Abstract
Cotranslational synthesis of proteins into the endoplasmic reticulum is preceded by targeting of the translating mRNA once a signal peptide emerges from the ribosome exit tunnel. Many mRNAs, however, are unlikely to be targeted by this process because they encode proteins that do not contain a signal peptide or because they are too short to be recognized by the signal recognition particle. Herein we tested the possible involvement of the 3′-UTR in the localization of an mRNA that encodes a very short Saccharomyces cerevisiae protein (Pmp1). We found by ribosome density mapping, sedimentation analysis, differential centrifugation, and fluorescent in situ hybridization that the 3′-UTR is essential for the association of the transcript with membrane compartments. Fusion of the 3′-UTR to heterologous open reading frames conferred on them a sedimentation and cellular localization pattern resembling that of PMP1. Mutation analysis revealed that a repeating UG-rich sequence within the 3′-UTR is important for membrane association. Taken together, our results reveal an essential role for elements within the 3′-UTR in the localization of an mRNA that is likely to be ignored by the standard signal-dependant mechanism.
Keywords: 3′-UTR, localization, plasma membrane, velocity sedimentation
INTRODUCTION
Targeting of plasma membrane and secreted proteins to their final destinations involves an initial step of insertion into the endoplasmic reticulum (ER) that is directed by the signal peptide (Hegde and Bernstein 2006). This transport may occur post-translationally, i.e., after complete translation of the entire protein. In such a case, the protein is fully synthesized in the cytosol and is targeted to a translocator complex in the ER membrane (the sec61 complex and its accessory proteins) (Panzner et al. 1995) with the assistance of various chaperones (Bukau and Horwich 1998). Alternatively, protein insertion may take place through a cotranslational mechanism, where the protein is inserted into the ER while being translated. Such a mechanism may prevent the premature or ectopic functioning of the synthesized protein.
Translation of proteins that are inserted cotranslationally begins in the cytosol, where, upon emergence of the signal peptide from the ribosome, translation is arrested through binding of the signal recognition particle (SRP) to the ribosome and the nascent chain (Wolin and Walter 1989, 1993; Mason et al. 2000; Halic et al. 2006). The SRP is then recognized by the SRP receptor on the ER membrane, and the ribosome is docked onto the sec61 translocator. At this stage, translation resumes and the polypeptide is inserted through the translocator channel (for review, see Pool 2003). As an outcome of the cotranslational process, mRNAs encoding for proteins that are targeted by this mechanism are associated with the ER membranes. This has served as a useful way to identify proteins that are targeted by this pathway (Diehn et al. 2000, 2006; Lerner et al. 2003).
An essential step in cotranslational targeting of mRNAs is the recognition of the signal peptide as it emerges from the ribosome's exit tunnel. This necessitates a minimal length of the encoded protein, which probably differs between proteins, yet was shown to be of at least 50 amino acids (Ibrahimi et al. 1986; Siegel and Walter 1988; Okun et al. 1990; Jungnickel and Rapoport 1995). This minimal length is consistent with the size of the ribosome exit tunnel and the length of the signal peptide. Thus, the requirement for translation of at least 50 amino acids is probably to allow for sufficient contact between the protruding signal peptide and the binding groove of the SRP complex. It is still unclear by what mechanism mRNAs encoding shorter plasma membrane polypeptides are targeted to the ER. Such polypeptides are unlikely to interact sufficiently with the binding groove and may require regions outside the coding region for correct targeting.
Elements in noncoding regions are known to be important for localization of various mRNAs. Such elements are more prevalent within the 3′-UTR and are usually present at several positions along the UTR (Chabanon et al. 2004). They usually serve as binding sites for specific RNA-binding proteins (RBPs) that interact with specific motor proteins that transport the mRNA to its destination via the cellular cytoskeleton (Lopez de Heredia and Jansen 2004). In some cases, mRNAs may simply diffuse within the cell and interact with anchoring proteins at their target sites. This anchoring will lead to a gradual increase in the local concentration of the mRNA, and thus to an apparent localization (Forrest and Gavis 2003). In recent years, 3′-UTRs of mRNAs encoding for mitochondrial proteins were shown to have a role in localization (Corral-Debrinski et al. 2000; Marc et al. 2002; Margeot et al. 2002; Sylvestre et al. 2003). These observations are surprising in light of the fact that these mRNAs also encode for functional mitochondrial targeting sequences, which are sufficient to target the mRNAs in a cotranslational manner. The 3′-UTR may allow for better targeting of the protein into the mitochondria; indeed, replacement of the 3′-UTR of ATP2 mRNA with a 3′-UTR of a cytosolic mRNA (ADH1) led to the mislocalization of the mRNA and inefficient translocation of the Atp2 protein (Margeot et al. 2002).
The Saccharomyces cerevisiae Pmp1 is a small plasma membrane proteolipid that copurifies with the major H+-ATPase protein (Pma1) and was shown to enhance its H+-ATPase activity (Navarre et al. 1992, 1994). It is encoded by a very short open reading frame (ORF) (41 codons) and has a single trans-membrane domain. Importantly, its mRNA was shown to be associated with membrane-bound polysomes (Diehn et al. 2000), implying a cotranslational targeting process.
In this study, we investigated the role of the 3′-UTR of PMP1 in its subcellular localization. We show by sedimentation analysis and fluorescent in situ hybridization (FISH) that PMP1 transcripts with deletions within the 3′-UTR become mislocalized. Fusion of the 3′-UTR to heterologous ORFs conferred to them a sedimentation and localization pattern similar to that of PMP1. Further mapping of regions within the 3′-UTR revealed elements in the 3′-UTR that are required for its targeting function. Our results reveal a critical role for the 3′-UTR in localization of an mRNA that encodes for a short plasma membrane protein.
RESULTS
Velocity sedimentation analysis of PMP1 mRNA in sucrose gradients
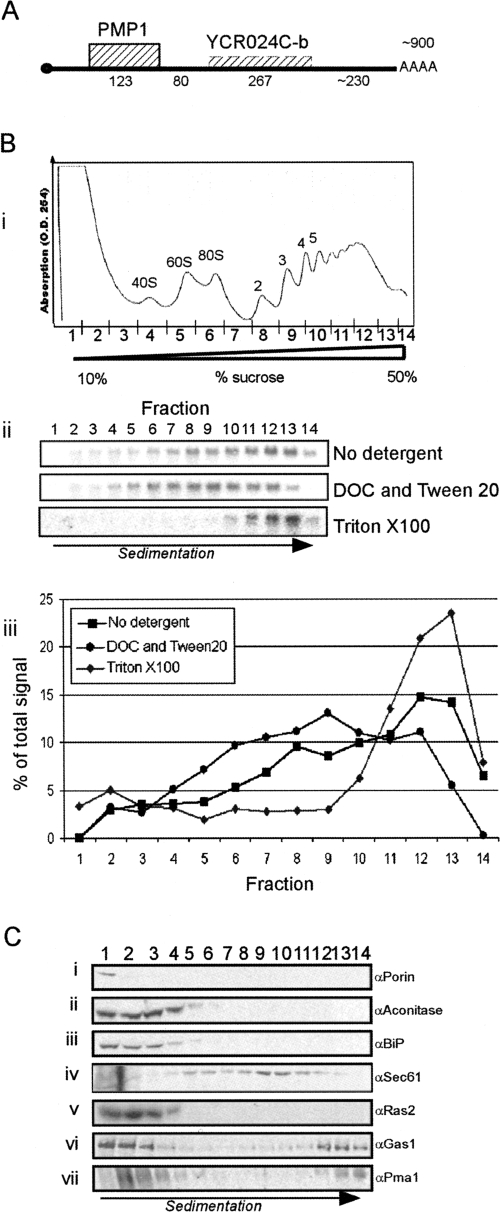
The PMP1 gene contains a short ORF (123 nucleotides [nt]) encoding for a small plasma membrane protein that is a regulatory subunit of the plasma membrane H+-ATPase (Navarre et al. 1994). The PMP1 mRNA was shown to be membrane-associated (Diehn et al. 2000) and was suggested to contain a relatively long 3′-untranslated region (3′-UTR) (∼600 nt) (Navarre et al. 1994). To confirm the length of the PMP1 3′-UTR, its mRNA was cleaved by RNase H and oligodeoxynucleotides (ODN) complementary to several regions downstream from the stop codon. The lengths of the reaction products were revealed by Northern analysis (Supplemental Fig. 1). The 3′-UTR of PMP1 appeared to be ∼600 nt, and the entire transcript was ∼900 nt long. The genomic sequence of S. cerevisiae in the Saccharomyces Genome Database (SGD) suggests the presence of a putative short ORF (YCR024C-B) 80 nt downstream from the PMP1 stop codon. Considering the length of the PMP1 3′-UTR, this putative ORF is included within it, consistent with recent observations (Fig. 1A; Miura et al. 2006).
