Abstract
In order to infect their hosts, many Gram-negative bacteria translocate agents of infection, called effector proteins, through the type III secretion system (TTSS) into the host cytoplasm. This process is thought to require at least partial unfolding of these agents, raising the question of how an effector protein might unfold to enable its translocation and then refold once it reaches the host cytoplasm. AvrPto is a well-studied effector protein of Pseudomonas syringae pv tomato. The presence of a readily observed unfolded population of AvrPto in aqueous solution and the lack of a known secretion chaperone make it ideal for studying the kinetic and thermodynamic characteristics that facilitate translocation. Application of Nzz exchange spectroscopy revealed a global, two-state folding equilibrium with 16% unfolded population, a folding rate of 1.8 s−1, and an unfolding rate of 0.33 s−1 at pH 6.1. TrAvrPto stability increases with increasing pH, with only 2% unfolded population observed at pH 7.0. The R1 relaxation of TrAvrPto, which is sensitive to both the global anisotropy of folded TrAvrPto and slow exchange between folded and unfolded conformations, provided independent verification of the global kinetic rate constants. Given the acidic apoplast in which the pathogen resides and the more basic host cytoplasm, these results offer an intriguing mechanism by which the pH dependence of stability and slow folding kinetics of AvrPto would allow efficient translocation of the unfolded form through the TTSS and refolding into its functional folded form once inside the host.
Keywords: AvrPto, Pseudomonas syringae, type III secretion system, pH-dependent stability, low stability, slow conformational exchange
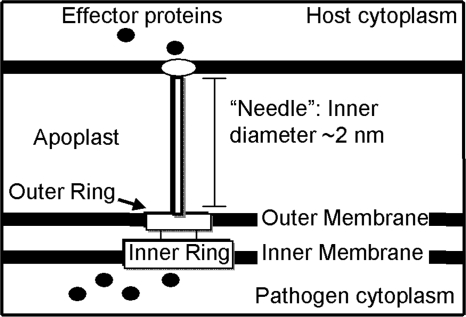
The type III secretion system (TTSS) is observed in a broad range of Gram-negative bacterial genera, including Salmonella, Shigella, Yersinia, and Pseudomonas (Preston et al. 1995). This syringe-like supramolecular assembly is composed of multiple copies of over 20 different proteins (Fivaz and van der Goot 1999; Galan and Collmer 1999) and can transport effector proteins from the bacterium in a process dependent upon both a putative ATPase in the inner membrane ring (Eichelberg et al. 1994; Woestyn et al. 1994; Tamano et al. 2000), and a proton gradient across the inner bacterial membrane (Fig. 1; Wilharm et al. 2004). Although effector proteins themselves vary widely among pathogens, the translocation machinery of the TTSS is a common feature found among many important pathogens (Jin et al. 2003).
Figure 1.
The type III secretion system (TTSS) provides a pathogen–host conduit. Schematic diagram of the TTSS used by pathogens to transport effector proteins from the bacterial cytoplasm into the host cytoplasm. These proteins are thought to pass through the hollow, needle-like structure (pilus) of the TTSS. Since the inner diameter of the pilus is ∼2–3 nm, most proteins must be at least partially unfold before secretion.
Partial or complete unfolding of effector proteins is thought to be necessary for translocation through the TTSS into the host (Johnson et al. 2005). A hollow, needle-like structure called the Hrp pilus extends from the basal body of the TTSS and has an inner diameter of 2–3 nm (Blocker et al. 2001; Cordes et al. 2003). It is known from immunogold-labeling experiments that the extracellular components of the TTSS and the effector proteins are conducted along the needle (Brown et al. 2001; Jin et al. 2003) and exit through the needle's tip (Li et al. 2002). In the current models for both TTSS needle elongation and effector translocation, these proteins pass through the inside of the needle in at least a partially unfolded form. This mechanism is analogous to the elongation model for the evolutionarily related bacterial flagellar assembly (Emerson et al. 1970; Yonekura et al. 2003).
The stability and folding kinetics of an effector protein have important effects on its ability to be secreted. These dependences are not simple, however. Fusion of ubiquitin to the C terminus of YopE, a Yersinia effector protein, prevented secretion of the fusion protein, whereas mutational destabilization of the ubiquitin fusion partner resulted in secretion (Lee and Schneewind 2002). Conversely, the RhoGAP domain of YopE and YopH effector proteins has reasonable stabilities of 5–6 kcal/mol (Zhang et al. 1992; Ghosh 2004), yet they are secreted. The importance of folding kinetics is also implicated by the finding that YopE secretion was prevented by fusion to rapidly folding domains such as dihydrofolate reductase (Sorg et al. 2005). Some, but not all, effector proteins associate with secretion chaperones that remain in the bacterium after effector secretion (Rosqvist et al. 1994; Frithz-Lindsten et al. 1995; Cheng and Schneewind 1999), which adds another dimension to the complexity of the secretion process. Proposed roles of secretion chaperones include assisting conjugate effector proteins to unfold (Stebbins and Galan 2001) or to compete for access to the TTSS (Luo et al. 2001; Wulff-Strobel et al. 2002; Ghosh 2004).The emerging picture is that the secretion efficiency of an effector protein appears to depend on some combination of its folding thermodynamics and kinetics.
The Pseudomonas syringae pv tomato effector protein AvrPto, which has no known chaperone and has been shown to have marginal stability (Wulf et al. 2004), offers a simple model system to probe the dependence of secretion and translocation on effector protein stability and kinetics. Upon delivery into the host cytoplasm, AvrPto has roles in promoting the infection of tomato leaf cells (Bogdanove and Martin 2000; Chang et al. 2000; Shan et al. 2000; Hauck et al. 2003) and, ironically, in the plant's defense against the pathogen (Scofield et al. 1996; Tang et al. 1999; Pedley and Martin 2003). The NMR structure of a truncated 105-residue version of AvrPto (TrAvrPto), in which the unstructured N and C termini were removed, reveals a ∼5-nm long and ∼2.5-nm wide three-helix bundle with a short orthogonal helix and a large Ω-loop (Wulf et al. 2004). These dimensions would necessitate at least partial unfolding for passage through the Hrp pilus.
Interestingly, both AvrPto and TrAvrPto have a significant unfolded population in slow exchange with the folded form in aqueous solution at pH 6 and 25°C (Wulf et al. 2004). The ability of AvrPto to unfold and refold in vitro without chaperone assistance, and the presence of an NMR-observable unfolded population, enable us to study relationships between the kinetics and thermodynamics of effector protein folding and chaperone-independent secretion. Of particular interest are the residue-specific folding and unfolding rates and their potential influence on the viability of a protein for TTSS secretion.
In this work, the equilibrium kinetics and thermodynamics of TrAvrPto folding were characterized using two-dimensional NMR techniques (Montelione and Wagner 1989; Farrow et al. 1994a,b; Mulder et al. 1996). Nzz exchange spectroscopy is sensitive to chemical exchange processes that occur at rates on the order of 0.1–10 s−1 (Palmer 2001). This NMR technique has the advantage of providing a multiple-site perspective that enables the elucidation of independent folding regions if present, or the accurate extraction of kinetic and thermodynamic parameters for a global two-state folding process. Here, Nzz exchange spectroscopy was applied to determine the folding and unfolding rates and stability of TrAvrPto at pH 6.1. Additional application of HSQC and longitudinal relaxation experiments showed that TrAvrPto stability increases with increasing pH and provided confirmation of the folding equilibrium parameters obtained from Nzz exchange spectroscopy. Given the acidic apoplast environment of the pathogen and the more basic host cytoplasm, the results presented here offer an intriguing mechanism by which the pH dependence of stability and slow folding kinetics of AvrPto would allow efficient translocation through the TTSS and refolding into its functional folded form once inside the host.
