products revealed that the 2.4-kbp fragment contained an intronic sequence, while the intronic sequence was spliced out in the 1.8-kbp fragment. (C) 5′ RACE analysis of pri-miR-194-2. 5′ RACE was performed using random primers instead of oligo dT primer. PCR was performed by primers indicated in A. PCR products were separated by 5% polyacrylamide gel electrophoresis and stained by EtBr. Consistent with B, two fragments were observed, and sequence analysis of these fragments revealed that difference was due to an intronic sequence. Both fragments had the same transcription start site.
FIGURE 4.
Identification of pri-miR-194-2 structure. (A) 3′ RACE analysis of pri-miR-194-2. Schematic representation of the primers used in the analysis is shown (left). PCR products were separated by 2% agarose gel electrophoresis and stained by EtBr (right). Two fragments are shown, designated as 1 and 2. (B) Schematic representation of pri-miR-194-2 structures. Sequence analysis revealed that difference in fragments 1 and 2 observed in A comes from the difference in length of the second exon. Both fragments had the same 3′ end, which also coincided with the 3′ end of AW207381. (C) Induced expression of pri-miR-194-2 during Caco-2 cell differentiation. To confirm the increased expression of the identified pri-miR-194-2 transcript during Caco-2 differentiation, RT-PCR was performed using primers represented in B. PCR products were separated by 2% agarose gel electrophoresis, and stained by EtBr.
Identification of the core promoter element for pri-miR-194-2
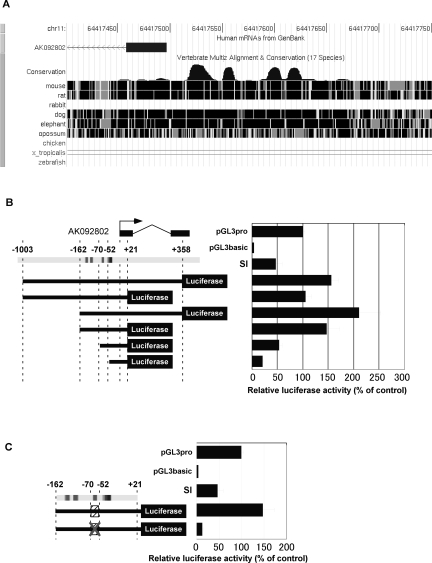
To know the transcriptional mechanism, we then sought to analyze the promoter region of pri-miR-194-2. Upstream genomic region close to the transcription start site of pri-miR-194-2 contains several highly conserved regions among human, mouse, rat, and dog (from −162 to +21 with respect to the transcription start site of AK092802) (Fig. 5A). To identify the promoter region, we constructed reporter plasmids carrying various genomic sequences around the transcription start site of pri-miR-194-2 (Fig. 5B) and subjected them to luciferase assay. Results demonstrated that the region from −162 to +21 had a high promoter activity in differentiated Caco-2 cells, comparable to that of the longest region from −1003 to +358 (Fig. 5B). In contrast, deletion of the conserved region dramatically reduced promoter activity.
FIGURE 5.
Potential HNF-1α binding element is located in the pri-miR-194-2 promoter. (A) Conserved sequences found within the pri-miR-194 promoter region. This figure is derived from the UCSC genome browser. (B) Schematic representation of human miR-194-2 promoter reporter constructs (left) and analysis of their promoter activity (right). Firefly luciferase reporter plasmids were constructed containing various regions of the putative pri-miR-194-2 promoter between positions −1003 to +358, designated with respect to the 5′end of AK092802. Arrow indicates transcription start site of pri-miR-194-2. pGL3 promoter (pGL3 pro) contains SV40 promoter, while pGL3basic has no promoter sequence upstream of the luciferase coding region. (C) Mutation of the region from −70 to −52 dramatically reduced pri-miR-194-2 promoter activity in Caco-2 cells. A scrambled sequence was introduced in the mutant pri-miR-194-2 promoter. Luciferase activities were normalized by Renilla luciferase activities.
Since conserved regions in a gene promoter are expected to contain regulatory elements, we focused on such region found within the pri-miR-194-2 promoter. Luciferase assay using a mutated construct revealed that the region from −70 to −52 is critically required for the pri-miR-194-2 promoter activity that is driven by the identified conserved region in differentiated Caco-2 cells, indicating that the corresponding region is the core element of pri-miR-194-2 promoter (Fig. 5C).