FIGURE 1.
PMP1 mRNA sedimentation is related to membrane subdomains. (A) Schematic representation of the PMP1 transcript. Hatched boxes indicate ORFs, and numbers indicate lengths in nucleotides. (B) Sedimentation pattern of PMP1 mRNA. Cells grown in rich media (YPD) to mid-logarithmic phase were harvested and lysed in a lysis buffer either without any detergent, with deoxycholate and Tween 20, or with Triton X-100. Complexes were then separated by velocity sedimentation in 10%–50% linear sucrose gradient. (B, i) OD254 profile of a representative gradient from which the sedimentation positions of the various polysomal complexes can be deduced. These profiles looked the same under all treatments (not shown). (B, ii) Northern analysis for the sedimentation of PMP1 mRNA upon lysis without detergent, with DOC and Tween 20 or with Triton X-100. (B, iii) Quantification results of the northern signals. (C) Western blot analysis for polysomal fractions. Cells treated with lysis buffer containing Triton X-100 were fractionated as above, and proteins in each fraction were resolved by PAGE-SDS. Western blot analyses were performed using the indicated antibodies.
Yeast cells contain another protein (Pmp2p) that is almost identical to Pmp1p (Navarre et al. 1994). However, the transcripts of PMP1 and PMP2 are of different lengths and differ substantially in their UTRs (only 38.8% sequence identity). Under our hybridization conditions, the PMP1 probe does not cross-react with the PMP2 mRNA, and vice versa (Supplemental Fig. 1B).
To characterize the ribosomal association of the PMP1 mRNA in cells, we mechanically broke the cells in a lysis buffer devoid of detergent, removed large complexes by centrifugation for 5 min at 8000g, and performed velocity sedimentation analysis in sucrose gradients (Fig. 1B). The PMP1 transcript appeared to sediment in the heavy region of the gradient, corresponding to an association with eight ribosomes or more (Fig. 1B, ii, “No detergent”). Such a high association with ribosomes is unlikely for an ORF of this size (123 nt) because the ribosome footprint is ∼40 nt (Wolin and Walter 1988).
Velocity sedimentation is routinely used to differentiate complexes based on size rather than density. Since the PMP1 mRNA is associated with membranes, we hypothesized that the heavy sedimentation was due to its association with large ribosome–membrane complexes (e.g., microsomes), which led to a dragging of the transcript to this position. To test this, we used conditions that are used to release membrane-bound polysomes from membranes (Stoltenburg et al. 1995; Diehn et al. 2000; see also Fig. 5 below). Following the mechanical breakage and removal of large complexes, extracts were incubated with detergents (0.2% deoxycholate [DOC] and 0.5% Tween 20) to release membrane-bound polysomes. The released complexes were then analyzed by velocity sedimentation. This treatment did not significantly change the overall OD254 polysomal profile or the sedimentation pattern of a nonmembrane mRNA (data not shown), yet led to a strong shift in the sedimentation of the PMP1 mRNA, with a peak signal in fraction 9 (Fig. 1B, ii, “DOC and Tween 20”). Fraction 9 represents the association with about three ribosomes, which is a reasonable number for the PMP1 ORF. Thus, PMP1 is associated with about three ribosomes, yet when bound to membranes it sediments as a much heavier complex. Interestingly, a commonly used extraction procedure (Kuhn et al. 2001; Arava et al. 2003), which uses 1% Triton X-100 in the lysis buffer, resulted in an increase in the amount sedimenting in the heavy region of the gradient (Fig. 1B, ii, “Triton X100”). This might be because cold Triton X-100 is known to stabilize, rather than dissolve, membrane subdomains known as detergent insoluble granules (DIGs) or lipid rafts. These stabilized particles, which in yeast start to assemble in the ER (Bagnat et al. 2001), might therefore sediment in the heavy region of the gradient with their associated polysomal mRNAs. Interestingly, the protein that interacts with Pmp1p (Pma1p) is a well-established marker of lipid rafts (Bagnat et al. 2001; Gong and Chang 2001; Malinska et al. 2003). Consistent with this, Western analyses for fractions collected along the gradient using various protein markers revealed that some of the lipid raft markers, Pma1 and Gas1, sediment in the heavy fractions of the gradient (Fig. 1C, vi,vii). The heavy sedimentation of lipid rafts under these experimental conditions is probably because they are associated with many polysomal mRNAs (not only PMP1 mRNA), which generates a very large complex. All further velocity sedimentation in sucrose gradient assays were performed in the presence of Triton X-100 because of the higher signals in the heavy fractions compared to those without detergent.
FIGURE 5.
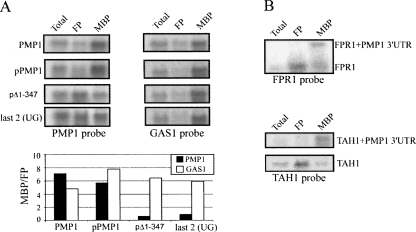
Effect of the 3′-UTR on the sedimentation of mRNAs as membrane-bound polysomes. (A) Cells expressing PMP1 from its genomic loci (PMP1), from a plasmid (pPMP1), or PMP1 with a deletion in its 3′-UTR (pΔ1-347) or point mutations within the 3′-UTR [two (UG) elements within the 3′-UTR, “last 2 (UG)”] were subjected to a protocol aimed at separation of free polysomes (FP) from membrane-bound polysomes (MBP) (Stoltenburg et al. 1995; Diehn et al. 2000). Equal amounts of unfractionated RNA (Total), FP, and MBP were resolved in a gel, and the localization of PMP1 mRNA or the control GAS1 mRNA was determined by hybridization with the appropriate probes. Histograms present the ratio between signals in the MBP, and the FP is presented for (closed bars) PMP1 variants and (open bars) GAS1 mRNA. (B) Cells expressing the fusion mRNAs FPR1-PMP1 3′-UTR or TAH1-PMP1 3′-UTR were subjected to fractionation analysis as in A, and a sample of each fraction was analyzed by Northern analysis with a probe for (top) FPR1 or (bottom) TAH1.