Results
The unfolding of TrAvrPto proceeds on the slow chemical exchange timescale across the entire protein
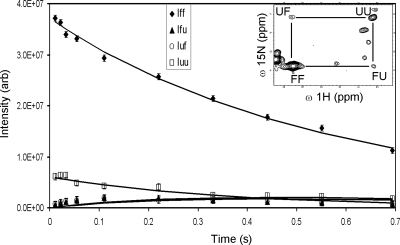
Nzz exchange spectroscopy (Montelione and Wagner 1989; Farrow et al. 1994b) was used to identify individual residues in slow chemical exchange and to quantify their folding and unfolding rate constants. For each spin in a slow two-state folding reaction, four peaks are expected: two auto-correlation peaks for the folded, FF, and unfolded, UU, resonances and two exchange cross-correlation peaks, FU and UF (Fig. 2, inset). The time-dependent changes in intensity of these four peaks—IFF(t), IFU(t), IUF(t), and IUU(t) (Fig. 2)—are modeled by the two-state solution of the Bloch-McConnell exchange equations (McConnell 1958; Cavanagh et al. 1996), which depends on the kinetic rate constants, kFU and kUF, and the rates of longitudinal relaxation in the absence of exchange for each conformation, R1F and R1U.
Figure 2.
Time-dependence of G95 peak intensities during Nzz experiment. The experimental data (symbols) and two-state exchange model best fits (solid lines) for G95 auto- and cross-correlation peak intensity are shown. Autopeak intensities for the folded (FF, filled diamonds) unfolded (UU, open squares) conformations, cross-correlation intensities, FU (filled triangles) and UF (open circles), are plotted vs. NZZ exchange spectroscopy mixing time. (Inset) G95 auto- and cross-correlation peaks at a mixing time of 0.1101 s.
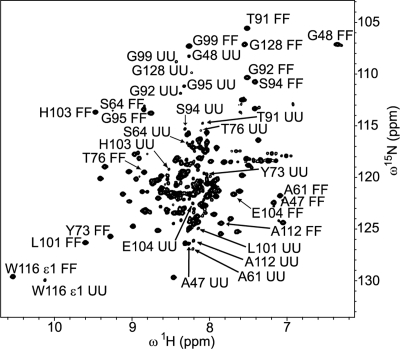
The presence of cross peaks in the Nzz exchange spectroscopy spectra of TrAvrPto immediately demonstrates that folding occurs on the slow chemical exchange timescale (circa (kFU + kUF) ≤ 100 s−1). Ninety-five potential unfolded TrAvrPto resonances were observed, suggesting that the entire 105-residue protein exchanges between its folded and unfolded conformations on this slow exchange timescale. The folded autopeaks of 41 N-H groups were linked to at least one of their corresponding cross peaks and, thereby, were identified as being in slow exchange (Fig. 3, Supplemental Table 1). Some of these residues (e.g., L37) displayed their folded autopeak and a single resolved cross peak. For other residues, three peaks (e.g., N105 and A47) or all four peaks (e.g., G92) were resolved. The unfolded autopeaks were well-resolved for a subset of 17 of these 41 residues (Fig. 4). Limited spectral resolution prevents further identification of backbone amides in slow exchange, especially in the center of the spectrum where resonances of unfolded TrAvrPto and α-helices congregate (Fig. 4). The distribution of residues in slow exchange across the structure (Fig. 3) and the good sampling of the well-resolved peaks (Fig. 4) indicate that exchange between folded and unfolded states occurs protein-wide.
Figure 3.
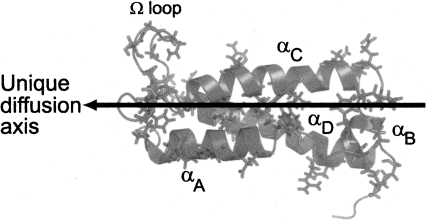
Prolate shape and global folding process of TrAvrPto. TrAvrPto is a prolate molecule, whose unique diffusion axis (shown as an arrow) is aligned with helices αA, αC, and αD. Forty-one slowly exchanging amide groups were identified and are shown mapped onto the structure of TrAvrPto as side chains. They are distributed across all secondary structural elements of TrAvrPto, implying that folding occurs globally: Q35, L37, H41, and E45 of helix αA; A47 and G48 of loop-AB; D52, H54, and E55 of αB; S58, S59, and A61 of loop-BC; Q63, S64, N67, L72, Y73, T76, R78, and L80 of αC; Q86, H87, M90, T91, G92, S94, G95, N97, G99, and L101 of the Ω-loop; H103, E104, N105, M109, R110, A112, W116 ε1, R120, and E121 of αC; and G128 and I129 on the C-terminal tail. Image rendered with PyMOL (DeLano Scientific) using NMR structure (PDB file 1R5E) (Wulf et al. 2004), ensemble-averaged via MOLMOL (Koradi et al. 1996).
Figure 4.
Folded and unfolded TrAvrPto populations are detected at pH 6.1. Two discrete sets of peaks are observed in a fully relaxed Nzz spectrum collected with zero mixing time, corresponding to the folded and unfolded conformations of TrAvrPto. The peaks for both unfolded TrAvrPto and the three parallel α-helices of folded TrAvrPto cluster near the center of the spectrum. The FF and UU peaks are labeled for the 17 residues with assigned UU resonances (A47, G48, A61, S64, Y73, T76, T91, G92, S94, G95, G99, L101, H103, E104, A112, W116 ε1, and G128). Nine of these UU peaks had been assigned previously (Wulf et al. 2004).
TrAvrPto is characterized by global two-state unfolding
A series of Nzz spectra were acquired, each with a different time interval, t, during which the 15N longitudinal magnetization decayed toward equilibrium and TrAvrPto exchanged between its folded and unfolded conformations. At each time t, the variation of IFU(t) and IUF(t) across TrAvrPto is comparable to the experimental uncertainty in peak volume, σ. Conversely, at a given time point, the autopeak intensities show more variation between residues than can be accounted for more by σ. The variation in IFF(t) was an order of magnitude greater than σ and, at early times, that of IUU(t) was approximately twice σ. To understand this difference in behavior between the autopeak and cross-peak intensities across the protein, it is informative to derive linear approximations of the Bloch-McConnell equations (McConnell 1958; Cavanagh et al. 1996) at short time intervals (t ≪ 1) using the Taylor series relation,
Using this approach, the intensities associated with each amide group in slow chemical exchange are given by
These simplified equations illustrate that, at early time points, the cross-peak intensities are sensitive to the kinetic rate constants only, while the autopeak intensities are sensitive to both the relaxation rates (R1F and R1U), and the kinetic rate constants (kFU and kUF). The observation that residue-to-residue variations of IFU(t) and IUF(t) are indistinguishable from experimental noise indicates that a single set of global kinetic rate constants, kFU and kUF, applies for all residues. Therefore, the larger variation in autopeak intensities is due to differences in R1F and R1U from residue to residue. As discussed in more detail below, folded TrAvrPto is a prolate molecule (Fig. 3), whose backbone amide R1Fs are expected to vary based on their placement in the protein. The smaller but significant variation in IUU(t) implies that there is some intramolecular variation in R1U, perhaps due to residual structure or to the amide group's proximity to the ends of the polypeptide chain.
