Abstract
Evolutionarily conserved triad glutamine amidotransferase (GAT) domains catalyze the cleavage of glutamine to yield ammonia and sequester the ammonia in a tunnel until delivery to a variety of acceptor substrates in synthetase domains of variable structure. Whereas a conserved hydrolytic triad (Cys/His/Glu) is observed in the solved GAT structures, the specificity pocket for glutamine is not apparent, presumably because its formation is dependent on the conformational change that couples acceptor availability to a greatly increased rate of glutamine cleavage. In Escherichia coli carbamoyl phosphate synthetase (eCPS), one of the best characterized triad GAT members, the Cys269 and His353 triad residues are essential for glutamine hydrolysis, whereas Glu355 is not critical for eCPS activity. To further define the glutamine-binding pocket and possibly identify an alternative member of the catalytic triad that is situated for this role in the coupled conformation, we have analyzed mutations at Gln310, Asn311, Asp334, and Gln351, four conserved, but not yet analyzed residues that might potentially function as the third triad member. Alanine substitution of Gln351, Asn311, and Gln310 yielded respective Km increases of 145, 27, and 15, suggesting that Gln351 plays a key role in glutamine binding in the coupled conformation, and that Asn311 and Gln310 make less significant contributions. None of the mutant k cat values varied significantly from those for wild-type eCPS. Combined with previously reported data on other conserved eCPS residues, these results strongly suggest that Cys269 and His353 function as a catalytic dyad in the GAT site of eCPS.
Keywords: glutamine, amidotransferase, carbamoyl phosphate, ammonia, hydrolytic triad
Because free ammonia is highly reactive and can readily diffuse across cell membranes, two types of glutamine amidotransferases (GAT) have evolved to avoid potential ammonia toxicity by catalyzing direct delivery of ammonia to acceptor substrates for the biosynthesis of purine and pyrimidine nucleotides, coenzymes, amino acids, and amino sugars. In both GAT types, the nucleophilic sulfhydryl side chain of a cysteinyl residue initiates the amide transfer by forming a thioester with the substrate glutamine. The reactive cysteine is part of a conserved triad of amino acid residues (Cys/His/Glu) in the triad GAT family, and is incorporated in a β-sheet in the N-terminal nucleophile (Ntn) GAT family (Zalkin and Smith 1998). The GAT domain has an α/β structure with the conserved residues clustered at one end of the core β-sheet, as demonstrated in all available crystal structures for triad GAT family members: GMP synthetase (Tesmer et al. 1996), carbamoyl phosphate synthetase (Thoden et al. 1997), anthranilate synthase (Knochel et al. 1999; Morollo and Eck 2001; Spraggon et al. 2001), imidazole glycerol phosphate synthetase (Douangamath et al. 2002; Korolev et al. 2002; Omi et al. 2002; Myers et al. 2003), aminodeoxychorismate synthase (McKinzie and Parsons 2002), CTP synthetase (Endrizzi et al. 2004; Hagihara et al. 2004), and formylglycinamidine synthetase (Hoskins et al. 2004). Although the overall fold of the GAT domain appears unrelated to that of any other enzyme with a catalytic triad, the conserved Cys/His/Glu residues assume a three-dimensional arrangement similar to that of the established catalytic triads for many hydrolytic enzymes, including serine and cysteine proteases, transglutaminases, and α/β hydrolases (Fig. 1; Tesmer et al. 1996; Nardini and Dijkstra 1999). The solved GAT structures each contain two additional features that are also observed in the other hydrolytic structural families, an “oxyanion hole” and “nucleophile elbow” location of the triad cysteine (Tesmer et al. 1996). Occurrence of this catalytic residue at the “elbow” that is both the last residue of a β-strand and the first residue of an α-helix yields a distinctive disallowed backbone conformation (ϕ = 55°, ψ = 110°).
Figure 1.
eCPS glutamine amidotransferase triad residues Cys269, His353, and Glu355 (PDB file 1jdbF) aligned with triad residues of papain (PDB file 9pap), trypsin (PDB file 1sgt), and subtilisin (PDB file 1sue).
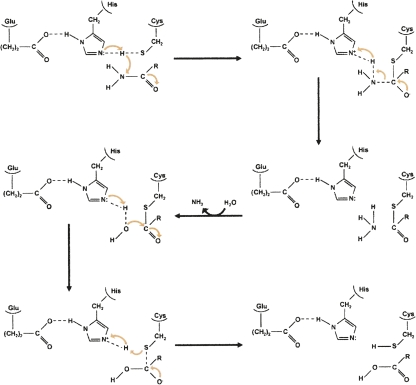
Escherichia coli carbamoyl phosphate synthetase (eCPS), which catalyzes the formation of the initial intermediate in the biosynthesis of both pyrimidine nucleotides and arginine, is one of the most extensively studied triad GATs. Two of the triad residues are known to be critical for the GAT activity of eCPS and to carry out the catalytic roles well established for the corresponding triad residues of serine proteases such as trypsin (Thoden et al. 1999). The triad cysteine (Cys269 in eCPS) forms a thiolate anion that attacks the carbonyl carbon of the substrate glutamine carboxamide group to yield a tetrahedral intermediate (Fig. 2). The transient negative charge on the glutamine amide oxygen in this intermediate is stabilized by the backbone amide hydrogens of the conserved “oxyanion hole.” Protonation of the amino group of the tetrahedral intermediate makes it a preferred leaving group. The intermediate then collapses to form free ammonia that departs via the internal channel and a glutamyl thioester intermediate that has been visualized by X-ray crystallography (Thoden et al. 1998). Finally, the glutamyl thioester is hydrolyzed to release the product glutamate and restore the triad cysteine residue, with acid/base catalysis increasing the nucleophilicity of the attacking water. The triad histidine (His353 in eCPS) appears to assist in forming the thiolate anion and/or acting as a general acid/base catalyst during hydrolysis of the thioester intermediate.