To exclude the possibility that the sedimentation of PMP1 was unrelated to ribosomes or to a translational process, we extracted the cells in a buffer either lacking or containing EDTA (Supplemental Fig. 2A). The EDTA treatment led to a strong shift in PMP1 sedimentation, as most of it sedimented in fractions 5 and 6, and as free of assembled ribosomes. In addition, cells were subjected to glucose deprivation for a short period, which is known to lead to immediate translational arrest (Ashe et al. 2000). This treatment led to a shift in PMP1 sedimentation to the lighter fractions, as associated with one ribosome (Supplemental Fig. 2B). The results of the EDTA treatment and the glucose deprivation experiment strongly suggest that the heavy sedimentation of PMP1 during optimal growth is related to the overall translation status.
PMP1 with premature stop codons sediments in heavy fractions
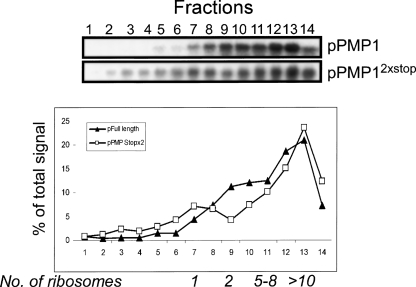
To determine if the sedimentation of PMP1 is dependent on its own translation, we used a yeast strain with both PMP1 and PMP2 genes deleted (kindly provided by Andre Goffeau, Catholic University of Louvain, Belgium) and transformed it by using either a plasmid expressing the natural PMP1 ORF or PMP1 with two consecutive stop codons inserted three codons downstream from the start codon (pPMP12xstop). Both constructs included the PMP1 promoter and untranslated regions. Sedimentation patterns of these two plasmid-encoded transcripts were analyzed by standard polysomal analysis (Fig. 2). Most transcripts of the normal PMP1 appeared in the heavy fractions, similar to the transcript expressed from its genomic loci (Fig. 1). pPMP12xstop also sedimented mainly in the heavy region of the gradient, similar to pPMP1 sedimentation. Thus, PMP1 mRNA sediments in the heavy region of the gradient even when its own translation is hindered, provided that the global translation is not impeded. This is consistent with the suggestion that the membrane compartments sediment in the heavy fractions because they are associated with many polysomal mRNAs; these polysomal mRNAs are affected by EDTA or glucose treatments, yet are not affected when only PMP1 mRNA is mutated.
FIGURE 2.
Effect of premature stop codons on PMP1 sedimentation. A yeast strain deleted of PMP1 and PMP2 genes was transformed with a plasmid expressing the entire PMP1 mRNA (pPMP1) or a plasmid expressing the same mRNA but with two consecutive stop codons at the beginning of PMP1 ORF (pPMP12xstop). Cell lysis and polysomal analysis were performed as in Figure 1, in the presence of Triton X-100.
A small peak of pPMP12xstop signal appeared in fractions 7 and 8, in the sedimentation position of mRNAs associated with a single ribosome. The mutations in pPMP12xstop were inserted three codons downstream from the start codon, thereby allowing for translation initiation and ribosome assembly at the start codon. Therefore, the peaks in fractions 7 and 8 represent pPMP12xstop transcripts with a single ribosome. These mRNAs probably escaped the membrane-targeting process, either due to the higher expression levels from a plasmid or because translation of the entire PMP1 protein has some contribution to targeting.
The heavy sedimentation of PMP1 is mediated by its 3′-UTR
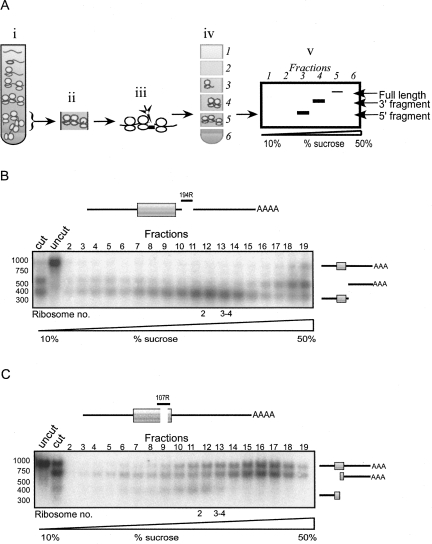
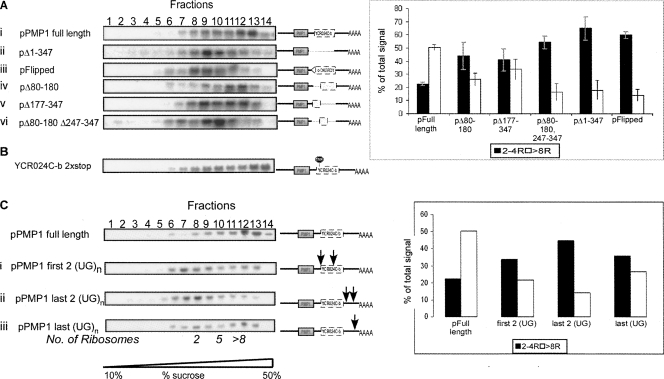
To test whether the 3′-UTR contributes to the heavy sedimentation of PMP1, we performed a ribosome density mapping (RDM) experiment (Arava et al. 2005), in which a sample of the heavy region of the gradient was collected and mixed with RNase H and antisense ODN complementary to the region near the PMP1 stop codon (see the scheme of the procedure in Fig. 3A). This led to cleavage of the PMP1 transcript into two fragments of distinct lengths, a 5′ fragment that contained the 5′-UTR and the ORF and a 3′ fragment that contained only the 3′-UTR. The cleavage products were then separated on a sucrose gradient, and Northern analysis with a probe recognizing PMP1 was performed to determine the sedimentation position of the cleavage products (Fig. 3B,C).
FIGURE 3.
Sedimentation pattern of the ORF or 3′-UTR of PMP1. (A) Scheme of the RDM procedure aimed at determining the sedimentation of different regions of the mRNA. (A, i) Yeast cells are collected and separated on a sucrose gradient. (A, ii) The fraction that contained the majority of the mRNA of interest (heavy fractions in the case of PMP1 mRNA) is isolated. (A, iii) RNase H and oligodeoxynucleotide (ODN) are added to the sample in order to cleave the PMP1 mRNA at the site of interest. (A, iv) The cleavage products are separated on a sucrose gradient, and multiple fractions are collected (six in the scheme). (A, v) The collected fractions are subjected to Northern blot analysis in order to determine the sedimentation positions of the cleavage products (that are identified according to their length). Usually not all transcripts are cleaved by RNase H. These uncleaved transcripts sediment in Step iv at the same position as in Step i. (B) RDM analysis of PMP1. The heavy fraction of a sucrose gradient was isolated and subjected to cleavage with RNase H and ODN complementary to the region 194 nt downstream from the start codon (indicated schematically in the cartoon). Cleavage reactions were separated on a sucrose gradient, and 19 fractions were collected. The indicated fractions were subjected to Northern analysis, and the sedimentation position of each cleavage product was determined using a radioactive probe. Cartoons to the right indicate the migration position of the two cleavage products and the uncut, full-length transcript. “Uncut” lanes indicate samples that were collected but were not mixed with RNase H or ODN. “Cut” lanes indicate samples that were collected and mixed with RNase H and ODN, but were not separated on a sucrose gradient. (C) RDM analysis is similar to the analysis presented in B, but with a different ODN added to the cleavage reaction.
Following cleavage of PMP1 at ∼194 nt downstream from the start codon, the fragment that contained the ORF sedimented mostly in fractions 11–14, which is consistent with the association of 2–4 ribosomes, whereas the 3′-UTR fragment sedimented in the heavy region of the gradient (Fig. 3B, fractions 17–19). To obtain a better resolution of the sedimentation of the 3′-UTR, we repeated the RDM assay with another ODN and a slightly shorter sedimentation time (Fig. 3C). Again, the fragment containing the ORF sedimented in fractions that corresponded to association with 2–4 ribosomes, and the 3′-UTR fragment sedimented at the heavy region of the gradient. Similar results were obtained with cleavage at position 166 (data not shown).