The above analysis motivates the modeling of TrAvrPto conformational exchange as a two-state process with global k FU and k UF and residue-specific R1F and R1U. The resulting global kinetic rate constants (extracted as described in Materials and Methods using data from the 17 residues with assigned IUU peaks) are k FU = 0.33 ± 0.04 s−1 and k UF = 1.8 ± 0.3 s−1, corresponding to ΔG = −RT*ln(k UF/k FU) = −1.0 ± 0.1 kcal/mol. Compared to the ambient thermal energy, RT = −0.6 kcal/mol, TrAvrPto is only modestly stable at pH 6.1. The population in the unfolded ensemble, pU, is
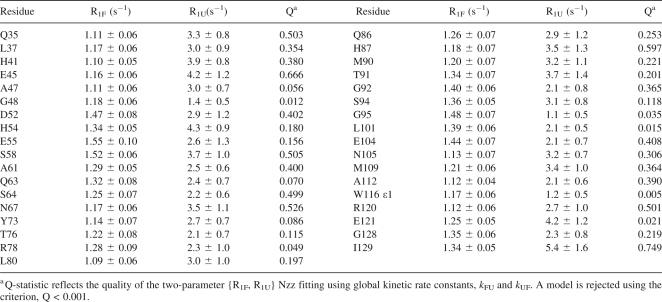
where pU + pF = 1 is the total population of TrAvrPto. The peak intensities for each of the 41 slowly exchanging residues were then individually fitted for R1F and R1U, using the global k FU and k UF as set constants. These fits yielded χ2 probability values, Q (Press et al. 1988), where the criterion for the rejection of a model was Q < 0.001. Thirty-five of the 41 residues had acceptable fits (Table 1; R1F values are plotted in Fig. 5A). The remaining residues (S59, L72, N97, G99, H103, and R110) had peaks that suffered from overlap, resulting in unacceptable fits. Although G99 and H103 were included in the global kinetic rate constants analysis, subsequent refitting without these residues yielded statistically equivalent rates. In general, with k FU and k UF as set constants, R1F and R1U could be acceptably fitted using, at minimum, well-resolved peaks including the folded autopeak and one of the cross peaks.
Table 1.
R1F and R1U extracted from pH 6.1 Nzz data
Figure 5.
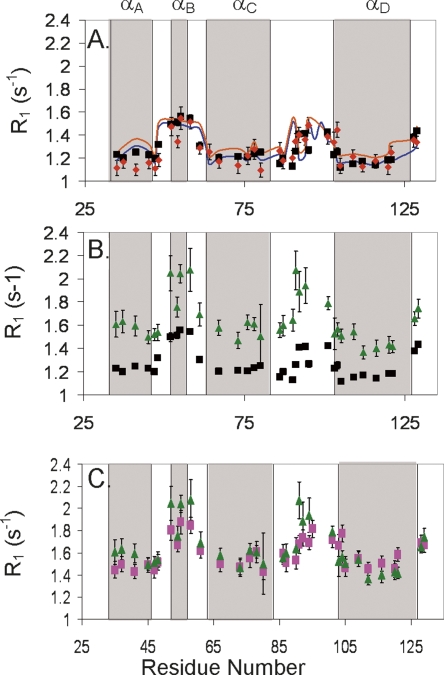
The effect of slow chemical exchange and global anisotropy on R1. TrAvrPto R1 relaxation rates are plotted vs. residue number (the four α-helices are shaded in gray boxes for reference) for three different comparisons: (A) Relaxation in the absence of TrAvrPto folding. R1 values for folded TrAvrPto in the absence of chemical exchange extracted from Nzz data measured at pH 6.1 (R1F Nzz, red diamonds) are comparable to those obtained from a T1 experiment at pH 7.0 (R1F(eff), black squares), where TrAvrPto is nearly completely folded. The R1F profiles were simulated by modeling TrAvrPto as a prolate monomer (blue line) and as a prolate monomer/oblate dimer equilibrium (orange line). (B) The pH dependence of R1F(eff) from T1 experiments: R1F(eff) values obtained from T1 experiments at pH 6.1 (green triangles) and at pH 7.0 (black squares) show nearly constant offset, with R1F(eff) at pH 6.1 shifted to greater values than at pH 7.0. (C) Nzz-extracted parameters accurately predict R1F(eff): calculated R1F(eff) values (pink squares), obtained using Equation 7 and parameters R1F Nzz and k FU extracted from the pH 6.1 Nzz data, are comparable to R1F(eff) values obtained from T1 experiments at pH 6.1 (green triangles). Error bars, experimental uncertainties for R1F extracted from Nzz and T1 data and propagated uncertainties for the calculated R1F(eff).
Longitudinal relaxation and HSQC data provide an independent view of TrAvrPto folding kinetics and pH dependence of stability
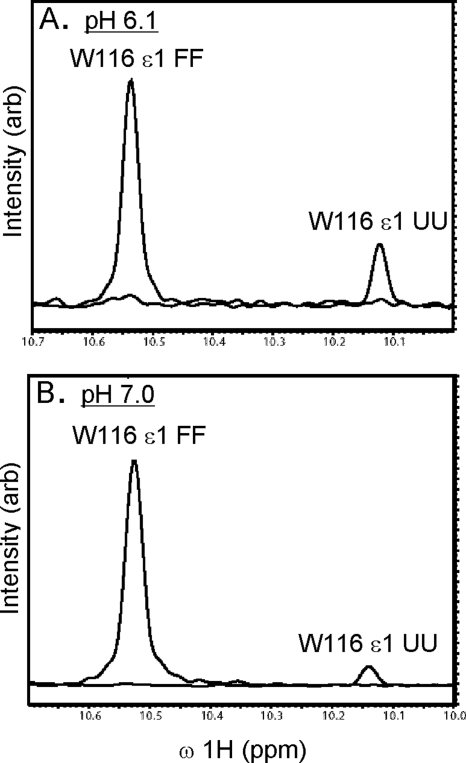
The stability of TrAvrPto is pH-dependent with greater stability at pH 7.0 than at pH 6.1 (Fig. 6). As determined from the Nzz data of 0.63 mM TrAvtPto, 16% of the population is unfolded at pH 6.1. At higher pH and higher concentration (pH 7.0 and 1.2 mM TrAvrPto), the unfolded population (quantified using a fully relaxed HSQC spectrum) decreased to 2%. As discussed in the next section, the increased relative intensity of the folded peaks at pH 7.0 is due purely to the pH dependence of TrAvrPto stability, not to self-association at the higher protein concentration.
Figure 6.
TrAvrPto is more stable at pH 7.0 than at pH 6.1: The relative volumes of peaks associated with folded and unfolded protein states depend on the stability of the protein. The TrAvrPto W116 ε1 side-chain amide group gives rise to well-resolved peaks in both folded (IFF) and unfolded (IUU) states. One-dimensional line shapes of the W116 ε1 IFF and IUU peaks extracted from a 2D 15N-1H HSQC spectrum at pH 6.1 (A) and pH 7.0 (B) illustrate the larger unfolded TrAvrPto population at pH 6.1 than at pH 7.0. Line shapes were generated using NMRDraw (Delaglio et al. 1995) and were overlaid using Adobe Illustrator.
In contrast to the R1F values extracted from Nzz data (R1F Nzz), which are the relaxation rates in the absence of exchange, the effective relaxation rate constants extracted from 15N T1 experiments (R1F[eff]) can contain contributions from the kinetic rates. Consequently, R1F(eff) values measured for TrAvrPto should reflect the folding equilibrium and be pH-dependent. R1F(eff) values at pH 6.1 are offset to greater values than those at pH 7.0 (Fig. 5B). The conserved profile of the R1F(eff) versus residue number curves indicates that the molecular shape of TrAvrPto does not dramatically change with changing pH. Rather, the offset between R1F(eff) profiles at pH 6.1 and 7.0 can be explained by the dependence of R1F(eff) on the rate of unfolding (k FU). For a single residue that interconverts between its folded and unfolded states on the slow exchange timescale, if we assume R1U ≫ R1F, p F ≫ p U (Leigh 1971), and R1U ≫ k UF, then the estimated effective longitudinal relaxation rate for the folded state will be
where k ex(= k FU + k UF) is the exchange rate constant and R1F is the longitudinal relaxation rate of the folded state independent of exchange. It is known from the Nzz fittings that R1U is generally greater than R1F (Table 1), and the population p F > 5p U. The final approximation (R1U ≫ k UF) is an oversimplification; however, the uncertainties in the fitted R1U prohibit the use of a more complete model that depends on this parameter and, as demonstrated below, the fits of the data to Equation 7 justify the use of this model.