HNF-1α regulates pri-miR-194-2 promoter
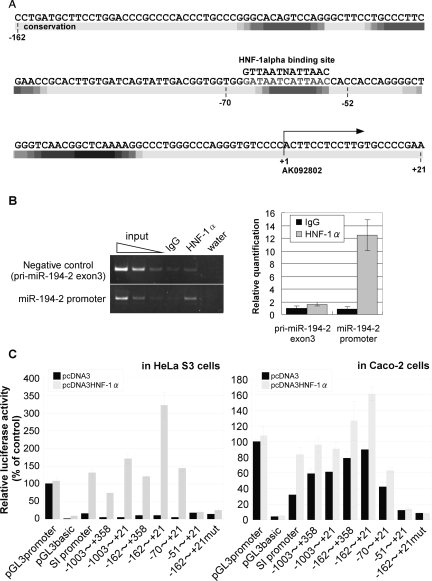
By a computational search for potential motifs of transcription factors, a putative binding site for HNF-1α was found in the conserved core element of pri-miR-194-2 promoter (Fig. 6A). HNF-1α is a member of a class of transcription factors that is distantly related to homeobox proteins, which contains a DNA binding domain. In addition, HNF-1α has been formerly described as a key transcriptional activator during Caco-2 differentiation (Wu et al. 1994; Mitchelmore et al. 1998; Boudreau et al. 2001, 2002; van Wering et al. 2002). Thus, we hypothesized that HNF-1α might bind to the core element of pri-miR-194-2 promoter and contribute to up-regulate pri-miR-194-2 transcription. Indeed, HNF-1α was shown to physically interact to the core element of the pri-miR-194-2 promoter in Caco-2 cells, as judged by ChIP assay (Fig. 6B). Furthermore, forced expression of HNF-1α activated the promoter of both pri-miR-194-2 and SI in HeLa S3 cells, which usually do not express miR-194 (Fig. 6C). Forced expression of HNF-1α in Caco-2 cells also showed an additive effect on the promoter activity of pri-miR-194-2. Mutation in the HNF-1α binding site resulted in almost complete loss of the pri-miR-194-2 promoter activity, confirming the requirement of HNF-1α binding to the corresponding site for pri-miR-194-2 transcription. Taken together, these data suggest that HNF-1α is a key transcriptional regulator of pri-miR-194-2 and activates its promoter activity by physically binding to the core promoter element.
FIGURE 6.
HNF-1α binds and regulates the miR-194-2 promoter in Caco-2 cells. (A) Sequence of pri-miR-194-2 promoter between −162 and +21. The region between −162 and +21 is partially conserved among human, mouse, and dog. Conserved sequences are indicated as density map (higher conservation becomes black). The transcription start site is indicated by an arrow. Consensus sequence of HNF-1α binding site is located at region between −67 and −55. (B) Chromatin immunoprecipitation (ChIP) analysis showing binding of HNF-1α to the miR-194-2 promoter in vivo in Caco-2 cells. Fixed chromatin from differentiated Caco-2 cells was prepared and immunoprecipitated either by anti HNF-1α antibody or normal goat IgG. ChIP primers were designed to amplify the region containing putative HNF-1α binding site in the pri-miR-194-2 promoter. Negative control primers were designed to amplify exon 3 of the pri-miR-194-2. PCR products were separated by acrylamide gel electrophoresis and stained by EtBr (left). Results of the quantitative PCR are shown as the relative amount of precipitated chromatin, in which chromatin precipitated by goat-IgG is set to 1. (C) Human pri-miR-194-2 promoter activity is up-regulated by forced expression of HNF-1α. Various luciferase reporter plasmids were cotransfected with either pcDNA3 mock vector (black bar) or HNF-1α expression vector (open bar) into HeLa S3 cells (left), or Caco-2 cells (right). SI-promoter contains promoter sequence of the sucrase-isomaltase (SI) gene, which is formerly reported to be up regulated by HNF-1α. Luciferase activities were normalized by Renilla luciferase activities.