In each RDM reaction, there appeared to be some remnants of full-length PMP1 that were not cut by RNase H and ODN (Fig. 3B,C), probably due to structural hindrance or the presence of proteins on some transcripts. The sedimentation position of these full-length mRNAs served as an “in-gradient” marker for the normal sedimentation of the full-length PMP1. Comparison of the sedimentation of the cleavage products to the full-length transcripts revealed that the sedimentation of the fragment that contains the ORF differed from the full-length transcript, while the 3′-UTR fragment has a sedimentation pattern identical to the full-length transcript. From this analysis, we conclude that the PMP1 ORF is associated with 2–4 ribosomes, and its apparent heavy sedimentation (i.e., membrane association) occurs only when it is associated with its 3′-UTR.
PMP1 3′-UTR affects the sedimentation of unrelated ORFs
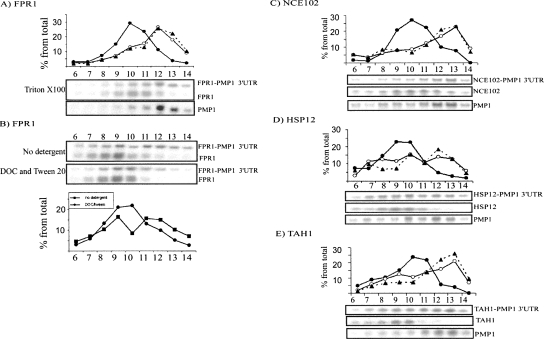
To examine if the PMP1 3′-UTR can affect the sedimentation of heterologous ORFs, we generated several fusion constructs in which the 3′-UTR of several unrelated mRNAs was replaced by the PMP1 3′-UTR (Fig. 4). These mRNAs included FPR1, HSP12, NCE102, and TAH1, each cloned with 500 nt upstream of its start codon, presumably containing the promoter and 5′-UTR. As shown in Figure 4, all fusion transcripts were in heavier complexes than their normal counterparts. This is easily seen in the strain containing both the normal FPR1 transcript and the one fused to the PMP1 3′-UTR, which are expressed at similar levels and are of distinct lengths, and therefore can be seen on the same image. The normal FPR1 sediments as associated with about three ribosomes, and the fused transcript sediments in the heavy fractions of the gradient (Fig. 4A). The heavy sedimentation of the FPR1-PMP1 3′-UTR is probably due to an association with membrane compartments since the treatment of the sample with DOC and Tween 20 led to a shift in the sedimentation of the heavy-sedimenting complex to lighter fractions (Fig. 4B). The analysis of all other mRNAs revealed in all cases that transcripts with the PMP1 3′-UTR sedimented significantly heavier than transcripts with their normal 3′-UTRs. The difference in sedimentation was not merely due to the difference in the length of the mRNA, as the transcript of the NCE102-PMP1 3′-UTR is of similar length to the natural NCE102 (∼1100 nt) (Fig. 4C). In all cases, the sedimentation of an unrelated mRNA (ADK1) was tested and was found to be similar (data not shown). Importantly, Northern blot analyses with a probe recognizing PMP1 confirmed that the mRNAs with the 3′-UTR of PMP1 sediment in the same fractions where PMP1 mRNA was found (Fig. 4). Thus, the PMP1 3′-UTR can shift the sedimentation of unrelated ORFs to a heavy region of the gradient.
FIGURE 4.
Sedimentation analysis of PMP1 3′-UTR fused to heterologous ORFs. Plasmids expressing the 3′-UTR of PMP1 cloned downstream from the ORFs of (A,B) FPR1, (C) NCE102, (D) HSP12, or (E) TAH1 were transformed into (A,B,E) wild-type cells or into (C,D) deletion strains. The cells were grown in selective media, subjected to polysomal separation on sucrose gradients, and resolved into 14 fractions. The sedimentation position of the mRNAs indicated to the right of each panel was determined by Northern analysis. In A and B, the expression levels and lengths of the mRNAs allowed for simultaneous detection of the fusion and natural transcripts on the same blot. In C–E, the sedimentation of the natural transcript was determined by similar analysis of wild-type cells. In the quantification graphs for panels A and C–E, the closed circles indicate signals of the normal transcripts, open circles represent the signals of the fusion transcripts, and triangles connected by dashed lines represent the sedimentation of the PMP1 transcript. In B, closed squares or circles represent the signals of FPR1-PMP1 3′-UTR in the assay without detergent or with DOC and Tween 20, respectively.
Further establishing the role of the 3′-UTR in membrane association
To substantiate the role of the 3′-UTR in membrane association, we used a protocol that allows for reliable isolation of membrane-bound polysomes (Stoltenburg et al. 1995; Diehn et al. 2000). This method employs several steps of differential centrifugation to free membrane-bound polysomes. The membrane-bound polysomes are then released from the membranes by treatment with DOC and Tween 20, and the unreleased material is removed by centrifugation at 20,000g for 20 min. By this method, the majority of PMP1 transcript, either expressed from its genomic loci (PMP1) or from a plasmid (pPMP1), appeared mostly in the fraction of membrane-bound polysomes (Fig. 5A). However, PMP1 transcript with a large deletion in its 3′-UTR (complete deletion of YCR024C-B, pΔ1–347; all numbers are relative to the PMP1 stop codon) appeared mostly in the free polysome fraction. The shift to the free polysomes is specific to the mutated PMP1, as is apparent from the analysis of another membrane-associated mRNA (GAS1).
Similar analyses for mRNAs in which the PMP1 3′-UTR was fused to an ORF that encoded a cytosolic protein (FPR1 or TAH1) revealed that the signal for the PMP1 3′-UTR-containing mRNA was mostly in membrane-bound polysomes, while the normal transcript was with free polysomes (Fig. 5B). Thus, it was also found to be the case by this assay that the PMP1 3′-UTR is effective when merged to another ORF.
The membrane distribution of PMP1 mRNA was also tested by Concanavalin A treatment (Con A) (Fig. 6; Kaiser et al. 2002). In this treatment, the plasma membrane was coated by Con A and precipitated by relatively low centrifugation [3000g, (P3)]. The rest of the membranes were then precipitated by high-speed centrifugation [100,000g (P100)] from the soluble material (S100). This protocol does not require the use of EDTA, therefore polyribosomes are not affected. As can be seen in Figure 6, PMP1 mRNA fractionated in the P3 and P100 fractions and not in the S100 fraction. This pattern was similar to the fractionation of an ER marker (Fig. 6, Sec63-RFP) and different from a plasma membrane marker (Fig. 6, Pma1-GFP). This further indicates that PMP1 mRNA is associated with cellular membranes, most likely the ER.
FIGURE 6.