The exchange process between folded and unfolded TrAvrPto explains the difference at pH 6.1 between R1F Nzz and R1F(eff) values. The R1F Nzz values are theoretically equivalent to the R1F in Equation 7. Experimental R1F(eff) can be simulated using the Nzz parameters (R1F Nzz and k FU) and Equation 7. The resulting simulated R1F(eff) values accurately predict the experimental R1F(eff) values measured at pH 6.1 (Fig. 5C; Supplemental Table 2). As a further validation test, R1F Nzz values should be approximately equal to R1F(eff) values measured at pH 7.0, where the effects of the conformational exchange are minimal due to the negligible (2%) unfolded population. Indeed, these values are in general within experimental uncertainty of each other (Fig. 5A). These results demonstrate that the variation of relaxation at different pH values is fully explained by the global folding exchange process and, importantly, provide independent verification of the Nzz-derived kinetic parameters.
The concentration-independent R1F rates of TrAvrPto are consistent with a prolate folded monomer
The exchange-independent R1F Nzz values differ across residues in a manner that can be explained by the prolate shape of folded TrAvrPto (Fig. 5A), where α-helices αA, αC, and αD are aligned along its unique diffusion axis and the Ω-loop and αB are approximately perpendicular to this axis (Fig. 3). A TrAvrPto monomer can rotate more rapidly about the long, unique axis than it can tumble end-over-end, a phenomenon that changes the relaxation rates, and so, an N–H bond has different R1 values depending on how it is oriented on the molecule. For folded TrAvrPto, R1F can be simulated using previously established theory (Woessner 1962; Lipari and Szabo 1982; Schurr et al. 1994) and the N–H bond vectors from the known structure of the protein (PDB file 1R5E) (Wulf et al. 2004). Using the monomeric, prolate TrAvrPto structure without a hydration layer yields a R1F profile that matches the fitted R1F Nzz profile but is shifted to higher values, reflecting a smaller molecule (data not shown). Expanding the effective dimensions of TrAvrPto to 6.1 nm long and 3.1 nm wide aligns the simulated R1F profile with the R1F Nzz curve (Fig. 5A). Although not necessarily a unique solution, the alignment of the simulated and experimental data is striking. The larger effective size could correspond to a hydration layer of ∼1.5 water molecules, drag from the loops and tails, or an internally hydrated and less compact average folded structure. A water-swollen structure is not implausible; the presence of internal waters has been demonstrated previously both experimentally and in MD folding simulations (Shea et al. 2002; Bezsonova et al. 2006; Nishiguchi et al. 2007). Since R1F Nzz rates are independent of conformational exchange, these values should be dominated by global tumbling. The above analysis demonstrates that the R1F Nzz values can in fact be reproduced based solely on the TrAvrPto monomer structure and further validates the global kinetic rates used in extraction of these values.
Previous work from this laboratory hypothesized that TrAvrPto is a transient dimer at pH 6 (Wulf et al. 2004). Indeed, it is also possible to reproduce the R1F Nzz values extracted from 0.63 mM Nzz data (Fig. 5A) as the population-weighted average R1F from monomer and side-by-side (oblate) dimer conformations in fast chemical exchange. Oblate dimers were modeled as 2.6 nm by 7.7 nm oblate ellipsoids and monomers were modeled as 5.6 nm by 2.6 nm prolate ellipsoids, dimensions that include one hydration layer. A reasonable fit of the R1F Nzz values was obtained by weighting the monomer population by pM = 0.7 and the oblate dimer subunit population by pD = (1 − pM) = 0.3. These populations correspond to weak self-association, described by the dissociation constant
In this simulation, [T] = 0.63 mM, the total TrAvrPto concentration during the Nzz experiments.
The monomer-dimer model fails to correctly predict the behavior of R1F(eff) data at other concentrations, however. Additional R1F(eff) values measured at pH 6.1 were obtained for total concentrations of 0.34 and 0.87 mM. Given K d = 2 mM and the previously estimated dimensions of the monomers and oblate dimers, the model predicts pM = 0.79 at 0.34 mM and pM = 0.65 at 0.87 mM. The predicted R1F values for 0.34 mM TrAvrPto were offset by an average of +0.18 s−1 relative to those for 0.87 mM TrAvrPto (Supplemental Table 3). Despite the contribution of k FU to R1F(eff) expected at pH 6.1 (see Equation 7), the average separation between experimental R1F(eff) data at 0.34 and 0.87 mM should be equivalent to +0.18 s−1 since k FU is a constant. However, the experimental 0.34 mM and 0.87 mM R1F(eff) data have negligible average separation, −0.04 s−1, with a standard deviation on the order of the experimental uncertainty (Supplemental Table 2). This inequality demonstrates that, while the 0.63 mM R1F Nzz profile can be coincidentally fit using a monomer-dimer equilibrium model, the observed concentration independence of R1F(eff) at pH 6.1 rules out this model. Additionally, measurements at pH 6.8 (0.92 mM) and 7.0 (1.2 mM) yielded R1F(eff) values within uncertainty of each other (Supplemental Table 2), further refuting the dimer model. The “dimers” observed in the previous DLS and native-gel studies most likely were unfolded protein, which would have a larger radius of gyration than the folded conformation (Piaggio et al. 2007). Altogether, these results argue against self-association and suggest that the dominant folded form of TrAvrPto is monomeric.
Discussion
The kinetic and thermodynamic properties of an effector protein may have important consequences on its ability to be efficiently translocated by the TTSS, a process requiring at least partial unfolding of the protein (Johnson et al. 2005). AvrPto is a well-characterized effector protein of P. syringae pv tomato and an excellent model system to probe requirements for efficient effector translocation. NMR spectroscopy allowed the folding of TrAvrPto to be probed at multiple locations across the protein. The multiple probes enrich the analysis, demonstrating that the entire TrAvrPto molecule, not just the core or the long Ω-loop, folds cooperatively with two-state kinetics. The low stability (16% unfolded population at pH 6.1), slow folding rate (k UF = 1.8 ± 0.3 s−1), and correspondingly long lifetime of the unfolded state (τ = 1/k UF = 0.6 ± 0.1 s) of TrAvrPto would allow a pool of readily secretable protein to be maintained, especially given that the full-length effector protein, AvrPto, is even less stable than TrAvrPto at pH 6 (Wulf et al. 2004).
After translocation into the host cell cytoplasm, an effector must be able to spontaneously fold into its functional form. As shown here, TrAvrPto is almost completely folded at pH 7.0, where the observed 2% unfolded population corresponds to a moderate folding free energy of -2.3 kcal/mol. Therefore, TrAvrPto should be predominately folded within the cytoplasm of the host cell.