Figure 2.
Mechanism for the generation of ammonia within the glutamine amidotransferase domain of eCPS. The carbonyl carbon of the glutamine side-chain carboxamide group is attacked by the thiolate anion of Cys269, yielding a tetrahedral intermediate. The collapse of this intermediate is facilitated by protonation of the amino group by His353, thereby forming a thioester intermediate and ammonia. The ammonia departs, after which the thioester intermediate is hydrolyzed by a water molecule that has been activated through its interaction with His353. Dashed lines represent H-bonds.
Whereas mutagenesis studies have verified the critical roles of Cys269 and His353 (Rubino et al. 1986; Miran et al. 1991), they have ruled out a significant role for the third eCPS canonical triad member, Glu355 (Huang and Raushel 1999). In the present work, we have carried out mutation analysis on potentially critical conserved residues in the eCPS GAT domain to determine whether any of these provide the significant rate enhancement expected for a residue that plays the expected role of the third triad member, i.e., increasing the nucleophilicity of Cys269 by increasing the polarity of His353 and/or accelerating the hydrolysis of the thioester intermediate. These studies were also intended to identify additional residues that contribute to the binding of glutamine and/or to the coupling of activity between the GAT and ammonia acceptor active sites of eCPS, and thereby contribute to the definition of the high-activity GAT conformation. For all GATs, glutamine hydrolysis activity is significantly stimulated when the GAT acceptor site is occupied by the substrate to which the ammonia moiety is transferred, with about 600-fold stimulation for eCPS (Zalkin and Smith 1998). This high-activity GAT conformation has thus far eluded crystal analysis, since only a single conformation, without an apparent glutamine specificity pocket, has been obtained for any GAT (Mouilleron and Golinelli-Pimpaneau 2007). Therefore, functional analysis, as reported in the present work, is essential for definition of the high-activity GAT conformation that occurs when acceptor substrate availability is coupled with glutamine availability.
Results
Identification of residues potentially critical for GAT domain activity
eCPS is a heterodimeric enzyme with a 42-kDa subunit containing the GAT domain and a 118-kDa subunit catalyzing the synthesis of CP from the ammonia channeled from the GAT domain utilizing bicarbonate and two molecules of MgATP (Fig. 3). Previous multisequence alignment analysis of the 200 residues within the eCPS GAT domain, as summarized in Figure 4A, revealed that 30 residues are invariant in 17 glutamine-utilizing CPSs, including eCPS (Saeed-Kothe and Powers-Lee 2003). Included among these 30 residues are the three triad residues (Cys269, His353, Glu355) and six additional residues (Gly241, Gly267, Gly271, Gln273, His312, Pro354) that are invariant in all triad GATs (Zalkin and Smith 1998). To identify candidate eCPS residues that might carry out the canonical proton-transfer role expected of the third catalytic triad residue, we have targeted the remaining GAT residues conserved in glutamine-utilizing CPSs (Fig. 4A). Site-directed mutagenesis of eCPS has demonstrated roles in binding glutamine for Leu270 (Saeed-Kothe and Powers-Lee 2003), Gln273 (Huang and Raushel 1999; Saeed-Kothe and Powers-Lee 2003), and His312 (Miran et al. 1991), as well as conserved Ser47 (Thoden et al. 1999) from the N-terminal domain of eCPS. As shown in Figure 4B and Table 1, the location of these residues in the solved crystal structure of eCPS is consistent with their involvement in glutamine binding. Additional mutagenesis analysis has suggested that Lys202 forms a salt bridge to Glu355 (Huang and Raushel 1999) and has ruled out a significant role for Asn240 in glutamine interaction (Saeed-Kothe and Powers-Lee 2003). Of the remaining residues conserved in glutamine-utilizing CPSs, only seven (Asp198, Lys285, Asn294, Gln310, Asn311, Asp334, Gln351) have side chains that could participate in proton transfer. As shown in Figure 4C and Table 1, Asp198, Lys285, Asn294, and Asp 334 are so distant from the glutamine site that even a significant conformational change is unlikely to allow their participation in catalysis. Therefore, we targeted for analysis the three conserved residues that are closer to the glutamine site: Gln310, Asn311, and Gln351 (Table 1; Fig. 4C). We also constructed Asp334Ala, based on an incorrect distance estimate, and included it in the study since it was available. It is noteworthy that most cysteine proteases have Asn or Gln as the third triad member in place of the Asp/Glu occurring in serine proteases (Brannigan et al. 1995).