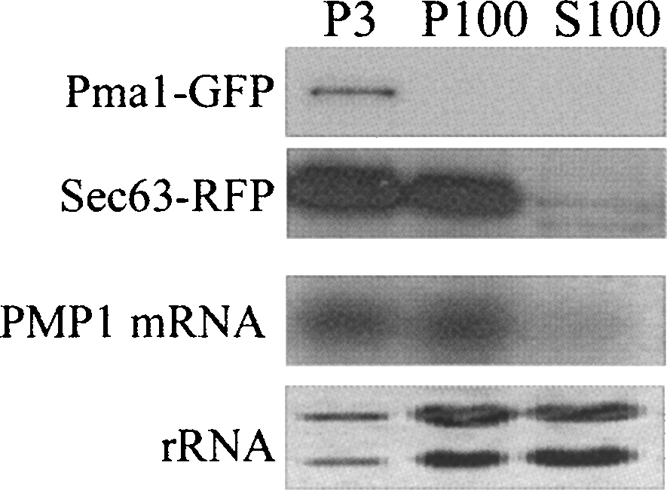
Fractionation of cellular membranes following Con A treatment. Yeast cells expressing Pma1-GFP and Sec63-RFP were converted to spheroplasts, mixed with Con A, and lysed, and Con A aggregates were collected by centrifugation at 3000g (P3). The supernatant was subjected to a high-speed centrifugation (100,000g) and separated into pellet (P100) and supernatant (S100). Protein samples from each fraction were taken for Western analysis using antibody to a plasma membrane marker (PMA1-GFP) or ER marker (Sec63-RFP). RNA samples were taken for Northern analysis using a probe for PMP1 mRNA or stained with methylene blue to visualize the rRNA.
The 3′-UTR is involved in cellular localization
To further test the role of the 3′-UTR in the compartmentalization of the PMP1 transcript, we analyzed its cellular localization by FISH. Cells deleted of the PMP1 and PMP2 genes and expressing PMP1 mRNA from a plasmid were subjected to FISH with a Cy3-labeled ODN complementary to a region from within the PMP1 ORF (Fig. 7A) or from within the 3′-UTR (Fig. 7B). A clear signal appeared at the cell periphery, which was absent in the control hybridizations of cells deleted of PMP1 or when a probe recognizing a mitochondria-localized mRNA was employed (Fig. 7E,F, respectively). In yeast, a substantial amount of the cellular ER is localized within the vicinity of the plasma membrane (hence termed “cortical ER”) (Prinz et al. 2000; Fehrenbacher et al. 2002). The FISH signals may suggest association of PMP1 mRNA with this compartment.
FIGURE 7.
Cellular localization of PMP1 variants by FISH. Yeast cells deleted of PMP1 and PMP2 genes were transformed with plasmids expressing either (A,B) full-length PMP1 transcripts, (C,D) the indicated deletions from within the 3′-UTR, (E) not transformed with a plasmid, or (H,I) the indicated fusion transcripts. Cells were subjected to fluorescent in situ hybridization with Cy3 labeled probes (red) complementary to either (A,C–E) the ORF region of PMP1 or (B,H,I) its 3′-UTR. (F) FISH analysis using a Cy3 labeled probe for an unrelated mRNA (ATM1). (G) Northern analysis to determine the steady-state mRNA levels obtained from the different plasmids. (−) Cells without any plasmid. Cells were costained with DAPI (blue) and mixed with cells expressing Pma1-GFP as a marker for the plasma membrane.
Importantly, the localization patterns in strains expressing PMP1 with a complete deletion of the YCR024C-B region of the 3′-UTR (Fig. 7C), or with partial deletions within it (Fig. 7D), were mostly cytoplasmic, displaying a clearly different localization when compared to the full-length mRNA. Northern blot analysis indicated that these localization differences cannot be attributed to lower expression of the modified mRNAs (Fig. 7G). These results indicate that the 3′-UTR of PMP1 is important for the localization of the transcript to the cell periphery.
FPR1 and TAH1 genes encode cytosolic proteins; therefore, their transcripts are expected to be in the cytoplasm (yet we were unable to obtain satisfactory FISH data for these). FISH analysis for the localization of FPR1 or TAH1 ORFs linked to PMP1 3′-UTR revealed a clear signal for these fusion transcripts in the cell periphery (Fig. 7H,I). This suggests that the PMP1 3′-UTR can affect the cellular localization of an unrelated ORF.
Mapping elements within the 3′-UTR
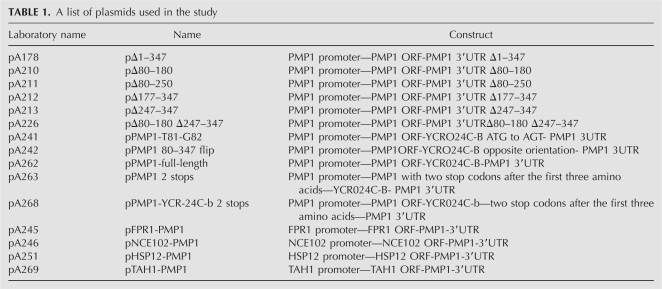
To narrow down the region within the PMP1 3′-UTR that is important for its heavy sedimentation, we generated a series of constructs carrying partial deletions and point mutations within the 3′-UTR. These constructs were inserted into a yeast strain deleted of the PMP1 and PMP2 genes, and the sedimentation of the transcript on sucrose gradients was determined.
The deletions within the 3′-UTR included a complete deletion of YCR024C-B (Δ1–347; all numbers are relative to the PMP1 stop codon) and partial internal deletions. In addition, we flipped the region that contains YCR024C-B, thus changing the sequence context without affecting the length. The sedimentation analysis of these mRNAs usually reveals a bi-modal sedimentation pattern, with mRNAs that sediment in fractions that correspond to association with 2–4 ribosomes, and mRNAs that sediment in the heavy fractions, as associated with many ribosomes (more than eight ribosomes). Based on the results presented in Figures 1–3, we considered PMP1 mRNAs that sedimented in the 2–4 ribosomes fractions as non-membrane-associated polysomes (i.e., free polysomes) and the mRNAs that sedimented in the heavy fractions as membrane-associated polysomes. The analyses presented in Figure 8 revealed that all deletions led to a decrease in the amount of PMP1 that sedimented in the heavy fractions (fractions 12–14) and an accumulation of transcripts in the fractions representing association with two to four ribosomes. It should be noted that sedimentation profiles may differ slightly between preparations due to biological or experimental variation. Because of this variation, the number of ribosomes that sediment in a particular fraction may change from one gradient to another. We therefore determined the relevant fractions for quantification (i.e., 2–4 ribosomes and more than eight ribosomes) by examining the OD254 profile of each experiment. Overall, relatively small deletions (pΔ80–-180 [Fig. 8A, iv], pΔ177–-347 [Fig. 8A, v], and pΔ80–250 or pΔ247–347 [data not shown]) had milder effects on the sedimentation, and ∼20% of the transcripts shifted from the heavy fraction to fractions representing association with two to four ribosomes (results are the averages of three independent experiments). Larger deletions (Fig. 8A, vi, pΔ80–180 Δ247–347) and reversal of YCR024C-B (Fig. 8A, iii) resulted in a much stronger effect (∼40% shift). This deletion analysis indicates that the apparent heavy sedimentation of the PMP1 transcript is due to elements spread along the 3′-UTR.
FIGURE 8.
Mapping elements within PMP1 3′-UTR that are important for its heavy sedimentation. Cells deleted of PMP1 and PMP2 were transformed with plasmids expressing various PMP1 transcripts. The sedimentation pattern of the various transcripts was determined as presented in Figure 1. (A) Northern blot results of the full-length transcript (A, i), various deletions, and a flip of the region between nucleotides 80 and 347 (the YCR024C-B region). The histogram presents the quantification results of signals in the fractions that contained (closed bars) two to four ribosomes and (open bars) eight or more ribosomes from at least three repeated experiments. (B) Insertion of two consecutive stop codons immediately downstream from the YCR024C-B start codon. (C) Mutagenesis of the UG-repeat into the AC-repeat. The histogram presents the quantification results of at least two repeats.