The pH dependence of TrAvrPto potentially explains at least part of the observed difference in full-length AvrPto's secretion efficiency under different culturing conditions. Expression of the TTSS and its associated proteins, including AvrPto, can be induced in P. syringae pv tomato using a hrp-inducing minimal medium (Rahme et al. 1992; Xiao et al. 1992). In this medium, AvrPto is expressed when the external pH, pHext, is 6 or 7; however, AvrPto is only secreted at pHext = 6 and not at pH 7 (van Dijk et al. 1999). The proton concentration inside a pathogen, pHint, can differ from pHext. Indeed, the rotation of the Escherichia coli flagellium in the evolutionally related flagellar apparatus (Jin et al. 2003) depends on the proton gradient, ΔpH, between the cytoplasm and the periplasm (Minamino et al. 2003). ΔpH is maintained even when pHext is varied between 5.5 and 8.0 with pHint remaining greater than pHext (Minamino et al. 2003). In Yersinia enterocolitica, secretion through the TTSS has also been found to be dependent on ΔpH, where again pHint is greater than pHext (Wilharm et al. 2004). Assuming that secretion by P. syringae requires a similar ΔpH, then pHint would exceed 7 if pHext is neutral. Our results indicate that TrAvrPto would be predominantly folded inside the pathogen under these conditions, providing an explanation for the lack of AvrPto secretion in hrp-inducing minimal medium at neutral pH (van Dijk et al. 1999).
The structure and low stability of TrAvrPto may have additional implications for its ability to compete for passage through the TTSS without chaperone assistance. One of the conserved proteins in the TTSS innermembrane ring is a putative ATPase (Ghosh 2004) required for translocation (Wilharm et al. 2004). The TTSS ATPases are unfoldases (Gorbalenya and Koonin 1993; Lupas and Martin 2002) whose functional similarity to the AAA+ ATPases has been suggested (Akeda and Galan 2005). For the AAA+ ATPase domain of the ClpX bacterial protease, the rate of catalyzed unfolding by the hexameric ring decreased with increasing substrate stability (Kenniston et al. 2003). The identity of the secondary structural element immediately after a degradation signal also affects the degradation efficiency (Lee et al. 2001). The greatest enhancement in efficiency occurs when this element is a surface loop or α-helix that can be removed relatively easily and whose removal destabilizes the protein core (Lee et al. 2001). By analogy, AvrPto appears to possess the correct architecture and stability that would allow for efficient action of the putative TTSS ATPase and subsequent translocation into the host. AvrPto's secretion signal is located on its N terminus, as in all effector proteins (Sory and Cornelis 1994; Anderson and Schneewind 1997), and the first secondary structural element is helix αA. The core of TrAvrPto is only slightly stable, and the removal of αA from the bundle would be expected to destabilize the fold. Therefore, an additional consideration for secretion and translocation efficiency may be the direction-specific mechanical force exerted on the N-terminal region of the effector protein structure by the TTSS ATPase (Matouschek 2003).
Two real-time confocal microscopy studies on the translocation rates for pathogen effector proteins into the host via the TTSS provide an in vivo functional context for our TrAvrPto folding kinetic rates. For Shigella flexneri, it was found that 50% of the populations of effector proteins IpaB and IpaC was translocated out of the bacterium in t1/2 ∼ 240 s (Enninga et al. 2005). Assuming a mono-exponential decay curve, the translocation rate is k = ln2/t1/2 ∼ 0.003 s−1. For Salmonella enterica, the translocation rate was estimated by the build-up of effector protein SipA within the host cell over time using a straight line approximation, y(t) = mt, where the observed slope was m = 7–60 molecules/s and the average total number of molecules translocated was y0 = 6*103 ± 3 * 103 molecules (Schlumberger et al. 2005). This is equivalent to a linear Taylor-series approximation of an exponential build-up, where y(t) = y0*{1 − exp(−kt)} is approximated by y(t) = y0*{1 − (1 − kt)} = y0kt. Hence, the corresponding mono-exponential translocation rate, k (estimated as m/y0), was on order of k ∼ 0.01 s−1. Based on these two microscopy studies, a rough estimate of the general TTSS translocation rate is on order of 10−3 to 10−2 s−1. Therefore, the k FU = 0.33 ± 0.04 s−1 unfolding rate of TrAvrPto observed here in solution would not represent a rate-limiting step of the secretion process.
Altogether, these results offer important insights into the folding and unfolding of TrAvrPto and, ultimately, into how the thermodynamics and kinetics of effector proteins affect their translocation efficiency. The folding kinetics, even while viewed in isolation without the TTSS or any putative ATPase unfolding machinery, are sufficient to sustain a translocation-competent pool of TrAvrPto. This observation is consistent with observed ability of AvrPto to be translocated through the TTSS (van Dijk et al. 1999). This application of NMR spectroscopy to detect and measure the pH-dependent stability and folding kinetics of TrAvrPto lays the groundwork for future studies of TrAvrPto mutants to determine the mechanism of a possible pH-triggered folding switch and the thermodynamic and kinetic thresholds that govern the translocation efficiency of effector proteins.
Materials and Methods
NMR sample preparation
The TrAvrPto expression vector encodes a fusion protein consisting of a N-terminal 6-His tag, an HRV 3c protease (3CPro) cut site (Walker et al. 1994), and TrAvrPto residues 29–133 cloned into the ampicillin-resistant plasmid pQEP (Qiagen) (gift from Pete Pascuzzi and Greg Martin, Cornell University, Ithaca, NY). The protein was expressed in kanamycin-resistant M15 [pREP4] E. coli cells (Qiagen) and cultured in M9 minimal media with 15NH4Cl (Isotec, Inc.), 1 mM thiamine HCl (Sigma), 100 μg/mL ampicillin (Sigma, sodium salt), and 30 μg/mL kanamycin monosulfate (Sigma). All cultures were grown at 37°C with shaking at ∼120 rpm. One liter cultures were started with a 50-mL overnight miniculture and grown to O.D.600∼0.8. Protein expression was induced with 1 mM IPTG (Acros Organics); the cells were grown for an additional 2 h and then pelleted. Pellets were washed in binding buffer (50 mM NaH2PO4•H2O [Mallinckrodt], 300 mM NaCl [Sigma], 10 mM imidazole [Sigma], pH 8), re-pelleted, and then resuspended in lysis buffer (20 mL binding buffer + 50 μL protease inhibitor) on ice. The cells were lysed with 1 mg/mL lysozyme (Sigma) and 0.75 mg/mL deoxylcholic acid (Sigma) and two rounds of sonication. After high-speed centrifugation, supernatant was filtered through a 0.8-μm syringe filter (Corning, Inc.) onto a Ni-NTA column (Qiagen) with a 3–5 mL bed volume. The column was washed with 20 bed volumes of binding buffer and then 20 bed of volumes wash buffer (50 mM NaH2PO4•H2O, 300 mM NaCl, 20 mM imidazole at pH 8). Cleavage of the 6His-TrAvrPto fusion protein to yield tag-free TrAvrPto was accomplished by adding ∼1 mg 6His-3CPro fusion protein (prepared as described elsewhere; Jayaraman and Nicholson 2007) to the Ni-NTA column and tumbling at 4°C overnight. The desired TrAvrPto was eluted with wash buffer and dialyzed into NMR buffer (10 mM NaH2PO4•H2O, 5 mM KH2PO4 [Sigma], 225 mM NaCl, pH 6.9). The final concentration was estimated via UV absorbance at 280 nm using the theoretical extinction coefficient of 9530 M−1cm−1 for TrAvrPto (Gasteiger et al. 2005). The pH values of the NMR samples were adjusted using HCl and NaOH.
NMR spectroscopy
The NMR experiments were performed at 25°C on a Varian Inova 600-MHz spectrometer using a {H,C,N} Z-axis gradient probe. The spectra were processed using NMRpipe and NMRDraw (Delaglio et al. 1995). The data for all experiments were apodized using a shifted sine-bell function and then zero-filled. The peak volumes were determined using nlinLS (Frank Delaglio, NIH/NIDDK) or Sparky (T.D. Goddard and D.G. Kneller, University of California, San Francisco), assuming a Gaussian line shape.