DISCUSSION
In the present study, we identified the structure of pri-miR-194-2 and determined both transcription start site and 3′ end using 5′ RACE and 3′ RACE, respectively. As pri-miRNAs could be either a protein-coding RNA or a noncoding RNA, we searched in silico for a candidate open reading frame (ORF) within a pri-miR-194-2, -192 transcript and did not find a single protein-coding ORF. In addition, conserved genomic region of pri-miR-194-2, -192 was restricted to the region encoding miRNA hairpin structures. Therefore, we suggest that pri-miR-194-2, -192 is a noncoding RNA.
We also identified that miR-194-2 and miR-192 are encoded within the intron but not in the exon (Fig. 4B). To assure this, we have shown that mature miR-194 surely arises from the miR-194-2, -192 cluster region, a partial sequence of the second intron (Fig. 4B), by forced expression of the corresponding region (Fig. 2B). This means that these miRNAs are processed by splicing and cropping of the pri-miRNA in the nucleus. Indeed, although the PCR product shown in Figure 3B contained the second intron, the PCR product shown in Figure 4C lacked the corresponding intron sequence. This difference may be attributed to the relatively short extension time (1.5 min) used for the PCR in Figure 4C, for detection of the transcript containing the intron (4.8 kb), but also suggests that the second intron containing the miR-194-2, -192 cluster is primarily transcripted along with the three exons but is rapidly spliced out from the pri-miR-194-2.
On the other hand, concerning these processing steps, a model of intronic miRNA processing was recently suggested (Kim and Kim 2007). According to their model, miRNA-harboring intron is detained while other introns are rapidly spliced out. In our study, RT-PCR analysis in Figure 3B showed 2.4-kbp and 1.8-kbp products, which might represent the nascent transcript and the partially spliced transcript for pri-miR-194-2, respectively. As the reverse primer used in the present RT-PCR is placed within the second intron, the result may indicate that the first intron is processed more rapidly, compared with the miRNA-harboring second intron. Therefore, pri-miR-194-2 might be processed through the proposed model.
Also in our study, we demonstrated that miR-194 is highly expressed in differentiated intestinal epithelial cells. Induced expression of miR-194 could be accounted by the regulatory mechanism of miR-194-2 gene in that a tissue-associated transcription factor, HNF-1α, plays a central role in its transcription. Although HNF-1α is well known for its regulatory roles of various genes specific for the intestine, its expression is not tightly restricted to the intestine but is also found in the liver or the kidney (Mendel and Crabtree 1991). However, mature miR-194 has been reported to appear not only in the intestine but also in the liver and in the kidney (Lagos-Quintana et al. 2003; Krutzfeldt et al. 2005; Wienholds et al. 2005; Kato et al. 2007). Our TaqMan miRNA analyses have also confirmed expression of miR-194 in these tissues (Fig. 1F). These findings are consistent with our present study describing the regulation of miR-194 expression by HNF-1α. Recent studies, however, have highlighted the regulation of mature miRNA generation at the post-transcriptional processing level (Thomson et al. 2006; Viswanathan et al. 2008) Although our data show consistent increase of mature miR-194 upon increase of its pri-miRNAs, there remains a possibility that expression of mature miR-194 might also be regulated at the processing level.
Our promoter analyses revealed that the consensus motif for HNF-1α found within the conserved region of the pri-miR-194-2 promoter plays a critical role in induction of miR-194-2 gene upon Caco-2 cell differentiation. HNF proteins are known to interact with other transcription factors to regulate the expression of various intestine-specific genes. For example, different members of the HNF family that are expressed in the intestine, such as HNF-1β and HNF-4, coordinately enhance target gene expression (Wu et al. 1994; Hu and Perlmutter 1999, 2002; Boudreau et al. 2001). Furthermore, GATA family and caudal related homeobox protein Cdx2 also coordinately enhance expression of intestine-specific genes (Krasinski et al. 2001; Boudreau et al. 2002; Wang et al. 2004). It is therefore speculated that the consensus motif for HNF-1α found within the core promoter region of pri-miR-194-2 may play a pivotal role in different aspects of regulation for this miRNA. Evolutional conservation of this consensus site among phylogenetically distant species further indicates a strong functional link between HNF-1α and miR-194-2 expression and also its significance in essential physiological events.