To test the role of the putative ORF (YCR024C-B) in the heavy sedimentation, we inserted two consecutive stop codons downstream from its start codon. As can be seen, the effects of these mutations on sedimentation were minor (Fig. 8B). A similar small effect was observed upon changing the start codon of this ORF to AGT (data not shown). These results suggest that translation of this ORF has little if any role in the sedimentation of PMP1. It should be noted that recent re-sequencing of all yeast ORFs revealed a nucleotide deletion at position 144 of YCR024C-B that led to a frameshift and substantial shortening of the reading frame (Hu et al. 2007).
The 3′-UTR was scanned for the presence of sequence motifs that might serve as binding sites for trans-acting factors. A sequence comprised of multiple repeats (four or more) of UG dinucleotide (UG)n appeared at four sites along the 3′-UTR, starting at positions 83, 261, 350, and 401 downstream from the stop codon. A similar sequence (UG/AUG/AUG/A) is implicated in the regulation of oskar mRNA, by serving as binding sites for Bruno (Kim-Ha et al. 1995). In addition, this sequence also appears at multiple sites within the 3′-UTR of the PMP2 mRNA, which also shows a heavy sedimentation pattern (Supplemental Fig. 1).
We mutated either the first (83 and 261) or the last two (UG)n sequences (positions 350 and 401) into AC repeats and tested the effects on PMP1 sedimentation (Fig. 8C). About 30% of the transcripts shifted their sedimentation to lighter fractions following mutations in positions 83 and 261, and a much larger change in sedimentation occurred following the mutation in the (UG)n elements at positions 350 and 401, with ∼50% PMP1 transcripts changing their sedimentation. Mutating only one UG motif (in position 401) (Fig. 8C) led to a much smaller effect, consistent with a necessity for multiple elements along the 3′-UTR.
To confirm that these mutations affected the membrane association of PMP1, we tested the fraction of membrane-bound polysomes of the transcript with the mutated last two UG-repeats by the method described by Stoltenburg et al. (1995) (Fig. 5). Indeed, this mutation led to a significant change in the amount of mRNA that appeared as membrane-bound, similar to the effect that occurred after complete deletion of a large region of the 3′-UTR (Fig. 5).
DISCUSSION
In agreement with previous work (Diehn et al. 2000), our study reveals that the PMP1 mRNA is associated with membrane compartments, probably the ER. Our data, obtained by several independent approaches, suggests that the 3′-UTR is essential for this association. First, we observed that the ORF region of PMP1, when cleaved of its 3′-UTR, sediments as associated with 2–4 ribosomes and not in a region of the gradient where large membrane-associated complexes sediment (Fig. 3). Second, PMP1 with a deletion within its 3′-UTR was isolated with cytosolic polysomes, while the normal PMP1 transcript was highly associated with membrane-bound polysomes (Fig. 5). Third, FISH analysis revealed that the PMP1 transcript was normally associated with the cell periphery, whereas deletions within the 3′-UTR resulted in a diffuse signal across the cytosol (Fig. 7). Finally, deletions and point mutations within the 3′-UTR led to a significant change in the sedimentation of the mRNA in a sucrose gradient, but insertion of two consecutive stop codons into the PMP1 ORF had almost no effect (Figs. 2, 8). Together, these findings indicate that the PMP1 transcript is localized in a membrane compartment that is in the cell periphery (probably the cortical ER), and this localization requires the 3′-UTR. It appears from our data that the membrane association of PMP1 is not essential for its translation because PMP1 transcript that is free of membranes (either because of detergent treatment or mutations in the 3′-UTR) sediments in sucrose gradient as associated with ribosomes.
The sedimentation analyses of the various PMP1 mutants usually reveal a bi-modal sedimentation pattern, with mRNAs associated with 2–4 ribosomes, and membrane-associated mRNAs that sediment in the heavy region of the gradient. The heavy sedimentation of the membrane-associated transcripts is probably because these specific membrane compartments are associated with multiple polysomal mRNAs (PMP1 and probably other mRNAs). We reason (based on Figures 1–3) that the PMP1 ORF is associated with 2–4 ribosomes, and the transcripts that sediment in this fraction of the gradient were released from their membrane association due to the mutations in the 3′-UTR. It appears from our data that large deletions or multiple mutations are necessary to confer a strong effect on sedimentation (i.e., significant shift from the heavy fractions to the 2–4 ribosome fraction) (Fig. 8). This may suggest the involvement of multiple binding sites from throughout the 3′-UTR (and probably several trans-factors) in targeting PMP1 to the membrane compartment.
The best studied pathway for targeting mRNA translation to the ER is the pathway mediated by the signal recognition particle (SRP). This targeting pathway involves the recognition of the nascent signal peptide as it emerges from the ribosome by the SRP54 subunit of SRP, which causes translation arrest and shuttling of the mRNA–ribosome–SRP complex to the SRP receptor at the ER. Translation then resumes, and the protein integrates into the ER and is transported to the appropriate compartment. Various studies attempted to identify the minimal length of a peptide that is needed to be synthesized in order to allow for SRP-dependant targeting (Ibrahimi et al. 1986; Siegel and Walter 1988; Wolin and Walter 1988, 1989; Okun et al. 1990; Wolin and Walter 1993; Jungnickel and Rapoport 1995). While lengths may vary depending on the protein and the experimental system, it appears that a minimal length of about 50 amino acids is necessary for proper targeting (Okun et al. 1990; Jungnickel and Rapoport 1995). PMP1 ORF, however, is only 41 codons long, which may explain the necessity for an alternative mechanism for its localization.
The deletion analysis revealed that a large region of the 3′-UTR is necessary for full activity (Fig. 8), as is common for many regulatory elements within the 3′-UTR (Chabanon et al. 2004). This may indicate the presence of multiple sites for RNA-binding proteins along the 3′-UTR, or that a large structural element is involved. We have identified a repeating UG-rich sequence that is located at the 3′-UTR of PMP1 and is important for its sedimentation. A similar sequence is present at the 3′-UTR of oskar mRNA and serves as a binding site for the Bruno protein (Kim-Ha et al. 1995). Interestingly, it was shown recently that a region containing this sequence (the AB BRE region) is capable of inducing oligomerization of oskar mRNA into large silencing complexes, thereby affecting its apparent sedimentation (Chekulaeva et al. 2006). Although there is no homolog for Bruno in yeast, it is possible that a related protein binds this sequence and leads to its heavy sedimentation. While our data suggest an involvement of specific membrane subdomains in the heavy sedimentation, we cannot exclude the possibility that oligomerization of PMP1 transcript also contributes to this sedimentation.
We have also identified a repeating UG-rich element in the 3′-UTR of PMP2, another mRNA that sediments in the heavy region of the gradient, which might suggest the involvement of this motif in targeting mRNAs encoding short plasma membrane polypeptides. However, a genome-wide search for the presence of the sequence UGUGUGUG (allowing one error) within all the 3′-UTRs of S. cerevisiae (Hurowitz and Brown 2003; Shalgi et al. 2005) identified more than 200 genes, with no significant enrichment to any specific group of genes. This indicates that additional elements are necessary in order to differentiate the PMP 3′-UTRs from the rest of the genes. To identify such elements, we performed an unbiased search for stable structural elements that contain any conserved sequence motif in their exposed loop (Robins et al. 2005; Wexler et al. 2007). Windows of 90 nt along the PMP1 3′-UTR were folded using the Vienna package folding tool (Hofacker 2003), and any 9-mers that appeared in exposed loops of any of the optimal structures or any of the top suboptimal structures (with a deviation of up to one kilocalorie from the minimal free energy [MFE]) were aligned (allowing one error) with exposed sequences that appear in all other yeast 3′-UTR sequences (∼6000 3′-UTRs) (Shalgi et al. 2005). The only exposed sequence within a stable hairpin structure (MFE < 20 Kcal) that was conserved between the 3′-UTRs of PMP1 and PMP2 (and three other genes of an unknown function) was “UGUGUGUGA.” Moreover, additional UG-rich subsequences appeared at multiple sites within the 3′-UTRs of PMP1 and PMP2, and the majority of them were also structurally exposed in MFE-predicted hairpins (although their structures are less stable than the original, strict MFE threshold we had applied in the genome-wide search). Thus, it might be that structure plays an important role in exposing UG-rich motifs and allowing their interaction with specific RNA-binding proteins. It remains to be determined experimentally whether these structural predictions are biologically relevant.