Figure 3.
Ribbon representation of one heterodimer of eCPS. A2 (purple) is the GAT domain and A1 (violet) is involved in communicating active site occupancy between the GAT and synthetase subunits. Domains B (green) and C (red) are regions of internal duplication, and each contains an ATP grasp fold. Domains D′ (yellow) and D (orange) contain the interfaces for eCPS self-association. In addition to this role, domain D is the site for allosteric regulation of eCPS. The intramolecular tunnel connecting the three active sites is illustrated with arrows. Also shown are the individual chemical reactions at each of the active sites that are synchronized in the overall reaction. Created from PDB file 1a9x.
Figure 4.
Identification of potentially critical eCPS GAT domain residues. (A) Invariant residues in the GAT domain of eCPS. Analysis of an amino acid sequence alignment revealed 30 residues that are invariant in the GAT domain of CPSs from 20 different organisms. The nine residues shown in blue are invariant in all triad GAT domains. The canonical Cys-His-Glu residues are indicated by ↓. The four residues shown in red are presently targeted for mutagenesis. (B) Conserved residues in the GAT domain of eCPS that have been subjected to previous mutation analysis. Residues in blue are members of the catalytic triad. Created from PDB file 1jdb. (C) Conserved residues in the GAT domain of eCPS with yet undefined function and with side chains that could participate in proton transfer. The residues in blue are those currently targeted for mutagenesis. Created from PDB file 1a9x.
Table 1.
Distances between canonical triad residues and other eCPS residues
Functional analysis of site-directed mutations
The four candidate residues for glutamine interaction, Gln310, Asn311, Asp334, and Gln351, were separately substituted by alanine. The functional effects of these mutations were identified by analyzing the overall and partial reactions catalyzed by eCPS. The synthesis of CP requires coordination of three eCPS active sites, with the GAT site occurring in domain A, and ATP cleavage occurring at duplicated ATP grasp domains B and C (Fig. 3). Individual contributions of the three active sites to the overall reaction are reflected in three partial reactions: (1) at the GAT site, hydrolysis of glutamine; (2) at ATPB, bicarbonate-dependent ATP hydrolysis that reflects formation of the intermediate carboxyphosphate and its hydrolysis in the absence of ammonia; and (3) at ATPC, the synthesis of ATP from CP and ADP that appears to reflect reversal of carbamate formation (Holden et al. 1999; Saeed-Kothe and Powers-Lee 2003). All three individual partial reactions proceed relatively slowly, but are conformationally coupled when all substrates are present (Miles and Raushel 2000). The rates of reaction at all three sites are increased and are synchronized so that one molecule of glutamine and two molecules of ATP are cleaved for each molecule of CP synthesized.
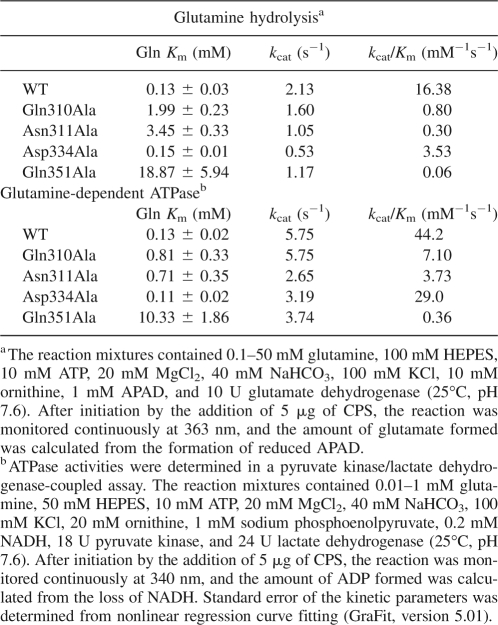
All four eCPS variants were able to effectively catalyze glutamine-dependent CP synthesis under standard assay conditions with all substrates present in excess. The specific activities, in micromoles CP/min/mg, were as follows: 0.855 for wild-type eCPS, 0.822 for Gln310Ala, 0.160 for Asn311Ala, 0.311 for Asp334Ala, and 0.200 for Gln351Ala. The kinetic parameters for glutamine utilization by wild-type and mutant eCPSs are shown in Table 2. None of the mutant k cat values varied significantly from those for wild-type eCPS. In both assays shown, the amount of glutamine was varied and all other substrates were present in excess. In the glutamine hydrolysis assay, the formation of glutamate was followed by coupling the CPS reaction with the glutamate dehydrogenase reaction. In the glutamine-dependent ATPase assay, the formation of ADP was followed by coupling the CPS reaction with that of pyruvate kinase and lactate dehydrogenase. Variations in the kinetic parameters obtained in the two assays presumably reflect the varying reaction mix components and/or the varying amounts of product formation, i.e., no glutamate buildup in the glutamine-hydrolysis assay and no ADP buildup or ATP depletion in the ATPase assay. Previous studies of eCPS-catalyzed glutamine hydrolysis (Miles et al. 1998) have established that the rate-limiting step is formation of the thioester intermediate when ATP and bicarbonate are present. In contrast, in the uncoupled conformation (i.e., only glutamine is present), the rate-determining step is the hydrolysis of the thioester intermediate. The observation of essentially unchanged ability to catalyze ammonium-dependent CP formation, where high levels of ammonium chloride can substitute for ammonia derived from the preferred substrate glutamine (Rubino et al. 1986), suggested the absence of any gross conformational change in the variants.