From the view of environmental regulation of miRNA, a recent study reported that miR-192 ectopically appears in diabetic renal glomeruli and that TGF-β is involved in the induction of this miRNA (Kato et al. 2007). As we have determined in the present study that miR-192 is the clustering partner of miR-194-2, it is possible that TGF-β may also regulate expression of miR-194. Interestingly, it is reported that TGF-β1 can modulate the differentiation process of Caco-2 cells in certain environments (Schroder et al. 1999). Therefore, it is of interest to examine the role of TGF-β upon regulation of miR-194 expression, as this cytokine shows diverse effects on intestinal physiology.
In conclusion, miR-194 is highly induced during intestinal epithelium differentiation, and pri-miR-194-2 expression is regulated by HNF-1α. As HNF-1α is one of the critical regulators of intestinal epithelial gene expression, control of miR-194 by this transcription factor adds miRNAs to the regulatory network of gene expression in intestinal epithelial cells. Therefore, the present work suggests that induced expression of miRNAs by tissue-specific transcription factors has an important role in intestinal epithelium maturation.
MATERIALS AND METHODS
Cell culture
Caco-2 cells were cultured in Minimal Essential Medium (Sigma) supplemented with nonessential amino acids and 10% heat-inactivated fetal bovine serum. For the differentiation assay, cells were seeded onto collagen-coated plate, and growth medium was changed every 3 d. HeLa S3 and 293 cells were cultured in Dulbecco's modified Eagle medium (Sigma) supplemented with 10% heat-inactivated fetal bovine serum.
RNA extraction, reverse transcription, and real-time quantitative PCR
Total RNA extraction was performed using miReasy mini kit with DNase I treatment (Qiagen). For detection of differentiation markers, LPH and SI, total RNAs were reverse transcribed by QuantiTect Reverse Transcription Kit (Qiagen). Semi-quantitative PCR was performed using LA Taq (Takara) with specific primer sets as follows:
LPH primer F 5′-TTTCTGTACGGACGGTTTCC-3′ and
LPH primer R 5′-AGAAAACGTGTCCCAAATGC-3′;
SI primer F 5′-AATCAGATGGCACAGGGTTC-3′ and
SI primer R 5′-TTCCTTTCCCCCATACATGA-3′;
GAPDH primer F 5′-GAAGGTCGGAGTCAACGGATTT-3′ and
GAPDH primer R 5′-ATGGGTGGAATCATATTGGAA-3′.
Quantification of mature miRNAs was performed by TaqMan miRNA assays Human Panel–Early Access Kit (Applied Biosystems) according to the manufacturer's instruction. As the kit did not contain probes for miR-192, data for miR-192 are not included in Figure 1B. Quantification of individual miRNAs (Fig. 2D) was performed by a TaqMan miRNA assay kit (Applied Biosystems).
Northern blot
To synthesize DIG-labeled RNA probe, vectors were constructed by inserting the following oligonucleotides into pcDNA3:
For mature miR-194 detection, 5′-TCCACATGGAGTTGCTGTTACA-3′;
For pre-miR-194-1 detection, 5′-AACTCCATGTGGACTGTGTACCAATTTCCAGTGGAGATGC-3′; and
For pre-miR-194-2 detection, 5′-AACTCCATGTGGAAGTGCCCACTGGTTCCAGTGGGGCTGC-3′.
DIG-labeled RNA probes were synthesized by DIG RNA labeling kit (Roche). For pre-miRNA detection, knockdown of Dicer was performed before RNA extraction, using siRNA. The Dicer siRNA was transfected into cells by LipofectAmine2000 (Invitrogen). The sequence of the Dicer siRNA was as follows:
Dicer siRNA sense, 5′-UGCUUGAAGCAGCUCUGGAdTdT; and
Dicer siRNA anti-sense, 5′-UCCAGAGCUGCUUCAAGCAdTdT.
The letters “dT” represent deoxythymidine. Thirty micrograms of total RNA was separated by 8 M urea PAGE and transferred to Nybond N+ (GE Healthcare). After UV cross-linking, the membrane was hybridized with DIG-labeled probes in hybridization buffer (50% formamide, 5× SSC, 0.1% SDS, 2× Denhardt's solution, and salmon sperm DNA). Detections were achieved by AP-conjugated anti-DIG antibody (Roche) and CDP-star (GE Healthcare).
