Abstract
A crystallization chaperone is an auxiliary protein that binds to a target of interest, enhances and modulates crystal packing, and provides high-quality phasing information. We critically evaluated the effectiveness of a camelid single-domain antibody (VHH) as a crystallization chaperone. By using a yeast surface display system for VHH, we successfully introduced additional Met residues in the core of the VHH scaffold. We identified a set of SeMet-labeled VHH variants that collectively produced six new crystal forms as the complex with the model antigen, RNase A. The crystals exhibited monoclinic, orthorhombic, triclinic, and tetragonal symmetry and have one or two complexes in the asymmetric unit, some of which diffracted to an atomic resolution. The phasing power of the Met-enriched VHH chaperone allowed for auto-building the entire complex using single-anomalous dispersion technique (SAD) without the need for introducing SeMet into the target protein. We show that phases produced by combining SAD and VHH model-based phases are accurate enough to easily solve structures of the size reported here, eliminating the need to collect multiple wavelength multiple-anomalous dispersion (MAD) data. Together with the presence of high-throughput selection systems (e.g., phage display libraries) for VHH, the enhanced VHH domain described here will be an excellent scaffold for producing effective crystallization chaperones.
Keywords: X-ray diffraction, crystallization, SeMet phasing, camelid single-domain antibody, VHH, yeast surface display
Structural biology has made a fundamental impact on our understanding of biological organization, function, and mechanism. Technical advances have fueled extraordinary progress toward attacking the most challenging and complex structural biology problems; however, currently we are realistically still only able to deal with a small subset of such problems. The major impediment for X-ray crystallography approaches still remains the production of high-quality diffracting crystals. This situation is not the result of any lack of effort to produce new tools or methods to assist in crystallization. In fact, this problem has been attacked on many fronts, from robotics to molecular biology (for reviews, see Weselak et al. 2003; McPherson 2004; Dauter 2006). Nevertheless, the process of crystallization is affected by many factors, including the chemical character of the protein and its inherent conformational heterogeneity that limit the number of productive lattice contacts that are available for crystallization.
An approach that holds promise especially for membrane proteins and large protein complexes is the use of crystallization chaperones. These chaperones are in the form of antibody fragments or other binding domains that have been engineered to bind specifically to a given protein target. The basis for the strategy is to increase the probability of getting well-ordered crystals by (1) increasing effective surfaces that can provide primary contact between molecules in the crystal lattice and (2) minimizing the conformational heterogeneity by binding to a specific conformation. An additional attribute inherent in the crystallization chaperone approach is that the chaperone can also provide initial model-based phasing information.
Much of the previous work employing crystallization chaperones involves application to crystallizing membrane proteins using antibody fragments (Fab and scFv) (for reviews, see Kovari et al. 1995; Hunte and Michel 2002; Venturi and Hunte 2003). These fragments are produced by proteolytic cleavage or recombinant reformatting of antibodies that have been raised to the particular protein target using animal immunization and the hybridoma technology. Unfortunately, this traditional approach is limited by its expense and throughput, which greatly reduce its potential as a broadly applicable method. Recent approaches based on recombinant technologies such as phage display libraries coupled with bacterial expression appear to have circumvented many of the issues limiting hybridoma-generated Fabs and thus greatly expand the potential of antibody fragments as crystallization chaperones (Brekke and Loset 2003; Bradbury and Marks 2004; Hoogenboom 2005; Sidhu and Fellouse 2006; Fellouse et al. 2007). Additionally, other non-antibody type scaffolds have been used such as ankyrin repeats (Schweizer et al. 2007) and fibronectin type 3 domains (Koide et al. 2007b).
A practical limitation for using Fabs for crystallization is their size, which is ∼550 amino acids, their heterodimeric architecture, and the low level of production in the bacterial systems. One might imagine that for some types of protein targets, especially the large class of proteins in the 100- to 300-amino-acid range, a smaller chaperone molecule would be preferable. In this regard, we introduce here the use of the antigen-binding domain of a camelid heavy-chain antibody as a viable alternative. The camels and other dromedaries have as a part of the antibody repertoire heavy-chain antibodies (the IgG2 and IgG3 classes) that lack the light chain and the CH domain found in the conventional IgG (Hamers-Casterman et al. 1993). The antigen-binding domains of the heavy-chain antibodies, termed VHH, include only the heavy-chain variable domain of the conventional antibody. These domains, despite their limited size (∼125 amino acids), possess binding loops that still can produce high-affinity interactions to protein targets (Muyldermans 2001). These domains are also structurally robust and are readily adapted to phage display systems to allow identification and affinity maturation in a similar fashion as has been exploited in the case of variable domains of conventional Fab antibody fragments (McCafferty 1990). The scaffold of a VHH domain has recently been engineered to optimize its use in phage display mutagenesis applications to produce highly specific binding partners to target proteins (Barthelemy et al. 2008). Therefore, the VHH domain is an attractive molecular scaffold with which one can develop a system to produce crystal chaperones.
When determining the structure of a target–chaperone complex, the quality of model-based phasing depends on the molecular mass of the chaperone relative to that in the total complex. Here, it would appear that the size of the Fab relative to VHH domains makes it the better phasing chaperone. However, the VHH domains have important advantages over Fabs in that they are amenable to facile misincorporation of SeMet into their scaffolds, facilitating the acquisition of complementary anomalous dispersion data. The number and spatial distribution of the SeMet sites and expression levels can also be optimized for the VHH scaffold. Importantly, by introducing SeMet exclusively into the chaperone molecule, the need to incorporate sites into the target molecules themselves is eliminated.
Although there are a number of crystal structures of antigen–antibody fragment complexes that can be viewed as successful examples of chaperone-assisted crystallography (CAC), there are a lack of systematic studies that address fundamental questions about the effectiveness of this approach. Do chaperones restrict the mode of crystal packing? How much phasing power can chaperones potentially produce? We present here a study that evaluates the chaperone-like crystallization and phasing capacities of a VHH domain. Our model system is a VHH domain (cAb-RN05 VHH) bound to its antigen, RNase A. It was originally isolated from an immunized camel and the crystal structure of the 1:1 antigen–antibody complex (Protein Data Bank [PDB] code 1BZQ) has been determined by Decanniere et al. (1999) at a 2.8 Å resolution.
In this work, we employed both structure-guided design and combinatorial library selection to produce VHH variants that exhibit enhanced levels of crystallizability and contain additional SeMet residues in the core. The engineered VHHs are capable of forming multiple crystal forms in the context of this complex, which can diffract up to atomic resolution. The addition of SeMet residues improved the phase quality significantly to such a level that high-quality electron density maps are produced by combining VHH model-based phases with single-wavelength anomalous dispersion (SAD) data. Together our results demonstrate that VHH domains have significant potential as crystallization chaperones and suggest the utility of the CAC approach.