Table 2.
Kinetic parameters for utilization of glutamine by WT and mutant eCPSs
The mutations Gln310Ala and Asn311Ala resulted in 15- and 27-fold greater K m values for glutamine, respectively, as gauged by the glutamine-hydrolysis assay. When determined in the glutamine-dependent ATPase assay, the glutamine K m values were increased, but to a lesser extent, with six- and fivefold increases, respectively, for Gln310Ala and Asn311Ala. The Asp334Ala mutation had essentially no effect on the K m for glutamine. However, the Gln351Ala mutation yielded a strikingly large effect on the glutamine K m with a 145-fold increase observed in the glutamine-hydrolysis assay and a 79-fold increase observed in the glutamine-dependent ATPase assay. For all constructs, the kinetic parameters for ATP utilization showed no significant differences from those for the wild-type enzyme (Table 3), again suggesting that the substitutions did not cause gross conformational changes.
Table 3.
Kinetic parameters for utilization of ATP by WT and mutant eCPSs
Synchronization of the eCPS catalyzed reaction
The synchronized overall reaction of wild-type eCPS is expected to yield products in a ratio of 1:1:2 for CP:Glu:ADP (Miles and Raushel 2000). Wild type and Gln310Ala showed the expected stoichiometry of glutamine cleavage to CP formation, with respective ratios of 1 CP, 0.9 Glu and 2.7 ADP and 1 CP, 1.3 Glu and 3.6 ADP, while Asp334Ala exhibited only slight uncoupling of glutamine cleavage, with a ratio of 1 CP, 2.2 Glu and 4.0 ADP. Both the Asn311Ala and Gln351Ala mutants showed more substantial uncoupling of glutamine cleavage from CP formation, with respective ratios of 1 CP, 4.6 Glu and 4.7 ADP and 1 CP, 2.8 Glu and 3.4 ADP.
Discussion
The present findings provide novel insight into the interaction of eCPS with glutamine. Although all members of the triad GAT family appear to undergo conformational changes that reciprocally signal ligand binding at the glutamine and acceptor sites, the crystal structure for only a single conformation has been determined for eCPS and the other GAT members with solved structures. In all cases, the solved glutamine-binding site is only incompletely formed (Mouilleron and Golinelli-Pimpaneau 2007). Thus, site-directed mutagenesis data are especially powerful in supplementing the available crystal structure data to yield an understanding of how eCPS interacts with glutamine in the fully formed glutamine site, with a turnover rate ∼600 times greater than that for glutamine in the absence of acceptor substrates. In the solved eCPS structure, specific side-chain interactions with glutamine were observed only for Ser47 and Gln273 (Thoden et al. 1999). The Ser47 hydroxyl group is within hydrogen-bonding distance of the carbonyl oxygen of the glutamyl side chain and the side-chain carboxamide of Gln273 is within hydrogen-bonding distance of the α-carboxyl group of glutamine. Additionally, backbone amide and/or carbonyl groups of Gly241, Gly243, Leu270, Asn311, Gly313, and Phe314 are positioned to hydrogen bond with the carboxamide, amino, and carboxyl moieties of glutamine. The present data strongly suggest that the side chain of Gln351 also plays a key role in glutamine binding, with a 145-fold higher K m for Gln351Ala, and that reorientation of this side chain is a component of the conformational change at the GAT domain that signals binding of ATP at the synthetase domain. The side chains of Asn311 and Gln310 appear to make less significant contributions to glutamine binding, making their reorientation as part of the activating conformation change possible, but not nearly as probable as Gln351 participation.
It is also notable that substitution of the smaller residue alanine for either Gln351 or Asn311 yields a three- to fivefold uncoupling of glutamine cleavage from CP formation. Inspection of the tunnel that channels ammonia from the GAT site to the acceptor site, where CP is formed in the eCPS crystal structure, revealed 10 amino residues that are located within 3.5 Å of the center of the molecular tunnel (Huang and Raushel 2000), with Asn311 (but not Gln351) included in the 10. Earlier studies aimed at blocking the tunnel (Huang and Raushel 2000) utilized only substitution of larger side chains for channel residues Ser35, Met36, Asp45, Gly293, Ala309, and Gly359. Substitution of leucine, tyrosine, or phenylalanine for Gly359 resulted in almost complete uncoupling of activities at the GAT and synthetase sites. The mutants Ser35Phe and Gly359Ser yielded, respectively, two- and fivefold uncoupling of glutamine cleavage from CP formation, values similar to those presently observed for Gln351Ala and Asn311Ala. The uncoupling observed in the present studies suggests that substitution of smaller residues can also cause a partial block in the tunnel that channels ammonia from the GAT site to the acceptor site, where CP is formed, presumably via a rearrangement of the normal molecular interactions underlying tunnel formation.