The pH 6.1 experiments were performed on 15N-labeled TrAvrPto samples at concentrations of 0.63 mM for the Nzz spectra (Montelione and Wagner 1989; Farrow et al. 1994b) and 0.34 mM and 0.87 mM for T1 experiments (Farrow et al. 1994a). The T1 experiments at 0.34 mM were taken at the timepoints 0.01, 0.01, 0.04, 0.08, 0.16, 0.32, 0.64, 1.0, 1.0, and 2.0 s. At 0.87 mM, the T1 experimental timepoints were 0.01, 0.01, 0.02, 0.04, 0.08, 0.08, 0.16, 0.32, 0.64, 1.0, and 2.0 s. T1 experiments (with timepoints at 0.01, 0.77, and 0.77 s) were recorded for a 1.2 mM, pH 7.0 15N-labeled sample. Additionally, a set of T1 data for pH 6.8, 0.92 mM TrAvrPto was collected for time points 0.01, 0.77, and 0.77 s. In the T1 experiments at each concentration, the time points were recorded out of sequential order to avoid bias.
The pH dependence of TrAvrPto stability was evaluated using 1H-15N 2D NMR spectroscopy. At pH 6.1, a fully relaxed (10-s delay interval) Nzz spectrum with zero mixing time was used. It was subsequently found that a 5-s delay was sufficient for complete recovery of equilibrium magnetization. The stability at pH 7.0 was determined using the fully relaxed (5-s delay interval) HSQC spectrum (Mulder et al. 1996) of 1.2 mM TrAvrPto.
Nzz exchange spectroscopy
Nzz exchange spectroscopy (Montelione and Wagner 1989; Farrow et al. 1994b) was performed on a 0.63 mM TrAvrPto solution at pH 6.1 and 25°C. Spectra were taken with mixing times of 0.01101, 0.02202, 0.03303, 0.04404, 0.05505, 0.1101, 0.2202, 0.3303, 0.4404, 0.5505, and 0.69363 s and replicate spectra at 0.01101, 0.02202, 0.1101, and 0.4404 s. As with the T1 experiments, the timepoint spectra were record out of sequential order to avoid bias. A delay interval of 3 s was used in all spectra.
Identification of sets of autopeaks and cross peaks was aided by the judicious use of overlaid spectra. The intensities in the frequency domain of all Nzz spectra were added together using NMRPipe (addNMR command) (Delaglio et al. 1995), creating a summed spectrum that contains all autopeaks and cross peaks. The Nzz spectrum with zero mixing time contains only the folded and unfolded autopeaks. The cross peaks and unfolded autopeaks have decayed to almost zero in the 0.69-s mixing time spectrum; only the folded resonances are seen at high contours (Fig. 2). Classification of the peaks was achieved by noting that the folded autopeaks are present in all three spectra, the unfolded autopeaks are in the zero mixing time and summed spectra, and the cross peaks are only observed in the summed spectrum.
Two types of standard deviations were used in the study. The first is the experimental uncertainty in peak volume, σ, which was estimated as the mean replication error:
 |
The term <(x − y)2> is the mean deviation squared of peak volumes from replicate spectra. The average is over all peaks, whether autopeak or cross peak, used in the Nzz analysis and is over the four sets of replicate spectra. The second type is specific to each timepoint t and to each kind of peak (IFF(t), IFU(t), IUF(t), or IUU(t)). These latter estimates (σFF(t), σFU(t), σUF(t), σUU(t)) measure the residue-to-residue variability in peak intensity within each spectrum.
The dependence of the peak intensity on mixing time (t) was modeled with the two-state solution of the Bloch-McConnell equations (McConnell 1958; Farrow et al. 1994b). The global kinetic rate constants {kFU, kUF} were extracted by simultaneously fitting {IFF(t), IFU(t), IUF(t), IUU(t)} data of multiple residues. Due to issues of computer memory and nonlinear minimization convergence, it was only possible to simultaneously fit data from a small subset of seven residues. Specifically, a total of 280 data points (4 curves per residue × 10 time points per curve × 7 residues = 280 data points) were fitted to 16 adjustable parameters (R1U and R1F for each of seven residues plus global kFU and kUF). It was critical to the robustness of the fitting that each member of the seven-residue fitting set possessed well-resolved IFF and IUU autopeaks. Since the cross-peak intensities are statistically similar for all residues, the average IFU(t) and IUF(t) were used in the fittings. These intensity data were fitted using Levenberg-Marquardt nonlinear least squares minimization (Levenberg 1944; Marquardt 1963), and the uncertainty in intensities were simulated via Monte Carlo with 100 repetitions. The value and uncertainty of the adjustable parameters were the mean and standard deviation over the 100 repetitions.
In order to prevent bias in the parameter extraction due to the choice of fitting set members, the data fitting was repeated multiple times, each round using intensities from a set of seven residues randomly selected from the 17 residues that have well-resolved IUU peaks at time zero (Fig. 4). Allowing the fitting set choice to vary in 10 rounds yields results statistically identical to results obtained using 100 rounds. The average extracted global kFU and kUF were then used as constants to extract R1F and R1U values by separately fitting the intensities of individual residues. All fittings were implemented using Matlab version R2006a.
The quality of fit for each individual residue {R1F, R1U} was evaluated with the χ2 probability, Q (Press et al. 1988). Due to the presence of experimental noise, the minimized χ2 for each fit can vary according to the χ2 distribution. The value Q is the probability that, if one refit the data, one would find a larger χ2 simply due to the experimental uncertainty. A model fitting is rejected if Q < 0.001.
Simulation of R1F relaxation rate constants
Anisotropic tumbling alters the relaxation rates of amide groups depending on how they are oriented within the macromolecule (Woessner 1962). TrAvrPto was modeled as various types of ellipsoids that are axially symmetric about their unique diffusion axis. The viscosity, 0.0010019 kg/s*m for H2O at 293.15 K (Weast 1982), and temperature, 298.15 K, were used in simulations of R1F.
R1F values were calculated for each residue with the aid of the structure of TrAvrPto (PDB file 1R5E) (Wulf et al. 2004), the atomic coordinates of which were ensemble-averaged using MOLMOL (Koradi et al. 1996) and then transformed into the inertial reference frame using the in-house program, NORMAdyn (Pawley et al. 2001, 2002). The principal axis in the inertial frame is aligned with the unique diffusion axis of a prolate ellipsoid and is perpendicular to the unique diffusion axis of an oblate ellipsoid. The spectral density function for completely rigid, axially symmetric anisotropic proteins (Lipari and Szabo 1982; Schurr et al. 1994) was used to calculate R1 using the standard approach (Abragam 1961), assuming Δσ = −163 ppm and <rNH> = 1.04 Å (Palmer 2001). For the simulation of folded monomer (M) and dimer (D) in fast exchange, the population-weighted R1F was calculated:
where pM = 1 − pD.
Electronic supplemental material
The assigned {FF, FU, UF, UU} chemical shifts for all 41 slowly exchanging residues are listed in Supplemental Table 1. Supplemental Table 2 contains, in numerical form, the R1F data shown in Figure 5. Supplemental Table 3 contains the R1F values simulated for monomer-dimer equilibria with varying monomer populations, pM. The pM = 0.7 R1F data from this table is also shown in Figure 5.
Acknowledgments
We thank Dr. Pete Pascuzzi and Professor Gregory Martin for reagents and helpful discussions, Kyriacos Leptos for construction of the 6His-3CPro expression vector, Kevin Tan for his contributions to the TrAvrPto purification protocol, and Soumya De, Bhargavi Jayaraman, Lea Michel, Valerie Anderson, Beth R. Dawson, and Alex Greenwood for helpful editing comments. This work was supported by NSF grant MCB-0641582 and the NIH Molecular Biophysics Training Grant T32GM08267.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Linda K. Nicholson, Department of Molecular Biology and Genetics, Cornell University, 239 Biotechnology Building, Ithaca, NY 14853, USA; e-mail: lkn2@cornell.edu; fax: (607) 255-6249.