Results
Introduction of additional Met residues into the VHH chaperone core
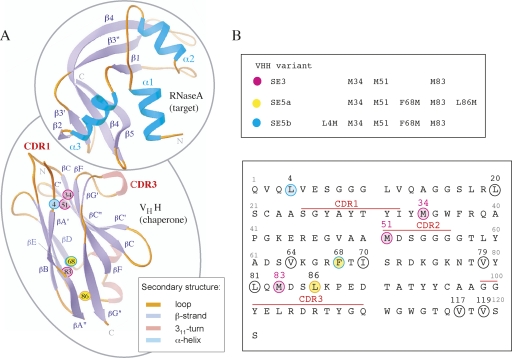
We have previously established phage display and yeast surface display systems for the cAb-RN05 VHH domain (Koide et al. 2007a). The native cAb-RN05 VHH domain contains three Met residues: M34, M51, and M83 (Fig. 1A,B). To evaluate and maximize the SeMet anomalous dispersion phasing capability of the VHH domain in the RNase A complex, additional Met residues were introduced into the VHH scaffold. Sites for Met incorporation that do not affect the protein folding and binding function were identified using a “shotgun methionine-scanning” selection strategy in a yeast surface display format. This strategy is similar to the shotgun mutagenesis technique developed by Sidhu and coworker for a phage display format (Weiss et al. 2000). A principal strength of the yeast surface display technology is in its ability to perform quantitative analysis and selection using fluorescence activated cell sorter (FACS). For each cell, the degree of surface display and that of target binding can be independently measured using specific, fluorescence-labeled detection reagents, allowing the acquisition of quantitative data on thermodynamics and kinetics of protein–target interaction (Boder and Wittrup 2000).
Figure 1.
(A) Ribbon diagrams of cAb-RN5 VHH and RNase A topology. The termini and the secondary structure elements are labeled. CDR1 and CDR3 residues are labeled in red. The position of the SeMet residues that were used for phasing are labeled and represented as small colored spheres. The Cα positions of the three native Met residues—34, 51, and 83—are shown in magenta. The Cα positions of two additional Met residues introduced into the SE5a variant (F68M and L86M) are shown in yellow, and those in the SE5b variant (L4M and F68M) are in cyan. (B) The amino acid sequence of cAb-RN05 VHH. Ten positions that were selected as potential sites for Met incorporation are circled. Positions at which SeMet is present in the proteins used in this study (shown in the upper panel) are colored in the same manner as in A.
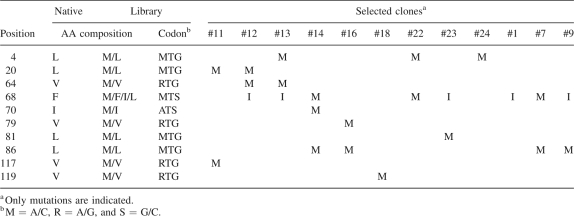
Ten spatially distributed positions in the hydrophobic core of the domain were chosen as potential methionine incorporation sites (Fig. 1B). A combinatorial library was constructed in which each of the 10 positions was mutated using an equal mixture of the native amino acid and Met, except for F68 where a combination of four amino acids were required to encode both Phe and Met, resulting in the total possible sequences of 2048 (Table 1). The yeast display library contains 2.5 × 104 independent clones, and thus, it is highly likely to include all sequences encoded by the library design. Only a small fraction of the naïve library was both expression and RNase A-binding positive as tested using FACS, indicating that some of the introduced mutations or combinations thereof are detrimental to the VHH function or stability. After two rounds of library sorting, the majority of the enriched library showed a FACS profile similar to that of the native (Supplemental Fig. S1).
Table 1.
The positions and amino acid compositions of the “methionine-scanning” library of the cAb-RN05 VHH and mutations found in selected clones
Sequence determination of 27 clones from the enriched library yielded 12 unique sequences with Met incorporation at each of the mutation sites (Table 1). Most clones contained no more than two or three additional methionine sites, indicating that there is an upper limit for substitution without destabilizing the protein. This observation is consistent with a large degree of destabilization of T4 lysozyme caused by multiple Met replacements (Gassner et al. 1996). The K d values were determined for three clones exhibiting the highest levels of binding in the yeast display format, all of which contained two additional methionine residues. The affinities for the clones 22 (L4M, F68M), 23 (F68I, L81M), and 7 (F68M, L86M) were 116 ± 33, 25 ± 8, and 20 ± 11 nM, respectively, similar to the value for native cAb-RN05 VHH (23 nM). Clones 7 and 22 that, respectively, contain two additional Met residues (Table 1) were chosen for the subsequent studies described below.
Multiple crystal forms of the VHH–RNase A complexes
The native and Met-enriched VHH proteins were expressed in Escherichia coli with the protein truncated at C-terminal residue 121, which removed a three native amino acids (RGR) and a His6 tag that were present in the original construct of Decanniere et al. (1999). We anticipated that the removal of the flexible C-terminal tail might facilitate crystallization (Derewenda 2004; Derewenda and Vekilov 2006).
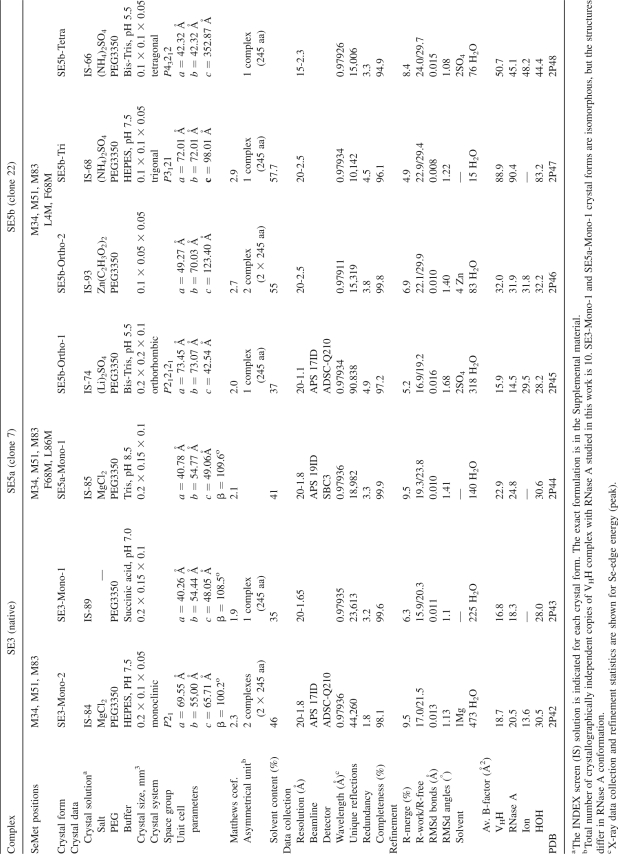
The native VHH (three SeMet sites) and the two selected VHH mutants (five SeMet sites) in complex with RNase A crystallized in several different space groups. From these VHH–RNase A complexes, six new crystal forms (Table 2), with the X-ray diffraction limits ranging from medium (∼2.5Å) to atomic resolution (1.1Å), were subsequently analyzed. Hereafter, these complexes are named based on the number of their SeMet sites: “SE3” refers to the native complex, “SE5a” to the mutant 7 complex, and “SE5b” to the mutant 22 complex (Fig. 1B). Generally, the solvent contents were relatively low (∼35%–45%), indicating that packing of the VHH–RNase A complex is very efficient in most of the space groups (Table 2). Interestingly, although the contacts between the N-terminal β-strands of the VHH are a common feature in several of the crystal forms, the other lattice contact interactions are generally quite distinct (Supplemental Figs. S2, S3).