To complement earlier studies (Huang and Raushel 1999) that demonstrated the substitution of the third eCPS canonical triad member, Glu355, does not significantly affect activity, we have performed mutation analysis on four other conserved eCPS residues that might potentially function as expected for the third member of a classical catalytic triad. Our present finding that alanine replacements for Gln310, Asn311, Asp334, and Gln351 do not significantly alter the maximal enzymatic rates of glutamine cleavage or CP formation disproves the hypothesis that any of these four residues are critically involved in catalysis of glutamine hydrolysis. Combined with previously reported data on other conserved eCPS residues, these results suggest that Cys269 and His353 function as a catalytic dyad in the GAT site of eCPS. Neither Glu355, the canonical third member of the GAT triad, nor any of the other conserved residues that occur near the glutamine site and have side chains capable of proton transfer, contribute significantly to the eCPS k cat value. In contrast, substitution of the corresponding triad residue of trypsin in the Asp102Asn mutant reduced the k cat to 0.003% of wild type (Corey et al. 1994). It must be noted that it is possible that a nonconserved residue might function as the third member of the GAT triad or that a large conformational change might position one of the distant conserved residues.
Although the apparent functioning of a catalytic dyad in eCPS behavior contrasts starkly with the requirement for an entire functional triad in the serine triad proteases, it is consistent with the observations for several cysteine triad proteases. Histidine is the only residue conserved in all triad proteases (Brannigan et al. 1995), with most cysteine proteases having Cys, His, and Asn as the triad in place of the serine protease triad Ser, His, and Asp/Glu. It has been proposed that the greater nucleophilicity of sulfur relative to oxygen allows the substitution of a third triad member with less ability to enhance the nucleophilicity (Brannigan et al. 1995). In the case of caspases (Chereau et al. 2003) and papain (Vernet et al. 1995), the third triad member Asn is conserved, but fails to act as an essential catalytic residue, presumably since the nucleophilicity of the Cys/His dyad is sufficient for effective catalysis. It is also notable that for the only member of the triad GAT family examined, p-aminobenzoate synthetase, replacement of the triad glutamyl residue had a minimal effect on glutamine hydrolysis, eliminating the possibility that the glutamyl residue was directly essential in catalysis (Roux and Walsh 1993). It thus appears that use of a catalytic dyad rather than a triad might well be a general feature of the “triad” GAT family. Additionally, γ-glutamyl hydrolase has been shown to have significant structural similarity to the GAT domain of eCPS (but not sufficient similarity to constitute a GAT family member) and to have a triad glutamyl residue that is not catalytically essential (Chave et al. 2000).
Our results suggest that Cys269 of eCPS, intrinsically a stronger nucleophile than the classic triad residue serine, does not require the assistance of a third triad residue for ionization. It is possible that a water molecule supplies the eCPS GAT site with some fraction of the acid/base catalysis normally supplied by the third triad residue in serine (and some cysteine) proteases. The eCPS crystal structure shows that a water molecule is hydrogen bonded to His353, as well as Asn311 and Gln351, and is well positioned to replace the canonical triad Glu/Asp salt bridge to His353 (Fig. 5). Although Glu355 does not provide the catalytic function of the canonical third triad member, it has been conserved in the eCPS GAT sequence throughout evolution and has been conserved in a triad-mimicking structure in the only solved CPS structure. Presumably, Glu355, possibly via the apparent salt bridge to the conserved Lys202 (Huang and Raushel 1999), has assumed a critical role in folding of nascent eCPS and/or in maintaining the folded structure that does not allow substitution.
Figure 5.
Orientation of eCPS residues Asn311, Gln351, and His353. Distances (in Å) are shown from the O of the hydrogen-bonded water. Created from PDB file 1a9x.