Perspective
In the present study, we have used the PMP1 transcript as a handle to identify unique cellular processes. We identified that the 3′-UTR is sufficient to target PMP1 and other ORFs to membrane-associated compartments. Involvement of untranslated regions in membrane targeting might be important for mRNAs encoding very short membrane proteins that cannot be recognized by the SRP, or the recently identified ER-associated mRNAs encoding for cytosolic proteins, which do not have a signal peptide (Lerner et al. 2003; Diehn et al. 2006). We have also identified a repetitive sequence that is involved in PMP1 sedimentation, and we speculate that this sequence is part of a larger structural element. The protein factors that are likely to bind this motif, as well as the importance of the structural features, are yet to be determined.
MATERIALS AND METHODS
Plasmid construction
Each plasmid (Table 1) was created using a PCR amplification reaction and specific primers, with either genomic DNA or other plasmids as the template. The PCR products were first cloned into p-GEM T easy vector (Promega) and then cut with the appropriate restriction enzymes and ligated into the multi-cloning site of pRS415 (Sikorski and Hieter 1989). All clones were verified by sequencing.
TABLE 1.
A list of plasmids used in the study
Yeast strains and growth conditions
The strain BY4741 was used in all polysomal, RDM, and Western blot analyses. The various constructs of PMP1 were inserted into strain W3031b deleted for PMP1 and PMP2 genes (a generous gift from Andre Goffeau at the Catholic University of Louvain, Belgium) (Table 2). All plasmid-containing strains were grown on SD (1.7 g/L Yeast Nitrogen Base, 5 g/L ammonium sulfate, 20 g/L glucose) with the following additions: 350 mg/L threonine, 40 mg/L methionine, 40 mg/L adenine, 50 mg/L lysine, 20 mg/L histidine, 50 mg/L tryptophan, and 20 mg/L uracil.
TABLE 2.
Yeast strains used in this study
Velocity sedimentation analysis
Velocity sedimentation analysis was performed as previously described with minor modifications (Arava 2003). Briefly, a 50 mL culture of yeast was harvested by centrifugation (4000 rpm, 4 min, 4°C) in the presence of 100 μg/mL cycloheximide. Following two washes in 4 mL of lysis buffer (20 mM HEPES at pH 7.4, 140 mM KAc, 1.5 mM MgAc, 0.5 mM dithiothreitol, 100 μg/mL cycloheximide, and 1 mg/mL heparin), cells were resuspended in 400 μL of lysis buffer, transferred to a screw-capped microfuge tube, supplemented with 1 mL of chilled glass beads, and lysed by vigorous vortexing using a bead beater (two rounds of 90 sec of vortexing). Lysate was transferred to a clean microfuge tube and centrifuged at 8000g for 5 min at 4°C. The supernatant was transferred to a clean microfuge tube, and one-half was directly loaded onto an 11 mL 10%–50% sucrose gradient (“No detergent”). To determine the effect of membrane on sedimentation, the other half was mixed with sodium deoxycholate to 0.2% final concentration and incubated on ice for 5 min, then Tween 20 was added to 0.5% final concentration, and the sample was incubated for an additional 5 min. The sample was then centrifuged for 20 min at 20,000g to remove unreleased membrane complexes, and the supernatant was loaded onto a sucrose gradient. Analysis in the presence of Triton X-100 was performed as described above, except that the lysis buffer included 1% Triton X-100 and Tris-Cl, KCl, and MgCl2 were used instead of HEPES, KAc, and MgAc, respectively (Arava et al. 2003). Gradients were centrifuged in an SW41 rotor (Beckman) at 35,000 rpm for 2.5 h, and the sedimentation positions of polysomal complexes were determined by monitoring RNA absorbance at 254 nm.
Western blot analysis and antibodies
Polysomal fractions were collected after centrifugation and precipitated by the addition of 2 volumes of 100% ethanol. After at least 30 min of incubation at −20°C, the samples were centrifuged for 25 min at 14,000g at 4°C, and the pellet was resuspended in 2× SDS sample buffer (4% SDS, 20% glycerol, 4 mM EDTA, 0.2 M Tris at pH 6.8, 4% β-mercaptoethanol, and 0.2 gr/mL Bromophenol blue), followed by incubation for 30 min at 40°C. The samples were run on 10% SDS-PAGE gel for 1–1.5 h and transferred to a nitrocellulose membrane. The membrane was then blocked with 1% milk in TBS-T and incubated with primary and secondary antibodies diluted in TBS-T (see Table 3). ECL reaction was performed using an ECL kit (Biological Industries) according to the manufacturer's instructions.
TABLE 3.
A list of the antibodies used in this study
Fractionation by Concanavalin A treatment
Con A fractionation was done as described previously (Kaiser et al. 2002) except that the coated spheroplasts were suspended in a lysis buffer that is more suitable for RNA assays (20 mM Tris-HCl at pH 7.4, 140 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.1 mg/mL cycloheximide, 1 mg/mL heparin, 1 mM PMSF, 0.5 μg/mL leupeptin, and 0.7 μg/mL pepstatin).
Fluorescent in situ hybridization
Fluorescent in situ hybridization was performed as previously described (Chartrand et al. 2000). Briefly, yeast cells were grown to the mid-log phase and fixed with 4% formaldehyde for 40 min, and the cell wall was removed by zymolase (25 μg/mL) treatment for 20 min (until ∼75% of the cells were spheroplasted). Cells were then adhered to poly-L-Lysine coated glass slides and stored in 70% ethanol at −20°C. For hybridization, ODN labeled by Cy3 at its 5′-end (30 pM) was added to the slides in 15 μL of hybridization buffer (10 mM NaHPO4 at pH 7.0, 40% formamide, 2× SSC, 40 μg of salmon sperm DNA, 40 μg of tRNA, 20 μg/μL BSA, 40 u/μL RNasin). After an overnight hybridization at 37°C, slides were washed twice with 2× SSC and 40% formamide for 15 min at 37°C, once with 2× SSC and 0.1% Triton X-100 for 15 min at room temperature, twice with 1× SSC for 15 min at room temperature, and finally with PBS. Slides were then mounted with 1 mg/mL anti-fade solution (p-phenylenediamine; Sigma) containing 1 μg/μL DAPI and imaged in a Leica DM IRE2 fluorescence microscope.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Amnon Harel of the Technion for the gift of α Porin; Dr. Ophry Pinnes of the Hebrew University, Israel for the gift of αBiP and αAconitase; Dr. Laura Popolo at the University of Milano, Italy for the gift of αGas1; Dr. Ramon Serrano at the University of Valencia, Spain for the gift of αPma1; Dr. R.P. Jansen at the Munich University, Germany for the gift of αSec61; Dr. Jeffery Gerst from the Weizmann Institute for the Sec63-RFP plasmid; and Dr. Andre Goffeau at the Catholic University of Louvain, Belgium for the gift of strain Δpmp1 Δpmp2. We also thank Dr. Hagai Abeliovich for technical advice and Dr. Dan Cassel for critically reading this manuscript. This work was supported by a grant no. 1096/05 from the Israel Science Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.867208.