Abbreviations: TrAvrPto, truncated AvrPto; 3CPro, 3c protease; TTSS, type three secretion system.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.034223.107.
References
- Abragam, A. 1st ed. Clarendon Press; Oxford: 1961. Principles of nuclear magnetism; pp. 1–599. [Google Scholar]
- Akeda, Y., Galan, J.E. Chaperone release and unfolding of substrates in type III secretion. Nature. 2005;437:911–915. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- Anderson, D.M., Schneewind, O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica . Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- Bezsonova, I., Korzhnev, D.M., Prosser, R.S., Forman-Kay, J.D., Kay, L.E. Hydration and packing along the folding pathway of SH3 domains by pressure-dependent NMR. Biochemistry. 2006;45:4711–4719. doi: 10.1021/bi060177r. [DOI] [PubMed] [Google Scholar]
- Blocker, A., Jouihri, N., Larquet, E., Gounon, P., Ebel, F., Parsot, C., Sansonetti, P., Allaoui, A. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secretion. Mol. Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J., Martin, G.B. AvrPto-dependent Pto-interacting proteins and AvrPto-inteacting proteins in tomato. Proc. Natl. Acad. Sci. 2000;97:8836–8840. doi: 10.1073/pnas.97.16.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, I.R., Mansfield, J.W., Taira, S., Roine, E., Romantschuk, M. Immunocytochemical localization of HrpA and HrpZ supports a role for the Hrp Ilus in the transfer of effector proteins from Pseudomonas syringae pv. tomato across the host plant cell wall. Mol. Plant-Microbe Interact. 2001;14:394–404. doi: 10.1094/MPMI.2001.14.3.394. [DOI] [PubMed] [Google Scholar]
- Cavanagh, J., Fairbrother, W.J., Palmer, A.G.I., Skelton, N.J. Academic Press; San Diego, CA: 1996. Protein NMR spectroscopy: Principles and practice; pp. 243–299. [Google Scholar]
- Chang, J.H., Rathjen, J.P., Bernal, A.J., Staskawicz, B.J., Michelmore, R.W. AvrPto enhances growth and necrosis caused by Pseudomonas syringae pv. tomato in tomato lines lacking either Pto or Prf. Mol. Plant-Microbe Interact. 2000;13:568–571. doi: 10.1094/MPMI.2000.13.5.568. [DOI] [PubMed] [Google Scholar]
- Cheng, L.W., Schneewind, O. Yersinia enterocolitica type III secretion. On the role of SycE in targeting YopE into HeLa cells. J. Biol. Chem. 1999;274:22102–22108. doi: 10.1074/jbc.274.31.22102. [DOI] [PubMed] [Google Scholar]
- Cordes, F.S., Komoriya, K., Larquet, E., Yang, S., Egelman, E.H., Blocker, A., Lea, S.M. Helical structure of the needle of the type III secretion system of Shigella flexneri . J. Biol. Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Eichelberg, K., Ginocchio, C.C., Galan, J.E. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: Homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 1994;176:4501–4510. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, S.U., Tokuyasu, K., Simon, M.I. Bacterial flagella: Polarity of elongation. Science. 1970;169:190–192. doi: 10.1126/science.169.3941.190. [DOI] [PubMed] [Google Scholar]
- Enninga, J., Mounier, J., Sansonetti, P., Tran Van Nhieu, G. Secretion of type III effectors into host cells in real time. Nat. Methods. 2005;2:959–965. doi: 10.1038/nmeth804. [DOI] [PubMed] [Google Scholar]
- Farrow, N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Forman-Kay, J.D., Kay, L.E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994a;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Farrow, N.A., Zhang, O., Forman-Kay, J.D., Kay, L.E. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR. 1994b;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- Fivaz, M., van der Goot, F.G. The tip of a molecular syringe. Trends Microbiol. 1999;7:341–343. doi: 10.1016/s0966-842x(99)01574-7. [DOI] [PubMed] [Google Scholar]
- Frithz-Lindsten, E., Rosqvist, R., Johansson, L., Forsberg, A. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensible for targeting to the secretion loci. Mol. Microbiol. 1995;16:635–647. doi: 10.1111/j.1365-2958.1995.tb02426.x. [DOI] [PubMed] [Google Scholar]
- Galan, J.E., Collmer, A. Type III secretion machines: Bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M.R., Appel, R.D., Bairoch, A. Protein identification and analysis tools on the ExPASy server. In: Walker John M., editor. The proteomics protocols handbook. Humana Press; Totowa, NJ: 2005. pp. 571–607. [Google Scholar]
- Ghosh, P. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 2004;68:771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A.E., Koonin, E.V. Helicases–amino-acid-sequence comparisons and structure-function-relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- Hauck, P., Thilmony, R., He, S.Y. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman, B., Nicholson, L.K. Thermodynamic dissection of the Ezrin FERM/CERMAD interface. Biochemistry. 2007;46:12174–12189. doi: 10.1021/bi701281e. [DOI] [PubMed] [Google Scholar]
- Jin, Q., Thilmony, R., Zwiesler-Vollick, J., He, S.Y. Type III protein secretion in Pseudomonas syringae . Microbes Infect. 2003;5:301–310. doi: 10.1016/s1286-4579(03)00032-7. [DOI] [PubMed] [Google Scholar]
- Johnson, S., Deane, J.E., Lea, S.M. The type III needle and the damage done. Curr. Opin. Struct. Biol. 2005;15:700–707. doi: 10.1016/j.sbi.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Kenniston, J.A., Baker, T.A., Fernandez, J.M., Sauer, R.T. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Lee, C., Schwartz, M.P., Prakash, S., Iwakura, M., Matouschek, A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Lee, V.T., Schneewind, O. Yop fusions to tightly folded protein domains and their effects on Yersinia enterocolitica type III secretion. J. Bacteriol. 2002;184:3740–3745. doi: 10.1128/JB.184.13.3740-3745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, J.S. Relaxation times in systems with chemical exchange: Some exact solutions. J. Magn. Reson. 1971;4:308–311. [Google Scholar]
- Levenberg, K. A method for the solution of certain problems in least squares. Q. Appl. Math. 1944;2:164–168. [Google Scholar]
- Li, C.M., Brown, I., Mansfield, J., Stevens, C., Boureau, T., Romantschuk, M., Taira, S. The Hrp pilus of Pseudomonas syringae elongates from its tip and acts as a conduit for translocation of the effector protein HrpZ. EMBO J. 2002;21:1909–1915. doi: 10.1093/emboj/21.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipari, G., Szabo, A. Model-free analysis to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- Luo, Y., Bertero, M.G., Frey, E.A., Pfuetzner, R.A., Wenk, M.R., Creagh, L., Marcus, S.L., Lim, D., Sicheri, F., Kay, C., et al. Structural and biochemical characterization of the type III secretions chaperones CesT and SigE. Nat. Struct. Biol. 2001;8:1031–1036. doi: 10.1038/nsb717. [DOI] [PubMed] [Google Scholar]
- Lupas, A.N., Martin, J. AAA proteins. Curr. Opin. Struct. Biol. 2002;12:746–753. doi: 10.1016/s0959-440x(02)00388-3. [DOI] [PubMed] [Google Scholar]
- Marquardt, D. An algorithm of least-squares estimation of nonlinear parameters. SIAM J. Appl. Math. 1963;11:431–441. [Google Scholar]
- Matouschek, A. Protein unfolding—an important process in vivo? Curr. Opin. Struct. Biol. 2003;13:98–109. doi: 10.1016/s0959-440x(03)00010-1. [DOI] [PubMed] [Google Scholar]