Materials and Methods
Strains, plasmids, and DNA methods
The expression host was E. coli strain L673, which lacks both CPS subunits and is defective in the Lon protease (Guillou et al. 1989). The expression plasmid pUCAB (8594 bp), encoding both the GAT and synthetase components of eCPS (carA and carB, respectively) under the control of the IPTG inducible trc promoter, was described previously (Saeed-Kothe and Powers-Lee 2003). XL1-Blue and Top10 E. coli were used for DNA manipulation. Bacterial transformations and recombinant DNA techniques were carried out as described in Sambrook et al. (2001). Site-directed mutants were created using nested PCR (Kwok and Higuchi 1989). Each mutagenesis cassette was sequenced to verify that no undesired changes were incorporated into the nucleotide sequence. Mutagenesis primers (mutated base pairs in bold) and outside non-mutagenic primers were as follows:
Q310A: 5′-CGTGGTAATGATAACCGCCGCGAACCACGGTTTTGCG-3′
N311A: 5′-CGTGGTAATGATAACCGCCCAGGCCCACGGTTTTGCG-3′
D334A: 5′-GCATAAATCCCTGTTCGCCGGTACCTTACAGGGCATTC-3′
Q351A: 5′-GCATTCAGCTTCGCCGGGCACCCCGAGGCCAGCCCTGGTC-3′
MPPF1: 5′-GACCTCTCTTCTTACCTGAAACGC-3′
MPPR3: 5′-TCTTTGTCGGTCAGCGTTTGGGC-3′.
eCPS purification and activity determination
The purification protocol for recombinant wild-type eCPS and for its site-directed mutants was as previously described (Saeed-Kothe and Powers-Lee 2003). The purity of all protein preparations was at least 95%, as assessed by Coomassie blue staining of SDS-PAGE gels (Laemmli 1970). Protein concentrations were determined either by the dye-binding assay of Bradford with bovine serum albumin as the standard (McGivan et al. 1976) or by measuring the absorbance at 280 nm (0.685 for 1 mg/mL eCPS) (Rubino et al. 1986). CP synthesis was determined in a two-step assay by coupling the CPS reaction to that of ornithine transcarbamoylase and then quantitating the resulting citrulline (Saeed-Kothe and Powers-Lee 2003). The reaction mixtures contained 50 mM HEPES, 100 mM KCl, 10 mM ATP, 20 mM MgCl2, 20 mM NaHCO3, 1 mM DTT, 5 mM ornithine, 0.2 units ornithine transcarbamoylase, and either 300 mM NH4Cl or 10 mM glutamine, final pH 7.6. ATPase activities were determined in a pyruvate kinase/lactate dehydrogenase coupled assay (Saeed-Kothe and Powers-Lee 2003). The reaction mixtures contained 10 mM ATP, 50 mM HEPES, 100 mM KCl, 20 mM MgSO4, 40 mM NaHCO3, 20 mM ornithine, 1 mM sodium phosphoenolpyruvate, 0.2 mM NADH, 18 units pyruvate kinase, and 24 units lactate dehydrogenase, final pH 7.6. To determine glutamine-dependent ATPase activity, 10 mM glutamine was included. For determination of bicarbonate-dependent ATPase activity, the assay was carried out in the absence of both NH4Cl and glutamine. ATP synthesis was assayed by following the rate of ATP formation from ADP and CP using a hexokinase/glucose-6-phosphate dehydrogenase coupling system (Saeed-Kothe and Powers-Lee 2003). The reaction mixtures contained 50 mM HEPES, 1 mM ADP, 20 mM MgCl2, 100 mM KCl, 1 mM NADP+, 20 mM glucose, 10 mM CP, 5 units hexokinase, and 2.5 units glucose-6-phosphate dehydrogenase, final pH 7.6. Glutamine hydrolysis was determined as previously described (Miles et al. 1998) by coupling glutamate formation to the glutamate dehydrogenase catalyzed reduction of 3-acetylpyridine dinucleotide (APAD, ε363 = 8.3 mM−1cm−1). The reaction mixtures contained 100 mM HEPES, 10 mM ATP, 20 mM MgCl2, 10 mM glutamine, 40 mM NaHCO3, 100 mM Kcl and 10 mM ornithine, 1 mM APAD, and 20 units glutamate dehydrogenase, final pH 7.6. Kinetic data were collected on a Beckman DU 640 spectrophotometer and were fit by nonlinear regression (Grafit, version 5.1) to the equation v = V max S/(K m + S), where v is the initial velocity, V max is the maximal velocity, S is the substrate concentration, and K m is the Michaelis-Menten constant.
Visualization of eCPS crystal structure
Figures that include representations of the eCPS crystal structure were created using the UCSF Chimera (Pettersen et al. 2004) and Friend (Abyzov et al. 2005) programs.
Acknowledgments
We thank Amna Saeed-Kothe for valuable discussions and Alexej Abyzov for bioinformatics expertise. This work was supported by National Institutes of Health Grant DK54423.
Footnotes
Reprint requests to: Susan G. Powers-Lee, Department of Biology, Northeastern University, 360 Huntington Avenue, Boston, MA 02115-5000, USA; e-mail: spl@neu.edu; fax: (617) 373-3724.
Abbreviations: APAD, 3-acetylpyridine dinucleotide; CP, carbamoyl-phosphate; CPS, carbamoyl-phosphate synthetase; eCPS, E. coli carbamoyl-phosphate synthetase; GAT, glutamine amidotransferase.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073428008.