REFERENCES
- Arava Y. Isolation of polysomal RNA for microarray analysis. Methods Mol. Biol. 2003;224:79–87. doi: 10.1385/1-59259-364-X:79. [DOI] [PubMed] [Google Scholar]
- Arava Y., Wang Y., Storey J.D., Liu C.L., Brown P.O., Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y., Boas F.E., Brown P.O., Herschlag D. Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Res. 2005;33:2421–2432. doi: 10.1093/nar/gki331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M.P., De Long S.K., Sachs A.B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M., Chang A., Simons K. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell. 2001;12:4129–4138. doi: 10.1091/mbc.12.12.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Horwich A.L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Chabanon H., Mickleburgh I., Hesketh J. Zipcodes and postage stamps: mRNA localization signals and their trans-acting binding proteins. Brief Funct. Genomic. Proteomic. 2004;3:240–256. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Bertrand E., Singer R.H., Long R.M. Sensitive and high-resolution detection of RNA in situ. Methods Enzymol. 2000;318:493–506. doi: 10.1016/s0076-6879(00)18072-3. [DOI] [PubMed] [Google Scholar]
- Chekulaeva M., Hentze M.W., Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M., Blugeon C., Jacq C. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 2000;20:7881–7892. [Google Scholar]
- Diehn M., Eisen M.B., Botstein D., Brown P.O. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- Diehn M., Bhattacharya R., Botstein D., Brown P.O. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2006;2:e11. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher K.L., Davis D., Wu M., Boldogh I., Pon L.A. Endoplasmic reticulum dynamics, inheritance, and cytoskeletal interactions in budding yeast. Mol. Biol. Cell. 2002;13:854–865. doi: 10.1091/mbc.01-04-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest K.M., Gavis E.R. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila . Curr. Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- Gong X., Chang A. A mutant plasma membrane ATPase, Pma1-10, is defective in stability at the yeast cell surface. Proc. Natl. Acad. Sci. 2001;98:9104–9109. doi: 10.1073/pnas.161282998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M., Blau M., Becker T., Mielke T., Pool M.R., Wild K., Sinning I., Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- Hegde R.S., Bernstein H.D. The surprising complexity of signal sequences. Trends Biochem. Sci. 2006;31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Rolfs A., Bhullar B., Murthy T.V., Zhu C., Berger M.F., Camargo A.A., Kelley F., McCarron S., Jepson D., et al. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae . Genome Res. 2007;17:536–543. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurowitz E.H., Brown P.O. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae . Genome Biol. 2003;5:R2. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi I.M., Cutler D., Stueber D., Bujard H. Determinants for protein translocation across mammalian endoplasmic reticulum. Membrane insertion of truncated and full-length prelysozyme molecules. Eur. J. Biochem. 1986;155:571–576. doi: 10.1111/j.1432-1033.1986.tb09526.x. [DOI] [PubMed] [Google Scholar]
- Jungnickel B., Rapoport T.A. A post-targeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Kaiser C.A., Chen E.J., Losko S. Subcellular fractionation of secretory organelles. Methods Enzymol. 2002a;351:325–338. doi: 10.1016/s0076-6879(02)51855-3. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Kerr K., Macdonald P.M. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- Kuhn K.M., DeRisi J.L., Brown P.O., Sarnow P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell. Biol. 2001;21:916–927. doi: 10.1128/MCB.21.3.916-927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner R.S., Seiser R.M., Zheng T., Lager P.J., Reedy M.C., Keene J.D., Nicchitta C.V. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Heredia M., Jansen R.P. mRNA localization and the cytoskeleton. Curr. Opin. Cell Biol. 2004;16:80–85. doi: 10.1016/j.ceb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Malinska K., Malinsky J., Opekarova M., Tanner W. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell. 2003;14:4427–4436. doi: 10.1091/mbc.E03-04-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc P., Margeot A., Devaux F., Blugeon C., Corral-Debrinski M., Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeot A., Blugeon C., Sylvestre J., Vialette S., Jacq C., Corral-Debrinski M. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 2002;21:6893–6904. doi: 10.1093/emboj/cdf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason N., Ciufo L.F., Brown J.D. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000;19:4164–4174. doi: 10.1093/emboj/19.15.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F., Kawaguchi N., Sese J., Toyoda A., Hattori M., Morishita S., Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre C., Ghislain M., Leterme S., Ferroud C., Dufour J.P., Goffeau A. Purification and complete sequence of a small proteolipid associated with the plasma membrane H+-ATPase of Saccharomyces cerevisiae . J. Biol. Chem. 1992;267:6425–6428. [PubMed] [Google Scholar]
- Navarre C., Catty P., Leterme S., Dietrich F., Goffeau A. Two distinct genes encode small isoproteolipids affecting plasma membrane H+-ATPase activity of Saccharomyces cerevisiae . J. Biol. Chem. 1994;269:21262–21268. [PubMed] [Google Scholar]
- Okun M.M., Eskridge E.M., Shields D. Truncations of a secretory protein define minimum lengths required for binding to signal recognition particle and translocation across the endoplasmic reticulum membrane. J. Biol. Chem. 1990;265:7478–7484. [PubMed] [Google Scholar]
- Panzner S., Dreier L., Hartmann E., Kostka S., Rapoport T.A. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Pool M.R. Getting to the membrane: How is cotranslational protein targeting to the endoplasmic reticulum regulated? Biochem. Soc. Trans. 2003;31:1232–1237. doi: 10.1042/bst0311232. [DOI] [PubMed] [Google Scholar]
- Prinz W.A., Grzyb L., Veenhuis M., Kahana J.A., Silver P.A., Rapoport T.A. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae . J. Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H., Li Y., Padgett R.W. Incorporating structure to predict microRNA targets. Proc. Natl. Acad. Sci. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R., Lapidot M., Shamir R., Pilpel Y. A catalog of stability-associated sequence elements in 3′-UTRs of yeast mRNAs. Genome Biol. 2005;6:R86. doi: 10.1186/gb-2005-6-10-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., Walter P. The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J. 1988;7:1769–1775. doi: 10.1002/j.1460-2075.1988.tb03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltenburg R., Wartmann T., Kunze I., Kunze G. Reliable method to prepare RNA from free and membrane-bound polysomes from different yeast species. Biotechniques. 1995;18:564–568. [PubMed] [Google Scholar]
- Sylvestre J., Margeot A., Jacq C., Dujardin G., Corral-Debrinski M. The role of the 3′-untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells. Mol. Biol. Cell. 2003;14:3848–3856. doi: 10.1091/mbc.E03-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler Y., Zilberstein C., Ziv-Ukelson M. A study of accessible motifs and RNA folding complexity. J. Comput. Biol. 2007;14:856–872. doi: 10.1089/cmb.2007.R020. [DOI] [PubMed] [Google Scholar]
- Wolin S.L., Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin S.L., Walter P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol. 1989;109:2617–2622. doi: 10.1083/jcb.109.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin S.L., Walter P. Discrete nascent chain lengths are required for the insertion of presecretory proteins into microsomal membranes. J. Cell Biol. 1993;121:1211–1219. doi: 10.1083/jcb.121.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]