- McConnell, H.M. Reaction rates by nuclear magnetic resonance. J. Chem. Phys. 1958;28:430–431. [Google Scholar]
- Minamino, T., Imae, Y., Oosawa, F., Kobayashi, Y., Oosawa, K. Effect of intracellular pH on rotational speed of bacterial flagellar motors. J. Bacteriol. 2003;185:1190–1194. doi: 10.1128/JB.185.4.1190-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelione, G.T., Wagner, G. 2D chemical-exchange NMR-spectroscopy by proton-detected heteronuclear correlation. J. Am. Chem. Soc. 1989;111:3096–3098. [Google Scholar]
- Mulder, F.A.A., Spronk, C.A.E.A., Slijper, M., Kaptein, R., Boelens, R. Improved HSQC experiments for the observation of exchange broadened signals. J. Biomol. NMR. 1996;8:223–228. doi: 10.1007/BF00211169. [DOI] [PubMed] [Google Scholar]
- Nishiguchi, S., Goto, Y., Takahashi, S. Solvation and desolvation dynamics in apomyoglobin folding monitored by time-resolved infrared spectroscopy. J. Mol. Biol. 2007;373:491–502. doi: 10.1016/j.jmb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Palmer, A.G.I. NMR probes of molecular dynamics: Overview and comparison with other techniques. Annu. Rev. Biophys. Biomol. Struct. 2001;30:129–155. doi: 10.1146/annurev.biophys.30.1.129. [DOI] [PubMed] [Google Scholar]
- Pawley, N.H., Wang, C., Koide, S., Nicholson, L.K. An improved method for distinguishing between anisotropic tumbling and chemical exchange in analysis of 15N relaxation parameters. J. Biomol. NMR. 2001;20:149–165. doi: 10.1023/a:1011249816560. [DOI] [PubMed] [Google Scholar]
- Pawley, N.H., Gans, J.D., Nicholson, L.K. Factors determining the reliable description of global tumbling parameters in solution NMR. J. Biomol. NMR. 2002;24:215–229. doi: 10.1023/a:1021664605995. [DOI] [PubMed] [Google Scholar]
- Pedley, K.F., Martin, G.B. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- Piaggio, M.V., Peirotti, M.B., Deiber, J.A. On the application of CZE to the study of protein denaturation. Electrophoresis. 2007;28:2223–2234. doi: 10.1002/elps.200600699. [DOI] [PubMed] [Google Scholar]
- Press, W.H., Flannery, B.P., Teukolsky, S.A., Vetterling, W.T. Cambridge University Press; Cambridge: 1988. Numerical recipes in C; pp. 521–522. [Google Scholar]
- Preston, G., Huang, H.C., He, S.Y., Collmer, A. The HrpZ proteins of Pseudomonas syringae pvs. Syringae, glycinea, and tomato are encoded by an operon containing Yersinia ysc homologs and elicit the hypersensitive response in tomato but not soybean. Mol Plant-Microbe Interact. 1995;8:717–732. doi: 10.1094/mpmi-8-0717. [DOI] [PubMed] [Google Scholar]
- Rahme, L.G., Mindrinos, M.N., Panopoulos, N.J. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J. Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosqvist, R., Magnusson, K.E., Wolf-Wantz, H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumberger, M.C., Müller, A.J., Ehrbar, K., Winnen, B., Duss, I., Stecher, B., Hardt, W.D. Real-time imaging of type III secretion: Salmonella SipA injection into host cells. Proc. Natl. Acad. Sci. 2005;102:12548–12553. doi: 10.1073/pnas.0503407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr, J.M., Babcock, H.P., Fujimoto, B.S. A test of the model-free formulas. Effects of anisotropic rotational diffusion and dimerization. J. Magn. Reson. B. 1994;105:211–224. doi: 10.1006/jmrb.1994.1127. [DOI] [PubMed] [Google Scholar]
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., Staskawicz, B.J. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- Shan, L., He, P., Zhou, J.M., Tang, X. A cluster of mutations disrupt the antivirulence but not the virulence function of AvrPto. Mol. Plant-Microbe Interact. 2000;13:592–598. doi: 10.1094/MPMI.2000.13.6.592. [DOI] [PubMed] [Google Scholar]
- Shea, J.E., Onuchic, J.N., Brooks C.L., III Probing the folding free energy landscape of the Src-SH3 protein domain. Proc. Natl. Acad. Sci. 2002;99:16064–16068. doi: 10.1073/pnas.242293099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg, J.A., Miller, N.C., Marketon, M.M., Schneewind, O. Rejection of impassable substrates by Yerinia type III secretion machines. J. Bacteriol. 2005;187:7090–7102. doi: 10.1128/JB.187.20.7090-7102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sory, M.P., Cornelis, G.R. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- Stebbins, C.E., Galan, J.E. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- Tamano, K., Aizawa, S., Katayama, E., Nonaka, T., Imajoh-Ohmi, S., Kuwae, A., Nagai, S., Sasakawa, C. Supramolecular structure of the Shigella type III secretion machinery: The needle part is changeable in length and essential for delivery of effectors. EMBO J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Xie, M., Kim, Y.J., Zhou, J., Klessing, D.F., Martin, G.B. Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell. 1999;11:15–29. doi: 10.1105/tpc.11.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk, K., Fouts, D.E., Rehm, A.H., Hill, A.R., Collmer, A., Alfano, J.R. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J. Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, P.A., Leong, L.E., Ng, P.W., Tan, S.H., Waller, S., Murphy, D., Porter, A.G. Efficient and rapid affinity purification of proteins using recombinant fusion proteases. Biotechnology (N. Y.) 1994;12:601–605. doi: 10.1038/nbt0694-601. [DOI] [PubMed] [Google Scholar]
- Weast, R.C. 63rd ed. CRC Press; Boca Raton, FL: 1982. CRC handbook of chemistry and physics. [Google Scholar]
- Wilharm, G., Lehmann, V., Krauss, K., Lehnert, B., Richter, S., Ruckdeschel, K., Heesemann, J., Trulzsch, K. Yersinia enterocolitica type III secretion depends on the proton motive force but not on the flagellar motor components MotA and MotB. Infect. Immun. 2004;72:4004–4009. doi: 10.1128/IAI.72.7.4004-4009.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner, D.E. Nuclear spin relaxation in ellipsoids undergoing rotational Brownian motion. J. Chem. Phys. 1962;37:647–654. [Google Scholar]
- Woestyn, S., Allaoui, A., Wattiau, P., Cornelis, G.R. YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol. 1994;176:1561–1569. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf, J., Pascuzzi, P.E., Fahmy, A., Martin, G.B., Nicholson, L.K. The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis. Structure. 2004;12:1257–1268. doi: 10.1016/j.str.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Wulff-Strobel, C.R., Williams, A.W., Straley, S.C. LcrQ and SycH function together a the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Mol. Microbiol. 2002;43:411–423. doi: 10.1046/j.1365-2958.2002.02752.x. [DOI] [PubMed] [Google Scholar]
- Xiao, Y., Lu, Y., Heu, S., Hutcheson, S.W. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J. Bacteriol. 1992;174:1734–1741. doi: 10.1128/jb.174.6.1734-1741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura, K., Maki-Yonekura, S., Namba, K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.Y., Clemens, J.C., Schubert, H.L., Stuckey, J.A., Fischer, M.W., Hume, D.M., Saper, M.A., Dixon, J.E. Expression, purification, and physiochemical characterization of a recombinant Yersinia protein tyrosine phosphatase. J. Biol. Chem. 1992;267:23759–23766. [PubMed] [Google Scholar]