References
- Abyzov, A., Errami, M., Leslin, C.M., Ilyin, V.A. Friend, an integrated analytical front-end application for bioinformatics. Bioinformatics. 2005;21:3677–3678. doi: 10.1093/bioinformatics/bti602. [DOI] [PubMed] [Google Scholar]
- Brannigan, J.A., Dodson, G., Duggleby, H.J., Moody, P.C., Smith, J.L., Tomchick, D.R., Murzin, A.G. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- Chave, K.J., Auger, I.E., Galivan, J., Ryan, T.J. Molecular modeling and site-directed mutagenesis define the catalytic motif in human γ-glutamyl hydrolase. J. Biol. Chem. 2000;275:40365–40370. doi: 10.1074/jbc.M007908200. [DOI] [PubMed] [Google Scholar]
- Chereau, D., Kodandapani, L., Tomaselli, K.J., Spada, A.P., Wu, J.C. Structural and functional analysis of caspase active sites. Biochemistry. 2003;42:4151–4160. doi: 10.1021/bi020593l. [DOI] [PubMed] [Google Scholar]
- Corey, J.L., Guastella, J., Davidson, N., Lester, H.A. GABA uptake and release by a mammalian cell line stably expressing a cloned rat brain GABA transporter. Mol. Membr. Biol. 1994;11:23–30. doi: 10.3109/09687689409161026. [DOI] [PubMed] [Google Scholar]
- Douangamath, A., Walker, M., Beismann-Driemeyer, S., Vega-Fernandez, M.C., Sterner, R., Wilmanns, M. Structural evidence for ammonia tunneling across the (βα)8 barrel of the imidazole glycerol phosphate synthase bienzyme complex. Structure. 2002;10:185–193. doi: 10.1016/s0969-2126(02)00702-5. [DOI] [PubMed] [Google Scholar]
- Endrizzi, J.A., Kim, H., Anderson, P.M., Baldwin, E.P. Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets. Biochemistry. 2004;43:6447–6463. doi: 10.1021/bi0496945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou, F., Rubino, S.D., Markovitz, R.S., Kinney, D.M., Lusty, C.J. Escherichia coli carbamoyl-phosphate synthetase: Domains of glutaminase and synthetase subunit interaction. Proc. Natl. Acad. Sci. 1989;86:8304–8308. doi: 10.1073/pnas.86.21.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara, S., Kumasawa, H., Goto, Y., Hayashi, G., Kobori, A., Saito, I., Nakatani, K. Detection of guanine-adenine mismatches by surface plasmon resonance sensor carrying naphthyridine-azaquinolone hybrid on the surface. Nucleic Acids Res. 2004;32:278–286. doi: 10.1093/nar/gkh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden, H.M., Thoden, J.B., Raushel, F.M. Carbamoyl phosphate synthetase: An amazing biochemical odyssey from substrate to product. Cell. Mol. Life Sci. 1999;56:507–522. doi: 10.1007/s000180050448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, A.A., Anand, R., Ealick, S.E., Stubbe, J. The formylglycinamide ribonucleotide amidotransferase complex from Bacillus subtilis: Metabolite-mediated complex formation. Biochemistry. 2004;43:10314–10327. doi: 10.1021/bi049127h. [DOI] [PubMed] [Google Scholar]
- Huang, X., Raushel, F.M. Deconstruction of the catalytic array within the amidotransferase subunit of carbamoyl phosphate synthetase. Biochemistry. 1999;38:15909–15914. doi: 10.1021/bi991805q. [DOI] [PubMed] [Google Scholar]
- Huang, X., Raushel, F.M. Role of the hinge loop linking the N- and C-terminal domains of the amidotransferase subunit of carbamoyl phosphate synthetase. Arch. Biochem. Biophys. 2000;380:174–180. doi: 10.1006/abbi.2000.1913. [DOI] [PubMed] [Google Scholar]
- Knochel, T., Ivens, A., Hester, G., Gonzalez, A., Bauerle, R., Wilmanns, M., Kirschner, K., Jansonius, J.N. The crystal structure of anthranilate synthase from Sulfolobus solfataricus: Functional implications. Proc. Natl. Acad. Sci. 1999;96:9479–9484. doi: 10.1073/pnas.96.17.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev, S., Skarina, T., Evdokimova, E., Beasley, S., Edwards, A., Joachimiak, A., Savchenko, A. Crystal structure of glutamine amidotransferase from Thermotoga maritima . Proteins. 2002;49:420–422. doi: 10.1002/prot.10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S., Higuchi, R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McGivan, J.D., Bradford, N.M., Mendes-Mourao, J. The regulation of carbamoyl phosphate synthase activity in rat liver mitochondria. Biochem. J. 1976;154:415–421. doi: 10.1042/bj1540415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinzie, P.B., Parsons, B.L. Detection of rare K-ras codon 12 mutations using allele-specific competitive blocker PCR. Mutat. Res. 2002;517:209–220. doi: 10.1016/s1383-5718(02)00077-3. [DOI] [PubMed] [Google Scholar]