Table 2.
Crystal data, X-ray data collection, and refinement statistics for cAb-RN05 VHH complexes with RNase A
In crystallization trials using commercially available screens, crystals appeared under multiple conditions containing PEG3350 (Table 2). SE5b was the most versatile complex, producing four different crystal forms without requiring any optimization of the crystallization conditions. Two forms are orthorhombic: SE5b-Ortho-1 crystals contain one molecule per asymmetric unit (ASU) and diffracted past 1.1 Å resolution; SE5b-Ortho-2, two molecules per ASU diffracted to 2.5 Å resolution. Additionally, the SE5b complex crystallized in a trigonal form (SE5b-Tri) and a tetragonal form (SE5b-Tetra) that diffracted to 2.5 Å and 2.3 Å resolution, respectively. We note that our objective was not to identify all possible crystal forms for this complex; our crystal screening strategy was relatively focused and did not involve an extensive search of crystallization space. Thus, it is probable that additional crystal forms could be obtained by a more expansive search strategy.
The identical monoclinic crystal form with one complex per ASU was identified for two complexes, SE3 and SE5a. The crystals diffracted to 1.65 Å (SE3) and 1.8 Å (SE5a) resolution and are referred to as SE3-Mono-1 and SE5a-Mono-1, respectively. Having isomorphous data for both the SE3 and SE5a complexes allowed us to make a direct comparison of the relative phasing capacity between chaperones containing three and five Se sites. In addition, the SE3 complex crystallized in a monoclinic space group with two molecules per ASU (SE3-Mono-2) that diffracts to 1.8 Å resolution. Diffraction data were collected, and selected crystallographic statistics are listed in Tables 2 and 3.
Phasing power of the VHH chaperones with different numbers of SeMet sites
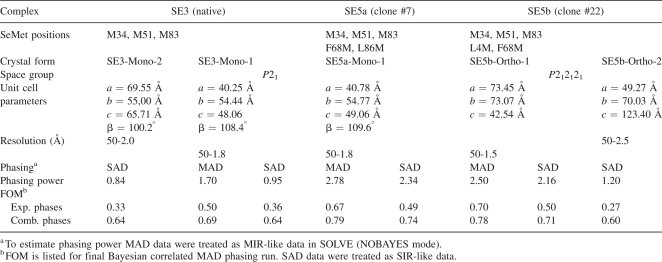
The relative phasing capacity of three vs. five SeMet sites and the contribution of the VHH model-based phases to the overall phasing potential of SAD and MAD data sets were evaluated using four different complexes: two with three SeMet sites in VHH (SE3-Mono-1, SE3-Mono-2) and two with five SeMet sites (SE5a-Mono-1, SE5b-Ortho-1). The SE5a and SE5b VHH variants differ by the location of one of the introduced SeMet groups: the C-terminal L86M in SE5a and N-terminal L4M in SE5b (Fig. 1).
Heavy atom searches using the Se anomalous signal in each crystal form indicated that all SeMet sites contribute to phasing. To compare the MAD and SAD approaches in the three and five SeMet cases, phases for two-wavelength (peak and inflection) and single-wavelength (peak) anomalous dispersion data sets were independently determined using SOLVE (Terwilliger and Berendzen 1999).
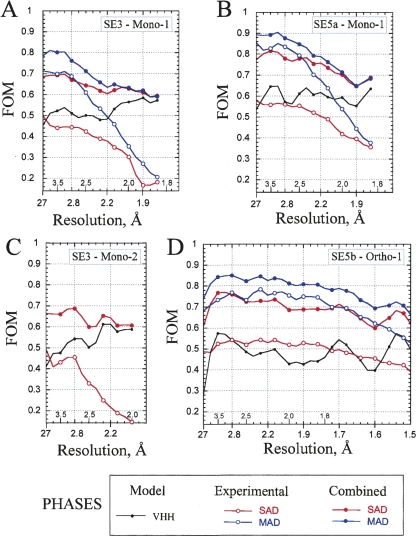
A comparison of phasing metrics from data collected in the isomorphous monoclinic space group for the three and five SeMet cases (SE3-Mono-1 vs. SE5a-Mono-1) provided a direct evaluation of the increased phasing capacity produced by the additional two SeMet groups (Table 3; Fig. 2). The phasing power from five SeMet MAD data (2.78) were much higher than that for the three SeMet case (1.70) (Table 3). The difference is even larger for SAD data, 2.34 vs. 0.95. Similar trends are seen for the figure-of-merit (FOM) values. SE3-Mono-1 gave FOMs of 0.50 and 0.36 for the experimental MAD and SAD data, respectively. The incorporation of two additional Se sites into SE5a variant resulted in increasing FOM value to 0.67 and 0.49 for MAD and SAD data, respectively. The quality of experimental phasing decreased for both SE3-Mono-1 and Se5a-Mono-1 significantly at last resolution shells (>2.5 Å), although not as precipitously for SE5a (Fig. 2A,B). A complementary analysis to the phasing metrics described below, based on auto-fitting electron density maps, confirms the importance of the additional SeMet groups and is provided in the Supplemental material.
Table 3.
MAD and SAD phasing statistics for cAb-RN05 VHH complexes with RNase A
Figure 2.
FOM for phasing based on SeMet-labeled VHH chaperones in the crystals of cAb-RN05 VHH–RNase A complex. Two-wavelength MAD (in blue) and SAD (in red) phasing statistics are plotted as a function of X-ray resolution bins. The value of the average resolution in each bin is indicated along the horizontal axis. Open circles represent the experimental phases derived by SOLVE. Filled circles represent the experimental phases combined with the VHH model-based phases. The phasing was performed for X-ray resolution range indicated in the parentheses in each case. The FOM for VHH model-based phases are included for reference and shown as a gray line. (A) SE3-Mono-1 (50–1.8 Å); (B) SE5a-Mono-1 (50–1.8 Å); (C) SE3-Mono-2 (50–2.0 Å); (D) SE5b-Ortho-1 (50–1.5 Å).
When the experimental phases were combined with the model-based phase contributions of the VHH domain (details are in Materials and Methods), the quality of SAD phasing became comparable to MAD phasing (Table 3). The FOM values for the SAD data are 0.64 for SE3 and 0.75 for SE5a, only slightly lower than those for the MAD data (0.69 and 0.79, respectively). A higher phasing capacity of a chaperone with five Se sites compared with three sites is similarly found in a comparison of SE5b-Ortho-1 and SE3-Mono-2 data sets (Table 3; Fig. 2C,D). We note that the drop-off in the FOM at the last resolution shells was significantly reduced when we combined the experimental phases with the VHH model-based phases for SE3-Mono-1, SE5a-Mono-1, and SE3-Mono-2 crystal forms that diffract to 1.65–1.8 Å resolution (Fig. 2A–C). In contrast, the phase combination marginally improved the phasing for the SE5b-Orto-1 crystal form (Fig. 2D) that diffracts to an atomic (1.1 Å) resolution.