RT-PCR for pri-miRNA
Pri-miRNA cluster detection PCR was performed using Quantitect SYBR PCR kit (Qiagen) with specific sets of primers as follows:
pri-miR-194-1 F, 5′-AGCGTTTCAAATCTACCAGT-3′;
pri-miR-194-1 R, 5′-TATCTTCTGTGTACCTGCCA-3′;
pri-miR-194-2 F, 5′-ATGATAAGAAGCCTCGGTGA-3′; and
pri-miR-194-2 R, 5′-GTGGGACCATGAGTGCTGCA-3′.
AK092802 and miRNA cluster RT-PCR (Fig. 3B) was performed with the following primers, and the sequence of the detected PCR products were confirmed by direct sequencing. Forward primer downstream of the transcription start site (DTSS F) was 5′-TTCCTCCTTGTGCCCCGAAG-3′, and pri-miR-194-2 R was used as the reverse primer (this primer is placed within the second intron). Template cDNA was prepared by reverse-transcription by SuperScript II (Invitrogen) using oligo dT primer.
RT-PCR for pri-miR-194-2 entire transcript (Fig. 4C) was performed with the following primers, Forward primer was DTSS F, and reverse primer was; 5′-CATCCCAGCCACAGAGCATC-3′.
RACE analysis
5′ RACE was performed using GeneRacer kit (Invitrogen) according to manufacturer's protocol, except for the use of random primer in reverse transcription. PCR amplification of 5′ end of pri-miR-194-2 was performed by touchdown PCR by LA Taq (Takara) using GeneRacer 5′ primer and reverse primer 5′-CAGCAGGCATTTTGGGAGAC-3′. Amplified 5′ RACE fragments were cloned into pGEM T-Easy (Promega) for sequence analysis. 3′ RACE was also performed using GeneRacer kit (Invitrogen). PCR Amplification of the 3′ end of pri-miR-194-2 was also performed using the gene-specific forward primer 5′-TTCCTCCTTGTGCCCCGAAG-3′ and GeneRacer 3′ primer. Amplified 3′ RACE fragments were also cloned into pGEM T-Easy (Promega) for sequence analysis.
Vector constructions
miRNA expression vectors were constructed by cloning miRNA coding region into pcDNA3.1(−) (Invitrogen). miRNA coding regions were amplified by Phusion DNA polymerase (New England Biolab) from Caco-2 genomic DNA using primers as follows:
miR-194-1cluster F, 5′-ATACTCGAGTAGAACATGAATAAATCGAGAC-3′;
miR-194-1cluster R, 5′-TATGAATTCTTACTCAATACATTTACATGGTAG-3′;
miR-194-2cluster F, 5′-ATACTCGAGCCTGGGGCCACGAAGACTGG-3′; and
miR-194-2cluster R, 5′-ATAGGATCCGGGGAATGAGACAGAGGGAGG-3′.
miR-194-2 promoter deletion variants were amplified by the following primers and cloned into pGL3basic between XhoI-MluI sites. Cloning primers were as follows:
−1003 primer F, 5′-ATGCACGCGTATGTCACCACCAGGGGTCGC-3′;
−162 primer F, 5′-ATGCACGCGTCCTGATGCTTCCTGGACCCG-3′;
−70 primer F, 5′-ATGCACGCGTTGGGATAATCATTAACCACC-3′;
−51 primer F, 5′-ATGCACGCGTCACCAGGGGCTGGGTCAACG-3′;
+21 primer R, 5′-TCGACTCGAGTTCGGGGCACAAGGAGGAAG-3′; and
+358 primer R, 5′-TCGACTCGAGACTCAGCCTGGCGGCCCTTC-3′.
Scrambled mutation was induced using Scrambled F 5′-GATACTAACGTAAGCCACCAGGGGCTGGGT-3′ and Scrambled R 5′-ACGTTAGTATCATAGCCACCGTCAATACTG-3′. HNF-1α expression vector was constructed by cloning HNF-1α cDNA into pcDNA3 (Invitrogen). HNF-1α primers were 5′-ATGCAAGCTTGCCACCATGGTTTCTAAACTGAGCCAGC-3′ and 5′-TAATGAATTCTTACTGGGAGGAAGAGGCCA-3′.