- Miles, B.W., Raushel, F.M. Synchronization of the three reaction centers within carbamoyl phosphate synthetase. Biochemistry. 2000;39:5051–5056. doi: 10.1021/bi992772h. [DOI] [PubMed] [Google Scholar]
- Miles, B.W., Banzon, J.A., Raushel, F.M. Regulatory control of the amidotransferase domain of carbamoyl phosphate synthetase. Biochemistry. 1998;37:16773–16779. doi: 10.1021/bi982018g. [DOI] [PubMed] [Google Scholar]
- Miran, S.G., Chang, S.H., Raushel, F.M. Role of the four conserved histidine residues in the amidotransferase domain of carbamoyl phosphate synthetase. Biochemistry. 1991;30:7901–7907. doi: 10.1021/bi00246a005. [DOI] [PubMed] [Google Scholar]
- Morollo, A.A., Eck, M.J. Structure of the cooperative allosteric anthranilate synthase from Salmonella typhimurium . Nat. Struct. Biol. 2001;8:243–247. doi: 10.1038/84988. [DOI] [PubMed] [Google Scholar]
- Mouilleron, S., Golinelli-Pimpaneau, B. Conformational changes in ammonia-channeling glutamine amidotransferases. Curr. Opin. Struct. Biol. 2007;17:653–664. doi: 10.1016/j.sbi.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Myers, R.S., Jensen, J.R., Deras, I.L., Smith, J.L., Davisson, V.J. Substrate-induced changes in the ammonia channel for imidazole glycerol phosphate synthase. Biochemistry. 2003;42:7013–7022. doi: 10.1021/bi034314l. [DOI] [PubMed] [Google Scholar]
- Nardini, M., Dijkstra, B.W. α/β Hydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Omi, R., Mizuguchi, H., Goto, M., Miyahara, I., Hayashi, H., Kagamiyama, H., Hirotsu, K. Structure of imidazole glycerol phosphate synthase from Thermus thermophilus HB8: Open–closed conformational change and ammonia tunneling. J. Biochem. 2002;132:759–765. doi: 10.1093/oxfordjournals.jbchem.a003284. [DOI] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., Ferrin, T.E. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Roux, B., Walsh, C.T. p-Aminobenzoate synthesis in Escherichia coli: Mutational analysis of three conserved amino acid residues of the amidotransferase PabA. Biochemistry. 1993;32:3763–3768. doi: 10.1021/bi00065a031. [DOI] [PubMed] [Google Scholar]
- Rubino, S.D., Nyunoya, H., Lusty, C.J. Catalytic domains of carbamyl phosphate synthetase. Glutamine-hydrolyzing site of Escherichia coli carbamyl phosphate synthetase. J. Biol. Chem. 1986;261:11320–11327. [PubMed] [Google Scholar]
- Saeed-Kothe, A., Powers-Lee, S.G. Gain of glutaminase function in mutants of the ammonia-specific frog carbamoyl phosphate synthetase. J. Biol. Chem. 2003;278:26722–26726. doi: 10.1074/jbc.M303774200. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., Maniatis, T. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Spraggon, G., Kim, C., Nguyen-Huu, X., Yee, M.C., Yanofsky, C., Mills, S.E. The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan. Proc. Natl. Acad. Sci. 2001;98:6021–6026. doi: 10.1073/pnas.111150298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer, J.J., Klem, T.J., Deras, M.L., Davisson, V.J., Smith, J.L. The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families. Nat. Struct. Biol. 1996;3:74–86. doi: 10.1038/nsb0196-74. [DOI] [PubMed] [Google Scholar]
- Thoden, J.B., Holden, H.M., Wesenberg, G., Raushel, F.M., Rayment, I. Structure of carbamoyl phosphate synthetase: A journey of 96 Å from substrate to product. Biochemistry. 1997;36:6305–6316. doi: 10.1021/bi970503q. [DOI] [PubMed] [Google Scholar]
- Thoden, J.B., Miran, S.G., Phillips, J.C., Howard, A.J., Raushel, F.M., Holden, H.M. Carbamoyl phosphate synthetase: Caught in the act of glutamine hydrolysis. Biochemistry. 1998;37:8825–8831. doi: 10.1021/bi9807761. [DOI] [PubMed] [Google Scholar]
- Thoden, J.B., Huang, X., Raushel, F.M., Holden, H.M. The small subunit of carbamoyl phosphate synthetase: Snapshots along the reaction pathway. Biochemistry. 1999;38:16158–16166. doi: 10.1021/bi991741j. [DOI] [PubMed] [Google Scholar]
- Vernet, T., Tessier, D.C., Chatellier, J., Plouffe, C., Lee, T.S., Thomas, D.Y., Storer, A.C., Menard, R. Structural and functional roles of asparagine 175 in the cysteine protease papain. J. Biol. Chem. 1995;270:16645–16652. doi: 10.1074/jbc.270.28.16645. [DOI] [PubMed] [Google Scholar]
- Zalkin, H., Smith, J.L. Enzymes utilizing glutamine as an amide donor. Adv. Enzymol. Relat. Areas Mol. Biol. 1998;72:87–144. doi: 10.1002/9780470123188.ch4. [DOI] [PubMed] [Google Scholar]