Packing arrangements
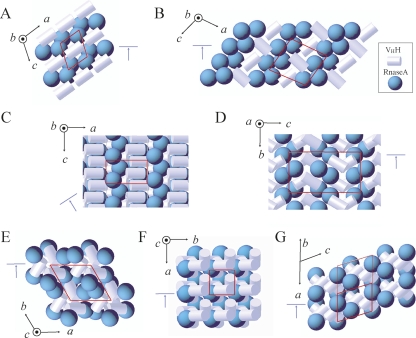
The diversity of the packing modes in the crystals of the VHH–RNase A complexes is rather extraordinary and demonstrates the potential versatility of the VHH domain to function as a crystallization chaperone. Figure 3 shows the schematic of the packing arrangements of VHH and RNase A molecules found in the six crystal forms analyzed and in the original 1BZQ structure. Selected packing interfaces are presented in Supplemental Figure S2.
Figure 3.
Schematics of the packing modes found in the crystals of the cAb-RN05 VHH–RNase A complexes. The unit cell is indicated in red. The orientation of the crystal axes is shown in black. The cyan arrow indicates the view shown in Supplemental Figure S2. The VHH and RNase A molecules are represented as white cylinders and cyan spheres, respectively. This representation is based on the calculation of molecular ellipsoids performed in MOLREP. (A) Monoclinic Mono-1 crystal form with one complex in ASU. This form was observed for the SE3 and SE5a complexes, crystallized isomorphously. (B) Monoclinic SE3-Mono-2 crystal form with two complexes in ASU. (C) Orthorhombic SE5b-Ortho-1 crystal form with one complex in ASU. (D) Orthorhombic SE5b-Ortho-2 crystal form with two complexes in ASU. (E) Trigonal SE5b-Tri crystal form with one complex in ASU. (F) Tetragonal SE5b-Tetra crystal form with one complex in ASU. (G) Original 1BZQ crystal structure crystallized in triclinic space group with four complexes in ASU.
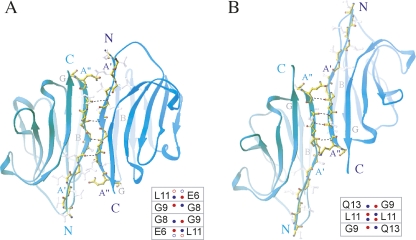
A key feature of the crystal packing is a twofold dimer interaction involving the N-terminal βA-strand of the VHH domain (Fig. 4). The intermolecular hydrogen bonds between main-chain atoms of the residues located in two adjacent βA-strands appear to nucleate formation of the dimer interface in SE5b (Fig. 4A) and 1BZQ (Fig. 4B) crystals. The residues that are involved in the dimer formation are different in SE5b and 1BZQ structures due to a shift of three amino acids along the β-strand. This shift is accompanied by a change in the relative orientation between the VHH molecules in the dimer. Similar arrangements have also been observed in a completely different VHH crystal system (PDB code 1F2X) (Decanniere et al. 2000).
Figure 4.
Favorable crystal contacts formed by dimerization of VHH through H-bonding in N-terminal β-strands. (A) VHH–VHH alignment in the SE5b dimer. Two VHH domains interact via their N-terminal βA′- and βA″-strands with the residues E6 (βA′), G8, G9, and L11 (βA″) being involved in the H-bonds (2.6–3.3 Å range). The SE5b-Tetra structure was selected for presentation. (Inset) Four H-bonding pairs found in SE5b dimers. The main-chain N- and O-atoms involved in H-bonds are shown as small circles colored in blue and red, respectively. There are four H-bonds in Se5b-Tri and Se5b-Tetra crystals (shown as filled circles). In Se5b-Ortho-2, the residues E6 and L11 contribute both N- and O-atoms in intermolecular H-bonding network, resulting in two additional H-bonds (shown as open circles). (B). VHH–VHH alignment in the 1BZQ dimer. Two VHH domains interact via their N-terminal βA″-strands with the residues G9, L11, and Q13 being involved in the H-bonds. (Inset) Three H-bonding pairs found in 1BZQ. L11 contributes both N- and O-atoms in H-bonding network, resulting in a total of four H-bonds.
Comparisons of X-ray crystal structures in different crystal forms
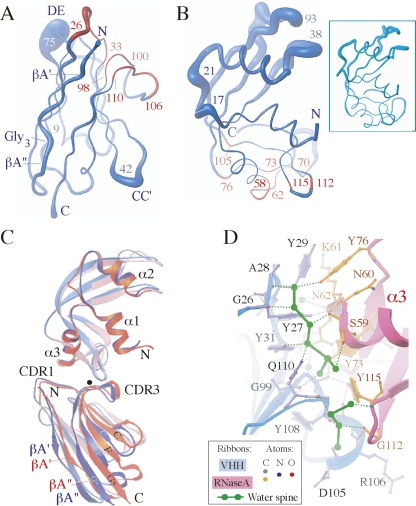
The six crystal forms studied in this work (Table 2) along with the original 1BZQ structure represent 14 crystallographically independent copies of the VHH complexed with RNase A. This number of independent copies of the complex provides a comprehensive database to evaluate the effects of crystal packing on the individual molecules. When the VHH main-chain atoms are superimposed, the average root mean square deviations (rmsd) between them are <1.1 Å. As expected for β-barrel scaffolds, the highest variability in the structure is observed for the loop regions connecting the β-strands. The CC′- and DE-loops are the most flexible in the VHH scaffold (Fig. 5A). A comparison of the seven available crystal forms reveals that among nine β-strands comprising the VHH scaffold (Fig. 1A), only the N-terminal βA-strand undergoes some structural changes (Fig. 5A) upon crystal packing. This strand consists of two short β-strands (βA′ and βA″) separated by the glycine residues 8–10. The conformational diversity of the βA-strand is presumably due to its extensive involvement in crystal packing as discussed above.
Figure 5.