Transfection and luciferase assay
Transfections were performed using LipofectAmine2000 (Invitrogen) according to the manufacturer's protocol. For examination of miRNA generation, miRNA expression vector, luciferase miRNA-sensor vector, and pRL-TK were mixed (10:9:1), and cotransfected into cells were cultured in 96-well plate. For examination of miRNA promoter activity, the HNF-1α expression vector, luciferase miRNA promoter vector, and pRL-TK were mixed (20:19:1) and cotransfected into cells cultured in 96-well plate. Luciferase activity was measured by the Dual luciferase assay kit (Promega).
ChIP assay
To cross-link chromatin, differentiated Caco-2 cells were treated with 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped by addition of 0.125 M glycine. After being washed twice with ice-cold PBS, cells were resuspended in NP-40 nuclear extraction buffer (10 mM HEPES at pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, and protease inhibitor) and centrifuged at 3000 rpm for 10 min. Crude nuclei were resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl at pH 8.0, and protease inhibitor), and extensively sonicated by Bioruptor. Sonicated chromatin was centrifuged at 15,000 rpm, and the supernatant was collected (input control). The supernatant was mixed with 9 vol of ChIP dilution buffer (11 mM Tris/HCl at pH 8.0, 154 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1.1% Triton X-100, 0.11% sodium deoxycholate, and protease inhibitors) and precleared with preblocked Protein G–Sepharose (GE Healthcare). Precleared chromatin was immunoprecipitated with 5 μg of anti-HNF-1α antibody (Santa Cruz, sc-6547) or 5 μg of normal goat IgG (Vector Laboratory, I-5000), and the immuno complexes were collected by preblocked Protein G–Sepharose. The beads were washed sequentially by RIPA, RIPA containing 500 mM NaCl, LiCl wash buffer, and twice by TE. Collected chromatin were eluted in ChIP elution buffer (1% SDS, 100 mM NaHCO3), adjusted to 200 mM NaCl, and incubated for at least 6 h at 65°C to reverse cross-link. After treatment with RNase A (Nippongene) and Proteinase K (Roche), DNA fragments were extracted by phenol/chloroform and ethanol precipitation. Quantitative PCR was performed with Quantitect SYBR PCR kit (Qiagen) using the following primers: miR-194-2 promoter ChIP primer F, 5′-TGATCAGTATTGACGGTGGTG-3′; primer R, 5′-AAGGAGGAAGTGGGGACAC-3′. Also used were negative control (exon3 of pri-miR-194-2) primer F, 5′-CCCACTGACCTGTGTCCTTT-3′; primer R, 5′-AGAGGGGTTGGAGGTGAGAC-3′. Detected PCR products were sequenced after cloning into pGEM T-Easy vector (Promega).
ACKNOWLEDGMENTS
This study was supported in part by grants-in-aid for Scientific Research, Scientific Research on Priority Areas, Exploratory Research, and Creative Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology; the Japanese Ministry of Health, Labor and Welfare; and the Japan Medical Association.
Footnotes
Abbreviations: miRNA, microRNA; SI, sucrase-isomaltase; kb, kilobases; LPH, lactase-phlorizin hydrolase; HNF-1α, hepatocyte nuclear factor-1 α; PCR, polymerase chain reaction; RT, reverse transcription; RACE, rapid analysis cDNA end; MEM, minimal essential medium; DMEM, Dulbecco's modified Eagle's Medium; PBS, phosphate buffered saline; FBS, fetal bovine serum.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.810208.