A comparison of different X-ray structures of the cAb-RN05 VHH–RNase A complexes. Fourteen crystallographically independent copies (those in Table 2 and the 1BZQ structure) were used for superposition and analysis. (A) Worm representations of VHH. The thickness of the worm is proportional to the deviations between the positions of corresponding Cα-atoms in the VHH structures superimposed using their main-chain atoms. The CC′- and DE-loops centered at the residues 42 and 75 exhibit the highest flexibility. CDR1 and CDR3 residues are colored in red. (B) Worm representations of RNase A structures. The thickness of the worm is proportional to the deviations between the positions of corresponding Cα-atoms in RNase A structures superimposed using their main-chain atoms. The residues involved in the interaction with VHH in the complex are indicated in red. (Inset) Six copies of RNase A molecule in the free form from four different crystal forms (PDB codes 7RSA, 1RTB, 1XPS, 1XPT) were superimposed using their main-chain atoms with rmsds <0.7 Å. (C) Superposition of VHH–RNase A complexes. The structures were superimposed using the main-chain atoms of RNase A. Two complexes where the VHH positions differ the most are shown: SE5b-Ortho-2 complex 2 (in blue) and SE3-Mono-2 complex 1 (in red). The VHH domains from two structures may be superimposed by ∼8° rotation around the axis running through the binding interface (close to CDR3) approximately perpendicularly to the page and represented as the black dot. (D) VHH–RNase A interface. Two spines of H-bonded water molecules are represented as connected green bonds. The H-bonds involving the protein residues are shown as dotted lines: water–protein, green; protein–protein, gray. The water spines are connected to the VHH residues G26(O), A28(N), Y31(OH), A98(O), G108(O), G109(O), Q110(NE2) and RNase A residues C58(O), S59(O, OG), Y73(OH), Y76(OH), D105(O), G112(O), Y115(O, N). The H-bond lengths are in 2.6–3.3 Å range. These H-bonds are preserved in all four high-resolution (1.8–1.1 Å) structures studied in this work (Table 2). The 1.1 Å X-ray structure SE5b-Ortho-1 was selected for presentation.
The rmsd between main-chain atoms of any two superimposed RNase A is <1.0 Å. The regions of RNase A flexibility are shown in Figure 5B. The relative orientation of the two RNase A domains differs slightly in the six crystal forms presented here. In the SE5b-Ortho-1 and SE5b-Tetra structures, the binding of sulfate ion to His12 and His119 results in the more compact conformation of RNase A. A comparison of these structures with the free form of RNase A crystallized alone or in complex with the small-molecule ligands revealed that VHH binding to RNase A produces only very minor changes in the RNase A structure (Figure 5B inset). Most of the RNase A residues interacting with VHH (colored red in Fig. 5B) maintain the backbone conformation found in the free form.
To assess the flexibility at the VHH-RNase A interface in the crystals, all 14 complexes were superimposed using the main-chain atoms of RNase A. Two complexes where the VHH positions differ the most are shown in Figure 5C. The VHH domain in the SE5b-Ortho-2 complex 2 (shown in blue) rotates by ∼8° from its position in the SE3-Mono-2 complex 1 (shown in red). The reorientation of VHH domains in the complexes is generally accompanied by small changes in the conformation of the CDR1 and CDR3 residues and the corresponding contact residues in RNase A (Fig. 5A,B).
The comparison of seven crystal forms shows that the hydrogen-bond pattern at the VHH–RNase A complex interface is preserved upon the small changes in the orientation of VHH (Fig. 5D). Four hydrogen bonds are formed across the interface through the carbonyl groups of G99, G100, R106, and T107 located in VHH CDR3. The CDR1 residues form three contacts to RNase A: One hydrogen bond is formed through the N-amino group of I32, and two hydrogen bonds involve the side-chain of Y27. A notable feature of the interaction is the extensive set of contacts mediated by a network of H-bonding water molecules (Fig. 5D) that complement VHH and RNase A interactions at the interface.
Discussion
We demonstrate here that the VHH domain can be successfully employed as a versatile crystallization chaperone. Similar to its Fab counterpart, the VHH domain has a highly structured β-sheet core, providing a rigid scaffold that can help stabilize potential crystal contacts. Because of its smaller size, VHH domains may be the chaperone of choice for protein targets of similar or smaller size. The example analyzed here with a VHH–RNase A complex, which crystallizes in at least six highly ordered crystal forms, suggests that one factor for the success is that these two protein molecules are well matched to initiate crystallization.
Another attribute of VHH domains is that they can be readily adapted to both phage and yeast display systems. This allows for the selection of a number of unique high-affinity VHH-binding domains to protein targets. Thus, crystallization trials are not limited to a single crystallization chaperone but can be expanded to include several different VHH chaperones, increasing the likelihood that high-quality crystals can be obtained from at least one of the VHH-target complexes.
The VHH scaffold nucleates crystallization
The six new crystal forms of the VHH–RNase A complexes displayed an array of space group symmetries, including two monoclinic and orthorhombic cell types as well as a trigonal and tetragonal cell type (Table 2; Fig. 3).
The examination of the six crystal forms shows that the contribution of VHH βA-strand is the primary element for the packing in most forms. This β-strand acts as a “hot spot” for the interactions that promote VHH dimerization (Fig. 4). In addition, it can interact with other VHH regions or with RNase A (Supplemental Fig. S2). The Gly3 segment in the middle of the βA-strand is likely to allow small adjustments upon crystal packing (Fig. 5A). The SE5b variant has the L4M mutation, which probably introduces an additional level of flexibility into the VHH N-terminal region. This variant is able to crystallize in four of a total of six crystal forms (Table 2). The local plasticity of the VHH–RNase A complex (Fig. 5C) is an additional factor that appears to play a significant role in crystallization, and the conservative water structure (Fig. 5D) may stabilize the VHH–RNase A interface upon small readjustments of VHH in the different crystal forms. In this regard, it is notable that SE5b has the lowest binding affinity among the VHH molecules tested in this study, suggesting that plasticity due to imperfect packing at the antigen–antibody interface may be sometimes beneficial for producing productive molecular contacts for lattice formation.
In addition, the VHH and RNase A molecules have comparable size, and their molecular surfaces have similar hydrophobic/hydrophilic composition. They potentially offer approximately the same number of residues (∼80) for packing interactions (Supplemental Figs. S3, S4). These characteristics may also have contributed to the formation of the many crystal forms.
Phasing power of SeMet-labeled VHH domain
The success of structure determination using phase information derived from molecular replacement (MR) alone is questionable when the sizes of the target and known chaperone portions in the complex become comparable and/or crystals exhibit limited diffraction quality. While the combination of multiple isomorphous replacement (MIR) and MR phasing has proven successful for chaperone complexes (Iwata et al. 1995; Ostermeier et al. 1995; Li et al. 1997; Ding et al. 1998; Kwong et al. 1998; Hunte et al. 2000), the requirement of producing a good heavy atom derivative is almost always problematic. Over the past decade, classical MIR approaches have generally been superseded by the use of anomalous dispersion data principally using SeMet incorporation (Hendrickson 1991, 1999). In fact, the anomalous experimental data using Se-edge energies generally provide superior and less biased phase information compared with either MIR or model-based MR phasing data.
In this regard, our analysis indicates that the phasing power provided by the three native SeMet sites in the VHH domain is substantial, but two additional sites increase it significantly. Introducing these additional Met residues into the protein scaffold was facilitated using a shotgun Met-scanning strategy that identified the most favorable candidate positions that did not disrupt the protein's structure. Candidate sites that failed based on grounds of introducing instability, deleterious effects on expression or binding, or a combination of these were eliminated in the library sorting step. Thus, the yeast display system allowed us to pick out the few “winners” in a sea of “losers” in a very efficient way. It also allowed us to choose candidate sites that optimize the spatial distribution of the sites to maximize the overall phasing capacity of the group.
Our analysis has established that combining MR and SAD can provide accurate initial phasing information to solve structures of the size reported here. Obviously, eliminating the need to collect multiple wavelength MAD data reduces significantly data collection times with, in this case, little loss in phasing quality. We note, however, that in the case of Fab and scFv chaperones, incorporating SeMet sites is challenging because expression levels of the native molecules themselves in E. coli are generally low. This is especially true for Fabs produced by hybridoma technology that need to be introduced into bacterial vectors for expression. In contrast, the VHH domains that generally express robustly are more suitable for SeMet incorporation. Maximizing the phasing potential of the chaperone molecule by introducing all the SeMet sites in the VHH chaperone molecule alone circumvents the need to incorporate SeMet sites in the target protein. This eliminates a major bottleneck, especially for membrane proteins and proteins that are difficult to express in microorganisms.
In summary, we have significantly enhanced the crystallizability and phasing power of the VHH scaffold and demonstrated the potential of its use in CAC. As fully recombinant libraries built on a single scaffold have shown promising capacities of producing highly functional binding interfaces, one can clearly envision that chaperones for a new target can be produced using a synthetic combinatorial library built on a VHH scaffold that already contains all of these SeMet sites.
Materials and Methods
Vector and library construction
Gene synthesis and construction of the yeast surface display system have been previously reported (Koide et al. 2007a). A yeast display library for shotgun Met-scanning was constructed by substituting 10 positions to have either a methionine or its original residue (Table 1). The mutations were introduced by PCR, and Saccharomyces cerevisiae EBY100 was transformed following Gietz's method (Gietz and Woods 2002).
Yeast surface display sorting and binding analysis
RNase A (Sigma) was biotinylated using EZ-Link NHS-LC-Biotin (Pierce). Yeast cells were first grown for 2 d in the SD-CAA media and then in media containing 2% galactose. The yeast cells were incubated with monoclonal anti-V5 antibody (Sigma) and biotinylated RNase A, pelleted down, and washed with BSS buffer (50 mM Tris-HCl, 100 mM NaCl, pH 8, 1 mg/mL BSA). The binding and washing steps were repeated with FITC-conjugated anti-mouse antibody and PE-conjugated streptavidin. Flow cytometry analysis was performed on a FACScan instrument (Beckton Dickinson) according to Boder and Wittrup (2000). The surface expression of the displayed VHH domain was found to reach a maximal level after 18 h of induction. The VHH domain was displayed robustly by incubating at 30°C, the standard incubation temperature for yeast, suggesting an efficient folding of the single-domain VHH molecule. From RNase A titration directly using yeast cells expressing the VHH, we determined the dissociation constant K d for the VHH–RNase A interaction to be 23 ± 3 nM, which is consistent with the previously published K d value of 35 nM as determined with surface plasmon resonance (Decanniere et al. 1999).
For library sorting, 5 × 106 yeast cells in BSS buffer were incubated with 1.2 μM of biotinylated RNase A and 4 μg/mL of monoclonal anti V5 antibody for 90 min. The cells were pellet down, washed with BSS, and incubated with 15 μg/mL of neutravidin-PE (Molecular Probes) and 25 μg/mL of anti-Mouse IgG-FITC (Sigma). After washing with BSS buffer, PE and FITC double-positive cells were recovered using a MoFlo High Speed Cell Sorter (Cytomation). After growing the recovered cells, the second round of sorting was performed using 8 nM or 20 nM biotinylated RNase A. Individual clones were then isolated and analyzed as described above.
Protein preparation
The genes for native and mutant VHHs were, respectively, subcloned into the expression vector pHFT1 (Huang et al. 2006) using PCR. This process generated an expression clone producing a protein with TEV cleavable His6-tag fused to the N terminus of the target protein and adds two artificial residues Gly-Ser on that end. E. coli BL21(DE3) (Novagen) harboring an VHH expression vector was grown in the LB media containing kanamycin (200 μg/mL). For SeMet labeling, cells were grown in the M9 media supplemented with SeMet (30 mg/L) and all other amino acids (50 mg/mL each). IPTG was added at a final concentration of 1 mM to initiate protein expression when OD at 600 nm was between 0.5 and 0.8. The culture was incubated for an additional 3–6 h at 30°C.
The cells were harvested by centrifugation and suspended in 10 mM Tris Cl (pH 8.0) and lysed by sonication. After centrifugation, the pellet containing VHH inclusion bodies was rinsed first with 50 mM Tris HCl buffer pH 8.0 containing 5 mM EDTA and 2% deoxycholic acid, second with 50 mM Tris (pH 8.0), and finally with distilled water. The pellet was dissolved in 50 mM sodium phosphate buffer (pH 7.4) containing 300 mM NaCl, 6 M guanidine HCl, and 2 mM reduced glutathione (GSH). The solution was applied to a HisTrap column (Amersham); equilibrated with 50 mM sodium phosphate buffer (pH 8.0) containing 300 mM NaCl, 6 M guanidine HCl, and 20 mM imidazole; and then eluted with the same buffer but containing 500 mM imidazole. The eluted protein was refolded in 20 mM Tris-HCl (pH 7.4) containing 2 mM reduced GSH and 0.2 mM oxidized GSH. The refolded protein was cleaved with TEV protease. The solution was passed through a HisTrap column to remove uncleaved protein and TEV protease.
RNase A was dissolved in 20 mM TBS at the concentration of 100 mg/mL. The purified SeMet-labeled VHH proteins were dialyzed against 20 mM TBS, concentrated to ∼20 mg/mL, and mixed with RNase A protein at a 1:2 ratio to form the complex. The SeMet-labeled VHH complexes were purified with a Superdex75 gel filtration column and eluted in 10 mM Tris-HCl, pH 8.0, 150 mM NaCl. The purified complexes were concentrated to 15–20 mg/mL using Amicon 5-kDa cutoff membranes.
Crystallization, X-ray data collection, and processing
Crystals of SeMet-labeled VHH complexes with RNase A (Table 2) were grown by vapor diffusion in hanging drops at 19°C. The crystallization screening was first performed with unlabeled complex using a Hampton Research INDEX screen. The crystals appeared under multiple conditions containing PEG3350. In further trials with SeMet-labeled VHH complexes, we used only solutions IS-49 through IS-96 of the INDEX screen. Drops were generated by mixing 1 μL of protein solution with 1 μL of reservoir solution and placed against 1 mL of reservoir solution. The crystals formed within 1–2 d and typically grew to the final dimensions in 1 wk. No optimization of the crystallization conditions was performed. The INDEX solutions for each crystal form are shown in Table 2. The full description is in the Supplemental material. The SE5b-Ortho-2, SE5b-Tri, and SE5b-Tetra crystals were grown as clusters and required special efforts for their separation in order to be used for X-ray data collection. The monoclinic crystal forms were not identified for SE5b complex from the initial crystallization screen.