REFERENCES
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Boudreau F., Zhu Y., Traber P.G. Sucrase-isomaltase gene transcription requires the hepatocyte nuclear factor-1 (HNF-1) regulatory element and is regulated by the ratio of HNF-1α to HNF-1β. J. Biol. Chem. 2001;276:32122–32128. doi: 10.1074/jbc.M102002200. [DOI] [PubMed] [Google Scholar]
- Boudreau F., Rings E.H., van Wering H.M., Kim R.K., Swain G.P., Krasinski S.D., Moffett J., Grand R.J., Suh E.R., Traber P.G. Hepatocyte nuclear factor-1α, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J. Biol. Chem. 2002;277:31909–31917. doi: 10.1074/jbc.M204622200. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila . Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A.M., Byrom M.W., Shelton J., Ford L.P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A.R. Wnt signalling in the mouse intestine. Oncogene. 2006;25:7512–7521. doi: 10.1038/sj.onc.1210065. [DOI] [PubMed] [Google Scholar]
- Crosnier C., Stamataki D., Lewis J. Organizing cell renewal in the intestine: Stem cells, signals and combinatorial control. Nat. Rev. Genet. 2006;7:349–359. doi: 10.1038/nrg1840. [DOI] [PubMed] [Google Scholar]
- Esau C., Kang X., Peralta E., Hanson E., Marcusson E.G., Ravichandran L.V., Sun Y., Koo S., Perera R.J., Jain R., et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gregorieff A., Clevers H. Wnt signaling in the intestinal epithelium: From endoderm to cancer. Genes & Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Perlmutter D.H. Regulation of α1-antitrypsin gene expression in human intestinal epithelial cell line caco-2 by HNF-1α and HNF-4. Am. J. Physiol. 1999;276:G1181–G1194. doi: 10.1152/ajpgi.1999.276.5.G1181. [DOI] [PubMed] [Google Scholar]
- Hu C., Perlmutter D.H. Cell-specific involvement of HNF-1β in α(1)-antitrypsin gene expression in human respiratory epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;282:L757–L765. doi: 10.1152/ajplung.00271.2001. [DOI] [PubMed] [Google Scholar]
- Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Kato M., Zhang J., Wang M., Lanting L., Yuan H., Rossi J.J., Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGFβ-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Kim V.N. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.K., Lee Y.S., Sivaprasad U., Malhotra A., Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasinski S.D., Van Wering H.M., Tannemaat M.R., Grand R.J. Differential activation of intestinal gene promoters: Functional interactions between GATA-5 and HNF-1α. Am. J. Physiol. 2001;281:G69–G84. doi: 10.1152/ajpgi.2001.281.1.G69. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with “antagomirs.”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Meyer J., Borkhardt A., Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel D.B., Crabtree G.R. HNF-1, a member of a novel class of dimerizing homeodomain proteins. J. Biol. Chem. 1991;266:677–680. [PubMed] [Google Scholar]
- Mitchelmore C., Troelsen J.T., Sjostrom H., Noren O. The HOXC11 homeodomain protein interacts with the lactase-phlorizin hydrolase promoter and stimulates HNF1α-dependent transcription. J. Biol. Chem. 1998;273:13297–13306. doi: 10.1074/jbc.273.21.13297. [DOI] [PubMed] [Google Scholar]
- Olsen P.H., Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Sancho E., Batlle E., Clevers H. Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- Schroder O., Hess S., Caspary W.F., Stein J. Mediation of differentiating effects of butyrate on the intestinal cell line Caco-2 by transforming growth factor-β1. Eur. J. Nutr. 1999;38:45–50. doi: 10.1007/s003940050045. [DOI] [PubMed] [Google Scholar]
- Stanger B.Z., Datar R., Murtaugh L.C., Melton D.A. Direct regulation of intestinal fate by Notch. Proc. Natl. Acad. Sci. 2005;102:12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.M., Newman M., Parker J.S., Morin-Kensicki E.M., Wright T., Hammond S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wering H.M., Huibregtse I.L., van der Zwan S.M., de Bie M.S., Dowling L.N., Boudreau F., Rings E.H., Grand R.J., Krasinski S.D. Physical interaction between GATA-5 and hepatocyte nuclear factor-1α results in synergistic activation of the human lactase-phlorizin hydrolase promoter. J. Biol. Chem. 2002;277:27659–27667. doi: 10.1074/jbc.M203645200. [DOI] [PubMed] [Google Scholar]
- Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin-28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Klopot A., Freund J.N., Dowling L.N., Krasinski S.D., Fleet J.C. Control of differentiation-induced calbindin-D9k gene expression in Caco-2 cells by cdx-2 and HNF-1α. Am. J. Physiol. 2004;287:G943–G953. doi: 10.1152/ajpgi.00121.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wu G.D., Chen L., Forslund K., Traber P.G. Hepatocyte nuclear factor-1α (HNF-1α) and HNF-1β regulate transcription via two elements in an intestine-specific promoter. J. Biol. Chem. 1994;269:17080–17085. [PubMed] [Google Scholar]
- Zamore P.D., Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]