For low temperature data collection, crystals were transferred to the cryoprotectant (reservoir solution plus 20% [v/v] glycerol) for a few seconds and flash-frozen in liquid nitrogen. Anomalous X-ray data for SeMet-labeled–VHH complexes were collected at Se-edge energy at 100K using the Advanced Photon Source (APS) at Argonne National Laboratory. Two-wavelength (peak and inflection) MAD data sets were collected for the SE3-Mono-1, SE5a-Mono-1, and SE5b-Ortho-1 crystals. SAD data (peak) were collected for the SE3-Mono-2, SE5b-Ortho-2, SE5b-Tri, and SE5b-Tetra crystals. The width of the oscillation frame was 0.5°, and exposure times were between 1 and 3 sec depending on the crystal size and beamline. Invert beam geometry data collections were performed to maximize the anomalous signal. We note that despite the higher symmetry, Se5b-Tri and SE5b-Tetra crystal forms were not able to provide us with the sufficient anomalous signal, presumably due to extensive spot overlaps which resulted from crystal twinning. Images were processed and scaled with HKL2000 (Otwinowski and Minor 1997). Crystal data and data collection statistics at Se-peak energy are listed in Table 2 for all six crystal forms obtained in this work.
X-ray structure determination
The SE3-Mono-1, SE3-Mono-2, SE5a-Mono-1, SE5b-Ortho-1, and SE5b-Ortho-2 structures were solved by anomalous dispersion using the SOLVE and RESOLVE suite of programs (Terwilliger and Berendzen 1999; Terwilliger 2000, 2002). The model building performed with the RESOLVE basic script (Table 3) was complete for SE5b-Ortho-1 but needed some adjustments for the other four structures. The SE3-Mono-1, SE3-Mono-2 models were completed with use of the ARP/wARP program (Lamzin et al. 2001). The ARP/wARP program did not build the complete RNase A model in the SE5a-Mono-1 (due to the partial disorder observed in the RNase A molecule even at 1.8 Å resolution) and SE5b-Orho-2 (2.5 Å X-ray data) cases. The model building for these two structures was completed manually in TURBO-FRODO (Cambillau and Roussel 1997). The SE5b-Tri and SE5b-Tetra structures were determined by MR using the SE5b-Ortho-1 structure as a starting model and the program MOLREP (Vagin and Teplyakov 1997).
All models were refined against X-ray data collected at Se-peak energy. Rigid-body, positional refinement, and stimulated annealing were performed in CNS1.1 (Adams et al. 1997). The TLS (translation, libration, and screw-rotation tensor) and bulk solvent parameters, restrained B-factor, and positional refinement were completed with REFMAC5 (Murshudov et al. 1997). Rfree was monitored by setting aside 5%–10% of the reflection as a test set. Selected refinement parameters are listed in Table 2. In the six crystal forms, the final VHH model includes all 121 residues. The final RNase A model is complete in five crystal forms. The RNase A residues 19–32 (∼10% from total of 124) are partially disordered in the SE5a-Mono-1 crystals. These residues are ordered in SE3-Mono-1 crystals obtained using a different crystallization buffer (Table 2). The Ramachandran plot calculated with PROCHECK (Laskowski et al. 1993) indicates that 99.5% of the non-Gly and non-Pro residues in the final models lie in the most favored and additional allowed regions. The VHH residue E102 lies in the generously allowed region and is well defined in the electron density. The electron density maps were calculated using CCP4 (Collaborative Computational Project, Number 4 1994). The calculations of crystal contacts and solvent accessible areas were performed in CNS1.1. The surface areas buried at the complex and packing interfaces were estimated using Protein–Protein Interaction Server (http://www.biochem.ucl.ac.uk/bsm/PP/server). Figures showing the three-dimensional structures, molecular surface, and electron density maps were prepared using RIBBONS (Carson 1996), MOLMOL (Koradi et al. 1996), and TURBO-FRODO (Cambillau and Roussel 1997).
MAD and SAD phase determination
Heavy atom searches were performed in SHELXD (Schneider and Sheldrick 2002) using 3.5 Å X-ray data resolution cut-off. Two-wavelength (peak and inflection, MAD) and single-wavelength (peak, SAD) anomalous dispersion phasing calculations were performed using SOLVE. Density modification and initial model building were done using a RESOLVE basic script. The overall phasing and auto-building statistics are shown in Table 3 and Supplemental Table S1, respectively. The resolution dependence of FOM value is shown in Figure 2. The examples of the SAD and MAD maps are demonstrated in Supplemental Figures S5 and S6, respectively.
The calculations of model phases from VHH part of the structure and phase combination were done in CNS.1.1. The VHH models derived by a RESOLVE basic script with starting phases from SOLVE were more than 80% complete for both MAD and SAD data in all analyzed crystal forms (SE3-Mono-1, SE3-Mono-2, SE5a-Mono-1, SE5b-Ortho-1, and SE5b-Ortho-2). Thus, there was no need to search for the VHH part of the structure by MR in order to calculate VHH model-based phases. Prior to phase combination, we completed the VHH models manually in TURBO-FRODO and refined them against the corresponding data set in CNS.1.1. The analysis of the relative contribution of SeMet anomalous dispersion and model to the phasing capability of the VHH chaperone is provided in the Supplemental material.
Protein Data Bank coordinates
Coordinates and structure factors have been deposited in the protein data bank (PDB accession codes are listed in Table 2).
Acknowledgments
This work was supported by NIH grants R01-GM72688 and U54 GM74946, and by the University of Chicago Cancer Research Center. We acknowledge the use of the Flow Cytometry and DNA sequencing core facilities at the University of Chicago. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38. Portions of this work were performed at the Industrial Macromolecular Crystallography Association (IMCA-CAT) and at the Structural Biology Center (SBC-CAT) located at Sectors 17ID and 19ID, respectively, of the Advanced Photon Source. Use of the IMCA-CAT was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago. SBC-CAT is supported by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract no. W-31-109-ENG-38.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Anthony A. Kossiakoff, Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA; e-mail: koss@bsd.uchicago.edu; fax: (773) 834-2777; or Shohei Koide, Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA; e-mail: skoide@uchicago.edu; fax: (773) 834-2777.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.034892.108.
References
- Adams, P.D., Pannu, N.S., Read, R.J., Brunger, A.T. Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl. Acad. Sci. 1997;94:5018–5023. doi: 10.1073/pnas.94.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelemy, P.A., Raab, H., Appleton, B.A., Bond, C.J., Wu, P., Wiesmann, C., Sidhu, S.S. Comprehensive analysis of the factors contributing to the stability and solubility of autonomous human VH domains. J. Biol. Chem. 2008;283:3639–3654. doi: 10.1074/jbc.M708536200. [DOI] [PubMed] [Google Scholar]
- Boder, E.T., Wittrup, K.D. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- Bradbury, A.R.M., Marks, J.D. Antibodies from phage antibody libraries. J. Immunol. Methods. 2004;290:29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Brekke, O.H., Loset, G.A. New technologies in therapeutic antibody development. Curr. Opin. Pharmacol. 2003;3:544–550. doi: 10.1016/j.coph.2003.05.002. [DOI] [PubMed] [Google Scholar]
- Cambillau, C., Roussel, A. Turbo Frodo, version OpenGL.1. University Aix-Marseille II; 1997. Marseille, France. [Google Scholar]
- Carson, M. Ribbons. Methods Enzymol. 1996;277:493–505. [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dauter, Z. Current state and prospects of macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2006;D62:1–11. doi: 10.1107/S0907444905034050. [DOI] [PubMed] [Google Scholar]
- Decanniere, K., Desmyter, A., Lauwereys, M., Ghahroudi, M.A., Muyldermans, S., Wyns, L. A single-domain antibody fragment in complex with RNaseA: Non-canonical loop structures and nanomolar affinity using two CDR loops. Struct. Fold. Des. 1999;7:361–370. doi: 10.1016/s0969-2126(99)80049-5. [DOI] [PubMed] [Google Scholar]
- Decanniere, K., Muyldermans, S., Wyns, L. Canonical antigen-binding loop structures in immunoglobulins: More structures, more canonical cases? J. Mol. Biol. 2000;300:83–91. doi: 10.1006/jmbi.2000.3839. [DOI] [PubMed] [Google Scholar]
- Derewenda, Z.S. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Derewenda, Z.S., Vekilov, P.G. Entropy and surface engineering in protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 2006;D62:116–124. doi: 10.1107/S0907444905035237. [DOI] [PubMed] [Google Scholar]
- Ding, J., Das, K., Hsiou, Y., Sarafianos, S.G., Clark A.D., Jr, Jacobo-Molina, A., Tantillo, C., Hughes, S.H., Arnold, E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- Fellouse, F.A., Esaki, K., Birtalan, S., Raptis, D., Cancasci, V.J., Koide, A., Jhurani, P., Vasser, M., Wiesmann, C., Kossiakoff, A.A., et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Gassner, N.C., Baase, W.A., Matthews, B.W. A test of the “jigsaw puzzle” model for protein folding by multiple methionine substitutions within the core of T4 lysozyme. Proc. Natl. Acad. Sci. 1996;93:12155–12158. doi: 10.1073/pnas.93.22.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Hamers-Casterman, C., Atarhouch, T., Muyldermans, S., Robinson, G., Hamers, C., Songa, E.B., Bendahman, N., Hamers, R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W.A. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W.A. Maturation of MAD phasing for the determination of macromolecular structures. J. Synchrotron Radiat. 1999;6:845–851. [Google Scholar]
- Hoogenboom, H.R. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- Hunte, C., Michel, H. Crystallisation of membrane proteins mediated by antibody fragments. Curr. Opin. Struct. Biol. 2002;12:503–508. doi: 10.1016/s0959-440x(02)00354-8. [DOI] [PubMed] [Google Scholar]
- Hunte, C., Koepke, J., Lange, C., Rossmanith, T., Michel, H. Structure at 2.3 Å resolution of the cytochrome bc1-complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- Huang, J., Koide, A., Nettle, K.W., Greene, G.L., Koide, S. Conformation-specific affinity purification of proteins using engineered binding proteins: Application to the estrogen receptor. Protein Expr. Purif. 2006;47:348–354. doi: 10.1016/j.pep.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Iwata, S., Ostermeier, C., Ludwig, B., Michel, H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans . Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- Koide, A., Tereshko, V., Uysal, S., Margalef, K., Kossiakoff, A.A., Koide, S. Exploring the capacity of minimalist protein interfaces: Interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope. J. Mol. Biol. 2007a;373:941–953. doi: 10.1016/j.jmb.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide, A., Gilbreth, R.N., Esaki, K., Tereshko, V., Koide, S. High-affinity single-domain binding proteins with a binary-code interface. Proc. Natl. Acad. Sci. 2007b;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., Wuthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kovari, L.C., Momany, C., Rossmann, M.G. The use of antibody fragments for crystallization and structure determinations. Structure. 1995;3:1291–1293. doi: 10.1016/s0969-2126(01)00266-0. [DOI] [PubMed] [Google Scholar]
- Kwong, P.D., Wyatt, R., Robinson, J., Sweet, R.W., Sodroski, J., Hendrickson, W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamzin, V.S., Perrakis, A., Wilson, K.S. The ARP/WARP suite for automated construction and refinement of protein models. In: Rossmann M.G., Arnold E., editors. International tables for crystallography: Crystallography of biological macromolecules. Vol. F. Kluwer Academic; Dordrecht, The Netherlands: 2001. pp. 720–722. [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Li, H., Dunn, J.J., Luft, B.J., Lawson, C.L. Crystal structure of Lyme disease antigen outer surface protein A complexed with an Fab. Proc. Natl. Acad. Sci. 1997;94:3584–3589. doi: 10.1073/pnas.94.8.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty, M.C. Immunity to Chlamydia psittaci with particular reference to sheep. Vet. Microbiol. 1990;25:87–99. doi: 10.1016/0378-1135(90)90096-e. [DOI] [PubMed] [Google Scholar]
- McPherson, A. Protein crystallization in the structural genomics era. J. Struct. Funct. Genomics. 2004;5:3–12. doi: 10.1023/B:JSFG.0000029199.43875.92. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E. Refinement of macromolecular structures by maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Muyldermans, S. Single domain camel antibodies: Current status. J. Biotechnol. 2001;74:277–302. doi: 10.1016/s1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]
- Ostermeier, C., Iwata, S., Ludwig, B., Michel, H. Fv fragment-mediated crystallization of the membrane protein bacterial cytochrome c oxidase. Nat. Struct. Biol. 1995;2:842–846. doi: 10.1038/nsb1095-842. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Schneider, T.R., Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Schweizer, A., Roschitzki-Voser, H., Amstutz, P., Briand, C., Gulotti-Georgieva, M., Prenosil, E., Binz, H.K., Capitani, G., Baici, A., Plückthun, A., et al. Inhibition of caspase-2 by a designed ankyrin repeat protein: Specificity, structure, and inhibition mechanism. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. Maximum likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;D56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. Automated main-chain model-building by template-matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 2002;D59:34–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C., Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, A.A., Teplyakov, A. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Venturi, M., Hunte, C. Monoclonal antibodies for the structural analysis of the Na+/H+ antiporter NhaA from Escherichia coli . Biochim. Biophys. Acta. 2003;1610:46–50. doi: 10.1016/s0005-2736(02)00713-7. [DOI] [PubMed] [Google Scholar]
- Weiss, D.J., Watanabe, C.K., Zhong, A., Goddard, A., Sidhu, S.S. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc. Natl. Acad. Sci. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weselak, M., Patch, M.G., Selby, T.L., Knebel, G., Stevens, R.C. Robotics for automated crystal formation and analysis. Methods Enzymol. 2003;368:45–76. doi: 10.1016/S0076-6879(03)68004-3. [DOI] [PubMed] [Google Scholar]